Abstract
As a key element in the cytoskeleton, actin filaments are highly dynamic structures that constantly sustain forces. However, the fundamental question of how force regulates actin dynamics is unclear. Using atomic force microscopy force-clamp experiments, we show that tensile force regulates G-actin/G-actin and G-actin/F-actin dissociation kinetics by prolonging bond lifetimes (catch bonds) at a low force range and by shortening bond lifetimes (slip bonds) beyond a threshold. Steered molecular dynamics simulations reveal force-induced formation of new interactions that include a lysine 113(K113):glutamic acid 195 (E195) salt bridge between actin subunits, thus suggesting a molecular basis for actin catch-slip bonds. This structural mechanism is supported by the suppression of the catch bonds by the single-residue replacements K113 to serine (K113S) and E195 to serine (E195S) on yeast actin. These results demonstrate and provide a structural explanation for actin catch-slip bonds, which may provide a mechanoregulatory mechanism to control cell functions by regulating the depolymerization kinetics of force-bearing actin filaments throughout the cytoskeleton.
Keywords: single-molecule force spectroscopy, mechanotransduction, mechanosensing, nemaline myopathy
The actin cytoskeleton, primarily a force-bearing structure, controls the morphology, motility, and adhesion of the cell (1–4). Its core filamentous component, assembled from actin monomers via noncovalent interactions (5), undergoes rapid and controlled polymerization and depolymerization, allowing the dynamic reorganization of the actin cytoskeleton (1, 2).
In cells, this dynamic process can be modulated by forces, and this is crucial to mechanosensitivity, mechanotransduction, and cellular adaptations to mechanical stresses (3, 6–8). For example, the assembly, stabilization, and reorganization of the actin stress fiber and the focal adhesion, where actin filaments constantly sustain tension, are induced by externally applied forces (9–12) dependent on myosin-generated contractility (4, 8, 13, 14) and sensitive to substrate rigidity (3, 15, 16). These observations led us to investigate the molecular mechanism by which actin dynamics are regulated by force.
The force-regulated kinetics of several molecular interactions important to adhesion and force-bearing functions of cells are governed by catch-slip bonds, in which the interaction is stabilized by tensile force in a low range and destabilized when force exceeds a threshold (17–22). Various mechanisms, such as the allosteric model based on intramolecular conformational change under forces (23, 24) and the sliding-rebinding model based on force-induced formation of new interactions due to intermolecular interface sliding (18, 25), have been proposed to provide structural explanations for catch-slip bonds in different molecular interactions.
Here we use atomic force microscopy (AFM) force-clamp experiments to determine how force regulates the off-rate of actin depolymerization and to elucidate the structural mechanism by a combination of steered molecular dynamics (SMD) simulations and yeast actin mutagenesis studies.
Results
Force-Dependent Bond Lifetimes of Actin Subunit Interactions.
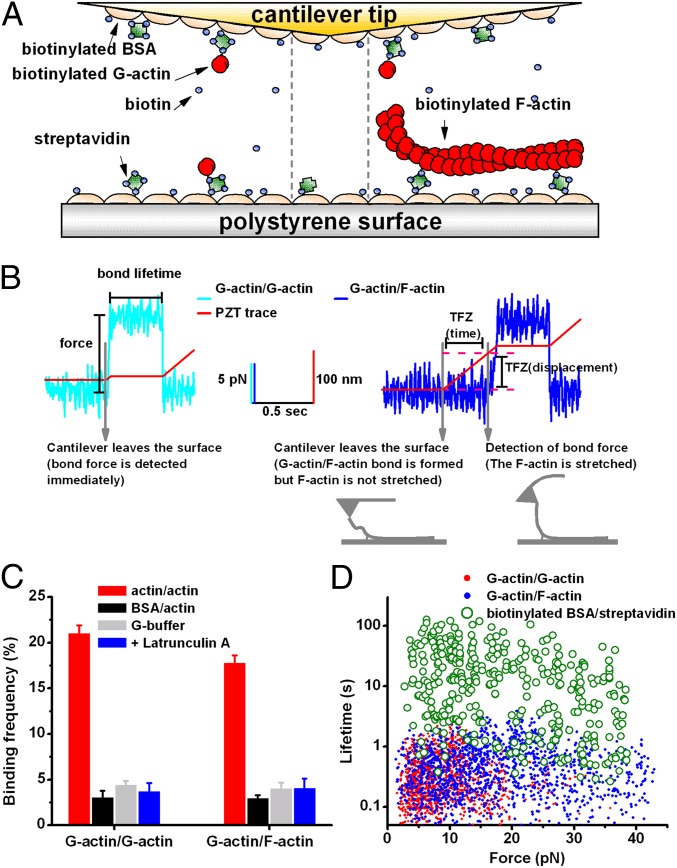
To study force-regulated actin dissociation kinetics at the single-bond level, we used biotin/streptavidin coupling to functionalize the AFM (Fig. 1A). The cantilever tip was coated with actin monomers and the polystyrene surface was coated with monomeric (Fig. 1A, Left) or filamentous (Fig. 1A, Right) actin for studying G-actin/G-actin or G-actin/F-actin interactions, respectively. The tip was driven close to the surface (10–20 and 20–35 nm for G-actin/G-actin and G-actin/F-actin interactions, respectively) and held for 0.5 s to allow bond formation, retracted to pull on the bond, and then held stationary for measurements of bond lifetime at a constant force (Fig. 1B and Fig. S1).
Fig. 1.
AFM experiment. (A) AFM cantilever tip and polystyrene surface functionalized for G-actin/G-actin (Left), biotin/streptavidin (Center) or G-actin/F-actin (Right) interactions. Actin (red), biotin (blue), streptavidin (green), and BSA (beige) are depicted. (B) Representative force traces for G-actin/G-actin (Left, cyan) and G-actin/F-actin (Right, blue) interactions illustrating bond lifetime measurements at clamped forces. The piezoelectric translator (PZT) (displacements of which are depicted as red traces) brought the cantilever tip close to the surface to allow bond formation, was retracted to pull on the bond, and then was held stationary to allow lifetime measurement at a constant force until rupture, signified by a drop of force to the baseline. A tension-free zone (TFZ, indicated)—i.e., a delay time (and displacement) from the onset of PZT retraction to the onset of force increase—was observed in G-actin/F-actin interactions but not in G-actin/G-actin interactions. As depicted, TFZ is interpreted as time (and displacement) required for the F-actin to be picked up and realigned along the direction of cantilever retraction until it was fully extended and stretched. (C) Binding frequencies of G-actin/G-actin (Left, red) or G-actin/F-actin (Right, red) interactions were significantly (P < 0.001) higher than those of the following respective controls that prevented actin/actin interactions: replacing G-actin by BSA (black), replacing F-buffer by G-buffer (gray), and adding 20 μM latrunculin A (blue). Data are presented as mean ± SEM of 10–30 binding frequencies, each estimated from 100 to 200 contacts in AFM experiments. (D) Individual lifetime vs. force scatter plots showing that lifetimes of biotin/streptavidin interactions (green; compare with A, Center) were several hundred-fold longer than those of G-actin/G-actin (red, compare with A, Left) and G-actin/F-actin (blue compare with A, Right) interactions.
A tension-free zone (TFZ) was characteristic of the force-scan traces of G-actin/F-actin interactions (Fig. 1B, Right and Fig. S1), but not of G-actin/G-actin interactions (Fig. 1B, Left and Fig. S1) or nonspecific binding controls. A TFZ has been observed in interactions between lengthy molecules because they need to be fully extended before they can resist any tensile forces (26, 27). In G-actin/F-actin interactions, the TFZ is interpreted as a period (and length of displacement) during which the G-actin/F-actin bond is formed but the F-actin filament is not fully extended (Fig. 1B, Right). It is terminated by detecting a tether force on the G-actin/F-actin bond presumably due to full extension of a segment of filament between the cantilever tip and the biotin–streptavidin anchoring site. This interpretation of TFZ is supported by data that show that the length distribution of TFZ is left-shifted when the ratio of biotinylated actin/unmodified actin used to prepare the F-actin is increased to reduce the average length of F-actin segments free of biotinylated actin (Fig. S2). Using TFZ to select and group data allowed us to confirm the binding specificity for G-actin/F-actin interactions and to obtain better and more uniformly oriented G-actin/F-actin bonds for lifetime measurement under tension. With a sufficiently long TFZ, the tether force must be applied along the axial direction of the extended segment of the actin filament; therefore, the bond lifetime is measured when the G-actin/F-actin bond is stressed in a proper orientation.
Binding was specific to the actin/actin interaction as its frequency was suppressed by coating the cantilever tip with BSA instead of G-actin, by using G-buffer instead of F-buffer (see Methods for the buffer contents) as the working buffer, or by adding 20 μM latrunculin A to prevent actin/actin interactions (Fig. 1C).
Lifetimes of the serial bonds (in which an actin–actin bond was sandwiched between biotin/streptavidin bonds) were determined by time-to-dissociation of the actin/actin bond, not the biotin/streptavidin bond, because the biotin/streptavidin bond alone lasted several hundred-fold longer (Fig. 1D). For G-actin/F-actin interactions, the actin/actin serial bonds most likely dissociated at the end, not in the middle of the F-actin, as stretching an F-actin segment from the middle via biotin/streptavidin coupling yielded lifetimes 10- to 100-fold longer than those of G-actin/F-actin interactions (Fig. S3). Also, rupture in the middle of an actin filament requires a force an order of magnitude higher than the force applied in our experiments (28).
Force Regulates Actin Subunit Dissociation Kinetics by Catch-Slip Bonds.
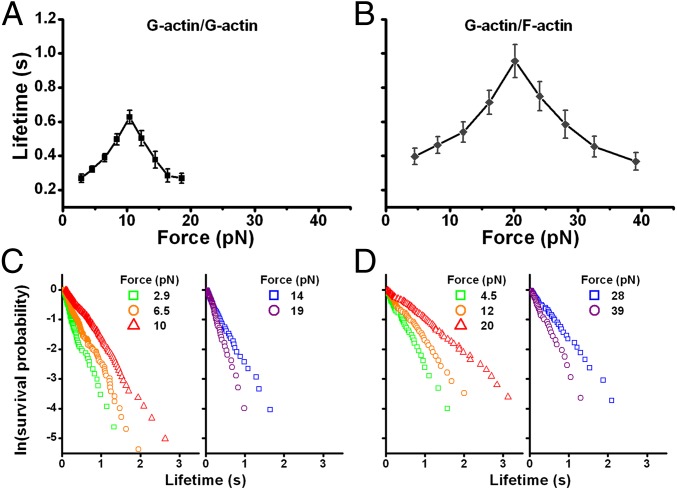
The G-actin/G-actin bond lifetime exhibited a biphasic catch-slip force dependence. It had a “catch” region characterized by increasing lifetimes as force increased, with a maximum of 0.63 s at 10 pN, followed by a “slip” region within which the lifetimes decreased as force further increased (Fig. 2A).
Fig. 2.
Experimental demonstration of actin catch-slip bonds. (A and B) Bond lifetimes exhibit biphasic force dependence, i.e., catch-slip bonds, for G-actin/G-actin (A: zero TFZ) and G-actin/F-actin (B: TFZ = 50–100 nm) interactions. Each point represents the mean ± SEM of >50 measurements. (C and D) Single exponential lifetime distributions exemplified by the linear semilog plots of survival frequency (i.e., fraction of bonds survived longer than a given time) versus lifetime for the G-actin/G-actin (C) and G-actin/F-actin (D) interactions at every other force bin presented in A and B, respectively.
G-actin/F-actin interactions also formed catch-slip bonds that were qualitatively similar to, but quantitatively different from, those of G-actin/G-actin interactions (Fig. 2B). Their maximum lifetime was 0.96 s [for data with TFZ = 50–100 nm, with ∼15–30 actin monomer subunits (5)] at 20 pN, and their lifetime was indifferent to the TFZ length (Fig. S4). The catch-slip phenotype does not result from premature dissociation of a partially formed actin bond due to lack of nucleotide hydrolysis because force-dependent lifetimes measured from bonds formed in 0.5- and 10-s contacts were indistinguishable (Fig. S5). The latter period should be sufficient for the conversion of a newly added ATP-actin to ADP-actin (1).
The bond survival probabilities at various force bins for G-actin/G-actin and G-actin/F-actin interactions decreased exponentially with increasing lifetime, as shown in semilog plots (Fig. 2 C and D). Such single exponential lifetime distributions suggest first-order dissociation kinetics from respective homogeneous states. Therefore, the dissociation rate constant (koff) at each force bin can be estimated from the reciprocal mean lifetime. At the smallest force measured (2.5 pN), the off-rates of G-actin/G-actin and G-actin/F-actin interactions were 3.6 s−1 (Fig. 2A) and 2.5 s−1 (Fig. 2B), respectively. The latter value is of the same order of magnitude as the previously reported off-rates at the ends of actin filaments (1). However, as the first measured off-rate for G-actin/G-actin interactions under force in a surface-based assay system, the former value is much smaller than the koff of actin dimers predicted by theoretical simulation with force-free solution-based assumptions (29).
CapZ and Tmod3 Isolate the Polar Activity in G-Actin/F-Actin Interactions.
F-actin is a polar molecule, and the barbed end has faster kinetics than the pointed end. It is likely that the two ends contribute to the G-actin/F-actin bond lifetime measurements differently. To isolate the activity of each end, we repeated the measurements in the presence of chicken actin capping protein muscle Z-line (CapZ) or tropomodulin3 (Tmod3) in solution, which specifically block actin filament turnover at the barbed or pointed end, respectively (30, 31) (Fig. 3A). Blocking the pointed end with Tmod3 lowered the binding frequency to 13%, whereas capping at the barbed end with CapZ reduced the G-actin/F-actin binding frequency from 18 to 8% (Fig. 3B, dark-gray and blue bars). In either situation, the measured lifetimes were largely mediated by specific G-actin/F-actin interactions, as binding frequencies were further diminished by conditions preventing these interactions (Fig. 3B, cyan and light-gray bars). Adding CapZ and Tmod3 together reduced the binding frequency to the level of nonspecific binding (3–4%), indicating complete blockade of both ends (Fig. 3B, green bar). This result suggests that G-actin binding to the filament side, if any, was undetectable compared with binding to the ends.
Fig. 3.
Effects of Tmod3 and CapZ on G-actin/F-actin interactions. (A) Tmod3 and CapZ block G-actin interaction with F-actin at the pointed and barbed ends, respectively. (B) G-actin/F-actin binding frequency was reduced by Tmod3 or CapZ and further lowered by both Tmod3 and CapZ to the level of controls in which actin/actin interactions were prevented (compare with Fig. 1C legend). Data are presented as mean ± SEM of 10–30 binding frequencies, each estimated from 100 to 200 contacts. (C) Catch-slip bonds of G-actin/F-actin interactions in the presence of Tmod3 or CapZ. Each point represents the mean ± SEM of >50 measurements.
The force-dependent lifetimes of G-actin bonds with the pointed and barbed ends of F-actin exhibited qualitatively similar catch-slip behaviors (Fig. 3C), with the slip bond portions being indistinguishable. Lifetimes at low forces in the catch bond regime were longer with CapZ than Tmod3; this result is consistent with previous studies showing that the barbed end has a faster koff than the pointed end in the force-free experiment (1).
SMD-Simulated Force-Induced Formation of New Interactions Between Actin Subunits.
To elucidate the structural mechanism of actin catch-slip bonds, we used SMD simulations to study force-induced actin dimer dissociation and actin filament depolymerization, by pulling, respectively, the subunits within an actin dimer apart and the ends of an actin 14-mer filament along its axial direction (Fig. 4).
Fig. 4.
SMD simulated actin dimer dissociation and filament depolymerization under force. (A–D) Sequential snapshots showing force-induced formation of new interactions within (A) long-pitch and (B) short-pitch actin dimers and an actin 14-mer filament pulled at the (C) barbed end or (D) pointed end. Each panel is representative of ≥ 3 simulations. For actin dimers, pulling caused a relative sliding between the pulled (cyan) and the constrained (green in A and purple in B) G-actins charged with ATP (orange), which induced the formation of new interactions between acidic (red) and basic (blue) residues that were far apart before pulling. (C and D) In the actin 14-mer filament (showing only three subunits) pulled at either the barbed (C) or the pointed (D) end, only the pulled terminal subunit (cyan) dissociated from the rest of actin filament. Pulling-induced relative sliding and formation of new interactions were also observed. (E–H) Time courses of distances between the indicated atoms of the residues identified in A–D, respectively. Interactions were absent before pulling (time 0), but noncovalent interactions formed when pulled, as signified by the decreases in the interatomic distances below the 3.5 Å threshold (horizontal dashed line in E–H).
For the actin dimer dissociation, simulations were performed with long-pitch (parallel, intrastrand) and short-pitch (antiparallel, interstrand) actin dimers, which were charged with either ATP or ADP. Pulling caused a relative sliding, enabling the formation of new interactions that tightened the binding between two interacting actin subunits (Fig. 4 A and B). For example, in the long-pitch dimer, a salt bridge formed between arginine (Arg)39 of the constrained G-actin and aspartic acid 286 of the pulled G-actin (Fig. 4 A and E). In the short-pitch dimer, salt bridges were observed to form between K113:E195 and Arg62:glutamic acid (Glu)270 (Fig. 4 B and F).
For F-actin depolymerization, simulations were performed by pulling the terminal actin monomer subunit of an actin 14-mer filament at either the barbed or pointed end while constraining the two actin subunits at the other end. In either case, the pulling-induced dissociation occurred at the G-actin/F-actin interface of the pulled end, but not at the middle of the filament. This further supports the assertion that in AFM experiments (Fig. 1A, Right) the lifetimes for G-actin/F-actin interactions corresponded to time-to-dissociation of the terminal actin monomer from the end of the filament.
Pulling-induced new interactions were also observed for actin filament depolymerization. For example, when pulling was applied to the barbed end, a salt bridge between E195 of the pulled actin subunit and the K113 of the neighboring interstrand subunit in the constrained filament was observed (Fig. 4 C and G). When pulling was applied to the pointed end, formation of two interstrand salt bridges (K113:E195 and E270:R39) was observed (Fig. 4 D and H).
Similar qualitative features were observed in simulations with actin subunits charged with ATP and ADP. New interactions were not observed in control simulations without the applied pulling force. No significant global conformational change in the actin subunits was observed during the pulling process (Fig. S6).
The sliding-rebinding model, based on previous SMD simulations of selectin/ligand (25) and glycoprotein Ibα/von Willebrand factor A1 domain (18) dissociations with results similar to the present observations, has been proposed to explain the catch bonds of these systems. This model may apply to actin subunit interactions. At low forces, the actin/actin bond can dissociate directly without forming any new interactions. Increasing force tilts the orientation of the interacting actins to allow sliding along their interface and induces the formation of new, strong interactions, which prolong the overall bond lifetime and give rise to catch bonds.
The pulling-induced formation of the K113:E195 salt bridge between interacting actin subunits was observed in the simulations for the short-pitch actin dimer dissociation and in those for the actin filament depolymerization at both barbed and pointed ends, suggesting the importance of this pair of residues in the structural basis of actin catch-slip bonds.
Yeast Actin Point Mutations K113S and E195S Suppress Actin Catch-Slip Bonds.
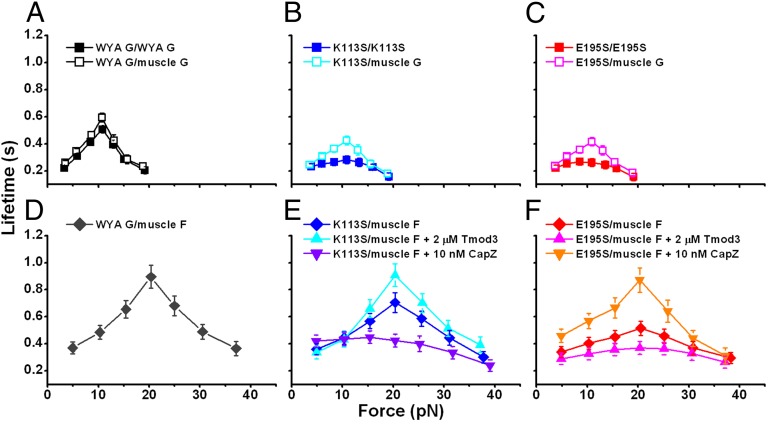
Because both K113 and E195 are conserved in rabbit skeletal muscle and yeast actins, we used the yeast actin mutants K113S and E195S to assess the importance of the K113:E195 ionic interaction in the formation of actin catch-slip bonds. The catch-slip phenotype similar to that of muscle actin was observed in the interaction of the wild-type yeast actin (WYA) monomer with either WYA monomer or muscle actin monomer (compare Figs. 2A and 5A). This observation indicates that the catch-slip phenotype is indifferent to the 13% sequence differences between rabbit skeletal muscle and yeast actins. Therefore, results obtained from the genetically mutable yeast actin regarding G-actin/G-actin catch-slip bonds may be generalized to other systems such as rabbit skeletal muscle actin.
Fig. 5.
Measured effects of K113S and E195S mutations on actin catch-slip bonds. (A) Catch-slip bonds between WYA monomers (solid square) and between WYA monomer and rabbit skeletal muscle actin monomer (open square). (B and C) Catch-slip bonds between yeast G-actin and muscle G-actin (open square) or yeast G-actin (solid square) were progressively suppressed by the mutation of K113S (B) or E195S (C) on one or both interacting partners. (D) Catch-slip bonds between WYA monomer and rabbit skeletal muscle actin filament. (E) The suppression effect of K113 mutation on yeast G-actin/muscle F-actin catch-slip bonds (blue diamond) was exacerbated by CapZ (purple down triangle) but diminished by Tmod3 (cyan up triangle). (F) The suppression effect of E195S mutation on yeast G-actin/muscle F-actin catch-slip bonds (red diamond) was exacerbated by Tmod3 (pink up triangle) but diminished by CapZ (orange down triangle). Each point represents the mean ± SEM of >50 measurements.
Substituting the neutral residue Ser for the cationic residue K113 (Fig. 5B and Fig. S7A) or the anionic residue E195 (Fig. 5C and Fig. S7B) similarly suppressed the G-actin/G-actin catch-slip bonds. The suppression was more prominent with the mutation of either K113S or E195S on both interacting G-actins than just one G-actin (Fig. 5 B and C). However, mutations of both K113S and E195S, with one on each of the two interacting G-actins, did not lead to greater suppression than that observed with only one mutation on either of the two interacting G-actins (Fig. S7). This finding suggests that the suppressive effect of mutating Lys113 or Glu195 on the G-actin/G-actin catch-slip bond resulted from elimination of the ionic interaction between these two residues.
The catch-slip bonds between the WYA monomer and skeletal muscle F-actin were quantitatively similar to those between the skeletal muscle G-actin and F-actin (compare Figs. 2B and 5D), again justifying the use of a mixed-species system in the study of G-actin/F-actin catch-slip bonds.
The G-actin/F-actin catch-slip bonds were similarly suppressed by the K113S or E195S mutations on the interacting yeast actin monomer (Fig. 5 E and F). For the K113S mutation, the suppression was exacerbated by CapZ and diminished by Tmod3 (Fig. 5E). For the E195 mutation, the suppression was exacerbated by Tmod3 and diminished by of CapZ (Fig. 5F). These findings are consistent with the polarity of F-actin in which residue 113 or 195 of the newly added monomer is accessible to only the F-actin barbed or pointed end, respectively. Together with the SMD simulations (Fig. 4), these results support the sliding-rebinding model (25) as a structural mechanism for actin catch-slip bonds in which the force-induced Lys113:Glu195 ionic interaction plays an important role.
Discussion
Actin filaments are the major force-bearing structures in the cytoskeleton and are highly dynamic. The actin filaments in stress fibers constantly sustain tensile force at focal adhesions and may undergo polymerization and depolymerization (32–35). Although rupture of actin filaments under tension (28) and the growth of an actin network under compression (36) have been studied, the effects of tensile force on actin depolymerization have not been investigated. The present work fills this gap by studying an in vitro model for the disassembly of the actin nucleus and the depolymerization of the actin filament under tensile force.
Depolymerization of the terminal actin subunit from the filament tip involves the dissociation of two G-actin/G-actin bonds (intrastrand long-pitch and interstrand short-pitch) arranged in parallel (5). This can be modeled by assuming that each component G-actin/G-actin bond at the G-actin/F-actin interface sustains half of the force applied to stretch the actin filament. This model is supported by the rightward and upward shifts of the lifetime versus force curve of the G-actin/F-actin interaction relative to that of the G-actin/G-actin interaction (compare Fig. 2 A and B). This is similar to the previously reported shifts in the lifetime versus force curve of dimeric interactions relative to that of monomeric interaction between P-selectin and its ligand (17).
The qualitative similarity of catch-slip bonds at the barbed and pointed ends of the actin filament (Fig. 3C) suggests that a common structural mechanism dominates the catch-slip phenotype at both ends. Among the residue pairs shown in our SMD simulations (Fig. 4) to form ionic bridges upon pulling, the K113:E195 ionic interaction was consistently observed at both ends (Fig. 4 C and D). Eliminating this interaction using actin mutants greatly suppressed catch-slip bonds at both ends (Fig. 5 E and F), experimentally confirming that the force-induced K113:E195 ionic interaction is a common mechanism.
The catch-bond regime where G-actin/F-actin interaction is stabilized by force (≤20 pN) corresponds to the estimated range of tensile forces physiologically sustained by a single actin filament, as summarized in Table S1. Thus, catch bonds may explain the tension-induced assembly and stabilization of the actin cytoskeleton (4, 8, 10, 12–14) and the fact that actin stress fibers are more developed in cells plated on rigid substrates than in cells plated on soft substrates (3, 15, 16). As F-actin likely bears intracellular forces transmitted across adhesion complexes from the extracellular matrix, the force-dependent actin depolymerization kinetics may contribute to mechanosensing by the cell (3, 37–39). Moreover, actin catch bonds could form a feedback mechanism controlling the length of F-actin in stress fibers where actin filaments constantly sustain tensions, as decreasing the length of the anchored F-actin could increase tension, which in turn would inhibit actin depolymerization, thus inducing filament growth to restore the length.
Actin catch-slip bonds could be important from a pathological standpoint. Key residues observed in our SMD simulations to be involved in the formation of actin catch-slip bonds (Fig. 4) are related to nemaline myopathy mutations in the human actin gene ACTA1 (40–42). These residues include R39, K113, E270, and D286. Among them, in particular, K113 has been verified experimentally to be a key residue contributing to the catch-slip bonds by forming a salt bridge with E195 upon pulling (Figs. 4 and 5). These results suggest that actin catch-slip bonds are important to the physiological function of actin.
Our study investigates the force dependence of actin dynamics at the single-bond level and suggests a possible mechanosensing mechanism for the force regulation of actin cytoskeleton dynamics.
Methods
AFM Force-Clamp Experiments.
Our custom-made AFM and force-clamped experimental procedures for measuring lifetimes of single bonds have been previously described (17, 18, 20). To functionalize the AFM for G-actin/G-actin interactions (Fig. 1A, Left), the cantilever tip and the polystyrene dish surface were incubated with 2 mg/mL biotinylated BSA (Sigma Aldrich) at 4 °C overnight, washed three times with PBS, and incubated with 1 mg/mL streptavidin (Sigma Aldrich) for 1 h at room temperature. After being washed three times with G-buffer (5 mM Tris⋅HCl, pH 8.0, 0.2 mM CaCl2, 0.2 mM ATP, 0.5 mM DTT), the cantilever tip and the polystyrene dish surface were incubated at 4 °C for 1 h with 1 μM biotinylated G-actin in G-buffer containing 0.00025% biotin to achieve the low G-actin coating density required for single-bond measurements. For G-actin/F-actin interactions (Fig. 1A, Right), the cantilever tip was functionalized in the same way, but the polystyrene surface was functionalized with sonicated F-actin instead of actin monomer. To prepare the sonicated F-actin, 4 μM of G-actin (biotinylated actin:nonmodified actin = 1:20 unless otherwise stated) was incubated in F-buffer (G-buffer + 50 mM KCl, 2 mM MgCl2, 1 mM ATP) for 15 min at room temperature and then sonicated for 1 min (5 s on, 5 s off), followed by two more cycles of 15-min incubation + 1-min sonication (Branson 1510). After the last sonication, the F-actin was immediately applied to the biotin/streptavidin-treated polystyrene surface with 0.00025% biotin. To measure interactions between actins, the G-buffer was replaced with F buffer containing 0.00025% biotin to block any nonsaturated binding sites of streptavidin. To measure biotin/streptavidin interactions (Fig. 1A, Center), only the polystyrene surface was further incubated with streptavidin after absorption with biotinylated BSA, and the working solution was F-buffer without soluble biotin.
The AFM was functionalized with an appropriate actin density to maintain a low binding frequency of ∼20% (Fig. 1C) to ensure ≥89% probability of single-bond formation as predicted by Poisson statistics (43). Only single-step dissociations were analyzed, to ensure that the lifetimes measured were time-to-dissociation of single bonds.
SMD Simulations.
The models for long- and short-pitch actin dimers and actin 14-mer filaments charged with ADP or ATP were constructed by using a recently developed F-actin model (44) as a template and taking the relevant G-actin structures into account (5, 45). The constructed models were equilibrated and the final structures were used for SMD simulations. In the simulations for the actin dimer, the Cα atom of a surface Lys residue of one actin monomer was harmonically constrained and a similar atom on the other monomer was pulled. For the simulations of the actin filament, the actin monomer subunit at the barbed or pointed end was pulled while the two subunits at the other end were constrained.
Similar to other SMD studies and limited by computational resources, the simulations here were performed under much larger forces and a much shorter timescale (18, 46, 47). Therefore, the SMD results should be interpreted only qualitatively rather than quantitatively by directly comparing the times and forces between simulations and experiments.
All methods are detailed in SI Methods.
Supplementary Material
Acknowledgments
We thank V. M. Fowler for reagents and F. Kong for help with AFM. This work was supported by National Institutes of Health Grants HL18672 and HL70537 (to L.V.M.); HL091020, HL093723, AI077343, and AI044902 (to C.Z.); AR48615 (to S.O.); and DC8803 (to P.A.R.); and by National Natural Science Foundation of China Grants 31070827, 31222022, and 81161120424 (to J.L.). The computational resources for the SMD simulations were provided by National Science Foundation Teragrid Large Resource Allocations Committee Grant MCA08X014 (to C.Z.) and by the Supercomputing Center of Chinese Academy of Sciences and National Supercomputing Center Tianjin Center (J.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. P.A.J. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1218407110/-/DCSupplemental.
References
- 1.Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112(4):453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- 2.Pollard TD, Cooper JA. Actin, a central player in cell shape and movement. Science. 2009;326(5957):1208–1212. doi: 10.1126/science.1175862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Geiger B, Spatz JP, Bershadsky AD. Environmental sensing through focal adhesions. Nat Rev Mol Cell Biol. 2009;10(1):21–33. doi: 10.1038/nrm2593. [DOI] [PubMed] [Google Scholar]
- 4.Parsons JT, Horwitz AR, Schwartz MA. Cell adhesion: Integrating cytoskeletal dynamics and cellular tension. Nat Rev Mol Cell Biol. 2010;11(9):633–643. doi: 10.1038/nrm2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holmes KC, Popp D, Gebhard W, Kabsch W. Atomic model of the actin filament. Nature. 1990;347(6288):44–49. doi: 10.1038/347044a0. [DOI] [PubMed] [Google Scholar]
- 6.Kaunas R, Usami S, Chien S. Regulation of stretch-induced JNK activation by stress fiber orientation. Cell Signal. 2006;18(11):1924–1931. doi: 10.1016/j.cellsig.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 7.Chien S. Mechanotransduction and endothelial cell homeostasis: The wisdom of the cell. Am J Physiol Heart Circ Physiol. 2007;292(3):H1209–H1224. doi: 10.1152/ajpheart.01047.2006. [DOI] [PubMed] [Google Scholar]
- 8.Galbraith CG, Sheetz MP. Forces on adhesive contacts affect cell function. Curr Opin Cell Biol. 1998;10(5):566–571. doi: 10.1016/s0955-0674(98)80030-6. [DOI] [PubMed] [Google Scholar]
- 9.Riveline D, et al. Focal contacts as mechanosensors: Externally applied local mechanical force induces growth of focal contacts by an mDia1-dependent and ROCK-independent mechanism. J Cell Biol. 2001;153(6):1175–1186. doi: 10.1083/jcb.153.6.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maniotis AJ, Chen CS, Ingber DE. Demonstration of mechanical connections between integrins, cytoskeletal filaments, and nucleoplasm that stabilize nuclear structure. Proc Natl Acad Sci USA. 1997;94(3):849–854. doi: 10.1073/pnas.94.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaunas R, Nguyen P, Usami S, Chien S. Cooperative effects of Rho and mechanical stretch on stress fiber organization. Proc Natl Acad Sci USA. 2005;102(44):15895–15900. doi: 10.1073/pnas.0506041102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kolega J. Effects of mechanical tension on protrusive activity and microfilament and intermediate filament organization in an epidermal epithelium moving in culture. J Cell Biol. 1986;102(4):1400–1411. doi: 10.1083/jcb.102.4.1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Watanabe N, Kato T, Fujita A, Ishizaki T, Narumiya S. Cooperation between mDia1 and ROCK in Rho-induced actin reorganization. Nat Cell Biol. 1999;1(3):136–143. doi: 10.1038/11056. [DOI] [PubMed] [Google Scholar]
- 14.Hirata H, Tatsumi H, Sokabe M. Dynamics of actin filaments during tension-dependent formation of actin bundles. Biochim Biophys Acta. 2007;1770(8):1115–1127. doi: 10.1016/j.bbagen.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 15.Yeung T, et al. Effects of substrate stiffness on cell morphology, cytoskeletal structure, and adhesion. Cell Motil Cytoskeleton. 2005;60(1):24–34. doi: 10.1002/cm.20041. [DOI] [PubMed] [Google Scholar]
- 16.Discher DE, Janmey P, Wang YL. Tissue cells feel and respond to the stiffness of their substrate. Science. 2005;310(5751):1139–1143. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- 17.Marshall BT, et al. Direct observation of catch bonds involving cell-adhesion molecules. Nature. 2003;423(6936):190–193. doi: 10.1038/nature01605. [DOI] [PubMed] [Google Scholar]
- 18.Yago T, et al. Platelet glycoprotein Ibalpha forms catch bonds with human WT vWF but not with type 2B von Willebrand disease vWF. J Clin Invest. 2008;118(9):3195–3207. doi: 10.1172/JCI35754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo B, Guilford WH. Mechanics of actomyosin bonds in different nucleotide states are tuned to muscle contraction. Proc Natl Acad Sci USA. 2006;103(26):9844–9849. doi: 10.1073/pnas.0601255103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kong F, García AJ, Mould AP, Humphries MJ, Zhu C. Demonstration of catch bonds between an integrin and its ligand. J Cell Biol. 2009;185(7):1275–1284. doi: 10.1083/jcb.200810002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thomas W, et al. Catch-bond model derived from allostery explains force-activated bacterial adhesion. Biophys J. 2006;90(3):753–764. doi: 10.1529/biophysj.105.066548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Akiyoshi B, et al. Tension directly stabilizes reconstituted kinetochore-microtubule attachments. Nature. 2010;468(7323):576–579. doi: 10.1038/nature09594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Le Trong I, et al. Structural basis for mechanical force regulation of the adhesin FimH via finger trap-like beta sheet twisting. Cell. 2010;141(4):645–655. doi: 10.1016/j.cell.2010.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen W, Lou J, Zhu C. Forcing switch from short- to intermediate- and long-lived states of the alphaA domain generates LFA-1/ICAM-1 catch bonds. J Biol Chem. 2010;285(46):35967–35978. doi: 10.1074/jbc.M110.155770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lou J, Zhu C. A structure-based sliding-rebinding mechanism for catch bonds. Biophys J. 2007;92(5):1471–1485. doi: 10.1529/biophysj.106.097048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sarangapani KK, Marshall BT, McEver RP, Zhu C. Molecular stiffness of selectins. J Biol Chem. 2011;286(11):9567–9576. doi: 10.1074/jbc.M110.196485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marshall BT, et al. Measuring molecular elasticity by atomic force microscope cantilever fluctuations. Biophys J. 2006;90(2):681–692. doi: 10.1529/biophysj.105.061010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kishino A, Yanagida T. Force measurements by micromanipulation of a single actin filament by glass needles. Nature. 1988;334(6177):74–76. doi: 10.1038/334074a0. [DOI] [PubMed] [Google Scholar]
- 29.Sept D, McCammon JA. Thermodynamics and kinetics of actin filament nucleation. Biophys J. 2001;81(2):667–674. doi: 10.1016/S0006-3495(01)75731-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zigmond SH, et al. Formin leaky cap allows elongation in the presence of tight capping proteins. Curr Biol. 2003;13(20):1820–1823. doi: 10.1016/j.cub.2003.09.057. [DOI] [PubMed] [Google Scholar]
- 31.Fischer RS, Fritz-Six KL, Fowler VM. Pointed-end capping by tropomodulin3 negatively regulates endothelial cell motility. J Cell Biol. 2003;161(2):371–380. doi: 10.1083/jcb.200209057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rossier OM, et al. Force generated by actomyosin contraction builds bridges between adhesive contacts. EMBO J. 2010;29(6):1055–1068. doi: 10.1038/emboj.2010.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Butler B, Gao C, Mersich AT, Blystone SD. Purified integrin adhesion complexes exhibit actin-polymerization activity. Curr Biol. 2006;16(3):242–251. doi: 10.1016/j.cub.2005.12.033. [DOI] [PubMed] [Google Scholar]
- 34.Hotulainen P, Lappalainen P. Stress fibers are generated by two distinct actin assembly mechanisms in motile cells. J Cell Biol. 2006;173(3):383–394. doi: 10.1083/jcb.200511093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goffin JM, et al. Focal adhesion size controls tension-dependent recruitment of alpha-smooth muscle actin to stress fibers. J Cell Biol. 2006;172(2):259–268. doi: 10.1083/jcb.200506179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parekh SH, Chaudhuri O, Theriot JA, Fletcher DA. Loading history determines the velocity of actin-network growth. Nat Cell Biol. 2005;7(12):1219–1223. doi: 10.1038/ncb1336. [DOI] [PubMed] [Google Scholar]
- 37.Bershadsky A, Kozlov M, Geiger B. Adhesion-mediated mechanosensitivity: A time to experiment, and a time to theorize. Curr Opin Cell Biol. 2006;18(5):472–481. doi: 10.1016/j.ceb.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 38.Geiger B, Bershadsky A. Exploring the neighborhood: Adhesion-coupled cell mechanosensors. Cell. 2002;110(2):139–142. doi: 10.1016/s0092-8674(02)00831-0. [DOI] [PubMed] [Google Scholar]
- 39.Moore SW, Roca-Cusachs P, Sheetz MP. Stretchy proteins on stretchy substrates: The important elements of integrin-mediated rigidity sensing. Dev Cell. 2010;19(2):194–206. doi: 10.1016/j.devcel.2010.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Feng JJ, et al. Direct visualisation and kinetic analysis of normal and nemaline myopathy actin polymerisation using total internal reflection microscopy. J Muscle Res Cell Motil. 2009;30(1–2):85–92. doi: 10.1007/s10974-009-9178-9. [DOI] [PubMed] [Google Scholar]
- 41.Laing NG, et al. Mutations and polymorphisms of the skeletal muscle alpha-actin gene (ACTA1) Hum Mutat. 2009;30(9):1267–1277. doi: 10.1002/humu.21059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sanoudou D, Beggs AH. Clinical and genetic heterogeneity in nemaline myopathy: A disease of skeletal muscle thin filaments. Trends Mol Med. 2001;7(8):362–368. doi: 10.1016/s1471-4914(01)02089-5. [DOI] [PubMed] [Google Scholar]
- 43.Zhu C, Long M, Chesla SE, Bongrand P. Measuring receptor/ligand interaction at the single-bond level: Experimental and interpretative issues. Ann Biomed Eng. 2002;30(3):305–314. doi: 10.1114/1.1467923. [DOI] [PubMed] [Google Scholar]
- 44.Oda T, Iwasa M, Aihara T, Maéda Y, Narita A. The nature of the globular- to fibrous-actin transition. Nature. 2009;457(7228):441–445. doi: 10.1038/nature07685. [DOI] [PubMed] [Google Scholar]
- 45.Graceffa P, Dominguez R. Crystal structure of monomeric actin in the ATP state. Structural basis of nucleotide-dependent actin dynamics. J Biol Chem. 2003;278(36):34172–34180. doi: 10.1074/jbc.M303689200. [DOI] [PubMed] [Google Scholar]
- 46.Lou J, et al. Flow-enhanced adhesion regulated by a selectin interdomain hinge. J Cell Biol. 2006;174(7):1107–1117. doi: 10.1083/jcb.200606056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sotomayor M, Schulten K. Single-molecule experiments in vitro and in silico. Science. 2007;316(5828):1144–1148. doi: 10.1126/science.1137591. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.