Abstract
Reported here is a piggyBac transposon-based expression system for the generation of doxycycline-inducible, stably transfected mammalian cell cultures for large-scale protein production. The system works with commonly used adherent and suspension-adapted mammalian cell lines and requires only a single transfection step. Moreover, the high uniform expression levels observed among clones allow for the use of stable bulk cell cultures, thereby eliminating time-consuming cloning steps. Under continuous doxycycline induction, protein expression levels have been shown to be stable for at least 2 mo in the absence of drug selection. The high efficiency of the system also allows for the generation of stable bulk cell cultures in 96-well format, a capability leading to the possibility of generating stable cell cultures for entire families of membrane or secreted proteins. Finally, we demonstrate the utility of the system through the large-scale production (140–750 mg scale) of an endoplasmic reticulum-resident fucosyltransferase and two potential anticancer protein therapeutic agents.
Keywords: glycoprotein, X-ray crystallography
The desire to produce eukaryotic proteins for applications in cell biology, structural biology, and biotechnology has greatly accelerated the use of mammalian cell expression systems during the past 10 to 15 years (1, 2). Among these, transient and stable approaches have been used, although those involving the latter are the most widely used for research laboratory- and industry-scale protein production. Typically, the transgene of interest is stably inserted into the mammalian genome by the classical integration method (i.e., the spontaneous integration of foreign DNA), an approach that often leads to significant transcriptional variation as a result of position effects (3–5). In addition, transgenic fragments integrated in this way are often found to be inserted as concatemers, genomic structures that can be inactivated by “repeat-induced gene silencing” (6, 7). As a result of these processes, time-consuming and laborious cloning steps are typically required to generate cell lines with acceptable protein expression levels.
To address the drawbacks associated with the classical integration method, a number of approaches have been developed. The use of dihydrofolate reductase/methotrexate–based gene amplification (8), for example, is used to increase expression levels, and chromatin-regulating elements, such as matrix attachment regions and insulators, have been used to limit gene silencing (9–11). Nevertheless, these methods do not necessarily eliminate the variation in specific cell productivity or the need for cell cloning steps. Finally, the site-specific recombinases Flp, Cre, and PhiC31 have been used to target the transgene to a single, defined, and actively transcribed site in the genome (12). Although this approach leads to uniform expression levels and eliminates the need for cell cloning, it is limited by the low efficiency of integration and the requirement of a host cell line engineered to contain the required recombination sequence.
The overexpression of a protein can lead to cell stress and slow growth, making it difficult to generate a cell line or bulk cell culture and/or to expand it for protein production, an important consideration in the design of an expression system. To address this issue, a number of inducible promoter systems have been developed and used for various applications (13–20). Although these systems differ with respect to their expression levels and the degree to which expression can be regulated, one or more exogenous inducer proteins must be expressed in the host cell, a requirement often necessitating the use of purpose-built cell lines.
The host factor-independent piggyBac (PB) transposon was recently introduced as a highly efficient method for inserting multiple copies of exogenous DNA fragments into mammalian genomes (21, 22). The PB transposase (PBase) recognizes a pair of terminal repeat sequences flanking the donor DNA fragment and inserts it into random TTAA sites on the chromosomal DNA in mammalian cells. It can insert fragments of as much as 9 kb with high efficiency and fragments of as much as 14 kb with reduced efficiency (22), and, most recently, it has been used for the insertion of a bacterial artificial chromosome into the genome of an embryonic stem cell (23). As many as 15 copies of the DNA fragment can be inserted per cell (24), and the individual copies are distributed over the genome (24, 25), a property expected to average out position effects.
Although use of the PBase for the generation of stably transfected mammalian cells for protein production holds much promise, the approach has seen only limited development and use to date (26). Reported here is a PB-based expression system for the generation of inducible, stably transfected mammalian cell cultures that has been in development and has been well tested in our laboratories during the past 3 years. The system takes advantage of the ability of the PBase to efficiently mediate the simultaneous integration of multiple DNA fragments (27), a property allowing for the concurrent stable integration of the target protein and the reverse tetracycline transactivator (rtTA) inducer (28) required for doxycycline-mediated expression from the tetracycline response element (TRE) promoter. As such, the system is not restricted to specially modified mammalian cell lines, and we demonstrate its use with adherent [Chinese hamster ovary (CHO)-K1 and human embryonic kidney 293T (HEK293T)] and suspension-adapted cell lines (Freestyle 293-F), as well as the HEK293S N-acetylglucosaminyltransferase I-deficient (GnTI−) cell line developed for applications in structural biology (29). By using a panel of 14 test proteins, we demonstrate the efficiency of the system, its performance relative to the classical integration method, and, as a proof of concept, the feasibility of its use for high-throughput stable cell culture generation in 96-well format. Finally, we report the use of the system for the production and purification of an endoplasmic reticulum (ER)-resident fucosyltransferase, a vascular endothelial growth factor (VEGF) Trap, and an anti-human epidermal growth factor receptor 2 (HER2) single-chain variable-domain antibody (Ab) in amounts ranging from 140 to 750 mg.
Results
Description of PB System.
The generation of stably transformed inducible cell cultures with this PBase-mediated system involves the use of three plasmids (one helper plasmid and two plasmids containing transposons; Fig. 1A and Fig. S1A). The first transposon-containing plasmid carries the TRE (30), which controls the expression of the target protein, and, in addition, a puromycin resistance gene driven by an attenuated simian vacuolating virus 40 (SV40) early promoter (PSV40Δ90). There are two plasmids of this type, one designed to supply an N-terminal Protein A fusion tag (PB-T-PAF), the other the Gateway Reading Frame cassette A (RfA; PB-T-RfA) to facilitate subcloning. The second transposon containing plasmid carries the rtTA gene and the blasticidin S (PB-RB) or neomycin (PB-RN) resistance gene. The helper plasmid, pCyL43 (24) (referred to as PBase plasmid hereafter; Fig. S1A), expresses the PBase. After cotransfection of the three plasmids into a mammalian cell, the transiently expressed PBase mediates the insertion of the transposons (PB-T-PAF/RfA and PB-RB/RN) into the host cell’s genome. Cells that have integrated the target protein (PB-T-PAF/RfA) and the rtTA (PB-RB/RN) transposons are selected by dual-drug resistance to puromycin and blasticidin S/neomycin. The integrated rtTA protein is constitutively expressed under the strong cytomegalovirus (CMV) promoter, and, in the presence of doxycycline, induces the expression of the target protein.
Fig. 1.
The inducible PB system is efficient and consistent. (A) The PB system is composed of a plasmid encoding the target protein (PB-T-PAF or PB-T-RfA) and a plasmid encoding the rtTA inducer (PB-RB or RN). The PB-T-PAF plasmid expresses an N-terminal Protein A (Pr-A) tagged target protein (Tr-P). The PB-T-RfA plasmid contains the Gateway RfA cassette (Invitrogen) to allow for simple insertion of the target protein DNA. (B) The domain structure of the 14 Protein A-fusion proteins from the PA-T-PAF vector. (C) Drug-resistant foci per 106 cells for each of the 14 test Protein A-fusion constructs using the PB method in three cell lines: CHO, HEK 293T, and HEK 293S GnTI−. The empty PB-T-PAF vector was used in the HEK293T cell as a control (“empty” column). (D) Drug resistant foci per 106 cells using the classical method in HEK 293T and HEK 293S GnTI− cell lines with the 14 pIRES constructs. The empty pIRES-puro3 vector was used in the HEK293T cell as a control (empty). (E) Drug resistant foci per 106 cells averaged over all 14 fusion protein constructs for each cell type tested. The HEK293S GnTI− PB/doxycycline (PB + dox) experiment was performed with the addition of 1 µg/mL doxycycline. Error bars represent SD (n = 3 in C and D and n = 42 in E; ***P < 0.001 and #P > 0.05).
PB System Is Simple and Efficient.
To test the efficiency of the PB system, we generated stably transfected HEK293T, HEK293S GnTI−, and CHO-K1 cell cultures for each of 14 Protein A fusion proteins (Fig. 1B and Table S1). All the proteins correspond to soluble fragments of ER/Golgi-resident glycosyltransferases or single-pass mammalian or viral membrane proteins. To allow a comparison between the PB system and that of the classical integration method, the genes encoding each protein were subcloned into the PB-T-PAF vector (Fig. 1A) and the pIRES-puro3 vector (Clontech; Fig. S1A). In all cases, the protein coding regions were identical in both vectors.
For all 14 PB-T-PAF constructs, the plasmid DNA mixture (0.4 µg PB-T-PAF, 0.05 µg PB-RB, and 0.05 µg PBase) was transfected into each of the three cell lines in 24-well plate format (50,000 cells per well) by using Lipofectamine 2000 (Invitrogen). On day 1 after transfection, each well was split, serially diluted, and cultured in the presence of blasticidin S and puromycin for 3 wk. Drug-resistant foci were then counted and stable cell transfection efficiencies were calculated. All 14 constructs produced large numbers of drug-resistant foci in each of the cell lines tested (Fig. 1C). On average, we found that 2.4 ± 0.5%, 3.8 ± 1.3%, and 6.4 ± 2.1% of the cells transfected generated drug-resistant foci for the HEK293S GnTI−, HEK293T, and CHO cell lines, respectively (Fig. 1E). In contrast, with the classical method and HEK293T cells (5 µg linearized pIRES plasmid DNA/600,000 cells per well in six-well plates), only 0.02% of the cells (averaged over the 14 constructs) generated drug-resistant foci, a value ∼100-fold lower than that obtained with the PB system (P < 0.001; Fig. 1 D and E). Similarly, 0.03% of the cells were rendered drug-resistant, using the empty vector as control (Fig. 1D). The efficiency was even worse in the case of HEK293S GnTI− cells, in which most constructs yielded no drug-resistant foci despite attempts to optimize the transfection protocol (Fig. 1 D and E). Taken together, these results clearly show that in contrast to the classical method, the PB approach is efficient and consistent regardless of the cell type or the nature of the protein construct.
To rationalize the higher efficiency and greater consistency observed with the PB system, we tested the effect of (i) the presence and absence of PBase and (ii) the effect of protein expression on the process of stable cell generation. To test the former, we transfected HEK293T cells with PB-T-PAF and compared the number of puromycin-resistant foci obtained in the presence of PBase with that obtained in the absence of PBase (Fig. S1B). The presence of PBase increased the foci yield by a factor of ∼1,000, demonstrating that the high efficiency obtained with the PB system was the result of PBase-mediated transposition. To test the effect of fusion protein production during the process of cell line generation, we repeated the generation of stable HEK293S GnTI− bulk cell cultures for all 14 PB-T-PAF constructs in the presence of 1 µg/mL doxycycline. As shown in Fig. S2, the presence of doxycycline resulted in the production of fewer drug-resistant foci. Moreover, induction led to a much greater variation in the number of drug-resistant foci generated for constructs 2, 5, 6, 7, 11, 12, and 14. These results show that expression of the fusion protein during the process of generating the stable bulk cell cultures reduces the number of drug-resistant foci, an observation highlighting one of the advantages of the inducible nature of this expression system.
Optimization of the PB System.
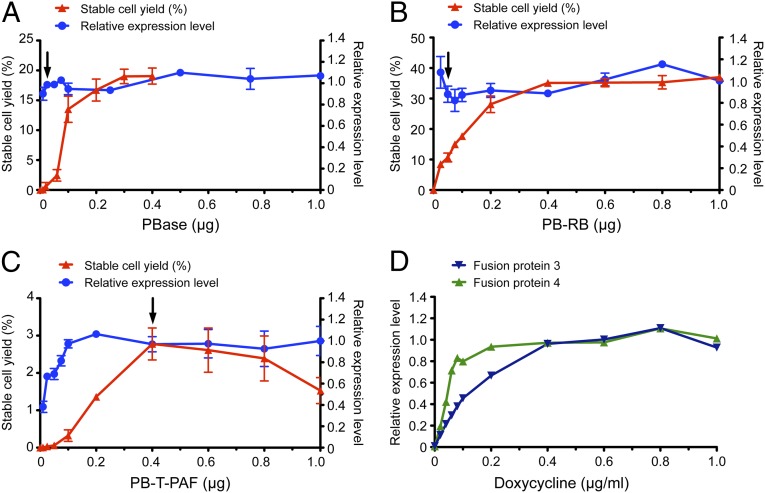
The absolute amount and the ratio of each of the three plasmids used for transfection were varied to assess the effect on foci yield and fusion protein production. As shown in Fig. 2A, as many as 20% of the transfected cells were rendered drug-resistant when large amounts of the PBase plasmid (>0.2 µg/50,000 cells per well) were used. Fig. 2 B and C also show that increasing the amount of the donor plasmids (PB-RB and PB-T-PAF) leads to an increase in drug-resistant foci. In contrast, protein expression levels were relatively insensitive to the amount of the PBase and PB-RB plasmids used (Fig. 2 A and B). Protein expression levels were, however, correlated with the amount of the PB-T-PAF plasmid used and reached a plateau at 0.2 µg plasmid/50,000 cells per well (Fig. 2C). However, given the potential cytotoxicity of the constitutively expressed rtTA inducer protein (31) and the possibility of PBase-mediated excision (22, 32), we routinely used the PB-T-PAF:PB-RB/RN:PBase plasmids at a mass ratio of 8:1:1. As shown in Fig. 2C, no puromycin-resistant foci were obtained when the PBase plasmid was transfected in the absence of PB-T-PAF (i.e., 0 µg/mL) even though the PBase plasmid possesses the puromycin resistance marker (Fig. S1A). This result clearly shows that the PBase plasmid itself was not efficiently integrated into the genome and would therefore not lead to stable cell cultures expressing PBase.
Fig. 2.
Plasmid and doxycycline dose–response curves. (A–C) The foci yield and protein expression levels obtained varying (A) PBase, (B) PB-RB, and (C) PB-T-PAF construct 4 while keeping the other two plasmids constant. Error bars represent SD (n = 3). In A, the cells were selected with puromycin and blasticidin S. In B, the cells were selected with blasticidin S only. In C, the cells were selected with puromycin only. The arrows indicate the standard condition (PB-T-PAF 0.4 µg, PBase 0.05 µg, PB-RB 0.05 µg). (D) Protein expression levels in response to the doxycycline concentration for PB-T-PAF constructs 3 and 4, showing the average value of two replicates.
As shown in Fig. S3, fusion protein expression was found to be low or undetectable in the absence of doxycycline and significantly induced in its presence (1.0 µg/mL doxycycline) in all cases. Dose–response studies with PB-T-PAF constructs 3 and 4 in HEK293S GnTI− cells showed that the former gave half-maximal expression levels at ∼0.2 µg/mL doxycycline, whereas the latter required only 0.04 to 0.06 µg/mL. The expression levels reached plateau at 0.4 and 0.2 µg/mL doxycycline, respectively (Fig. 2D).
Clonal Analysis and Use of Bulk Cell Cultures.
To further compare the PB and classical integration methods, we measured the amount of fusion protein produced by clones generated by the two methods for constructs 3 and 4 in HEK293S GnTI− cells (Fig. 3). All 20 clones examined for each of the two constructs showed robust protein expression levels using the PB approach (Fig. 3 A and B and Fig. S4). In contrast, a number of the clones derived by the classical integration method (using the pIRES vector), for both constructs, were found to produce very little protein. In the case of protein 3, all PB-derived clones showed higher expression levels than the best clone derived by the pIRES method (Fig. 3A), and, with protein 4, 50% of the PB-derived clones were better than the best pIRES clone (Fig. 3B). The PB approach allowed for four- and twofold higher average protein production compared with the classical (i.e., pIRES) method for fusion proteins 3 and 4, respectively (Fig. 3 C and D). For both proteins, the average expression level of the PB clones exceeded or was comparable to that of the best pIRES clone, an observation that allowed us to routinely use bulk cell cultures with the PB method. By eliminating the time-consuming cloning steps required by traditional approaches, the PB method leads to the simple and rapid generation of high-expressing stable cell cultures.
Fig. 3.
Protein expression levels for clones generated by the PB and classical integration methods. Twenty clones from each of the PB and classical (pIRES) methods for two Protein A constructs, 3 and 4, in HEK293S GnTI− cells were analyzed for protein production levels by dot blot analysis. (A and B) Expression of the individual clones (dashed lines mark the average expression levels for the PB clones). (C and D) Averaged expression level for each construct. Error bars represent SD (n = 20; ***P < 0.001).
High-Throughput Generation of Stable Bulk Cell Cultures.
The efficiency with which cells can be stably transfected by using the PB approach prompted us to test its use in a high-throughput format. Eight replicas of each of the 14 PB-T-PAF constructs were cotransfected with the rtTA and PBase plasmids in an 8:1:1 ratio (0.08 µg plasmid DNA in total for each well) into HEK293S GnTI− cells on 96-well plates. Three weeks after the start of simultaneous puromycin and blasticidin S selection, all wells were found to be densely populated by drug-resistant cells. When induced with doxycycline, expression of the Protein A fusion proteins was observed in all wells as shown by dot blot analysis (Fig. S5). Notably, little variation in protein expression was observed among replicas of the same construct, an observation that showed the robustness of the system even in 96-well format (Fig. 4).
Fig. 4.
High-throughput generation of stable bulk cell cultures by using the PB system. Each column corresponds to the average expression level of eight replicas of each of the 14 Protein A-fusion protein constructs expressed in HEK293S GnTI− cells. Error bars represent SD (n = 8).
Example 1: Stable Transfection and Liter-Scale Protein Production from Adherent Bulk Cell Culture.
A slightly truncated (3 aa shorter) version of the PB-T-PAF construct 4 was used to demonstrate the utility of the PB system using adherent cells. Plasmid DNA (20 µg PB-T-PAF plus 2.5 µg PB-RB plus 2.5 µg PBase) was used to transfect a 50% confluent 10-cm dish containing HEK293S GnTI− cells grown in 10% (vol/vol) fetal bovine serum (FBS) by using the calcium phosphate method. On day 3, the cells were transferred to a single T175 flask with medium containing 10% (vol/vol) FBS, 10 µg/mL puromycin, and 5 µg/mL blasticidin S. The surviving cell number reached 108 on day 15 post transfection, at which point the cells were seeded into a 4-L spinner flask containing 1 L Dulbecco's Modified Eagle Medium (DMEM)/F12 [with 3% (vol/vol) FBS] and 3 g Fibra-Cel disks (New Brunswick Scientific) as cell support. Doxycycline induction was started on day 23 post transfection and continued for ∼2 mo in the absence of drug selection. A total medium exchange was performed weekly. As shown by Western blot analysis, robust protein production was observed for the entire 2-mo period (Fig. 5 and Fig. S6). In total, 140 mg of the fusion protein was obtained from 10 L medium before the experiment was stopped. The protein production phase for this construct has also been performed in pleated 1,450-cm2 roller bottles, a technique we have routinely used for HEK293T and HEK293S GnTI− cells grown in the presence of 3% (vol/vol) FBS. Our medium generation rates in this mode are 600 to 800 mL/bottle/wk.
Fig. 5.
Liter-scale expression from a PB-generated adherent HEK2393S GnTI− bulk cell culture. Expression levels of the Protein A–4 fusion protein were assayed by anti-Protein A Western blot analysis. The 100% level was defined by the average production after induction (i.e., samples taken on days 6–74).
Example 2: Stable Transfection and Liter-Scale Protein Expression from Suspension-Adapted Bulk Cell Culture.
The utility of the inducible PB system for use with shake-flask suspension cell culture was demonstrated with a VEGF Trap and a single-chain variable-domain antibody fragment recognizing HER2 (Fig. 6A). Freestyle 293-F cells (1 × 106 cells/mL in 35 mL of Freestyle 293 Expression Medium; Invitrogen) were transfected with plasmid DNA (25 µg PB-T-RfA plus 5 µg PB-RN plus 5 µg PBase). Two days post transfection, selection for neomycin resistance (geneticin 500 µg/mL) was started and continued for 2 wk. To test the possibility that cells expressing multiple proteins could be obtained by selecting for one protein only, we selected for the rtTA inducer protein plasmid (PB-RN) alone in this example. During the drug selection process, media were renewed every 3 d by using centrifugation at 550 rpm (60 × g) to remove dead cells. In both cases, the derivation of stable bulk cell cultures was completed within the 2-wk period following transfection. The resultant cultures were then expanded in the presence of geneticin. Doxycycline induction (2 µg/mL) was performed when the cells reached a density of 0.8 to 1.2 × 106 cells/mL in the absence of geneticin. During this protein production phase, media from 25% of the culture was renewed every 1 or 2 d. Induction was typically performed for 8 to 10 d for each 1-L flask containing 400 mL medium. Fig. 6 B and C show the total cell number, cell viability, and glucose concentration over the 8-d period for several flasks. As shown in Fig. 6D, protein expression levels were relatively uniform among cultures initiated from different frozen vials and independent of whether the media was renewed every day or every other day. By using this protocol, the stable cell cultures were expanded to generate ∼25 L within 6 to 8 wk from the day of transfection for both constructs. In total, ∼250 mg of the VEGF Trap (∼10 mg/L) and 750 mg of the HER2 Ab fragment (∼30 mg/L) were produced. As shown in Fig. S7, both the VEGF Trap and the HER2 Ab fragment produced showed high affinity protein ligand binding and cell growth inhibition properties. In addition to these molecules, we have produced eight other Fc-fusion proteins (5–30 mg/L) with therapeutic potential by using this system. Further details for examples 1 and 2 are provided in SI Materials and Methods.
Fig. 6.
Expression of the VEGF Trap and HER2 Ab from PB-generated suspension Freestyle 293-F bulk cell cultures. (A) Domain structure of the VEGF Trap and HER2 Ab constructs. (B and C) The cell density, survival rates, and glucose levels for different shake flasks during an 8-d expression course for VEGF (B) and HER2 Ab (C). (D) Quantification of the expression levels from different flasks after purification. Each column represents a replica starting from a vial of frozen cells derived from a common stock. For columns without hatch marks, media renewal was performed every day; for the hatched columns, it was performed every other day. Error bars represent SD (n = 4 for VEGF Trap and n = 2 for HER2 Ab).
Discussion
The generation of stably transfected mammalian cells for protein production has traditionally been an arduous and time-consuming process. Low DNA integration efficiencies and position effects have typically necessitated cell cloning steps that can take months to complete. Moreover, toxicity effects can make it difficult to generate, expand, and maintain stable cell cultures produced with vectors using constitutive promoters. To address these issues, we have developed a stable mammalian cell expression system based on the recently described PB transposon (21, 22, 24, 32, 33). The high efficiency of PB-mediated integration has allowed us to develop a doxycycline inducible system that is cell line-independent and requires only a single transfection/selection step. As clones derived by the PB method show uniform and high protein expression levels, the approach has also eliminated the cell cloning steps typically required with the classical integration method.
In practical terms, the PB system is robust and simple to use and employs techniques typical of small research laboratories. The high efficiency ensures that each transfection/selection experiment results in a robust drug-resistant bulk cell culture that is ready for large-scale expansion 2 to 3 wk after transfection. Moreover, our protocols for liter-scale production from adherent and suspension-adapted cell lines are straightforward. Significantly, the expression levels observed by using the PB system exceeded that obtained by the classical method (Fig. 3), and, even under continuous doxycycline induction, expression levels were found to be stable for at least 2 mo. Taken together, these results clearly establish the superiority of the PB approach relative to that of the classical integration method.
The PB system has also allowed us to easily work with cell lines, such as HEK293S GnTI−, that are more difficult to stably transfect (Fig. 1D). Glycoproteins produced in this cell line are of value in protein crystallography (29, 34, 35), and the ability to rapidly generate high-productivity stable bulk cell cultures from this cell line is expected to greatly facilitate protein domain/fragment screening. In addition to the 14 targets described here, we have also used the PB system to produce more than 30 other protein constructs in the HEK293S GnTI− cell line. Our secreted fusion protein yields are typically ∼5 mg/L (ranging from 2 to 50 mg/L), and several of the target proteins have been crystallized, including the PB-T-PAF construct 4 described here. The significance of these results is underscored by the fact that all these proteins correspond to secreted glycoproteins or fragments of mammalian or viral membrane glycoproteins ranging in size from 15 to 150 kDa.
The high efficiency of PB-mediated DNA integration also promises to drive the development of new applications that would be difficult to implement with other approaches. The ability to simultaneously integrate a number of different DNA fragments (27, 32), for example, is expected to facilitate the coexpression of interacting protein partners as well as the expression of multisubunit proteins, including antibodies. Our work with the VEGF Trap and the HER2 Ab shows that the target proteins themselves need not even be selected for, as high expression levels were obtained with selection for the rtTA inducer alone. We have also demonstrated the feasibility of rapidly generating bulk cell cultures in 96-well format, a result that will facilitate the expression of entire classes of mammalian membrane or secreted proteins for novel applications in structural genomics and membrane protein research. Given the demonstrated utility of the approach and the potential for future development, we anticipate that this PB-based expression system will make a significant impact on a wide range of basic and applied research disciplines.
Materials and Methods
Plasmids and Cell Culture.
Further details pertaining to the plasmids and cell culture are provided in SI Materials and Methods. The complete and annotated sequences and maps of the plasmids are provided in Dataset S1. The experiments concerning the adherent HEK293T, HEK293S GnTI−, and CHO-K1 cells were performed in DMEM or DMEM/F12 medium supplemented with FBS. The FreeStyle 293-F suspension cultures (catalog no. R790-07; Invitrogen) were grown in Freestyle 293 Expression Medium (catalog no. 12338; Invitrogen).
Stable Transfection with the PB Transposon and Classical Integration Methods.
A total of 14 identical pairs of cDNAs encoding the Protein A-fusion proteins (listed in Table S1) were subcloned into the PB-T-PAF and pIRES-puro3 vectors. For the classical integration method, 5 µg of each of the 14 pIRES plasmids were linearized and transfected into cells on six-well plates (∼600,000 cells per well). For the PB method, a mixture of the PB-T-PAF, PB-RB, and PBase plasmids was transfected at a ratio of 8:1:1 (0.4 µg:0.05 µg:0.05 µg) into cells plated on 24-well plates (∼50,000 cells per well). One day after transfection, the cells were trypsinized and distributed by serial dilution into fresh tissue culture vessels. Drug selection started on day 3 and was continued for 2 to 3 wk until the foci became visible. The foci were then counted to determine the stable transfection yield. When testing the effect of induction during stable cell construction, 1 µg/mL doxycycline was present in the media in all stages starting 1 d before transfection. For the high-throughput generation of stable bulk cell cultures, HEK293S GnTI− cells were plated at 25% confluency (∼10,000 cells per well) in 96-well plates. The following day, each well was transfected with plasmid DNA (0.064 µg PB-T-PAF, 0.008 µg PB-RB, and 0.008 µg PBase). Three days later, dual drug selection was started by using 10 µg/mL puromycin and 5 µg/mL blasticidin S. Further details are provided in SI Materials and Methods.
Western Blot, Dot Blot, and Protein Purification.
The Western blot and dot blot analyses of the expression medium samples were performed with a monoclonal anti-Protein A antibody (catalog no. P2921; Sigma). The Protein A-tagged proteins were purified from 20-fold concentrated media by using IgG Sepharose 6 Fast Flow resin (catalog no. 17-0969-01; GE Healthcare). The VEGF Trap and HER2 Ab were purified from the concentrated media by immobilized-metal affinity chromatography on Ni-NTA columns. More details are provided in SI Materials and Methods.
Statistical Analysis.
Statistical significances were assessed by Student t test or one-way ANOVA followed by the Newman–Keuls test, by using PRISM software (version 5.0; GraphPad).
Supplementary Material
Acknowledgments
This work was funded by grants from the Canadian Institutes of Health Research (to J.M.R.); and grants from the Natural Sciences and Engineering Research Council of Canada and Canadian Cancer Society Research Institute (all to A.N.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1218620110/-/DCSupplemental.
References
- 1.Wurm FM. Production of recombinant protein therapeutics in cultivated mammalian cells. Nat Biotechnol. 2004;22(11):1393–1398. doi: 10.1038/nbt1026. [DOI] [PubMed] [Google Scholar]
- 2.Durocher Y, Butler M. Expression systems for therapeutic glycoprotein production. Curr Opin Biotechnol. 2009;20(6):700–707. doi: 10.1016/j.copbio.2009.10.008. [DOI] [PubMed] [Google Scholar]
- 3.Henikoff S. Position effect and related phenomena. Curr Opin Genet Dev. 1992;2(6):907–912. doi: 10.1016/s0959-437x(05)80114-5. [DOI] [PubMed] [Google Scholar]
- 4.Martin DI, Whitelaw E. The vagaries of variegating transgenes. Bioessays. 1996;18(11):919–923. doi: 10.1002/bies.950181111. [DOI] [PubMed] [Google Scholar]
- 5.Narlikar GJ, Fan HY, Kingston RE. Cooperation between complexes that regulate chromatin structure and transcription. Cell. 2002;108(4):475–487. doi: 10.1016/s0092-8674(02)00654-2. [DOI] [PubMed] [Google Scholar]
- 6.Garrick D, Fiering S, Martin DI, Whitelaw E. Repeat-induced gene silencing in mammals. Nat Genet. 1998;18(1):56–59. doi: 10.1038/ng0198-56. [DOI] [PubMed] [Google Scholar]
- 7.McBurney MW, Mai T, Yang X, Jardine K. Evidence for repeat-induced gene silencing in cultured Mammalian cells: Inactivation of tandem repeats of transfected genes. Exp Cell Res. 2002;274(1):1–8. doi: 10.1006/excr.2001.5443. [DOI] [PubMed] [Google Scholar]
- 8.Kaufman RJ. Selection and coamplification of heterologous genes in mammalian cells. Methods Enzymol. 1990;185:537–566. doi: 10.1016/0076-6879(90)85044-o. [DOI] [PubMed] [Google Scholar]
- 9.Kwaks TH, et al. Identification of anti-repressor elements that confer high and stable protein production in mammalian cells. Nat Biotechnol. 2003;21(5):553–558. doi: 10.1038/nbt814. [DOI] [PubMed] [Google Scholar]
- 10.West AG, Fraser P. Remote control of gene transcription. Hum Mol Genet. 2005;14(spec no 1):R101–R111. doi: 10.1093/hmg/ddi104. [DOI] [PubMed] [Google Scholar]
- 11.Harraghy N, Buceta M, Regamey A, Girod PA, Mermod N. Using matrix attachment regions to improve recombinant protein production. Methods Mol Biol. 2012;801:93–110. doi: 10.1007/978-1-61779-352-3_7. [DOI] [PubMed] [Google Scholar]
- 12.Birling MC, Gofflot F, Warot X. Site-specific recombinases for manipulation of the mouse genome. Methods Mol Biol. 2009;561:245–263. doi: 10.1007/978-1-60327-019-9_16. [DOI] [PubMed] [Google Scholar]
- 13.Friedman HM, et al. Use of a glucocorticoid-inducible promoter for expression of herpes simplex virus type 1 glycoprotein gC1, a cytotoxic protein in mammalian cells. Mol Cell Biol. 1989;9(6):2303–2314. doi: 10.1128/mcb.9.6.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci USA. 1992;89(12):5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weber W, et al. Gas-inducible transgene expression in mammalian cells and mice. Nat Biotechnol. 2004;22(11):1440–1444. doi: 10.1038/nbt1021. [DOI] [PubMed] [Google Scholar]
- 16.No D, Yao TP, Evans RM. Ecdysone-inducible gene expression in mammalian cells and transgenic mice. Proc Natl Acad Sci USA. 1996;93(8):3346–3351. doi: 10.1073/pnas.93.8.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rivera VM, et al. A humanized system for pharmacologic control of gene expression. Nat Med. 1996;2(9):1028–1032. doi: 10.1038/nm0996-1028. [DOI] [PubMed] [Google Scholar]
- 18.Beerli RR, Schopfer U, Dreier B, Barbas CF., 3rd Chemically regulated zinc finger transcription factors. J Biol Chem. 2000;275(42):32617–32627. doi: 10.1074/jbc.M005108200. [DOI] [PubMed] [Google Scholar]
- 19.Fussenegger M, et al. Streptogramin-based gene regulation systems for mammalian cells. Nat Biotechnol. 2000;18(11):1203–1208. doi: 10.1038/81208. [DOI] [PubMed] [Google Scholar]
- 20.Weber W, et al. Macrolide-based transgene control in mammalian cells and mice. Nat Biotechnol. 2002;20(9):901–907. doi: 10.1038/nbt731. [DOI] [PubMed] [Google Scholar]
- 21.Wilson MH, Coates CJ, George AL., Jr PiggyBac transposon-mediated gene transfer in human cells. Mol Ther. 2007;15(1):139–145. doi: 10.1038/sj.mt.6300028. [DOI] [PubMed] [Google Scholar]
- 22.Ding S, et al. Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell. 2005;122(3):473–483. doi: 10.1016/j.cell.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 23.Rostovskaya M, et al. Transposon-mediated BAC transgenesis in human ES cells. Nucleic Acids Res. 2012;40(19):e150. doi: 10.1093/nar/gks643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang W, et al. Chromosomal transposition of PiggyBac in mouse embryonic stem cells. Proc Natl Acad Sci USA. 2008;105(27):9290–9295. doi: 10.1073/pnas.0801017105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rad R, et al. PiggyBac transposon mutagenesis: A tool for cancer gene discovery in mice. Science. 2010;330(6007):1104–1107. doi: 10.1126/science.1193004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matasci M, Baldi L, Hacker DL, Wurm FM. The PiggyBac transposon enhances the frequency of CHO stable cell line generation and yields recombinant lines with superior productivity and stability. Biotechnol Bioeng. 2011;108(9):2141–2150. doi: 10.1002/bit.23167. [DOI] [PubMed] [Google Scholar]
- 27.Kahlig KM, et al. Multiplexed transposon-mediated stable gene transfer in human cells. Proc Natl Acad Sci USA. 2010;107(4):1343–1348. doi: 10.1073/pnas.0910383107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gossen M, et al. Transcriptional activation by tetracyclines in mammalian cells. Science. 1995;268(5218):1766–1769. doi: 10.1126/science.7792603. [DOI] [PubMed] [Google Scholar]
- 29.Reeves PJ, Callewaert N, Contreras R, Khorana HG. Structure and function in rhodopsin: High-level expression of rhodopsin with restricted and homogeneous N-glycosylation by a tetracycline-inducible N-acetylglucosaminyltransferase I-negative HEK293S stable mammalian cell line. Proc Natl Acad Sci USA. 2002;99(21):13419–13424. doi: 10.1073/pnas.212519299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Agha-Mohammadi S, et al. Second-generation tetracycline-regulatable promoter: Repositioned tet operator elements optimize transactivator synergy while shorter minimal promoter offers tight basal leakiness. J Gene Med. 2004;6(7):817–828. doi: 10.1002/jgm.566. [DOI] [PubMed] [Google Scholar]
- 31.Morimoto M, Kopan R. rtTA toxicity limits the usefulness of the SP-C-rtTA transgenic mouse. Dev Biol. 2009;325(1):171–178. doi: 10.1016/j.ydbio.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Woltjen K, et al. piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature. 2009;458(7239):766–770. doi: 10.1038/nature07863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu SC, et al. piggyBac is a flexible and highly active transposon as compared to sleeping beauty, Tol2, and Mos1 in mammalian cells. Proc Natl Acad Sci USA. 2006;103(41):15008–15013. doi: 10.1073/pnas.0606979103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao Y, et al. Automation of large scale transient protein expression in mammalian cells. J Struct Biol. 2011;175(2):209–215. doi: 10.1016/j.jsb.2011.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chaudhary S, Pak JE, Gruswitz F, Sharma V, Stroud RM. Overexpressing human membrane proteins in stably transfected and clonal human embryonic kidney 293S cells. Nat Protoc. 2012;7(3):453–466. doi: 10.1038/nprot.2011.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.