Abstract
Throughout their life cycle, plants produce new organs, such as leaves, flowers, and lateral roots. Organs that have served their purpose may be shed after breakdown of primary cell walls between adjacent cell files at the site of detachment. In Arabidopsis, floral organs abscise after pollination, and this cell separation event is controlled by the peptide INFLORESCENCE DEFICIENT IN ABSCISSION (IDA), which signals through the leucine-rich repeat receptor-like kinases HAESA (HAE) and HAESA-LIKE2 (HSL2). Emergence of new lateral root primordia, initiated deep inside the root under the influence of auxin, is similarly dependent on cell wall dissolution between cells in the overlaying endodermal, cortical, and epidermal tissues. Here we show that this process requires IDA, HAE, and HSL2. Mutation in these genes constrains the passage of the growing lateral root primordia through the overlaying layers, resulting in altered shapes of the lateral root primordia and of the overlaying cells. The HAE and HSL2 receptors are redundant in function during floral organ abscission, but during lateral root emergence they are differentially involved in regulating cell wall remodeling genes. In the root, IDA is strongly auxin-inducible and dependent on key regulators of lateral root emergence—the auxin influx carrier LIKE AUX1-3 and AUXIN RESPONSE FACTOR7. The expression levels of the receptor genes are only transiently induced by auxin, suggesting they are limiting factors for cell separation. We conclude that elements of the same cell separation signaling module have been adapted to function in different developmental programs.
Keywords: root development, peptide signaling, pectin degradation, polygalacturonases, propidium iodide
More than 200 genes encoding leucine-rich repeat receptor-like kinases (1) and more than 1,000 genes encoding putative secreted peptides have been identified in Arabidopsis thaliana (2), suggesting that peptide ligand–receptor interactions are important for cell-to-cell communication in plants. However, fewer than a dozen signaling modules, including INFLORESCENCE DEFICIENT IN ABSCISSION (IDA)-HAESA (HAE)/HAESA-LIKE2 (HSL2) controlling the separation step of floral organ abscission, have been identified by genetic and/or biochemical methods (3). Both the ida mutant and the hae hsl2 double mutant retain their floral organs indefinitely owing to lack of breakdown of the middle lamella between cell layers of the abscission zone (AZ) at the base of organs to be shed (4–6). IDA is expressed in the AZ region of the flowers (4), whereas both HAE and HSL2 expression is confined specifically to the specialized AZ cells (5, 6). Overexpression of IDA leads to premature and ectopic abscission, but not in hae hsl2 mutant background, consistent with IDA being the ligand of these receptors (5, 6).
The abscission process involves initial cell wall loosening by enzymes like xyloglucan endotransglucosylase/hydrolases (XTHs) and expansins (EXPs) (7, 8). The loosening facilitates the access of cell wall degradation enzymes like polygalacturonases (PGs) that hydrolyze pectins, which are major components of plant cell walls (7, 8). Mutational analyses indicate that PGs are of particular importance in cell separation events (8). Microarray data suggest that the IDA-HAE/HSL2 signaling module is involved in the regulation of cell wall remodeling (CWR) genes (7). However, XTHs, EXPs, and PGs are expressed not only in the floral AZs but also at other sites of cell wall remodeling and breakdown, including the dehiscence zone (DZ) of the siliques, which undergo cell separation to allow seed shedding, and the cells overlaying emerging lateral roots (7–10). IDA and HAE are also found expressed in DZs of mature siliques (11). We therefore hypothesized that IDA-HAE/HSL2 signaling might control other cell-separation processes than just floral organ abscission.
Here we demonstrate that the IDA-HAE/HSL2 signaling module is important for lateral root emergence (LRE), a process mainly regulated by auxin accumulating in the central cells of early-stage lateral root primordia (LRP) and later at the tip of the LRP (12). We show that IDA-HAE/HSL2 is part of the genetic network regulated by auxin, where IDA and HAE control expression of PGs and pectin degradation in the overlaying endodermal (EN), cortical (CO), and epidermal (EP) cells. In single mutants as well as the hae hsl2 double mutant, morphogenesis of the LRP is altered owing to constraints imposed by the overlaying layers. Hence, the IDA-HAE/HSL2 signaling module has been adapted to function in different root and shoot cell separation processes.
Results
IDA, HAE, and HSL2 Are Required for Normal Progression of LRP Through the Overlaying Layers.
The involvement of the IDA peptide and the HAE and HSL2 receptor-like kinases during LRE was initially indicated by significant lower densities of emerged LRs for the ida mutant [ida-2 in Col-0 background (5)] and the hae hsl2 double mutant compared with WT (ecotype Col-0) (Fig. 1A and Fig. S1A). The single mutants of the receptor genes show no abscission defect (5, 6), but interestingly both showed a lateral root (LR) phenotype with significantly reduced LR density (Fig. S1 A and B), suggesting that they influence LR development in different ways.
Fig. 1.
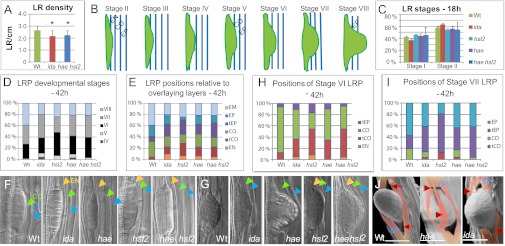
Mutations in IDA, HAE, and HSL2 delay LR emergence. (A) LR density (number of LRs per cm root) for ida (n = 21), hae hsl2 (n = 22), and WT plants (n = 20) 12 d after stratification. *Significant deviation from WT based on Student t test. (B) Schematic presentation of the stages of LR development. (C) Percentage of LRP at stages I and II for ida (n = 68), hae (n = 68), hsl2 (n = 81), hae hsl2 (n = 81), and WT plants (n = 69), 18 h after LR induction. (D) Percentage of LRP at stages IV to VIII (emerged) for ida (n = 68), hae (n = 59), hsl2 (n = 58), hae hsl2 (n = 56), and WT plants (n = 62), 42 h after LR induction. (E) Percentage of LRP with tips positioned in the overlaying EN, CO, and EP tissues or touching the CO or EP layer (tCO and tEP, respectively) for WT and mutants, as indicated. EM, emerged LRs. Numbers as in D. (F and G) WT and mutant LRP, as indicated, at stage V (F) and stage VI to VII (G). Overlaying EN, CO, and EP cells are indicated in orange, green, and blue arrowheads, respectively. (H) Percentage of stage VI LRP with tips positioned in the overlaying EN, CO, and EP tissues or touching the CO or EP layer (tCO and tEP, respectively) for WT and mutants, as indicated. (I) Percentage of stage VII LRP with tips positioned in the overlaying CO and EP tissues or touching the CO or EP layer (tCO and tEP, respectively) for WT and mutants, as indicated. (J) Cryo SEM images of emerged WT, hae, and ida LRs. The EP cells on each side of the LR have been colored. Arrows, symmetrical openings between EP cells in WT. Arrowheads, rifts in flattened and ruptured EP cells of mutant. (Scale bar: 50 μM.)
To monitor whether reduced LR density reflected differences in rates of LRP initiation and/or emergence, we used an assay exploiting the ability of a root gravitropic response to induce LRP organogenesis (13–17). Before emergence (stage VIII), LRP go through seven developmental stages, defined by the number of cell layers, and have to pass through overlaying layers of EN, CO, and EP cells (Fig. 1B) (18). Eighteen hours after gravitropic induction of LR development approximately 40% of LRP were at stage I and the remaining 60% at stage II in both WT and mutants (Fig. 1C), indicating that LR initiation was not affected. However, after 42 hours, 40% of the WT but only 20–24% of mutant LRs had emerged, with the highest percentage in hsl2 (Fig. 1D). More stage IV and V LRP were present in mutant lines; stage VII was in particular overrepresented for ida and hae, stage VI for hsl2, and stages VI and VII for hae hsl2, compared with WT (Fig. 1D). For all of the mutants the distribution of stages deviated significantly from a distribution expected if the mutants equaled WT distribution (P < 0.03). As anticipated if IDA and the two receptors were functioning in the same pathway, the ida and hae hsl2 distributions of LRP stages were not significantly different (P = 0.2), whereas the hsl2 distribution deviated significantly from that of ida (P = 0.04).
The size and number of cells of LRP at a given stage varies in the WT, but the number of cell layers and the shape of the LRP are characteristic for each stage [reported in companion article by Lucas et al. (19)]. The delayed emergence in the mutants suggested that defects in the IDA-HAE/HSL2 module delayed the passage of LRP through EN, CO, and/or EP layers. This could be due to restricted growth of the LRP and/or to constraints imposed by the overlaying tissues. Therefore, we registered the position of each LRP tip relative to the overlaying layers, a relationship that has been described as very consistent for WT LRP (Fig. 1B) (18, 20). The distribution of positions of the mutant LRP deviated highly significantly from WT distribution (P < 10−10) (Fig. 1E). The passage of the LRP from EN to CO involves the breaking of the Casparian strip, a lignified network forming an impermeable barrier between the EN and the outer layers (21, 22), and WT LRP stage IV–V changes from “flat-topped” to “dome-shaped” (compare LRP in Fig. 1 F vs. G) (19). In the mutants more and older-staged LRP seemed to be trapped in the EN layer, displaying an extraordinary flattened shape (Fig. 1 E and F). In accordance with previous observations (18), WT stage VI primordia were predominantly (>80%) positioned in the CO, whereas in the mutants a substantial fraction of stage VI LRP were only touching the CO (tCO) and had not yet penetrated the CO layer (Fig. 1H). At stage VII the majority of the WT LRP had, as expected, entered the EP layer, whereas most mutant stage VII LRP were only touching this layer (tEP) (Fig. 1I), often pushing intact overlaying cell files outwards (Fig. 1G). Additionally, mutant LRP with the size and shape of just-emerged WT LRs were observed covered with one or two cell layers of primary root tissues (Fig. S2A).
As for the distribution of stages, the ida and hae hsl2 LRP distribution of positions were statistically equivalent. The distribution of positions of hsl2 stage VI and VII LRP deviated most from the WT and the other mutants (Fig. 1 H and I). However, the distribution of stages and positions in the hae hsl2 double mutant was compatible with an additive effect of the two receptor single mutants: the double mutant displayed a percentage of emerged roots similar to that in hae and lower than that in hsl2 (Fig. 1D), and the percentage of stage VI LRP not yet positioned in the CO layer was similar to that in hsl2 and higher than that for hae (Fig. 1H).
We concluded that the three genes encoding the IDA-HAE/HSL2 signaling module were required for normal progression of LRP through the overlaying layers. Cryo scanning electron microscopy (SEM) allowed a more detailed inspection of emerged LRP. In the WT, EP cells on each side of emerged LRs were nicely rounded and curved like a pair of lips, with triangular equal sized openings between the separated cells above and below the emerged WT LR (Fig. 1J), as previously shown with environmental SEM (23). In contrast, in 9 of 10 ida and hae mutant roots, these openings, if present, were of unequal size, and the cells surrounding emerged roots appeared as flattened and torn sheets clinging to the LR itself (Fig. 1J and Fig. S2B). In the most affected cases no normal opening was present at all (Fig. 1J and Fig. S2B), indicating LRP penetration of cells impeded in cell separation.
IDA, HAE, and HSL2 Belong to an Auxin-Dependent Network of Genes Expressed in Cells That Separate During LRE.
The expression patterns of IDA, HAE, and HSL2 during LRE were investigated using promoter–reporter gene constructs. In the root, other members of the IDA gene family are mainly expressed in the vasculature (Fig. S3A). In contrast, expression of the IDA promoter β-glucuronidase (GUS) reporter construct pIDA:GUS (4) was detected in cells overlaying new LRP (Fig. 2A and Fig. S3B), initially in single EN cells overlaying stage I–IV LRP. Subsequently, pIDA:GUS expression was observed in the CO layer before the entrance of the LRP. Likewise, pIDA:GUS was active in the EP from stages IV to V before the LRP had entered this layer. Moreover, the GUS signal persisted in these cells after LRE (stage VIII) (Fig. 2A).
Fig. 2.
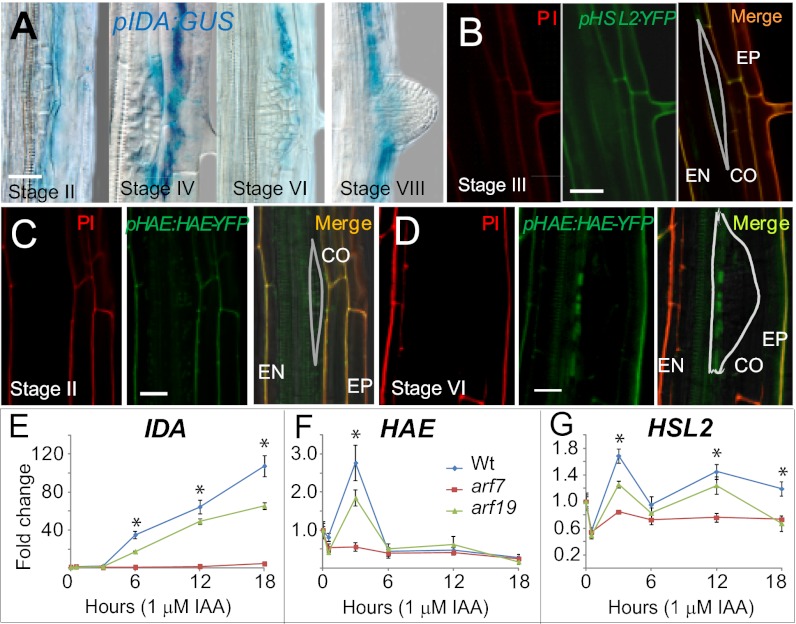
IDA, HAE, and HSL2 are expressed in cells overlaying new LRP and induced by auxin. (A) pIDA:GUS expression shown for the stages indicated. (B) Confocal images of pHSL2:YFP expression at stage III. (Left) PI-stained root; (Center) YFP signal; (Right) merge. The outline of the LRP is shown. Note that PI does not penetrate the EN or the LRP and therefore does not stain the inner EN wall and LR cell walls. (C and D) Confocal images of pHAE:HAE-YFP expression in hae hsl2 mutant (stage II) and WT background (stage IV), respectively. Note that the pHAE:HAE-YFP construct rescued the hae hsl2 mutants phenotypes (Fig. S5 A and B). Annotation as in B. (E–G) Relative expression levels of IDA, HAE, and HSL2 in roots in the presence of 1 μM IAA in WT, arf7, and arf19 background determined by RT-qPCR. The WT expression level before IAA addition was set to 1. SDs are shown. *Significant differences between WT and both mutants (P < 0.05).
Plants transformed with a promoter:YFP construct for HSL2 (pHSL2:YFP) revealed expression in the EN, CO, and EP from stage II to III, and a weak expression in the LRP (Fig. 2B and Fig. S4A). The expression in the LRP increased from stage IV to stage VI and further in emerged LR (Fig. S4 B–D). Interestingly, at the same time expression was reduced in separated CO and EP cells surrounding the emerging LRP (Fig. S4 C and D). After emergence, prominent expression was seen in the tip of the LR, similar to the expression pattern of pHSL2:YFP in the primary root tip (Fig. S4 E and F). Thus, the late-stage LRP expression could be related to a subsequent function in the tip of the emerged root.
HAE expression was investigated using a functional translational YFP fusion construct under the control of the HAE promoter (pHAE:HAE-YFP). In agreement with the hypothesized involvement of HAE in IDA-dependent cell separation, HAE-YFP was detected in cells overlaying LRP from stage II with stronger expression in the epidermis at later stages (Fig. 2 C and D). From stage VI an intensified HAE-YFP signal was found at the base of the LR (Fig. 2D).
LRP outgrowth and cell separation in overlaying tissues are coordinated by auxin derived from the tip of the primordium reprogramming EN, CO, and EP cells. In the overlaying CO and EP cells, the auxin influx carrier LIKE AUX1-3 (LAX3) is induced, resulting in preferential auxin accumulation (10). Auxin-induced degradation of the SOLITARY ROOT1 (SLR1)/IAA14 repressor releases the AUXIN RESPONSE FACTOR (ARF) 7 and ARF19 transcription factors, leading to the induction of CWR gene expression (10, 24). We have investigated the relation between IDA and this auxin-controlled gene regulatory network. pIDA:GUS expression was first observed at stage IV in lax3 mutant background and was clearly delayed compared with WT (Fig. S3B). Thus, IDA expression in the root seems dependent on auxin. Consistent with this, the pIDA:GUS transgene was activated in the root by exogenous indole-3-acetic acid (IAA) or naphthaleneacetic acid (NAA) (1 µM) (Fig. S3C), and real-time quantitative RT-PCR (RT-qPCR) demonstrated a more than 100-fold increase in IDA expression level from 6 to 18 h after addition of IAA (Fig. 2E). In the arf19 background the induction level was significantly reduced, whereas in the arf7 mutant normal IDA induction was blocked. HAE and HSL2 induction by IAA was only transient and weak, especially for HSL2; however, the expression peaks after 3 h were strongly dependent on ARF7 and also significantly reduced in arf19 background (Fig. 2 F and G). We conclude that the expression patterns of IDA, HAE, and HSL2 and the strong ARF7-dependent IDA induction by auxin are consistent with a role for IDA signaling through these receptors in the process of LRE.
IDA and HAE Are Involved in Control of Pectin Degradation.
The pectin-rich middle lamella at the interphase of adjacent plant cells must be broken down for cell separation to occur. We developed an assay for pectin breakdown based on propidium iodide (PI), commonly used to stain root cell walls. PI binds the negatively charged carboxyl and hydroxyl groups on homogalacturonans, which constitute a major component of pectic polysaccharides in the middle lamella, and it was recently demonstrated that cell wall degradation by pectinase reduces PI fluorescence (25). In the WT, reduced PI staining of cell walls between EN and CO cells directly overlaying LRP could be observed already at stage I (Fig. 3 A and B), consistent with an expected pectin dissolution during cell separation (7). Interestingly, diminished PI fluorescence was also seen in hsl2 roots (Fig. 3C), indicating that the pectin degradation occurred as in the WT. In contrast, the PI intensity of the EN/CO cell wall in front of the primordia in the ida, hae, and hae hsl2 mutants was equal to the intensity at positions above and below the LRP (Fig. 3 D–G), suggesting a defect in pectin degradation in these mutants. Three-dimensional images indicated that the pectin component of the middle lamella between the EN and CO layers overlaying the hae, ida, and hae hsl2 LRP was intact (Fig. 3E and Fig. S5 A and C). The pHAE:HAE-YFP construct rescued this hae hsl2 phenotype as well as the abscission defect (Fig. S5 A and B). Likewise, introduction of the WT IDA gene into ida background (4) reduced PI staining to WT levels (Fig. S5C). Taken together, the results of the PI assay suggest that IDA regulates LR emergence by signaling through the HAE receptor to control breakdown of cell wall components such as pectin.
Fig. 3.
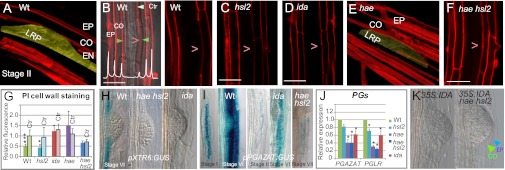
IDA, HAE, and HSL2 influence cell wall remodeling and degradation. (A) 3D Z-stack confocal image of PI-stained WT root at the site of a stage I WT LR. The LRP has been colored yellow. (B) 2D confocal image showing scan (white graph) of the fluorescence from PI-stained EN, CO, and EP cell walls at the site of WT LRP (green arrowheads). Control scans were taken below and above the LRP (light green arrowhead). >, position of LRP. (C–F) Confocal images of PI-stained roots at the site of a stage I LRs for hsl2 (2D), ida (2D), and hae (3D, yellow colored LRP) and hae hsl2 (2D). (G) PI fluorescence of LRP-overlaying cell walls between EN and CO relative to the reference point at the opposite side based on scans for six to nine roots. Bars indicate SD. Asterisks denote significant differences from fluorescence at control positions relative to the same reference point (Ctr) (*P < 0.05; ***P < 0.001, Student t test). (H and I) Expression of pXTR6:GUS and pPGAZAT:GUS and in WT, ida, and hae hsl2 background at the LR stages indicated. (J) Expression levels of PGAZAT and PGLR in mutant 7-d-old roots relative to WT determined by RT-qPCR. Bars indicate SD. *Significant deviation from WT (P < 0.05). (K) Stage VII LRP of 35S:IDA and 35S:IDA hae hsl2, as indicated. Arrowheads, cell walls of overlaying CO and EP cells. (Scale bars: 30 μM in 3D images, 20 μM in 2D images.)
IDA, HAE, and HSL2 Regulate Expression of CWR Genes.
Because the mutant phenotypes indicated deficiencies in cell wall degradation, we investigated the expression levels of representative members of the XTH, EXP, and PG gene families encoding CWR enzymes expressed in cell layers overlaying LRP [i.e., XTH 23/XYLOGLUCAN ENDOTRANSGLYCOSYLASE6 (XTR6), EXP17, PG LATERAL ROOT (PGLR), and PG ABSCISSION ZONE ARABIDOPSIS THALIANA (PGAZAT)] (9, 10, 24, 26). The latter, also called ARABIDOPSIS DEHISCENCE ZONE POLYGALACTURONASE2 (ADPG2), is regulated by IDA in the AZ and implicated in cell separation during dehiscence and floral organ abscission (8, 26).
The involvement of IDA, HAE, and HSL2 in control of expression of these CWR genes was demonstrated by reduced transcript levels and delayed expression of promoter:GUS reporter constructs in ida and hae hsl2 mutant background (Fig. 3 H–J and Fig. S6 A–C). In the WT, pXTR6:GUS expression was detected in LRP-overlaying tissues (10) and additionally in EN cells neighboring primordia and later at the base of LRP (Fig. 3G and Fig. S6B), where both HAE and HSL2 are expressed (Fig. 2 B–D). RT-qPCR, used to investigate the role of each of the receptor genes, revealed that the receptors were functionally redundant in regulating XTR6 and also EXP17, because substantial reduction in transcript levels was seen in the hae hsl2 double mutant but not single mutants (Fig. S6A). In contrast, the PGAZAT and PGLR levels were very similar in hae and hae hsl2 background (Fig. 3J), indicating that the expression levels of these two genes are influenced mainly by IDA signaling through the HAE receptor. Interestingly, pXTR6:GUS, pPGLR:GUS, and pPGAZAT:GUS expression was controlled by IDA-HAE/HSL2 during floral organ abscission (Fig. S6 B–D) (26).
Plant lines overexpressing IDA using the 35SCaMV promoter (35S:IDA) in WT and hae hsl2 background have been used to demonstrate that IDA signals through HAE and HSL2 in above-ground organs (6). These lines were used to substantiate that this is also the case in the root. 35S:IDA produce few seeds (6) with low germination rate, developmentally arrested seedlings, and restricted root growth and development, including very low production of LRs (Fig. S7A). Deficiencies in seed germination and root development were rescued in 35S:IDA hae hsl2 plants (Fig. S7B), which displayed the characteristic hae hsl2 LR phenotype with delayed penetration of the overlaying layers. This phenotype was not seen in the few lateral roots produced by 35S:IDA plants (Fig. 3I).
Discussion
In this work we have demonstrated that the IDA-HAE/HSL2 signaling module is required to facilitate the passage of LRP through overlaying root tissues. Lateral roots in Arabidopsis originate exclusively from pericycle cells located deep within the primary root tissues (18). As cells within new LRP divide, they push against overlaying EN, CO, and EP tissues (summarized in Fig. 4). Auxin originating from the tips of new LRP is likely to promote LR emergence by generating different chemical and mechanical signals (19) and reprogram overlaying cells to separate by controlling the expression of aquaporin water channels (16) and cell wall remodeling enzymes (10). We report that the peptide signal IDA and its receptor-like kinases HAE and HSL2 are important components in this auxin-regulated pathway by controlling cell separation. The reduced expression of PG genes, consistent with the persistently high fluorescence level in the PI assay in the ida, hae, and hae hsl2 mutants, argues for direct involvement of IDA and HAE in control of pectin degradation in walls of LRP overlaying cells. Moreover, where the receptors have overlapping expression patterns, they may both relay the IDA signal and be functionally redundant in controlling other CWR genes.
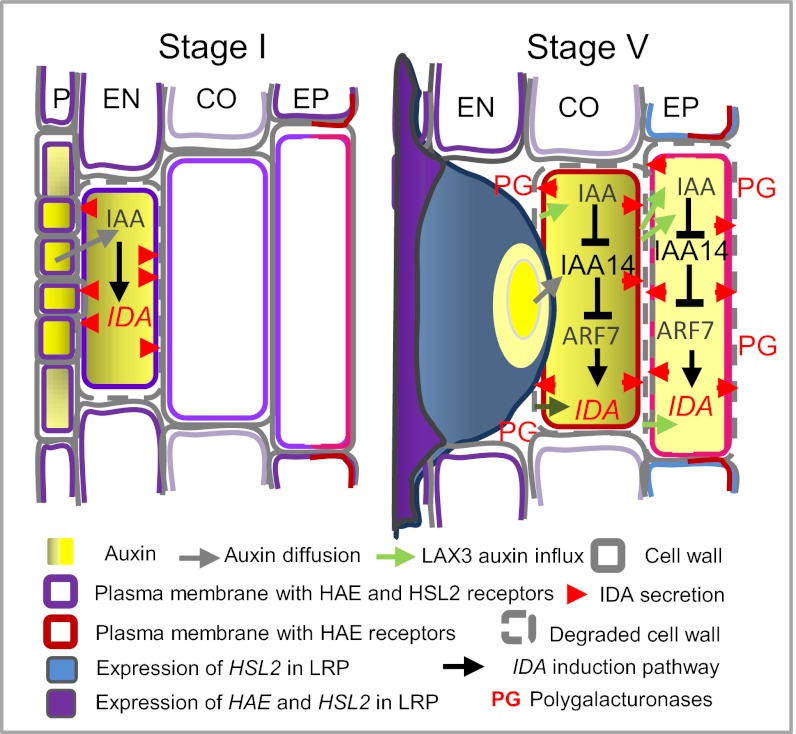
Fig. 4.
Model of IDA-HAE/HSL2 signaling in LR emergence. IDA expression and signaling to open a gateway for the LRP is hypothesized to occur in a wave-like fashion in the cells directly overlaying the developing LRPs. IDA is suggested induced in the EN by auxin diffusing from the young LRP and secreted to interact with HAE and HSL2 receptors already present, to trigger expression of CWR genes, leading to loosening and breakdown of the cell wall of the EN cells directly overlaying LRPs. Expression of IDA in the CO and EP overlaying the LRP is dependent on auxin influx via LAX3 and the auxin-dependent cascade leading to activation of ARF7, which also transiently induces expression of the receptor genes. HAE and HSL2 function redundantly to up-regulate some CWR genes like XTR6, for instance at the base of the LRP, whereas IDA signaling, mainly through the HAE receptor, in overlaying tissues will induce downstream genes encoding cell wall degradation enzymes like PGs, leading to cell separation.
We propose a model for auxin-IDA-HAE/HSL2 regulated LRP emergence based on the spatiotemporal distribution of these signaling components and their downstream targets (Fig. 4): at stages I and II, auxin derived from the LRP induces IDA expression in overlaying EN cells. Next, IDA signals through HAE and HSL2 receptors already present in the membrane of these cells, resulting in up-regulation of CWR genes like XTR6 and EXP17. Subsequently, LAX3-mediated auxin influx into overlaying CO and thereafter EP cells induces IDA expression and transiently increases the expression of the receptors in an ARF7-dependent manner. In the cells directly overlaying the LRP, IDA signaling through HAE triggers the expression of CWR enzymes like PGAZAT and PGLR that dissolve cell walls and open a gateway for the developing LR. Thus, the concerted but differential regulation of IDA, HAE, and HSL2 in the auxin-governed network of regulatory and enzymatic factors facilitates fine-tuned control of cell wall remodeling and cell wall degradation during LRE.
IDA, HAE, and HSL2 have previously been reported to regulate another cell separation process in Arabidopsis, floral organ abscission (4). Our analyses of expression patterns, mutant phenotypes, and genetic relationships allow a comparison of the role of IDA, HSL2, and HAE during both cell separation processes. In both cases, peptide signaling cross-talks with classic hormone signals. Interestingly, auxin has a promoting effect on IDA expression and LRE, but exogenous auxin commonly delays organ shedding and counteracts the abscission-promoting effect of ethylene (27, 28). AUXIN RESPONSE FACTORS, including ARF7 and ARF19, are involved in the timing of abscission (29), but it is not known whether auxin is involved in the regulation of IDA or its receptors in the floral organ AZs. This indicates organ-specific regulation of key components common to different cell separation processes.
Exogenous auxin induced IDA expression 100-fold in root tissues, but HAE and HSL2 mRNAs were only up-regulated transiently, suggesting that these receptors, and not the peptide, are the limiting factors controlling cell separation. This may explain why a general separation of cells is not observed at the used auxin concentration. With substantially higher concentrations, abnormal sloughing of root cells has been observed (30, 31). The HAE and HSL2 receptors are also limiting factors for above-ground organ shedding. Overexpression of IDA does not lead to general cell separation, only ectopic organ abscission at sites with vestigial AZ where HAE and HSL2 are found expressed (5, 6, 32, 33). Here we report that overexpression of IDA affects root growth, that this phenotype is dependent on functional HAE and HSL2 receptors, and that overexpression of IDA cannot override the delay in LRE of hae hsl2.
In floral organ AZs, HAE and HSL2 exhibit identical expression patterns overlapping with that of IDA, and single mutants display no aberrant phenotypes (5, 6). In the root, the patterns are more complex, because both single mutants show a phenotype (i.e., reduced LR density and delayed emergence). Differential expression patterns and mutant stage and position profiles indicate involvement of HAE and HSL2 in different aspects of the emergence process. Strong expression of pHSL2:YFP from stage VI and the particular accumulation of stage VI hsl2 LRP may indicate that HSL2 directly influences LRP growth. We cannot exclude that IDL genes expressed in the LRP, which in part can substitute for IDA function (6), may signal through HSL2.
The modest but significant difference in the distribution of LRP stages between WT and ida and the receptor mutants could also suggest that the IDA-HAE/HSL2 signaling module directly affects the development of the LRP. However, both receptors are expressed in the overlaying layers where IDA is expressed. Furthermore, the most significant difference between the WT and the mutants was the delayed passage through these layers, evident from the highly significant difference in distribution of LRP positions. This suggests that the primary effect of the mutations is a defect in the passaging of the LRP through these layers. The simplest explanation for this would be that the IDA peptide, expressed in the EN, CO, and EP, signals through the receptors present in the very same layers to induce CWR genes executing cell separation, consistent with the documented function of IDA-HAE/HSL2 in cell separation in the floral organ AZ (4–6). The delay in LRP development could be an effect of a feedback to the primordia, similar to mutants with increased or changed deposition of material to the Casparian strip of the endodermis that are delayed at different stages of the LRE process (19).
Materials and Methods
Plant Material and Constructs.
The arf7, arf19, lax3, ida-1, ida-2 (SALK_133209), hae, hsl2, and hae hsl2 mutants, pIDA:GUS, pXTR6:GUS, pPGLR:GUS, and pPGAZAT:GUS lines, and the 35S:IDA line have been described in refs. 6, 9, 10, and 34). Plants were cultivated in growth chambers at 22 °C for 8 h of dark and 16 h of light (100 μE·m−2·s−1). For root experiments sterilized seeds were plated on square plates with 0.5× MS medium (35), stratified 2–3 d in the dark at 4 °C, and thereafter placed vertically under the above conditions. For auxin treatment, seedlings were transferred 5 d after germination onto plates containing 1 µM auxin. Emerged LRs were counted 9 or 12 d after stratification. For staging of LR development from initiation to emergence, seedlings were grown for 10 d on vertical plates and then rotated 90° to induce LR formation (13–15). The χ2 test was used to test whether the distribution of stages and positions were statistically different between mutants and WT and among the mutants, using the WT, ida, or hae hsl2 distribution as alternative null hypotheses.
A single-locus pIDA:GUS line in ecotype C24 (11) was introgressed into Col-0 WT and the lax3 mutant in Col-0 background. The construct pHSL2:YFP with 2,300 bp upstream of the start codon in vector pHGY (RIKEN Plant Science Center), pHAE:HAE-YFP with 1,601 bp upstream region in vector pMDC111, were generated using Gateway technology.
RT-qPCR.
cDNA was prepared with SuperScript II reverse transcriptase (Invitrogen) from RNA isolated with Spectrum Plant Total RNA Kit or the RNeasy kit (Qiagen), and expression levels for selected genes were determined using LightCycler 480, using four replicates. Expression levels were normalized to At1G04850 and At5g18800 and analyzed for significant differences using REST 2009 Softeware (Qiagen).
Cryo SEM.
Five- to seven-day-old Arabidopsis seedlings were frozen in liquid nitrogen and analyzed with an accelerating voltage of 5–10 kV using a Hitachi S-4800 SEM microscope. Seven to ten roots were inspected per genotype. The “Gatan-CRYO chamber” sputter coated the nonsupplemented samples with paladium-gold. In Cryo SEM images EP cells neighboring the LRP were colored using Adobe Photoshop.
Staining Procedures.
GUS assays were performed and inspected using differential interference contrast optics as described previously (18, 36). Root cell walls were stained with 10 µM PI for YFP analyses and 20–30 µM PI for 30 s for analyses of cell wall degradation. PI fluorescence emission at 550–680 nm with 488-nm excitation was captured using an inverted LSM 710 META confocal microscope (Carl Zeiss) equipped with 40- and 63-fold Plan Apochromat oil immersion objectives (Carl Zeiss). The fluorescence signals at the point of highest fluorescence in front of stage I LRP (LR position), at the opposite side of the LRP (reference position), and below and above the LRP (control positions) were analyzed using Zen2011 (Zeiss) software. PI fluorescence for the overlaying cells and for the control positions was calculated relative to the reference fluorescence using 6–10 roots per genotype.
Primers.
The sequences of all primers used can be found in Table S1.
Accession Numbers.
Sequence data from the genes investigated can be found in the GenBank/European Molecular Biology Laboratory (EMBL) data libraries under accession numbers AT1G68765 (IDA), AT4G28490 (HAE), AT5G65710 (HSL2), AT5G14650 (PGLR), AT2G41850 (PGAZAT), AT4G25810 (XTR6), and AT4G01630 (EXP17).
Supplementary Material
Acknowledgments
We thank Roy Falleth and Solveig H. Engebretsen for technical assistance; Tara Holman [Centre for Plant Integrative Biology (CPIB)] for the reference genes for qPCR; Zinnia Gonzalez-Carranza for the pPGAZAT:GUS line; Mari Wildhagen for root length measurements; the Electron Microscopy Laboratory at the Department of Biosciences, University of Oslo, where the cryo SEM was performed; and the Confocal Laser Scanning Microscopy Core Facility at Oslo University Hospital (http://core.rr-research.no/index.php?section=141). This work was supported by Research Council of Norway Grants 175238/S10 (to C.-L.S. and R.B.A.) and 178049 (to M.A.B. and R.B.A.); Belgian Scientific Policy Contract (Belgian Arabidopsis Root Network) grant (to A.L. and M.J.B.); and by Biotechnology and Biological Sciences Research Council and Engineering and Physical Sciences Research Council funding to the CPIB (to A.L. and M.J.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1210835110/-/DCSupplemental.
References
- 1.Shiu SH, Bleecker AB. Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc Natl Acad Sci USA. 2001;98(19):10763–10768. doi: 10.1073/pnas.181141598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lease KA, Walker JC. The Arabidopsis unannotated secreted peptide database, a resource for plant peptidomics. Plant Physiol. 2006;142(3):831–838. doi: 10.1104/pp.106.086041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Butenko MA, Vie AK, Brembu T, Aalen RB, Bones AM. Plant peptides in signalling: Looking for new partners. Trends Plant Sci. 2009;14(5):255–263. doi: 10.1016/j.tplants.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 4.Butenko MA, et al. INFLORESCENCE DEFICIENT IN ABSCISSION controls floral organ abscission in Arabidopsis and identifies a novel family of putative ligands in plants. Plant Cell. 2003;15(10):2296–2307. doi: 10.1105/tpc.014365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cho SK, et al. Regulation of floral organ abscission in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2008;105(40):15629–15634. doi: 10.1073/pnas.0805539105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stenvik G-E, et al. The EPIP peptide of INFLORESCENCE DEFICIENT IN ABSCISSION is sufficient to induce abscission in Arabidopsis through the receptor-like kinases HAESA and HAESA-LIKE2. Plant Cell. 2008;20(7):1805–1817. doi: 10.1105/tpc.108.059139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cai S, Lashbrook CC. Stamen abscission zone transcriptome profiling reveals new candidates for abscission control: Enhanced retention of floral organs in transgenic plants overexpressing Arabidopsis ZINC FINGER PROTEIN2. Plant Physiol. 2008;146(3):1305–1321. doi: 10.1104/pp.107.110908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ogawa M, Kay P, Wilson S, Swain SM. ARABIDOPSIS DEHISCENCE ZONE POLYGALACTURONASE1 (ADPG1), ADPG2, and QUARTET2 are Polygalacturonases required for cell separation during reproductive development in Arabidopsis. Plant Cell. 2009;21(1):216–233. doi: 10.1105/tpc.108.063768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.González-Carranza ZH, Elliott KA, Roberts JA. Expression of polygalacturonases and evidence to support their role during cell separation processes in Arabidopsis thaliana. J Exp Bot. 2007;58(13):3719–3730. doi: 10.1093/jxb/erm222. [DOI] [PubMed] [Google Scholar]
- 10.Swarup K, et al. The auxin influx carrier LAX3 promotes lateral root emergence. Nat Cell Biol. 2008;10(8):946–954. doi: 10.1038/ncb1754. [DOI] [PubMed] [Google Scholar]
- 11.Butenko MA, et al. Ethylene dependent and -independent pathways controlling floral abscission are revelaed to converge using promoter:reporter gene constructs in the ida abscission mutant. J Exp Bot. 2006;58:3719–3730. doi: 10.1093/jxb/erl130. [DOI] [PubMed] [Google Scholar]
- 12.Péret B, et al. Arabidopsis lateral root development: An emerging story. Trends Plant Sci. 2009;14(7):399–408. doi: 10.1016/j.tplants.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 13.De Smet I, et al. Auxin-dependent regulation of lateral root positioning in the basal meristem of Arabidopsis. Development. 2007;134(4):681–690. doi: 10.1242/dev.02753. [DOI] [PubMed] [Google Scholar]
- 14.Lucas M, Guédon Y, Jay-Allemand C, Godin C, Laplaze L. An auxin transport-based model of root branching in Arabidopsis thaliana. PLoS ONE. 2008;3(11):e3673. doi: 10.1371/journal.pone.0003673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moreno-Risueno MA, et al. Oscillating gene expression determines competence for periodic Arabidopsis root branching. Science. 2010;329(5997):1306–1311. doi: 10.1126/science.1191937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Péret B, et al. Auxin regulates aquaporin function to facilitate lateral root emergence. Nat Cell Biol. 2012;14(10):991–998. doi: 10.1038/ncb2573. [DOI] [PubMed] [Google Scholar]
- 17.Péret B, et al. AUX/LAX genes encode a family of auxin influx transporters that perform distinct functions during Arabidopsis development. Plant Cell. 2012;24(7):2874–2885. doi: 10.1105/tpc.112.097766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Malamy JE, Benfey PN. Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development. 1997;124(1):33–44. doi: 10.1242/dev.124.1.33. [DOI] [PubMed] [Google Scholar]
- 19.Lucas M, et al. Lateral root morphogenesis is dependent on the mechanical properties of the overlaying tissues. Proc Natl Acad Sci USA. 2013;110:5229–5234. doi: 10.1073/pnas.1210807110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Casimiro I, et al. Dissecting Arabidopsis lateral root development. Trends Plant Sci. 2003;8(4):165–171. doi: 10.1016/S1360-1385(03)00051-7. [DOI] [PubMed] [Google Scholar]
- 21.Naseer S, et al. Casparian strip diffusion barrier in Arabidopsis is made of a lignin polymer without suberin. Proc Natl Acad Sci USA. 2012;109(25):10101–10106. doi: 10.1073/pnas.1205726109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roppolo D, et al. A novel protein family mediates Casparian strip formation in the endodermis. Nature. 2011;473(7347):380–383. doi: 10.1038/nature10070. [DOI] [PubMed] [Google Scholar]
- 23.Péret B, Larrieu A, Bennett MJ. Lateral root emergence: A difficult birth. J Exp Bot. 2009;60(13):3637–3643. doi: 10.1093/jxb/erp232. [DOI] [PubMed] [Google Scholar]
- 24.Laskowski M, Biller S, Stanley K, Kajstura T, Prusty R. Expression profiling of auxin-treated Arabidopsis roots: Toward a molecular analysis of lateral root emergence. Plant Cell Physiol. 2006;47(6):788–792. doi: 10.1093/pcp/pcj043. [DOI] [PubMed] [Google Scholar]
- 25.Rounds CM, Lubeck E, Hepler PK, Winship LJ. Propidium iodide competes with Ca(2+) to label pectin in pollen tubes and Arabidopsis root hairs. Plant Physiol. 2011;157(1):175–187. doi: 10.1104/pp.111.182196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.González-Carranza ZH, et al. A novel approach to dissect the abscission process in Arabidopsis. Plant Physiol. 2012;160(3):1342–1356. doi: 10.1104/pp.112.205955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aalen RB. Flower and floral organ abscission—control, gene expression and hormone interaction. In: Yaish MW, editor. The Flowering Process and Its Control in Plants: Gene Expression and Hormone Interaction. Kerala, India: Research Signpost/Transworld Research Network; 2011. pp. 307–327. [Google Scholar]
- 28.Sexton R, Roberts JA. Cell biology of abscission. Annu Rev Plant Physiol. 1982;33:133–162. [Google Scholar]
- 29.Ellis CM, et al. AUXIN RESPONSE FACTOR1 and AUXIN RESPONSE FACTOR2 regulate senescence and floral organ abscission in Arabidopsis thaliana. Development. 2005;132(20):4563–4574. doi: 10.1242/dev.02012. [DOI] [PubMed] [Google Scholar]
- 30.Boerjan W, et al. superroot, a recessive mutation in Arabidopsis, confers auxin overproduction. Plant Cell. 1995;7(9):1405–1419. doi: 10.1105/tpc.7.9.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laskowski MJ, Williams ME, Nusbaum HC, Sussex IM. Formation of lateral root meristems is a two-stage process. Development. 1995;121(10):3303–3310. doi: 10.1242/dev.121.10.3303. [DOI] [PubMed] [Google Scholar]
- 32.Jinn TL, Stone JM, Walker JC. HAESA, an Arabidopsis leucine-rich repeat receptor kinase, controls floral organ abscission. Genes Dev. 2000;14(1):108–117. [PMC free article] [PubMed] [Google Scholar]
- 33.Leslie ME, Lewis MW, Youn JY, Daniels MJ, Liljegren SJ. The EVERSHED receptor-like kinase modulates floral organ shedding in Arabidopsis. Development. 2010;137(3):467–476. doi: 10.1242/dev.041335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fukaki H, Nakao Y, Okushima Y, Theologis A, Tasaka M. Tissue-specific expression of stabilized SOLITARY-ROOT/IAA14 alters lateral root development in Arabidopsis. Plant J. 2005;44(3):382–395. doi: 10.1111/j.1365-313X.2005.02537.x. [DOI] [PubMed] [Google Scholar]
- 35.Murashige T, Skoog F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant. 1962;15(3):473–497. [Google Scholar]
- 36.Beeckman T, Engler G. An easy technique for the clearing of histochemically stained plant tissue. Plant Mol Biol Rep. 1994;12:37–42. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.