Abstract
Ribosome stalling on eukaryotic mRNAs triggers cotranslational RNA and protein degradation through conserved mechanisms. For example, mRNAs lacking a stop codon are degraded by the exosome in association with its cofactor, the SKI complex, whereas the corresponding aberrant nascent polypeptides are ubiquitinated by the E3 ligases Ltn1 and Not4 and become proteasome substrates. How translation arrest is linked with polypeptide degradation is still unclear. Genetic screens with SKI and LTN1 mutants allowed us to identify translation-associated element 2 (Tae2) and ribosome quality control 1 (Rqc1), two factors that we found associated, together with Ltn1 and the AAA-ATPase Cdc48, to 60S ribosomal subunits. Translation-associated element 2 (Tae2), Rqc1, and Cdc48 were all required for degradation of polypeptides synthesized from Non-Stop mRNAs (Non-Stop protein decay; NSPD). Both Ltn1 and Rqc1 were essential for the recruitment of Cdc48 to 60S particles. Polysome gradient analyses of mutant strains revealed unique intermediates of this pathway, showing that the polyubiquitination of Non-Stop peptides is a progressive process. We propose that ubiquitination of the nascent peptide starts on the 80S and continues on the 60S, on which Cdc48 is recruited to escort the substrate for proteasomal degradation.
Keywords: quality control, Saccharomyces cerevisiae
Cytoplasmic quality control mechanisms prevent the accumulation of potentially toxic translation products by recognition and elimination of nonconforming mRNAs and their associated nascent peptides stalled on ribosomes. Three quality control pathways are known, each targeting a specific type of aberrant mRNA: NonSense-Mediated mRNA Decay (NMD) (1), Non-Stop mRNA Decay (NSD) (2, 3), and NoGo mRNA Decay (NGD) (4), eliminating transcripts with a premature termination codon, lacking a STOP codon, or carrying an element causing translational arrest, respectively.
NSD constitutes an important quality control process especially in the case of the [PSI]+ prion state, a physiological context that promotes stop codon readthrough, thus generating extended proteins (5). Furthermore, NSD substrates can also result from transcription termination at cryptic polyadenylation [poly(A)] sites located in the coding region of the mRNA. A global map of poly(A) sites recently generated by direct RNA sequencing revealed that many cryptic poly(A) sites exist in yeast as well as in the human genome (6).
It has been shown that ribosome stalling and NGD can be induced by the synthesis of highly positively charged amino acid sequences, such as a polyarginine or polylysine stretches. The positive charges of these basic tracts would electrostatically interact with the negative charges within the exit tunnel (7), resulting in ribosome stalling and translation arrest. Such a polybasic tract can arise from the translation of a poly(A) tail into polylysine, which occurs during translation of a Non-Stop mRNA (8). Hence, the boundary between these two quality control pathways remains unclear (9). NSD and NGD would both be activated by a stalled ribosome with an empty A site recognized by Dom34/Hbs1, homologs proteins of the eRF1/eRF3 translation termination heterodimer, which triggers ribosome dissociation and peptidyl-tRNA release (10). In Saccharomyces cerevisiae, Ski7 protein, another homolog of the release factor eRF3, can also trigger NSD (2, 3). In the case of NGD, a yet unknown endonuclease cleaves the aberrant mRNA. The 5′ cleavage product is degraded upon binding of the SKI complex and the exosome whereas the 3′ fragment is degraded by Xrn1 (4). It was recently proposed that, in the case of NSD, an endonucleolytic cleavage also occurs, upstream of the ribosome stalled at the poly(A) tail (11). In addition, in both NGD and NSD, the nascent peptide is targeted to the proteasome and rapidly degraded. In the case of ribosome stalling induced by the synthesis of consecutive basic amino acid residues, the E3 ubiquitin ligase Not4 was shown to be required for the rapid degradation of the nascent peptide (12), whereas the E3 ligase Ltn1 was found to be required for the ubiquitination and proteasomal degradation of the aberrant peptide synthesized from a Non-Stop mRNA (13).
Therefore, during NGD and NSD, the degradation processes of the aberrant mRNA and of the associated nascent peptide are coupled. The absence of the SKI complex combined with the deletion of LTN1 results in synthetic slow growth (13), showing that a combination of mutations in both Non-Stop mRNA decay and aberrant nascent peptide degradation is detrimental to the cell.
In many cases, such as in endoplasmic-reticulum–associated protein degradation (ERAD), the AAA-ATPase Cdc48 recognizes polyubiquitinated proteins and is required for their efficient targeting to the proteasome (14, 15). The binding of Cdc48 to specific targets is determined by its physical interaction with a large number of cofactors. Here we identified two factors, translation-associated element 2 (Tae2) and ribosome quality control 1 (Rqc1), which, together with Ltn1 and Cdc48, form a complex associated to the 60S and required for the degradation of nascent peptides synthesized from aberrant Non-Stop mRNAs. We also show that ubiquitination of the nascent peptides starts on the stalled 80S ribosome and continues on the 60S after ribosomal dissociation.
Results
New Set of Proteins Functionally Linked to the SKI Complex.
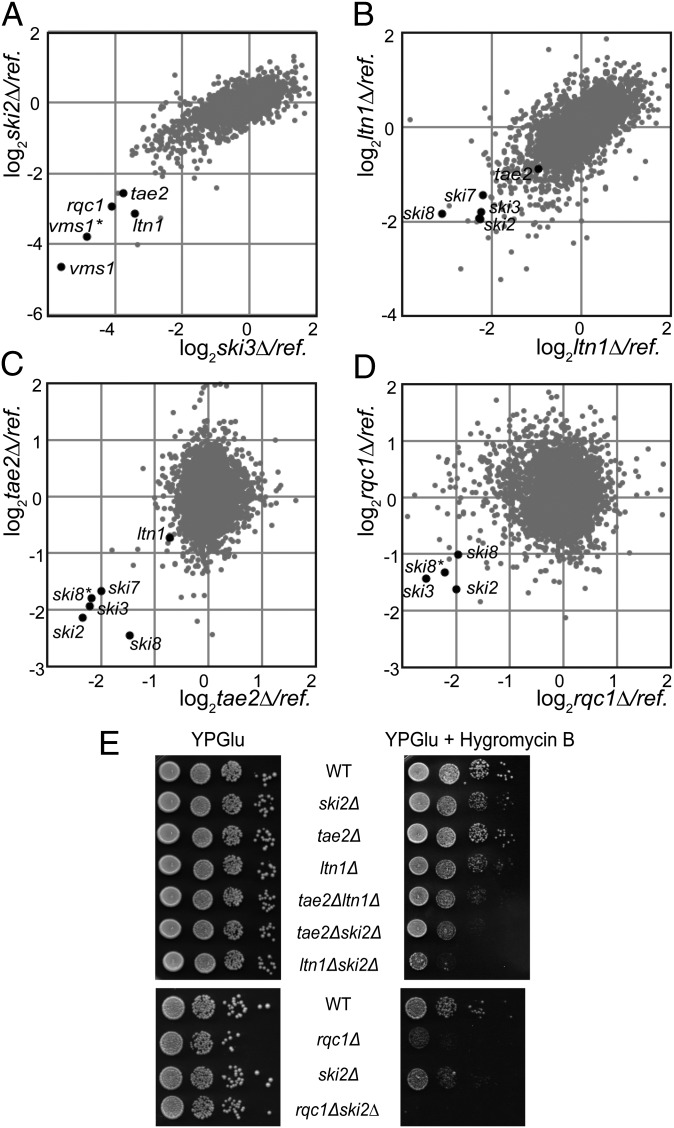
To identify additional factors involved in the recognition and degradation of aberrant mRNAs or associated nascent peptides, we performed genetic screens, using the genetic interaction mapping (GIM) strategy (16) with mutants of the SKI complex (ski2∆, ski3∆, and ski8∆) and with ltn1∆. GIM genome-wide genetic screens enable the identification of functional interactions, whether synthetic lethal, synthetic slow growth, or epistatic, by combining a query mutation with a collection of mutant strains: either complete gene deletions (17) or DamP mutants for essential genes (18). Among the 20 mutants having the strongest growth defect when combined with gene deletions of the SKI complex, we identified four gene deletions that were common in all of the screens: ltn1∆, vms1∆, ypl009c∆ (tae2∆), and ydr333c∆ (rqc1∆) (Dataset S1). These candidates were also selected in GIM screens performed with ski7∆ (Fig. S1). Fig. 1A illustrates the overlap of genetic interactions between ski2∆ and ski3∆. The genetic interactions between the SKI complex and the E3-Ubiquitin ligase Ltn1 confirmed the synthetic slow-growth phenotype reported earlier (13). Vms1 binds Cdc48 and is involved in diverse cellular functions, such as mitochondrial protein turnover (19), ERAD (20), or Cdc13 degradation (21). The deletion of the dubious ORF YDR048C was marked vms1∆* because its deletion overlaps with VMS1 and thus constitutes an independent mutation for this gene. YPL009C was named TAE2, following the identification of this gene deletion in a screen as conferring sensitivity to cycloheximide, a translation inhibitor (22). YDR333C has been recently named RQC1 (23). Remarkably, the GIM screen performed with ltn1∆ selected the four mutants of the SKI complex (ski2∆, ski3∆, ski8∆, and ski7∆) (Fig. 1B).
Fig. 1.
Tae2, Rqc1, and Ltn1 are functionally linked to the SKI complex. (A) GIM screens with ski2∆ and ski3∆ query strains. Each dot indicates a deletion mutant strain. In each screen, we compared the relative changes in given mutant levels in combination with the query or a reference mutation after 22 generations of a mixture of double-mutant populations. Microarray estimation of changes was normalized and the log2 of the ratio of query to reference was plotted. (B) GIM screens with the ltn1∆ query strain. Details are as in A. (C) GIM screens with the tae2∆ query strain. Details are as in A. (D) GIM screens with the rqc1∆ query strain. Details are as in A. (E) Serial dilutions of wild-type (BY4741) or deletion mutant strains were spotted on rich medium plates with or without hygromycin B (40 µg/mL) and incubated for 2 d at 30 °C.
In the course of this study, we further investigated the cellular role of the two factors Tae2 and Rqc1, which could explain the functional links with the SKI complex. First, we confirmed the functional interactions between TAE2 and RQC1 and the SKI complex by GIM screens with tae2∆ and rqc1∆. Similarly to the ltn1∆ screen, the ski2∆, ski3∆, ski7∆, and ski8∆ mutants exhibited the strongest growth defects in association with tae2∆ or rqc1∆, demonstrating the reciprocity and the specificity of the functional interactions between TAE2, RQC1, LTN1, and the SKI complex (Fig. 1 C and D and Fig. S1).
We next validated the functional links between the SKI genes and TAE2 or RQC1 by testing the growth of deletion mutant strains on rich medium plates, including ltn1∆ mutant strains as a control. Surprisingly, the growth of single- or double-mutant strains was not significantly affected in these conditions (Fig. 1E). Bengtson and Joazeiro also observed that the deletion of LTN1 had no obvious phenotype on rich medium plates, but demonstrated the hypersensitivity of the ltn1∆ strain to hygromycin B, an antibiotic that decreases translation fidelity and causes stop codon readthrough (13). This observation prompted us to test the viability of the single- and double-mutant strains in the presence of hygromycin B. The growth of each single-mutant strain ltn1∆, ski2∆, or tae2∆ was weakly affected but a stronger growth phenotype was observed for rqc1∆. Interestingly, the double-deletion strains were clearly more sensitive to hygromycin B compared with the corresponding simple mutant strains. The ski2∆ mutant exhibited a synthetic slow growth defect in combination with tae2∆ and ltn1∆ and a synthetic lethal effect in combination with rqc1∆ (Fig. 1E). These growth defects correlated with those observed under competitive growth in the GIM screens, which are performed in the presence of G418 and nourseothricin (Fig. 1 A–D and Fig. S1).
Tae2, Rqc1, Ltn1, and Cdc48 Are Bound to the Same 60S Ribosomal Subunits.
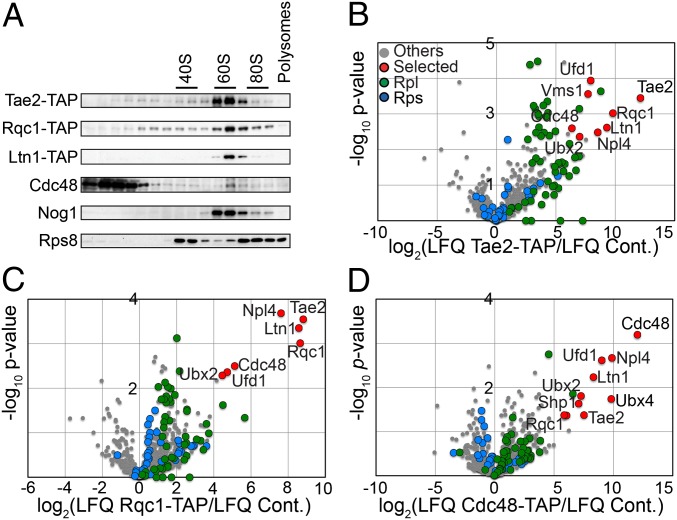
The overlap of genetic interaction patterns between TAE2, RQC1, and LTN1 suggested that these factors could be involved in similar biological processes. Because the deletion of any of these genes caused sensitivity to translation-affecting antibiotics such as hygromycin B and cycloheximide (24), and because it has been shown that Ltn1 binds 60S ribosomal particles and is involved in translational quality control (13), we suspected that Tae2 and Rqc1 could be involved in a translation-associated pathway. We therefore analyzed the localization of Tae2 and Rqc1 in a polysome gradient to see whether they also cosedimented with the 60S ribosomal particles. We used strains expressing TAP-tagged versions of Tae2, Rqc1, and Ltn1 to monitor the sedimentation of each of these factors in a polysome gradient (Fig. 2A). We observed that Tae2, Rqc1, and Ltn1 were localized in the same fractions as Nog1, a pre60S-binding-specific factor here used as a marker for 60S-containing fractions, suggesting that all these factors are associated with 60S ribosomal subunits (25).
Fig. 2.
Tae2, Rqc1, Ltn1, and Cdc48 are associated to 60S ribosomal particles. (A) Total cellular extracts were prepared from each strain expressing Tae2-TAP or Rqc1-TAP or Ltn1-TAP and separated on a sucrose gradient (10–30%). Fractions of the gradient were analyzed by Western blot, using antibodies against TAP, Cdc48, Nog1, and Rps8, a ribosomal protein of the small subunit. (B) Affinity purification of the Tae2-TAP–associated complex was subjected to LC-MS/MS identification and quantification. Tae2-associated complex was compared with the BY4741 strain. Volcano plot shows the fold change (log2 LFQ cpx/LFQ ref.) (LFQ, label-free quantification) on the x axis and the P-value distribution (−log10 P value) on the y axis for the proteins identified in the TAP purification. Each circle indicates an identified protein (in red, for the most significant enriched candidates in the TAP purification; in green, for RPLs; in blue, for the RPSs). (C) TAP purification with Rqc1-TAP compared with the BY4741 strain. Details are as in B. (D) TAP purification with Cdc48-TAP compared with the BY4741 strain. Details are as in B.
To see whether Tae2, Rqc1, and Ltn1 were bound together to the same 60S particles, we performed tandem affinity purification coupled with label-free mass spectrometry (LC-MS/MS), using strains expressing, respectively, the Tae2-TAP (Tandem affinity Purification) and Rqc1-TAP fusions as baits and a wild-type strain (BY4741) as a negative control. Statistical analysis of label-free quantitative MS data (26) revealed significant enrichment of proteins of the large ribosomal subunit (RPL) in both Tae2-TAP and Rqc1-TAP purifications, confirming the binding of these two factors to 60S particles (Fig. 2 B and C). In addition, we observed that Tae2, Rqc1, and Ltn1 were enriched specifically in both purifications, indicating that they are present together on the same 60S subunits. Moreover, these purifications revealed a strong enrichment of an additional component, Cdc48, and several of its known partners, Ufd1, Npl4, Vms1, Ubx2, Ubx4, and Shp1.
Affinity purifications using Cdc48-TAP as bait with the same wild-type strain as a negative control showed a strong enrichment of known Cdc48 partners (Ufd1, Npl4, Ubx2, and Ubx4) and of Ltn1, Tae2, and Rqc1, which confirmed the presence of Cdc48 in complexes that also contain Tae2, Rqc1, and Ltn1 (Fig. 2D and Dataset S2). In parallel, we examined the localization of Cdc48 in a polysome gradient. We observed that, even if most of the Cdc48 hexamers were localized in lighter fractions of the gradient, a subset of Cdc48 sedimented in the 60S fractions (Fig. 2A). This subset is likely to correspond to Cdc48 physically interacting with 60S particles associated with Tae2, Rqc1, and Ltn1.
Ltn1 and Rqc1 Are Essential for the Recruitment of Cdc48 to Ribosomal Subunits.
Cdc48 (called p97 in mammals) is a highly conserved member of the AAA-ATPase family involved in a large number of cellular functions and forming homohexamers able to interact with diverse partners that confer specificity toward various processes (14). One of the main functions of Cdc48 is to recognize polyubiquitinated molecules, extract them from a complex, and make these substrates accessible to the proteasome for degradation (27). Therefore, the ubiquitination activity of Ltn1 could be an important step for the recruitment of the Cdc48-Ufd1-Npl4 complex to 60S particles associated with Tae2 and Rqc1.
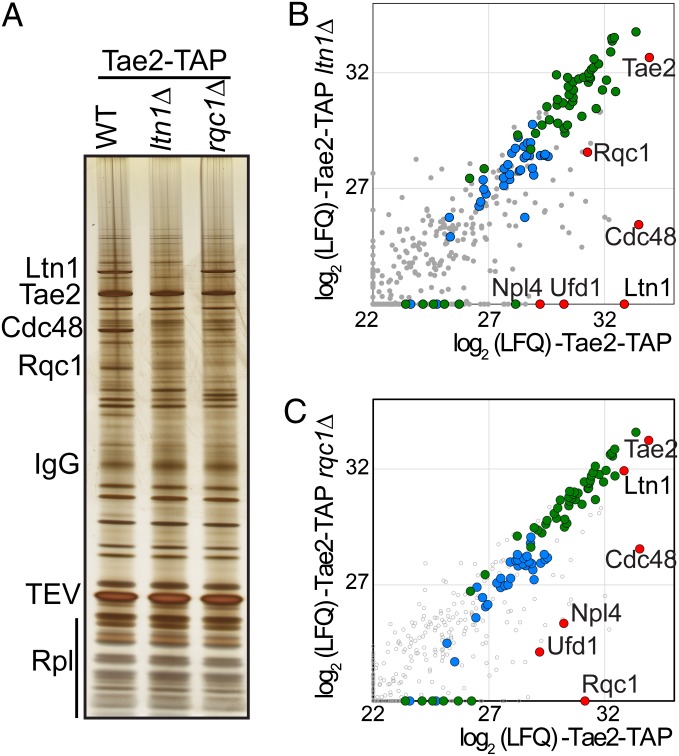
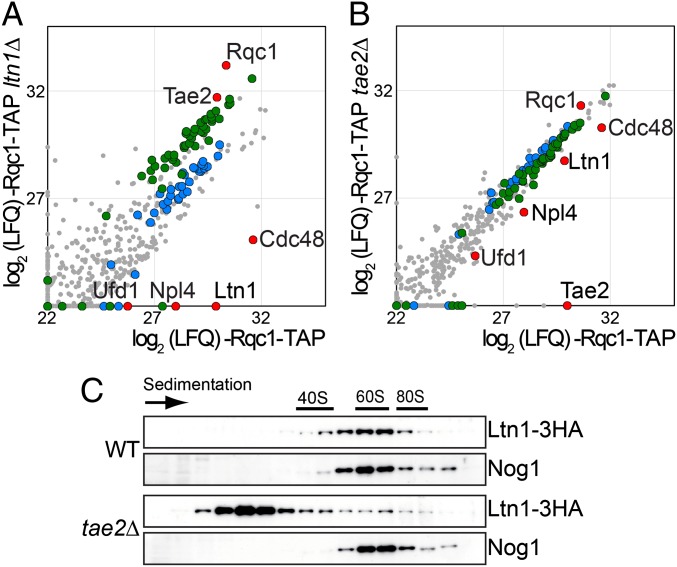
We thus tested whether the absence of Ltn1 affected the binding of Cdc48 to the Tae2- and Rqc1-associated complexes. We observed that the deletion of LTN1 resulted in a decrease in the amount of Cdc48 and its partners Ufd1 and Npl4 from Tae2-TAP (Fig. 3 A and B and Dataset S3)- and Rqc1-TAP (Fig. 4A and Dataset S4)-associated complexes. In addition, we examined the effect of the absence of Rqc1 in Tae2-TAP purifications and noticed a similar Cdc48 decrease (Fig. 3C and Dataset S3).
Fig. 3.
Cdc48 association with Tae2 depends on Ltn1 and Rqc1. (A) Affinity purification of the Tae2-TAP–associated complex in WT or in the absence of Ltn1 or in the absence of Rqc1. The associated proteins were separated on a polyacrylamide gel and revealed by silver staining. (B) Affinity purification of Tae2-TAP–associated complex in presence or absence of Ltn1. Graph shows the comparison between the intensities (LFQ) for each protein identified by LC-MS/MS (log2 scale). (Color legend is as in Fig. 2). (C) Affinity purification of the Tae2-TAP–associated complex in the presence or absence of Rqc1 as in B.
Fig. 4.
Ltn1 association to 60S ribosomal particles is affected by Tae2. (A) Affinity purification of the Rqc1-TAP–associated complex in the presence or absence of Ltn1. Graph shows the comparison of the intensities of each protein identified by LC-MS/MS (log2 scale) as in Fig. 3B. (Legend for the dots is as in Fig. 2A.) (B) Affinity purification of the Rqc1-TAP–associated complex in the presence or absence of Tae2 as in Fig. 3B. (C) Cellular extracts from the tagged strains Ltn1-3HA and Ltn1-3HA tae2∆ were separated on a sucrose gradient (10–30%) as in Fig. 2. The tagged proteins were revealed by Western blot as in Fig. 2, using HA antibody.
These data were strengthened by an independent experiment showing that the absence of Ltn1 or, to a somewhat lesser extent, Rqc1 efficiently reduced the amount of Cdc48 found in the 60S fractions of a polysome gradient (Fig. S2).
The amount of Cdc48 was also decreased to some extent from the Rqc1-TAP complex in the absence of Tae2 (Fig. 4B and Dataset S4), an observation also corroborated by the diminution of the amount of Cdc48 sedimenting with the 60S in a polysome gradient when Tae2 is absent (Fig. S2). This decrease was paralleled by a similar diminution of Ltn1 (Fig. 4B), suggesting that the decrease of Cdc48 might be indirect, resulting from the lower stability or recruitment of Ltn1 on the complex. In addition, we observed that the Ltn1-3HA fusion, which localizes with the 60S in a polysome gradient in a wild type, is shifted for a large part toward the lighter fractions in the absence of Tae2 (Fig. 4C). This confirms the importance of Tae2 for the stable association of Ltn1 with the complex.
In summary, the Cdc48-Ufd1-Npl4 complex requires both Ltn1 and Rqc1 to efficiently bind 60S particles, which suggests that ubiquitination by Ltn1 is essential for Cdc48 recruitment. Whether Rqc1 has a role in the ubiquitination process as well or whether it has a more structural role for Cdc48 binding remains to be determined.
Tae2, Rqc1, and Cdc48 Are Involved, Together with Ltn1, in Non-Stop Protein Decay.
The new 60S-associated complex described above contains Cdc48, Rqc1, Tae2, and the E3 ubiquitin ligase Ltn1, which has been previously shown to ubiquitinate aberrant nascent peptides synthesized from Non-Stop mRNAs to ensure their proteasomal degradation (13). In parallel, components of the SKI complex are essential for the recruitment of the cytoplasmic exosome and thus aberrant mRNA clearance (2). Because Tae2 and Rqc1 are functionally linked with the SKI complex and physically associated with Cdc48 and Ltn1 on the 60S, we tested whether Tae2, Rqc1, and Cdc48 were also involved in the degradation of aberrant Non-Stop mRNAs and the associated nascent peptides.
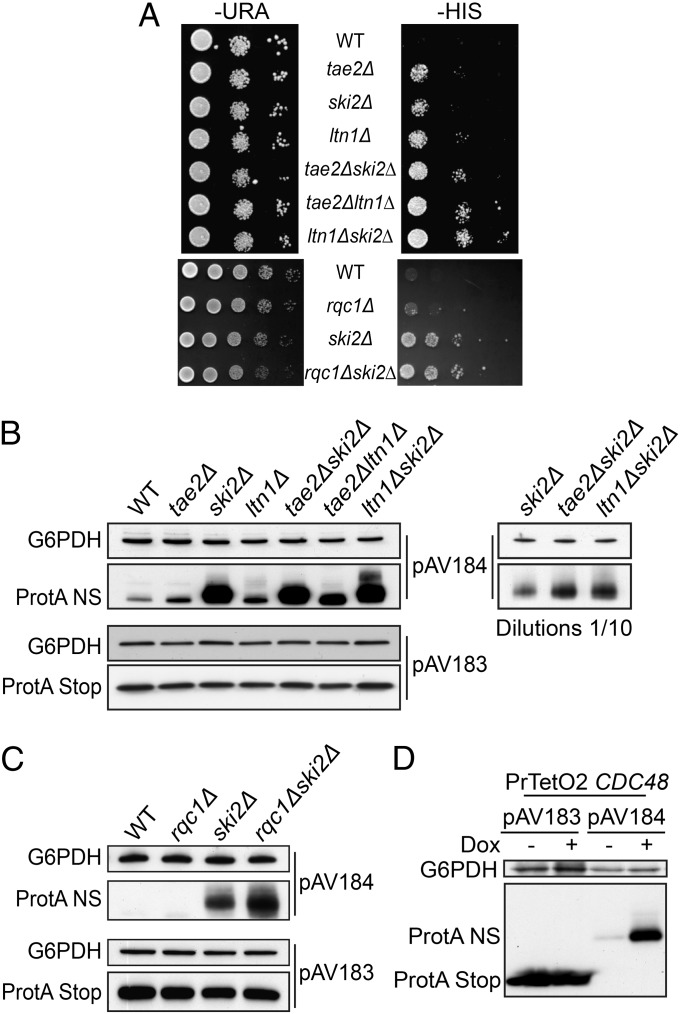
The HIS3 Non-Stop reporter gene (pAV188 plasmid) (2, 3) was used to test whether the Non-Stop degradation pathway was still active in the absence of Rqc1 or Tae2. In a wild-type strain, his3-Non-Stop mRNAs are efficiently degraded; therefore cells from a parental his3∆100 strain do not grow on media without histidine (Fig. 5A). In contrast, mutations of the SKI complex hinder Non-Stop mRNA decay activity that stabilizes aberrant Non-Stop mRNAs, thus enabling the growth of cells transformed with pAV188 even in the absence of histidine. Wilson et al. showed that a deletion of LTN1 also allowed pAV188-transformed cells to grow in the absence of histidine, revealing that the stabilization of proteins synthesized from Non-Stop mRNAs can be sufficient to restore a functional level of His3 activity (28). We performed drop tests, using deletion strains for either TAE2 (Fig. 5A, Upper) or RQC1 (Fig. 5A, Lower) and observed that both deletions allowed growth in the absence of histidine with pAV188. In addition, combinations of TAE2 or RQC1 deletions with ski2∆ (and ltn1∆ in the case of Tae2) revealed a synergistic effect on growth rate on this medium. These results showed that the absence of Tae2 or Rqc1 caused an accumulation of functional His3-Non-Stop proteins and that this accumulation was even stronger in combination with the absence of Ski2 or Ltn1.
Fig. 5.
Absence of Tae2, Rqc1, and Ltn1 as well as Cdc48 leads to the accumulation of Non-Stop proteins. (A) Absence of Tae2 (Upper) or Rqc1 (Lower) enables the production of Non-Stop His3. Wild-type (BY4741) or deletion mutant strains were transformed with the Non-Stop HIS3-containing plasmid (pAV188). Serial dilutions of the strains were spotted on SC −Ura and on SC −His and the plates were incubated for 3 d at 30 °C. (B) The same strains as in A were transformed with the pAV184 plasmid containing the Non-Stop protA gene under the control of the GAL10 promoter and with the pAV183-containing protA gene with a Stop codon as a control. Nondiluted and 10-fold–diluted total cellular extracts were separated on a 10% SDS/PAGE polyacrylamide gel and the protein A was revealed by Western blot, using PAP antibody. A loading control of the total protein amounts was done using anti-G6PDH antibody. (C) As in B with the strains related to rqc1∆, ski2∆, and rqc1∆ ski2∆. (D) As in B with the strain PrTetO2:CDC48. The depletion of Cdc48 was obtained by incubation of the strain for 12 h in the presence of Doxycyclin (10 µg/mL).
To address whether Tae2 and Rqc1 are involved in Non-Stop mRNA clearance or specifically in the degradation of aberrant nascent peptides, we used the pAV184 reporter that carries a gene coding for Protein A from an mRNA that lacks a termination codon. The same gene but with a normal termination codon was used as a control (pAV183) (2). We performed Western blots and Northern blots with total extracts from mutant cells transformed with pAV184 (Non-Stop Protein A) or pAV183 (Stop Protein A). As previously shown, deletion of SKI complex components caused an accumulation of Non-Stop mRNAs and therefore restored expression of aberrant proteins (Fig. S3 A and B). The ltn1∆ mutant exhibited an accumulation of aberrant proteins without affecting the levels of aberrant Non-Stop mRNAs (Fig. S3A) (13). We noticed that in the absence of Tae2, the aberrant Non-Stop Protein A was stabilized compared with the wild type (Fig. 5B, Upper) whereas Stop Protein A showed a similar abundance in all tested mutants (Fig. 5B, Lower). We showed that the absence of Tae2 did not affect the level of aberrant Non-Stop mRNA or its stability (Fig. S3 A and D). We did not see any clear effect of the RQC1 deletion on Non-Stop Protein A, however, as for the HIS3 Non-Stop reporter, combined deletions of TAE2 or RQC1 with the deletion of SKI2 revealed a synergistic effect on aberrant protein accumulation (Fig. 5 B and C). We tested in parallel the effect of CDC48 depletion and also observed a strong accumulation of Non-Stop Protein A (Fig. 5D).
The absence of Tae2, Rqc1, or Cdc48 did not affect the steady-state levels of the Non-Stop Protein A mRNA (Fig. S3 A–C), suggesting that like Ltn1, these three factors are specifically involved in the degradation of nascent peptides synthesized from Non-Stop mRNAs but have no role in Non-Stop mRNA decay.
Aberrant Nascent Peptides Remain Bound to the Ribosome in the Absence of Tae2, Rqc1, or Cdc48.
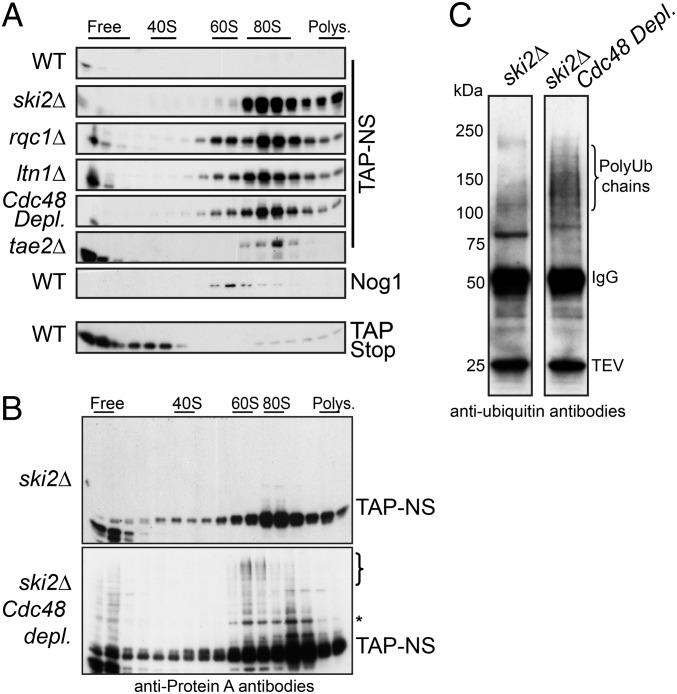
To understand how Tae2, Rqc1, and Cdc48 act on the recognition or the degradation of aberrant nascent peptides, we analyzed the sedimentation profile of the Non-Stop peptide in a polysome gradient in the absence of Tae2, Rqc1, or Cdc48 (in this experiment, we used a TAP-Non-Stop reporter derived from the Non-Stop Protein A reporter pAV184) (SI Materials and Methods). It has been previously shown that in mutants of the SKI complex or in the absence of Ltn1, aberrant nascent peptides accumulated in the 80S fractions of a polysome gradient, suggesting that although Ltn1 sediments with the 60S fractions, the recognition process of aberrant proteins could start on the stalled ribosome and trigger the dissociation of the two ribosomal subunits (13). Whereas the control TAP-Stop protein was localized at the top of the gradient in the free fractions, the TAP-Non-Stop protein accumulated in the 80S fractions in the absence of Tae2, Rqc1, or Cdc48 as in the absence of Ski2 or Ltn1 (Fig. 6A). Interestingly, in the absence of Ltn1 and to a lesser extent in the absence of Rqc1 or Cdc48, we could observe a subset of aberrant proteins sedimenting with the 60S fractions in addition to the 80S fractions. This is consistent with the hypothesis that aberrant nascent peptides could remain blocked in the 60S exit tunnel even after dissociation of the stalled 80S. This subset of TAP-Non-Stop proteins localized in the 60S fractions could thus represent intermediates of degradation between ribosome dissociation and ubiquitination of the aberrant nascent peptide shown by Bengtson and Joazeiro (13). However, whether the ubiquitination of aberrant nascent peptides starts before or after ribosome dissociation remains unclear.
Fig. 6.
Stabilized Non-Stop proteins accumulate on 80S and 60S particles. (A) Cellular extracts from wild-type or deletion mutant strains bearing the TAP Non-Stop reporter were separated on sucrose gradients. The sedimentation profile of the TAP-stop protein expressed in the wild-type strain is shown as a control. The fractions were analyzed by Western blot, using antibodies against TAP. In addition, an antibody against Nog1 was used for the WT to mark the 60S position. (B) Polysome gradient of cellular extracts from ski2∆ single-mutant and ski2∆ Cdc48-depleted strains that express TAP-Non-Stop construct, detected as in A. (C) Immunoprecipitation of TAP-Non-Stop reporter in ski2∆ single-mutant and ski2∆ Cdc48-depleted strains. The cellular extracts from the strains in B were purified on IgG beads and eluted with TEV protease and the associated proteins were analyzed by Western blot, using monoclonal anti-Ubiquitin antibodies (clone P4D1).
Cdc48 Is Involved in the Proteasomal Targeting of Polyubiquitinated Substrates Stalled on 60S Subunits.
No high-molecular-weight species of the TAP-Non-Stop protein, which could correspond to ubiquitinated forms, were readily apparent in these single-mutant conditions. Given its documented function described in other pathways (14, 15), the Cdc48-Ufd1-Npl4 complex could be expected to play a role in the recognition and proteasomal targeting of polyubiquitinated nascent peptides. We therefore examined the polysome sedimentation profile of TAP-Non-Stop proteins in the absence of Ski2 and Cdc48, to stabilize TAP-Non-Stop mRNAs and the polyubiquitinated forms of the TAP-Non-Stop proteins, respectively. In addition to its strong accumulation in both the 80S and the 60S fractions (Fig. 6B), we observed that upon depletion of Cdc48, a subset of TAP-Non-Stop proteins migrated as high-molecular-weight proteins, forming a ladder of increasing size from the 80S fractions to the 60S fractions. Affinity purifications of the TAP-Non-Stop reporter and its analysis by Western blot using antiubiquitin antibodies revealed the presence of a smear of polyubiquitinated species upon Cdc48 depletion (Fig. 6C), suggesting that the high-molecular-weight ladder formed by the TAP-Non-Stop protein in the 60S fractions indeed corresponds to polyubiquitinated forms of this peptide.
Taken together, these results show that (i) ubiquitination of the aberrant nascent peptide might start at the 80S level, because some TAP-Non-Stop protein-shifted species of intermediate size are already visible in the 80S fractions; (ii) high-molecular-weight polyubiquitinated forms of the aberrant peptide accumulate on 60S ribosomal particles in the absence of Cdc48; and (iii) Cdc48 is essential for proteasomal targeting of these polyubiquitinated aberrant substrates, because a depletion of Cdc48 strongly stabilized polyubiquitinated forms of the TAP-Non-Stop protein.
Discussion
Performing GIM genetic screens, we identified two factors, Tae2 and Rqc1, which, similarly to Ltn1, become essential during translation stress when the SKI complex is absent (Fig. 1). Remarkably, biochemical purifications revealed that these three factors, together with Cdc48, are part of a common complex in association with the 60S ribosomal subunit (Fig. 2). We found that these factors are all required for the efficient degradation of aberrant nascent peptides synthesized from mRNA lacking a stop codon (NSPD) (Fig. 5).
During the completion of this paper, another study was published (23), showing the existence of the same Rqc1-Tae2-Ltn1-Cdc48 complex in association with 60S particles, coined the ribosome quality control (RQC) complex. This work characterized the RQC complex as a trigger for degradation of stalled polypeptides and as a signal for translational stress, which is fully consistent with our findings.
The presence of a stalled ribosome that does not have a stop codon in its A site was proposed to be the signal that triggers the degradation of Non-Stop or NoGo mRNAs. Dom34/Hbs1 would then be recruited instead of the canonical termination factor eRF1/eRF3, peptidyl-tRNA hydrolysis would not be activated, and the nascent peptide, in the form of a peptidyl-tRNA, would remain within the exit tunnel and the P site when Rli1 drives the dissociation of the stalled ribosome. However, the absence of Ltn1 was reported to result in the stabilization of the aberrant peptide on the 80S (13), suggesting that the ubiquitination process starts before ribosome dissociation. It has been also shown that Ltn1 is localized on the 60S, an observation strengthened by our observation that Ltn1, Tae2, Rqc1, and Cdc48 are present on the same 60S particles, supporting the existence of a hypothetical transient 60S-peptidyl-tRNA intermediate. Indeed, we could detect a Non-Stop peptide bound to the 60S particle (Fig. 6A) and show that polyubiquitinated forms of this peptide accumulate on the 60S in the absence of Cdc48 (Fig. 6 B and C). This observation, together with the fact that Cdc48 does not associate with the RQC particle in the absence of Ltn1 (Figs. 3B and 4A), strongly suggests that the Cdc48-Ufd1-Npl4 complex recognizes polyubiquitinated forms of the aberrant peptide at a late stage of the process. Considering the general role of Cdc48, it might extract the polyubiquitinated peptides from the dissociated 60S and escort them to the proteasome for degradation.
In addition, we observed that Cdc48 binding to the complex is also impeded by the absence of Rqc1 (Fig. 3C), whereas Ltn1 binding is not affected. Whether this reflects the requirement of Rqc1 for peptide ubiquitination or a more structural role of this factor for Cdc48 binding remains to be determined. In addition, the absence of Tae2 weakly affected Cdc48 binding to the complex; this could be an indirect effect due to the decreased binding efficiency of Ltn1 in this context (Fig. 4 B and C).
Finally, Cdc48 was implicated in the degradation of nonfunctional 60S subunits (25S NRD for nonfunctional rRNA decay) by promoting the dissociation of stalled 80S, thus allowing the recruitment of the proteasome to stalled 60S to perform ribosomal proteins proteolysis (29). These results are consistent with the hypothesis that after dissociation of stalled ribosomes, the 60S particles are not recycled to perform new rounds or translation but targeted for proteasomal degradation, together with the aberrant nascent peptide.
Materials and Methods
Yeast Strains, Plasmids, and GIM Screens.
The yeast strains used in this study are listed in Tables S1 and S2. They were constructed by high-fidelity PCR transformation. Serial dilutions were spotted on rich medium plates with or without hygromycin B (40 µg/mL) and incubated for 2 d at 30 °C. Details of plasmid constructions and GIM screens (16) can be found in SI Materials and Methods.
Polysome Gradients and Western Blotting.
Polysome gradients analyses were conducted as described in SI Materials and Methods. Collected fractions were precipitated and separated on a 10% (wt/vol) acrylamide gel. The proteins were detected by hybridization with the appropriate antibodies (Table S3).
Tandem Affinity Purifications Mass Spectrometry and Data Analysis.
Biochemical purifications were performed using strains expressing TAP fusion proteins. A first-step purification was done with magnetic beads coupled with IgG (SI Materials and Methods). An aliquot of the Tobacco Etch Virus protease (TEV) eluate was separated on acrylamide gels and silver stained. The rest of the eluates were analyzed by mass spectrometry. Briefly, after digestion, peptides were identified on an LTQ-Orbitrap and raw MS data were analyzed using the MaxQuant software as described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank A. Van Hoof, A. Buchberger, and G. Dieci for providing plasmids and antibodies against Cdc48 and Rps8, respectively. We thank the proteomics platform of the Pasteur Institute for the availability of the Orbitrap. Q.D. and Y.Y. were supported by a fellowship from the Ministère de l’Enseignement Supérieur et de la Recherche. This work was supported by grants from the Agence Nationale de la Recherche (ANR), ANR-2011-BSV6-011-02 and ANR-08-JCJC-0019-01.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1221724110/-/DCSupplemental.
References
- 1.Kervestin S, Jacobson A. NMD: A multifaceted response to premature translational termination. Nat Rev Mol Cell Biol. 2012;13(11):700–712. doi: 10.1038/nrm3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Frischmeyer PA, et al. (2002) An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science 295:2258–2261. [DOI] [PubMed]
- 3.van Hoof A, Frischmeyer PA, Dietz HC, Parker R. Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science. 2002;295(5563):2262–2264. doi: 10.1126/science.1067272. [DOI] [PubMed] [Google Scholar]
- 4.Doma MK, Parker R. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature. 2006;440(7083):561–564. doi: 10.1038/nature04530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilson MA, Meaux S, Parker R, van Hoof A. Genetic interactions between [PSI+] and nonstop mRNA decay affect phenotypic variation. Proc Natl Acad Sci USA. 2005;102(29):10244–10249. doi: 10.1073/pnas.0504557102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ozsolak F, et al. Comprehensive polyadenylation site maps in yeast and human reveal pervasive alternative polyadenylation. Cell. 2010;143(6):1018–1029. doi: 10.1016/j.cell.2010.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu J, Deutsch C. Electrostatics in the ribosomal tunnel modulate chain elongation rates. J Mol Biol. 2008;384(1):73–86. doi: 10.1016/j.jmb.2008.08.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ito-Harashima S, Kuroha K, Tatematsu T, Inada T. Translation of the poly(A) tail plays crucial roles in nonstop mRNA surveillance via translation repression and protein destabilization by proteasome in yeast. Genes Dev. 2007;21(5):519–524. doi: 10.1101/gad.1490207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shoemaker CJ, Green R. Translation drives mRNA quality control. Nat Struct Mol Biol. 2012;19(6):594–601. doi: 10.1038/nsmb.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shoemaker CJ, Eyler DE, Green R. Dom34:Hbs1 promotes subunit dissociation and peptidyl-tRNA drop-off to initiate no-go decay. Science. 2010;330(6002):369–372. doi: 10.1126/science.1192430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsuboi T, et al. Dom34:hbs1 plays a general role in quality-control systems by dissociation of a stalled ribosome at the 3′ end of aberrant mRNA. Mol Cell. 2012;46(4):518–529. doi: 10.1016/j.molcel.2012.03.013. [DOI] [PubMed] [Google Scholar]
- 12.Dimitrova LN, Kuroha K, Tatematsu T, Inada T. Nascent peptide-dependent translation arrest leads to Not4p-mediated protein degradation by the proteasome. J Biol Chem. 2009;284(16):10343–10352. doi: 10.1074/jbc.M808840200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bengtson MH, Joazeiro CA. Role of a ribosome-associated E3 ubiquitin ligase in protein quality control. Nature. 2010;467(7314):470–473. doi: 10.1038/nature09371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stolz A, Hilt W, Buchberger A, Wolf DH. Cdc48: A power machine in protein degradation. Trends Biochem Sci. 2011;36(10):515–523. doi: 10.1016/j.tibs.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 15.Meyer H, Bug M, Bremer S. Emerging functions of the VCP/p97 AAA-ATPase in the ubiquitin system. Nat Cell Biol. 2012;14(2):117–123. doi: 10.1038/ncb2407. [DOI] [PubMed] [Google Scholar]
- 16.Decourty L, et al. Linking functionally related genes by sensitive and quantitative characterization of genetic interaction profiles. Proc Natl Acad Sci USA. 2008;105(15):5821–5826. doi: 10.1073/pnas.0710533105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giaever G, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418(6896):387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 18.Schuldiner M, et al. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell. 2005;123(3):507–519. doi: 10.1016/j.cell.2005.08.031. [DOI] [PubMed] [Google Scholar]
- 19.Heo J-M, et al. A stress-responsive system for mitochondrial protein degradation. Mol Cell. 2010;40(3):465–480. doi: 10.1016/j.molcel.2010.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tran JR, Tomsic LR, Brodsky JL. A Cdc48p-associated factor modulates endoplasmic reticulum-associated degradation, cell stress, and ubiquitinated protein homeostasis. J Biol Chem. 2011;286(7):5744–5755. doi: 10.1074/jbc.M110.179259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baek GH, Cheng H, Kim I, Rao H. The Cdc48 protein and its cofactor Vms1 are involved in Cdc13 protein degradation. J Biol Chem. 2012;287(32):26788–26795. doi: 10.1074/jbc.M112.351825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alamgir M, Erukova V, Jessulat M, Azizi A, Golshani A. Chemical-genetic profile analysis of five inhibitory compounds in yeast. BMC Chem Biol. 2010;10:6. doi: 10.1186/1472-6769-10-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brandman O, et al. A ribosome-bound quality control complex triggers degradation of nascent peptides and signals translation stress. Cell. 2012;151(5):1042–1054. doi: 10.1016/j.cell.2012.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hillenmeyer ME, et al. The chemical genomic portrait of yeast: Uncovering a phenotype for all genes. Science. 2008;320(5874):362–365. doi: 10.1126/science.1150021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saveanu C, et al. Sequential protein association with nascent 60S ribosomal particles. Mol Cell Biol. 2003;23(13):4449–4460. doi: 10.1128/MCB.23.13.4449-4460.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cox J, et al. Andromeda: A peptide search engine integrated into the MaxQuant environment. J Proteome Res. 2011;10(4):1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 27.Meyer HH, Wang Y, Warren G. Direct binding of ubiquitin conjugates by the mammalian p97 adaptor complexes, p47 and Ufd1-Npl4. EMBO J. 2002;21(21):5645–5652. doi: 10.1093/emboj/cdf579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilson MA, Meaux S, van Hoof A. A genomic screen in yeast reveals novel aspects of nonstop mRNA metabolism. Genetics. 2007;177(2):773–784. doi: 10.1534/genetics.107.073205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fujii K, Kitabatake M, Sakata T, Ohno M. 40S subunit dissociation and proteasome-dependent RNA degradation in nonfunctional 25S rRNA decay. EMBO J. 2012;31(11):2579–2589. doi: 10.1038/emboj.2012.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.