Abstract
Newly synthesized cellular proteins can be tagged with a variety of metabolic labels that distinguish them from preexisting proteins and allow them to be identified and tracked. Many such labels are incorporated into proteins via the endogenous cellular machinery and can be used in numerous cell types and organisms. Though broad applicability has advantages, we aimed to develop a strategy to restrict protein labeling to specified mammalian cells that express a transgene. Here we report that heterologous expression of a mutant methionyl-tRNA synthetase from Escherichia coli permits incorporation of azidonorleucine (Anl) into proteins made in mammalian (HEK293) cells. Anl is incorporated site-selectively at N-terminal positions (in competition with initiator methionines) and is not found at internal sites. Site selectivity is enabled by the fact that the bacterial synthetase aminoacylates mammalian initiator tRNA, but not elongator tRNA. N-terminally labeled proteins can be selectively conjugated to a variety of useful probes; here we demonstrate use of this system in enrichment and visualization of proteins made during various stages of the cell cycle. N-terminal incorporation of Anl may also be used to engineer modified proteins for therapeutic and other applications.
Keywords: protein synthesis, proteomics, translational profiling, protein engineering
To ensure proper function, cells must make the right proteins at the right times and in the correct amounts. The protein synthesis program of the cell depends on many factors, including environment, developmental stage, and cell type. As cells encounter stresses or other stimuli, protein synthesis is adjusted, allowing cells to accommodate changing conditions and respond to external signals. Because changes in protein synthesis can occur rapidly (1), time-resolved methods are needed to track newly synthesized proteins on timescales of a few minutes. Such measurements are challenging because new proteins are often far outnumbered by their older counterparts (e.g., the amount of protein produced in 1 h by a growing HeLa cell accounts for less than 1% of the total protein content of the cell) (2).
Newly synthesized proteins can be tagged with metabolic labels that allow them to be identified, quantified, or visualized. Current strategies use isotopic amino acids (which can be tracked by mass spectrometry) (3) or bioorthogonal labels that can be selectively conjugated to dyes and affinity probes (4). Labels such as “heavy” lysine, azidohomoalanine (Aha) (5), or O-propargyl-puromycin (OP-puro) (6) can be introduced to cells in a pulse during which actively translated proteins are tagged. Incorporation of these labels requires only the endogenous cellular machinery, making them broadly applicable across cell types and organisms. However, global incorporation of the label can be disadvantageous. For instance, elucidating cell-specific programs of protein synthesis in complex cellular systems (e.g., cocultures, tissues, and multicellular organisms) is challenging because proteins derived from different cell types are chemically indistinguishable. Though heavy lysine, Aha, and OP-puro are powerful tools for labeling proteins in time-resolved fashion, they lack the cell selectivity needed to understand cell-to-cell variation in heterogeneous systems (7). Thus, cell-selective tools for the study of protein synthesis are also required.
In 2009, we reported a strategy for cell-selective metabolic labeling of proteins in complex cellular mixtures (8). Expression of the L13N/Y260L/H301L mutant form of the Escherichia coli methionyl-tRNA synthetase (referred to as NLL-EcMetRS) enables cells to use the methionine (Met) surrogate azidonorleucine (Anl; Fig. 1A; SI Appendix, Fig. S1) in protein synthesis (8, 9). Proteins made in cells that express the NLL-EcMetRS can be labeled with Anl during a pulse in which the noncanonical amino acid competes with Met in the decoding of AUG codons. Anl-labeled proteins can be selectively conjugated to affinity probes (for enrichment) (10) or fluorescent dyes (for visualization) (11, 12). Expression of the mutant synthetase in a subset of cells in a cellular mixture restricts labeling to that subset. Proteins synthesized in cells that do not express the synthetase are not labeled. In multicellular environments, this approach simplifies the protein population and permits identification of the cellular origins of labeled proteins (8, 13).
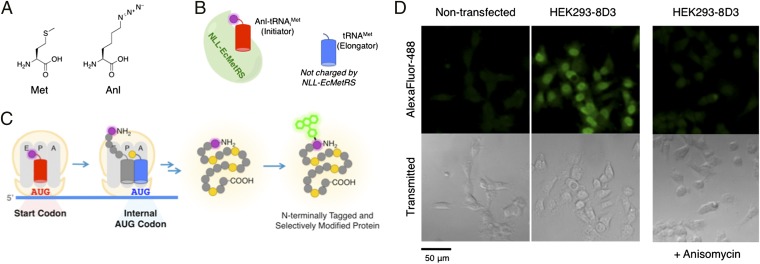
Fig. 1.
NLL-MetRS permits incorporation of Anl into proteins expressed in mammalian cells. (A) Structures of Met and Anl. (B) Selective aminoacylation by NLL-EcMetRS charges Anl to the initiator tRNA but not elongator tRNAMet. (C) Anl-tRNAiMet (red cylinder) initiates protein translation with Anl (purple dot). Internal AUG codons are interpreted only as Met (yellow dot). N-terminally labeled proteins can be modified with probes through bioorthogonal ligation with Anl. (D) HEK293 cells stably expressing the NLL-MetRS (HEK293-8D3) and nontransfected cells were pulsed with Anl and labeled proteins were conjugated to Alexa Fluor 488-alkyne. Fluorescence microscopy revealed labeled proteins in HEK293-8D3 cells, whereas proteins were not labeled in nontransfected cells. HEK293-8D3 cells treated with Anl in the presence of the protein synthesis inhibitor anisomycin exhibited significantly diminished levels of fluorescence.
Recently we sought to extend cell-selective labeling to proteins made in mammalian systems. Previous work had shown that wild-type EcMetRS catalyzes aminoacylation of mammalian tRNA (14), and that it is functional when expressed in COS-1 cells (15). Thus, we pursued heterologous expression of NLL-EcMetRS as an initial strategy. Because Anl is not used by the endogenous mammalian translational machinery (8), we anticipated that labeling would be limited to cells that express the mutant bacterial MetRS.
Here we demonstrate that heterologous expression of NLL-EcMetRS enables incorporation of Anl into proteins expressed in HEK293 cells. Furthermore, we show that Anl is incorporated site selectively at N-terminal positions and is not introduced at internal sites. Site selectivity is a consequence of the fact that NLL-EcMetRS catalyzes aminoacylation only of the mammalian initiator tRNA (tRNAiMet), as does the wild-type bacterial MetRS (14–19). Elongator Met tRNAs (tRNAMet) are not recognized by the NLL-EcMetRS and are charged only with Met. The use of tRNAiMet only in translational initiation (16, 18–21) directs Anl to N-terminal positions and prohibits its insertion at internal sites. Here we describe the use of this system in labeling, detection, and visualization of newly synthesized proteins in mammalian cells, and demonstrate its utility by examining cell cycle-dependent changes in translation.
Results
NLL-EcMetRS Selectively Aminoacylates Mammalian Initiator tRNA.
We first determined whether the NLL-EcMetRS retained selectivity for eukaryotic tRNAiMet. Synthetic tRNAiMet and tRNAMet sequences were prepared by the run-off transcript method (22) and treated in vitro with purified NLL-EcMetRS. tRNAiMet and tRNAMet from Caenorhabditis elegans were compared with the equivalent bacterial sequences. Formation of Anl-tRNAs was monitored by strain-promoted conjugation of the Anl side chain to difluorinated cyclooctyne (DIFO)-functionalized Alexa Fluor 488 (DIFO-Alexa Fluor 488) (23). Fluorescently labeled tRNAs were separated on 2% agarose gels and detected by in-gel fluorescence scanning. Though we observed aminoacylation of eukaryotic tRNAiMet, we found no evidence of charging of eukaryotic tRNAMet by the bacterial synthetase (SI Appendix, Fig. S2).
Expression of NLL-EcMetRS Enables Incorporation of Anl into Proteins Made in Mammalian Cells.
A stable HEK293-derived cell line constitutively expressing NLL-EcMetRS (referred to as HEK293-8D3) was generated by transfection. We confirmed that the bacterial synthetase was expressed as a soluble and intact protein in the HEK293-8D3 cytoplasm (SI Appendix, Fig. S3). To determine whether HEK293-8D3 cells could incorporate Anl into proteins, cells were treated with 1 mM Anl for 4 h. Because the specificity constant (kcat/Km) of the NLL-EcMetRS is higher for Anl than for Met (9), we anticipated that incorporation of Anl would occur even in the presence of high concentrations of Met. Thus, cells were not depleted of Met, and Anl was added to standard growth medium (Materials and Methods). After 4 h, Anl-labeled proteins were detected by Cu (I)-catalyzed azide-alkyne ligation (24) to Alexa Fluor 488-alkyne in fixed (permeabilized) cells. Imaging of cells by fluorescence microscopy revealed prominent labeling of proteins in HEK293-8D3 cells (Fig. 1D), whereas nontransfected cells exhibited only background levels of fluorescence. HEK293-8D3 cells treated with Anl and the protein synthesis inhibitor anisomycin were only dimly fluorescent, indicating that fluorescence arises predominantly from proteins made during the Anl pulse.
We tested various pulse times and Anl concentrations, and estimated the relative levels of Anl tagging after treatment of cell lysates with a dibenzoazacyclooctyne (DIBAC)-functionalized tetramethylrhodamine dye (25) (DIBAC-TAMRA; SI Appendix, Fig. S1). Treatment with DIBAC-TAMRA enables efficient dye-labeling of Anl-tagged proteins via the strain-promoted azide-alkyne cycloaddition; prior alkylation of cysteine residues with iodoacetamide was necessary to eliminate the background reaction of DIBAC with protein thiols (26) (SI Appendix, Fig. S4). After separation by 1D SDS/PAGE, TAMRA-labeled proteins were quantified by in-gel fluorescence scanning. Labeled proteins could be detected after a 15-min pulse with 1 mM Anl (shorter times were not tested; SI Appendix, Fig. S5A). The degree of labeling varied linearly with the pulse length up to 4 h (SI Appendix, Fig. S5A). Protein labeling was detected at Anl concentrations as low as 15 μM (SI Appendix, Fig. S5B; lower concentrations were not tested), and the degree of labeling was saturable in Anl with an apparent Km of 0.5 mM (SI Appendix, Fig. S5B). The value of Km determined previously for the purified NLL-EcMetRS was 2.2 mM (9).
Enrichment of Labeled Proteins and Analysis by Tandem Mass Spectrometry.
To determine whether Anl was incorporated into cellular proteins only at N-terminal positions, we subjected labeled proteins to analysis by mass spectrometry. First, Anl-tagged proteins were labeled with a DIBAC-functionalized biotin reagent (DIBAC-biotin; SI Appendix, Fig. S1) and enriched by binding to immobilized streptavidin (Materials and Methods). The specificity of the enrichment step was examined by comparing lysates from transfected and nontransfected cells (SI Appendix, Fig. S6A). Though numerous proteins were enriched from the lysate of transfected cells, proteins were not detected in the elution fraction of the mock enrichment experiment (i.e., from nontransfected cells treated with Anl). To estimate the extent of enrichment, purified GFP containing a single Anl residue was mixed with unlabeled lysate and recovered by the same procedure. By comparison of Coomassie-stained proteins in the initial mixture to those released from streptavidin, we estimate that GFP was enriched by at least 500-fold (SI Appendix, Fig. S6B).
Proteins enriched from cell lysates were digested with trypsin, and the tryptic peptides were analyzed by tandem mass spectrometry. The collected spectra were used to direct a peptide identification search in which the masses of Anl and DIBAC-biotin–modified Anl were included as variable modifications (SI Appendix, Fig. S7). In an initial analysis we identified 1,690 peptides (Table 1 and Dataset S1). Most notably, the N-terminal tryptic fragment of polypyrimidine tract binding protein-1 (PTBP-1; a housekeeping protein) was detected with an Nα-acetylated, DIBAC-biotin–modified Anl in place of Met (SI Appendix, Fig. S8). The mass of the parent ion matched the expected mass of the substituted peptide, and the modification was directly observed in both the b- and y-ion spectra upon fragmentation. Including fragments identified from internal regions of the protein, PTBP-1 was detected with 43% sequence coverage and 15 unique peptides (three of which contained one or more internal Met residues).
Table 1.
Summary of detected peptides from enriched proteins
| Species detected | DIBAC-biotin* | DIBAC-biotin† | DIBAC-S-S-biotin | Combined |
| Peptides | 1,690 | 1,538 | 5,956 | 9,184 |
| Peptides containing ≥1 Met | 354 | 186 | 1,131 | 1,671 |
| Total Met residues detected | 410 | 217 | 1,538 | 2,165 |
| Internal Met residues | 403 | 217 | 1,531 | 2,151 |
| N-terminal Anl residues | 1 | 0 | 11 | 12 |
| Internal Anl residues | 0 | 0 | 0 | 0 |
Detected peptides from cells pulsed with Anl only.
Detected heavy lysine-containing peptides from cells copulsed with Anl and heavy lysine.
In addition to the PTBP N-terminal peptide, 15 N-terminal peptides that did not contain Anl were also identified (Table 2 and Dataset S1). Eight peptides contained Met at the initiator position; the other seven were derived from proteins from which the initiator amino acid had been removed. Though it is possible that such peptides could have been derived from proteins tagged with Anl at internal positions, we suspected that they were fragments of contaminating proteins present in the elution fraction at low concentrations. Several of these peptides were derived from highly abundant proteins (such as the 70-kDa heat shock protein and γ-actin), which may have not been removed completely during enrichment. The remaining peptides were derived from proteins that were probably enriched unintentionally, such as endogenously biotinylated proteins or proteins covalently attached to Anl-tagged proteins (such as ubiquitin). To eliminate the ambiguity associated with contaminating proteins, we analyzed an enriched proteome collected from cells labeled simultaneously with Anl and [13C6, 15N2]lysine (heavy lysine). When we limited our analysis to peptides that contained the isotopic label, only a single N-terminal peptide lacking Anl was identified (Dataset S1). The N terminus of cofilin-1 was detected without Anl, but with the isotopic label, indicating that it was synthesized during the Anl pulse. Cofilin-1 is known to form intermolecular disulfide bonds (27), so it may have been carried through the enrichment step while bound to an Anl-tagged partner. None of the other unmodified N termini observed in our initial analysis were detected in the heavy lysine experiment.
Table 2.
Summary of heavy lysine-labeled peptides derived from enriched proteins
| Species detected | Detected peptides | Detected peptides containing heavy lysine |
| Total detected peptides | 1,690 | 1,538 |
| Unmodified N-terminal peptides | 15 (0.89%) | 1 (0.00065%) |
| C-terminal peptides | 25 (1.47%) | 23 (1.50%) |
To facilitate detection of additional sites of Anl incorporation, we used a cleavable enrichment probe in which a disulfide bond is positioned between the DIBAC and biotin moieties (DIBAC-S-S-biotin; SI Appendix, Fig. S1). The selectivity of DIBAC-S-S-biotin was similar to that of DIBAC-biotin with respect to labeling of cysteine-free proteins (SI Appendix, Fig. S9); for proteomic analyses, protein samples were reduced and alkylated before treatment with DIBAC-S-S-biotin. Following enrichment, reduction of the disulfide bond releases a fragment of the probe from modified proteins. The fragment that remains appended to the site of modification is more than 200 mass units smaller than that obtained upon treatment with DIBAC-biotin, and is more readily detected by MS analysis. We enriched proteins from labeled cell lysates by using the cleavable probe and analyzed them as before, with the fragment derived from reduced and alkylated DIBAC-S-S-biotin as an additional variable substitution (SI Appendix, Fig. S7). Eleven additional Anl modifications were identified by using the cleavable probe, each of which corresponded to a unique N-terminal peptide (Table 1; SI Appendix, Figs. S8B and S10; Dataset S1).
In sum, 12 peptides provided direct evidence of Anl at N-terminal positions, indicating that Anl is charged to tRNAiMet and that Anl-tRNAiMet can be used by the cell to initiate translation. Though we identified 1,671 peptides containing one or more Met residues and sampled 2,165 Met positions (2,151 of which were internal), we did not find evidence of Anl incorporation at internal positions (Table 1).
N-Terminal Anl Is Cleaved by Methionyl Aminopeptidase at an Attenuated Rate.
N-terminal Met residues are subject to excision from proteins by methionyl aminopeptidases (MetAPs) (28). Noncanonical amino acids at N-terminal sites can also be subject to N-terminal excision, although the rate of cleavage is typically reduced (29, 30). To investigate removal of Anl, we measured the activity of recombinant human MetAP-2 (cytoplasmic form) using Anl-7-amino-4-methylcoumarin (Anl-AMC; SI Appendix, Fig. S11A) and Met-7-amino-4-methylcoumarin (Met-AMC; SI Appendix, Fig. S11A) as substrates in a fluorometric assay. Though the enzyme cleaved both Anl-AMC and Met-AMC, kinetic analysis revealed that cleavage of Anl-AMC was approximately threefold slower (SI Appendix, Fig. S11B). We suspect that the rate of Anl excision inside cells may be even further attenuated, because treatment of cells with fumagillin (a MetAP-2 inhibitor) led to only a slight increase in the amount of Anl-labeled protein detected in cell lysates (SI Appendix, Fig. S11C). Several known MetAP substrates were detected with Anl residues at N-terminal positions (SI Appendix, Table S1).
N-Terminal Anl Is Recognized by Nα-Acetyltransferases.
Proteins are subject to modification by Nα-acetyltransferases (NATs), which catalyze the addition of acetyl groups to the N termini of substrate proteins (28). Nα-acetylation occurs primarily cotranslationally as nascent polypeptides emerge from the ribosome. Substrate selection is governed by N-terminal sequence determinants (31); it is estimated that more than 80% of human proteins are NAT substrates (32). Of the 12 peptides identified with N-terminal modifications, seven were derived from known NAT substrates; all seven were detected in Nα-acetyl-Anl form (SI Appendix, Table S2). The remaining five fragments were detected with unmodified terminal amines and were derived from proteins that do not normally contain Nα-acetyl-Met.
Distribution of Newly Synthesized Proteins Is Cell-Cycle Dependent.
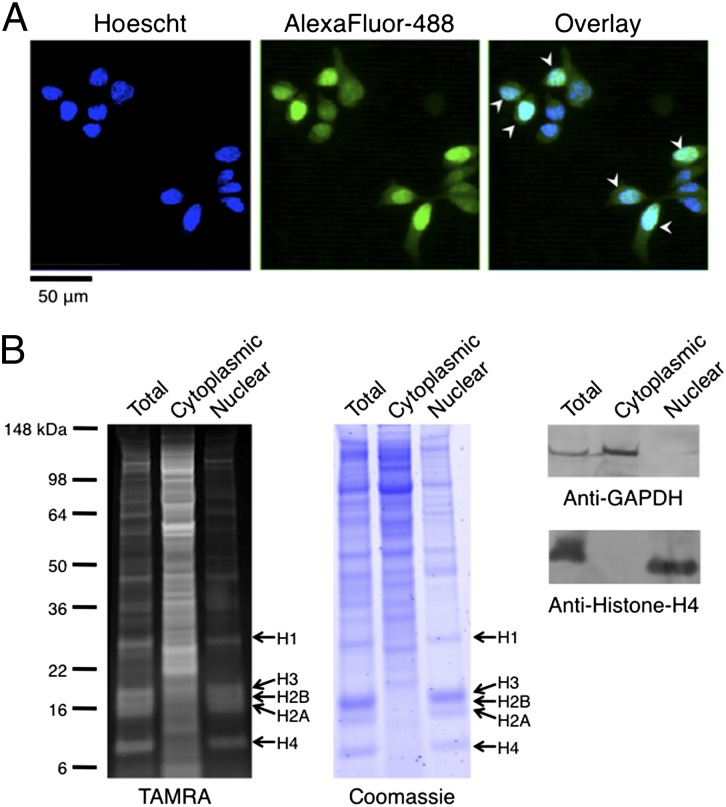
Fluorescence imaging of dye-labeled HEK293-8D3 cells revealed striking cell-to-cell variation in the localization of newly synthesized proteins. Though many cells exhibited nearly uniform spatial distributions of labeled proteins, a portion of the cell population (approximately one-third of all cells) displayed prominent intensities that colocalized with the nuclear stain Hoechst 33342 (Fig. 2A), suggesting that in these cells, much of the newly synthesized protein was translocated to the nucleus. To identify these proteins, we analyzed subcellular protein fractions by treatment with DIBAC-TAMRA, electrophoretic separation, and in-gel fluorescence scanning. In gels prepared from the nuclear fraction of Anl-treated HEK293-8D3 cells, we observed TAMRA-labeled proteins that migrated in a pattern characteristic of the core histone proteins (Fig. 2B). Histone labeling is consistent with the imaging results discussed previously; histones are abundant nuclear proteins synthesized predominantly during S phase of the cell cycle (33). Because we labeled growing, asynchronous cells, a fraction of the population was undoubtedly in S phase. Thus, the prominent nuclear intensities we saw in a fraction of cells were likely the result of labeled histones synthesized and translocated to the nucleus by cells undergoing DNA replication.
Fig. 2.
Detection and visualization of newly synthesized proteins localized to cell nuclei. (A) Labeled proteins in pulsed HEK293-8D3 cells were modified with Alexa Fluor 488-alkyne and visualized by fluorescence microscopy. Prominent intensities that colocalized with nuclear staining (Hoechst 33342) were observed in a fraction of cells. (B) Total protein, as well as cytoplasmic and nuclear fractions from pulsed HEK293-8D3 cells, was treated with DIBAC-TAMRA, and labeling was detected by in-gel fluorescence scanning. Labeling in the nuclear fraction was highest at bands corresponding to the core histone proteins (H3, H2B, H2A, and H4) and the adaptor histone (H1). Western blot detection of marker proteins for the cytoplasm (GAPDH) and the nucleus (histone H4) verified the quality of fractionation.
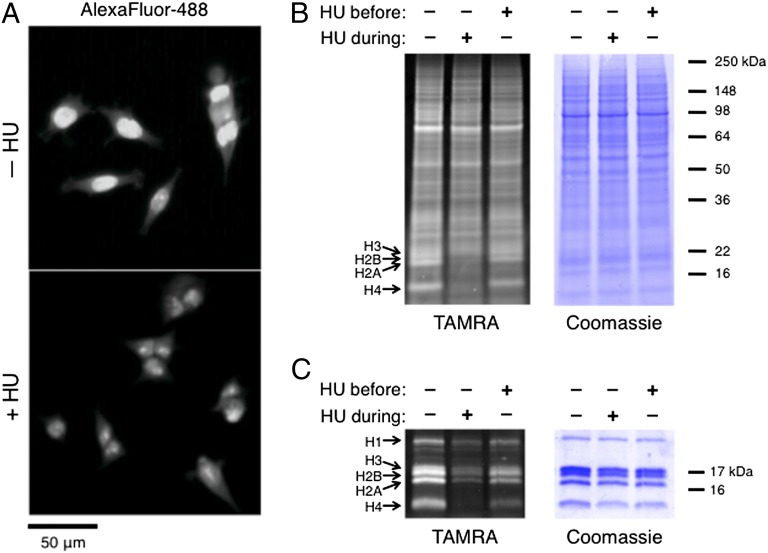
To verify that the observed intensities did in fact represent newly synthesized histones, we treated HEK293-8D3 cells with Anl in the presence of hydroxyurea (HU), which slows histone synthesis by reducing the levels of histone messenger RNAs in the cell (34). Imaging confirmed that the localized intensities did indeed represent new histones, because these intensities were not observed in HU-treated cells (Fig. 3A). We also analyzed total protein and histone extracts from Anl-treated cells by DIBAC-TAMRA labeling and in-gel fluorescence scanning. Though the majority of the labeled proteins appeared to be unchanged, HU treatment led to significantly diminished TAMRA intensity at bands corresponding to the core histone proteins (H3, H2B, H2B, H4) and the adaptor histone (H1; Fig. 3 B and C). The decrease in TAMRA intensity (83% decrease; SI Appendix, Fig. S12) agreed with previous measurements of the effect of HU on histone synthesis determined by using [3H]lysine (80–85% decrease) (35). In cells that were treated with HU before the pulse and released from the agent shortly before adding Anl, an intermediate level of histone synthesis was observed. Notably, none of the effects of HU treatment on histone synthesis could be detected in conventional Coomassie-stained gels (Fig. 3 B and C).
Fig. 3.
Visualization and quantification of the effect of HU on histone synthesis. (A) Cells treated with Anl in the absence or presence of HU were labeled with Alexa Fluor 488-alkyne and visualized by fluorescence microscopy. The prominent nuclear intensities seen in a portion of untreated cells were not observed in HU-treated cells, confirming that the observed nuclear intensities represented labeled histones synthesized by cells in S phase. (B) Proteins from Anl-treated cells were modified by DIBAC-TAMRA and detected by in-gel fluorescence scanning. Proteins from untreated and HU-treated cells, as well as from cells treated and released from HU before the pulse, were compared. Though the labeling of most proteins appears unchanged, TAMRA intensities at bands corresponding to histones were significantly reduced for HU-treated cells. (C) Histone extracts were analyzed, and quantification of TAMRA intensity indicated that histone synthesis was reduced by 82% during HU treatment. Histone synthesis was recovered in cells released from HU.
Detection of p53 Stabilization in Response to Stalled DNA Replication.
HU acts by quenching the tyrosyl radical of ribonucleotide reductase, thus limiting the pool of nucleotides available for DNA synthesis (36). Because histone and DNA synthesis are tightly coupled, HU also inhibits histone translation. In addition, HU elicits the genotoxic stress response and leads to stabilization of the p53 tumor suppressor (37). p53 is a transcription factor that is continually synthesized, but normally targeted for rapid degradation; the transcription factor is activated by phosphorylation, which inhibits its ubiquitination and, in turn, its degradation. As a result, newly synthesized p53 accumulates and enters the nucleus, where it facilitates transcription of target genes that mediate cell-cycle arrest.
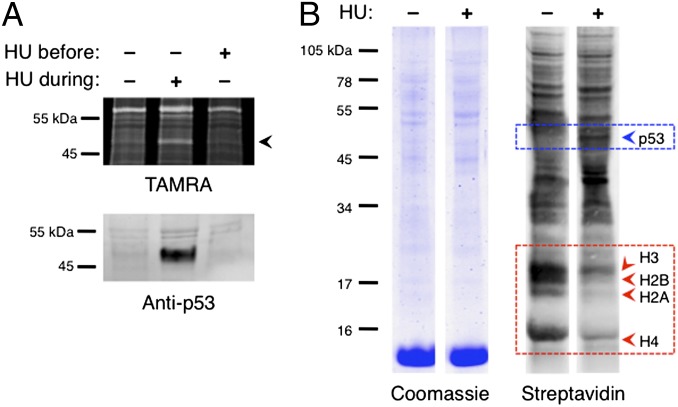
In analysis of Anl-labeled proteins, we detected a band with an approximate mass of 53 kDa that was present in HU-treated samples and absent from untreated and HU-released samples (Fig. 4A). Western analysis with anti-p53 antibody confirmed stabilization of p53 in HU-treated cells; the position of the antibody-labeled band matched that of the 53-kDa TAMRA-labeled band induced by HU. Both the HU-mediated decrease in histone translation and stabilization of p53 were observed in direct analysis of enriched proteins (Fig. 4B).
Fig. 4.
Detection of p53 stabilization and destabilization. (A) A protein labeled with an approximate mass of 53 kDa was labeled in HU-treated cells but not in untreated or HU-released cells (black arrowhead). The band represents p53 tumor suppressor, a protein that is continually synthesized yet rapidly degraded. HU treatment of cells stabilizes newly synthesized copies of p53. Western analysis confirmed stabilization of p53 during HU treatment. (B) Coomassie staining and streptavidin detection of proteins enriched from untreated and HU-treated cell lysates using DIBAC-biotin. Decreased histone translation (red arrowheads) and p53 stabilization seen during analysis of cell lysates were also apparent following enrichment (blue arrowhead).
Discussion
Noncanonical amino acids have been used to measure the kinetics of protein and nucleosome turnover (38, 39), visualize local protein synthesis in neuronal subcompartments (40, 41), and identify the products of stimulus-induced protein synthesis in axons and dendrites (42, 43). Here we introduce an additional level of control in the metabolic labeling of proteins made in mammalian cells, by using a noncanonical amino acid that requires a mutant aminoacyl-tRNA synthetase for activation. We believe that this system will enable cell-selective interrogation of protein synthesis in cocultures of mammalian cells or in virally transfected tissues. Use of Aha to examine protein synthesis in zebrafish has recently been described (44); extension of the strategy introduced here should permit cell-selective analysis in vivo and provide a valuable complement to existing methods for cell-selective isolation of mRNAs (45–47). Use of promoters to limit expression of mutant synthetases to specific cell states (such as specific developmental stages) (48) may allow further refinement of labeling specificity. It is important to note that NLL-EcMetRS may not prove to be the synthetase of choice for such experiments; our preliminary efforts to prepare transgenic C. elegans strains that express NLL-EcMetRS under control of cell-selective promoters have thus far been unsuccessful.
Expression of NLL-EcMetRS enables incorporation of Anl specifically at the N termini of proteins made in mammalian cells. The bacterial synthetase accepts the mammalian initiator tRNA, but none of the mammalian elongator tRNAs, as a substrate. Discrimination against mammalian elongator tRNAs is an intrinsic property of the bacterial MetRS; the enzyme recognizes Met tRNAs with seven-member anticodon loops (such as bacterial Met tRNAs and mammalian tRNAiMet) but not those with nine-member anticodon loops (such as the mammalian cytoplasmic elongator tRNAMet) (17). Selective charging in this manner prohibits Anl incorporation at internal positions. We reasoned that a confident determination of selective N-terminal incorporation required (i) direct evidence of translational initiation with Anl and (ii) a reliable inference of the absence of Anl at internal positions. We identified 12 peptides that provided direct evidence of Anl at N-terminal positions. These peptides provided confirmation that Anl was indeed charged to tRNAiMet and that Anl-tRNAiMet could be used to initiate protein synthesis.
Evidence of Anl incorporation at internal sites was not observed despite sampling a total of 2,143 Met positions in 1,671 peptides. This result is especially significant considering that N-terminal peptides are substantially outnumbered by internal Met-containing peptides in any digested proteome. For instance, trypsin digestion of the E. coli and human proteomes generates on average 28 and 45 peptides per protein, respectively (49). The E. coli genome encodes 8.2-fold more internal Mets than initiator Mets, and the human genome encodes 9.2-fold more [average values calculated from the codon distribution of the protein coding genes of E. coli and Homo sapiens genomes from National Center for Biotechnology Information (NCBI) GenBank Release 160]. Thus, even if Anl were incorporated 10× more slowly at internal sites than at N-terminal sites, one would expect to detect comparable numbers of N-terminal and internal modifications. Consistent with this argument are the results of Nessen et al. (50), who reported an analysis of peptides isolated from proteome-wide labeling in E. coli and found 10-fold more Aha modifications at internal sites than at N termini.
We found that the rate of MetAP-catalyzed excision of N-terminal Anl residues was attenuated in comparison with that observed for N-terminal Met. However, because Anl is incorporated selectively at N-terminal positions, proteins that are subject to cleavage of N-terminal signal sequences would not retain their Anl labels. In an unbiased analysis of N-terminal peptides collected from Jurkat cells, 32% of the identified N termini indicated a non-MetAP proteolytic cleavage site or an alternative translational initiation site (51). These “neo” N termini may have been generated following cleavage by signal peptidases (52), mitochondrial processing peptidase (52), or other intracellular proteases. Development of mutant mammalian synthetases that would permit incorporation of labels at internal positions would complement the labeling method introduced here, by allowing enrichment and identification of proteins that lack N-terminal Met residues.
Finally, we anticipate that the approach introduced here will be useful in preparing proteins containing site-specific modifications for biotechnological applications. Selective aminoacylation by EcMetRS has been previously used to produce proteins containing N-terminal biotinylated or fluorescent Met derivatives in cell-free systems (53–55). Adaptation of the methods introduced here to suspension culture of mammalian cells would complement existing strategies (56–58) for preparation of site-specific protein conjugates.
Materials and Methods
Full details regarding experimental procedures and the compounds used in this study can be found in SI Appendix, SI Materials and Methods.
Anl Labeling Procedure.
HEK293 cells were grown in Dulbecco’s Modified Eagle Medium containing 10% (vol/vol) FBS, penicillin, and streptomycin, and 50 μg⋅mL−1 Geneticin at 37 °C, with 5% CO2 using standard procedures. The culture medium was exchanged with fresh medium either several hours or the night before each experiment. Cells at 70–80% confluence were labeled by addition of Anl to the culture medium at the indicated concentrations and incubated for the indicated times at 37 °C under 5% CO2. Labeling was terminated by aspiration of the culture medium, and cells were washed with PBS and collected by trypsinization and centrifugation. Pelleted cells were washed with PBS to remove trypsin and stored at −20 °C until further processing.
Protein Modification and Detection with DIBAC-Functionalized Probes.
DIBAC probe stocks (5–20 mM) were prepared in DMSO, and conjugation reactions were initiated by dilution of probe into solutions of alkylated proteins to a final concentration of 20 μM. Reactions with DIBAC-TAMRA were incubated at room temperature for 20 min with gentle mixing and protection from light. Reactions with DIBAC-biotin and DIBAC-S-S-biotin were mixed gently at room temperature for 1 h. Reactions were quenched by addition of a fivefold excess (with respect to probe) of free Anl and immediately mixed by vortexing.
Supplementary Material
Acknowledgments
We thank Michael Sweredowski, Robert Graham, and Sonja Hess of the Proteome Exploration Laboratory (Beckman Institute, Caltech) for their help and advice in collecting and analyzing tandem mass spectrometry data; Angela Ho and Jost Vielmetter of the Protein Expression Center (Beckman Institute, Caltech) for their help in generating stable cell lines; Carolyn Bertozzi and her laboratory (University of California, Berkeley) for their gift of DIFO-Alexa Fluor 488; and John Phillips (Caltech) and members of D.A.T.’s laboratory for helpful discussions. Support for this work was provided by National Institutes of Health Grant R01 GM062523, the Joseph J. Jacobs Institute for Molecular Engineering for Medicine, and the Institute for Collaborative Biotechnologies through Grant W911NF-09-0001 from the US Army Research Office.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1216375110/-/DCSupplemental.
References
- 1.Gray NK, Wickens M. Control of translation initiation in animals. Annu Rev Cell Dev Biol. 1998;14:399–458. doi: 10.1146/annurev.cellbio.14.1.399. [DOI] [PubMed] [Google Scholar]
- 2.Eagle H, Piez KA, Fleischman R, Oyama VI. Protein turnover in mammalian cell cultures. J Biol Chem. 1959;234(3):592–597. [PubMed] [Google Scholar]
- 3.Schwanhäusser B, Gossen M, Dittmar G, Selbach M. Global analysis of cellular protein translation by pulsed SILAC. Proteomics. 2009;9(1):205–209. doi: 10.1002/pmic.200800275. [DOI] [PubMed] [Google Scholar]
- 4.Ngo JT, Tirrell DA. Noncanonical amino acids in the interrogation of cellular protein synthesis. Acc Chem Res. 2011;44(9):677–685. doi: 10.1021/ar200144y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dieterich DC, Link AJ, Graumann J, Tirrell DA, Schuman EM. Selective identification of newly synthesized proteins in mammalian cells using bioorthogonal noncanonical amino acid tagging (BONCAT) Proc Natl Acad Sci USA. 2006;103(25):9482–9487. doi: 10.1073/pnas.0601637103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu J, Xu Y, Stoleru D, Salic A. Imaging protein synthesis in cells and tissues with an alkyne analog of puromycin. Proc Natl Acad Sci USA. 2012;109(2):413–418. doi: 10.1073/pnas.1111561108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ong C-T, Corces VG. Enhancer function: New insights into the regulation of tissue-specific gene expression. Nat Rev Genet. 2011;12(4):283–293. doi: 10.1038/nrg2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ngo JT, et al. Cell-selective metabolic labeling of proteins. Nat Chem Biol. 2009;5(10):715–717. doi: 10.1038/nchembio.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tanrikulu IC, Schmitt E, Mechulam Y, Goddard WA, 3rd, Tirrell DA. Discovery of Escherichia coli methionyl-tRNA synthetase mutants for efficient labeling of proteins with azidonorleucine in vivo. Proc Natl Acad Sci USA. 2009;106(36):15285–15290. doi: 10.1073/pnas.0905735106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Szychowski J, et al. Cleavable biotin probes for labeling of biomolecules via azide-alkyne cycloaddition. J Am Chem Soc. 2010;132(51):18351–18360. doi: 10.1021/ja1083909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beatty KE, Xie F, Wang Q, Tirrell DA. Selective dye-labeling of newly synthesized proteins in bacterial cells. J Am Chem Soc. 2005;127(41):14150–14151. doi: 10.1021/ja054643w. [DOI] [PubMed] [Google Scholar]
- 12.Beatty KE, et al. Fluorescence visualization of newly synthesized proteins in mammalian cells. Angew Chem Int Ed Engl. 2006;45(44):7364–7367. doi: 10.1002/anie.200602114. [DOI] [PubMed] [Google Scholar]
- 13.Grammel M, Zhang MM, Hang HC. Orthogonal alkynyl amino acid reporter for selective labeling of bacterial proteomes during infection. Angew Chem Int Ed Engl. 2010;49(34):5970–5974. doi: 10.1002/anie.201002050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blanquet S, Petrissant G, Waller JP. The mechanism of action of methionine-tRNA synthetase: 2. Interaction of the enzyme with specific and unspecific tRNAs. Eur J Biochem. 1973;36(1):227–233. doi: 10.1111/j.1432-1033.1973.tb02904.x. [DOI] [PubMed] [Google Scholar]
- 15.Drabkin HJ, Estrella M, Rajbhandary UL. Initiator-elongator discrimination in vertebrate tRNAs for protein synthesis. Mol Cell Biol. 1998;18(3):1459–1466. doi: 10.1128/mcb.18.3.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gupta NK, Chatterjee KK, Bose S, Chung A. Roles of methionine transfer RNAs in protein synthesis in rabbit reticulocytes. J Mol Biol. 1970;54(1):145–154. doi: 10.1016/0022-2836(70)90452-3. [DOI] [PubMed] [Google Scholar]
- 17.Meinnel T, Mechulam Y, Fayat G, Blanquet S. Involvement of the size and sequence of the anticodon loop in tRNA recognition by mammalian and E. coli methionyl-tRNA synthetases. Nucleic Acids Res. 1992;20(18):4741–4746. doi: 10.1093/nar/20.18.4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stanley WM., Jr Specific aminoacylation of the methionine-specific tRNA’s of eukaryotes. Methods Enzymol. 1974;29:530–547. doi: 10.1016/0076-6879(74)29049-9. [DOI] [PubMed] [Google Scholar]
- 19.Kozak M. Comparison of initiation of protein synthesis in procaryotes, eucaryotes, and organelles. Microbiol Rev. 1983;47(1):1–45. doi: 10.1128/mr.47.1.1-45.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith AE, Marcker KA. Cytoplasmic methionine transfer RNAs from eukaryotes. Nature. 1970;226(5246):607–610. doi: 10.1038/226607a0. [DOI] [PubMed] [Google Scholar]
- 21.Dettman GL, Stanley WM., Jr Recognition of eukaryotic initiator tRNA by an initiation factor and the transfer of the methionine moiety into peptide linkage. Biochim Biophys Acta. 1972;287(1):124–133. doi: 10.1016/0005-2787(72)90336-x. [DOI] [PubMed] [Google Scholar]
- 22.Pestova TV, Hellen CUT. Preparation and activity of synthetic unmodified mammalian tRNAi(Met) in initiation of translation in vitro. RNA. 2001;7(10):1496–1505. doi: 10.1017/s135583820101038x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baskin JM, et al. Copper-free click chemistry for dynamic in vivo imaging. Proc Natl Acad Sci USA. 2007;104(43):16793–16797. doi: 10.1073/pnas.0707090104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hong V, Presolski SI, Ma C, Finn MG. Analysis and optimization of copper-catalyzed azide-alkyne cycloaddition for bioconjugation. Angew Chem Int Ed Engl. 2009;48(52):9879–9883. doi: 10.1002/anie.200905087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Debets MF, et al. Aza-dibenzocyclooctynes for fast and efficient enzyme PEGylation via copper-free (3+2) cycloaddition. Chem Commun (Camb) 2010;46(1):97–99. doi: 10.1039/b917797c. [DOI] [PubMed] [Google Scholar]
- 26.van Geel R, Pruijn GJ, van Delft FL, Boelens WC. Preventing thiol-yne addition improves the specificity of strain-promoted azide-alkyne cycloaddition. Bioconjug Chem. 2012;23(3):392–398. doi: 10.1021/bc200365k. [DOI] [PubMed] [Google Scholar]
- 27.Bernstein BW, Bamburg JR. ADF/cofilin: A functional node in cell biology. Trends Cell Biol. 2010;20(4):187–195. doi: 10.1016/j.tcb.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bradshaw RA, Brickey WW, Walker KW. N-terminal processing: The methionine aminopeptidase and N alpha-acetyl transferase families. Trends Biochem Sci. 1998;23(7):263–267. doi: 10.1016/s0968-0004(98)01227-4. [DOI] [PubMed] [Google Scholar]
- 29.Wang A, Winblade Nairn N, Johnson RS, Tirrell DA, Grabstein K. Processing of N-terminal unnatural amino acids in recombinant human interferon-beta in Escherichia coli. ChemBioChem. 2008;9(2):324–330. doi: 10.1002/cbic.200700379. [DOI] [PubMed] [Google Scholar]
- 30.Wiltschi B, Merkel L, Budisa N. Fine tuning the N-terminal residue excision with methionine analogues. ChemBioChem. 2009;10(2):217–220. doi: 10.1002/cbic.200800605. [DOI] [PubMed] [Google Scholar]
- 31.Pestana A, Pitot HC. Acetylation of nascent polypeptide chains on rat liver polyribosomes in vivo and in vitro. Biochemistry. 1975;14(7):1404–1412. doi: 10.1021/bi00678a010. [DOI] [PubMed] [Google Scholar]
- 32.Arnesen T, et al. Proteomics analyses reveal the evolutionary conservation and divergence of N-terminal acetyltransferases from yeast and humans. Proc Natl Acad Sci USA. 2009;106(20):8157–8162. doi: 10.1073/pnas.0901931106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marzluff WF, Duronio RJ. Histone mRNA expression: Multiple levels of cell cycle regulation and important developmental consequences. Curr Opin Cell Biol. 2002;14(6):692–699. doi: 10.1016/s0955-0674(02)00387-3. [DOI] [PubMed] [Google Scholar]
- 34.Sittman DB, Graves RA, Marzluff WF. Histone mRNA concentrations are regulated at the level of transcription and mRNA degradation. Proc Natl Acad Sci USA. 1983;80(7):1849–1853. doi: 10.1073/pnas.80.7.1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Senshu T, Ohashi M. Fate of newly synthesized histones shortly after interruption of DNA replication. J Biochem. 1979;86(5):1259–1267. doi: 10.1093/oxfordjournals.jbchem.a132641. [DOI] [PubMed] [Google Scholar]
- 36.Atkin CL, Thelander L, Reichard P, Lang G. Iron and free radical in ribonucleotide reductase. Exchange of iron and Mössbauer spectroscopy of the protein B2 subunit of the Escherichia coli enzyme. J Biol Chem. 1973;248(21):7464–7472. [PubMed] [Google Scholar]
- 37.Meek DW. Tumour suppression by p53: A role for the DNA damage response? Nat Rev Cancer. 2009;9(10):714–723. doi: 10.1038/nrc2716. [DOI] [PubMed] [Google Scholar]
- 38.Zhang MM, Tsou LK, Charron G, Raghavan AS, Hang HC. Tandem fluorescence imaging of dynamic S-acylation and protein turnover. Proc Natl Acad Sci USA. 2010;107:8623–8627. doi: 10.1073/pnas.0912306107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deal RB, Henikoff JG, Henikoff S. Genome-wide kinetics of nucleosome turnover determined by metabolic labeling of histones. Science. 2010;328(5982):1161–1164. doi: 10.1126/science.1186777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tcherkezian J, Brittis PA, Thomas F, Roux PP, Flanagan JG. Transmembrane receptor DCC associates with protein synthesis machinery and regulates translation. Cell. 2010;141(4):632–644. doi: 10.1016/j.cell.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dieterich DC, et al. In situ visualization and dynamics of newly synthesized proteins in rat hippocampal neurons. Nat Neurosci. 2010;13(7):897–905. doi: 10.1038/nn.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoon BC, et al. Local translation of extranuclear lamin B promotes axon maintenance. Cell. 2012;148(4):752–764. doi: 10.1016/j.cell.2011.11.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hodas JJ, et al. Dopaminergic modulation of the hippocampal neuropil proteome identified by bioorthogonal noncanonical amino acid tagging (BONCAT) Proteomics. 2012;12(15-16):2464–2476. doi: 10.1002/pmic.201200112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hinz FI, Dieterich DC, Tirrell DA, Schuman EM. Non-canonical amino acid labeling in vivo to visualize and affinity purify newly synthesized proteins in larval zebrafish. ACS Chem Neurosci. 2012;3(1):40–49. doi: 10.1021/cn2000876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heiman M, et al. A translational profiling approach for the molecular characterization of CNS cell types. Cell. 2008;135(4):738–748. doi: 10.1016/j.cell.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sanz E, et al. Cell-type-specific isolation of ribosome-associated mRNA from complex tissues. Proc Natl Acad Sci USA. 2009;106(33):13939–13944. doi: 10.1073/pnas.0907143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miller MR, Robinson KJ, Cleary MD, Doe CQ. TU-tagging: Cell type-specific RNA isolation from intact complex tissues. Nat Methods. 2009;6(6):439–441. doi: 10.1038/nmeth.1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ngo JT, Babin BM, Champion JA, Schuman EM, Tirrell DA. State-selective metabolic labeling of cellular proteins. ACS Chem Biol. 2012;7(8):1326–1330. doi: 10.1021/cb300238w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cagney G, Amiri S, Premawaradena T, Lindo M, Emili A. In silico proteome analysis to facilitate proteomics experiments using mass spectrometry. Proteome Sci. 2003;1(1):5–20. doi: 10.1186/1477-5956-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nessen MA, et al. Selective enrichment of azide-containing peptides from complex mixtures. J Proteome Res. 2009;8(7):3702–3711. doi: 10.1021/pr900257z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Staes A, et al. Selecting protein N-terminal peptides by combined fractional diagonal chromatography. Nat Protoc. 2011;6(8):1130–1141. doi: 10.1038/nprot.2011.355. [DOI] [PubMed] [Google Scholar]
- 52.Paetzel M, Karla A, Strynadka NCJ, Dalbey RE. Signal peptidases. Chem Rev. 2002;102(12):4549–4580. doi: 10.1021/cr010166y. [DOI] [PubMed] [Google Scholar]
- 53.Olejnik J, Gite S, Mamaev S, Rothschild KJ. N-terminal labeling of proteins using initiator tRNA. Methods. 2005;36(3):252–260. doi: 10.1016/j.ymeth.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 54.Mamaev S, Olejnik J, Olejnik EK, Rothschild KJ. Cell-free N-terminal protein labeling using initiator suppressor tRNA. Anal Biochem. 2004;326(1):25–32. doi: 10.1016/j.ab.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 55.Gite S, Mamaev S, Olejnik J, Rothschild K. Ultrasensitive fluorescence-based detection of nascent proteins in gels. Anal Biochem. 2000;279(2):218–225. doi: 10.1006/abio.1999.4472. [DOI] [PubMed] [Google Scholar]
- 56.Wang L, Xie J, Schultz PG. Expanding the genetic code. Curr Opin Chem Biol. 2005;9:548–554. doi: 10.1016/j.cbpa.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 57.Davis L, Chin JW. Designer proteins: Applications of genetic code expansion in cell biology. Nat Rev Mol Cell Biol. 2012;13(3):168–182. doi: 10.1038/nrm3286. [DOI] [PubMed] [Google Scholar]
- 58.Carrico IS, Carlson BL, Bertozzi CR. Introducing genetically encoded aldehydes into proteins. Nat Chem Biol. 2007;3(6):321–322. doi: 10.1038/nchembio878. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.