Abstract
Aims
The aim of this study was to quantify the collective effect of common lipid-associated single nucleotide polymorphisms (SNPs) on blood lipid levels, cardiovascular risk, use of lipid-lowering medication, and risk of coronary heart disease (CHD) events.
Methods and results
Analysis was performed in two prospective cohorts: Whitehall II (WHII; N = 5059) and the British Women’s Heart and Health Study (BWHHS; N = 3414). For each participant, scores were calculated based on the cumulative effect of multiple genetic variants influencing total cholesterol (TC), low-density lipoprotein cholesterol (LDL-C), high-density lipoprotein cholesterol (HDL-C), and triglycerides (TG). Compared with the bottom quintile, individuals in the top quintile of the LDL-C genetic score distribution had higher LDL-C {mean difference of 0.85 [95% confidence interval, (CI) = 0.76–0.94] and 0.63 [95% CI = 0.50–0.76] mmol/l in WHII and BWHHS, respectively}. They also tended to have greater odds of having ‘high-risk’ status (Framingham 10-year cardiovascular disease risk >20%) [WHII: odds ratio (OR) = 1.36 (0.93–1.98), BWHHS: OR = 1.49 (1.14–1.94)]; receiving lipid-lowering treatment [WHII: OR = 2.38 (1.57–3.59), BWHHS: OR = 2.24 (1.52–3.29)]; and CHD events [WHII: OR = 1.43 (1.02–2.00), BWHHS: OR = 1.31 (0.99–1.72)]. Similar associations were observed for the TC score in both studies. The TG score was associated with high-risk status and medication use in both studies. Neither HDL nor TG scores were associated with the risk of coronary events. The genetic scores did not improve discrimination over the Framingham risk score.
Conclusion
At the population level, common SNPs associated with LDL-C and TC contribute to blood lipid variation, cardiovascular risk, use of lipid-lowering medications and coronary events. However, their effects are too small to discriminate future lipid-lowering medication requirements or coronary events.
Keywords: Lipid genetic score, Lipid medication, Framingham
See page 949 for the editorial comment on this article (doi:10.1093/eurheartj/ehs439)
Introduction
The causal relevance of low-density lipoprotein cholesterol (LDL-C) in coronary disease has been established by numerous trials of LDL-C-lowering interventions.1 In Britain, ∼50% of all coronary events have been attributed to elevated cholesterol level2 and over 7 million people now use statins to reduce cholesterol for the prevention of coronary heart disease (CHD). However, cholesterol levels identify patients at risk of future coronary events only moderately well.3 Many individuals have a cholesterol concentration sufficient to raise the risk of coronary events, but the strength of the association of LDL-C with coronary events is only modest, with about a three-fold relative difference in the risk of coronary events among those at the extremes of the population LDL-C distribution.4
On the basis of observations from prospective studies and randomized trials on statin drugs, UK, European, and Australasian guidelines on the primary prevention of cardiovascular disease (CVD) recommend prescription of statins on the basis of absolute CVD risk rather than solely on LDL-C thresholds. The recommended methods for evaluating absolute CVD risk are based on multiple cardiovascular risk factors. The Framingham 10-year CVD risk equation,5 for example, incorporates information on lipid levels as well as age, sex, blood pressure, smoking habit, and diabetes status. Other risk scores used in Europe include QRISK,6 Euroscore,7 or Prospective Cardiovascular Münster (PROCAM) risk score.8 Despite guidelines, doctors may be persuaded in their therapeutic decisions by high absolute values of total cholesterol (TC) or LDL-C.
All the principal blood lipid fractions: TC, LDL-C, high-density lipoprotein cholesterol (HDL-C), and triglycerides (TG), have both environmental and genetic determinants, with a reported heritability of 40–70%.9 Recently, whole genome and our own previous dense gene-centric array analysis have identified numerous common single nucleotide polymorphisms (SNPs) associated with TC, LDL-C, HDL-C, and TG concentration. In the two cohorts studied here, we previously reported that, though individually these SNPs had a small average effect, lipid concentration differences between individuals carrying few vs. many such variants may be substantial.10 However, there is little information on (nor did we previously examine) the population effect of multiple lipid-associated SNPs on clinically relevant healthcare outcomes such as estimates of cardiovascular risk, prescription of lipid-lowering drug therapies, and subsequent clinical events. There is interest in lipid-associated genotypes as a potential health technology for predicting adverse health outcomes, including the risk of clinical events and the need for later preventative treatments. Potential advantages of genotypes include being fixed from conception, indexing long-term differences in blood lipid values without the biological variation that affects assays of blood lipids themselves, and being assayed at low cost and with very high fidelity.
We therefore studied the influence of common SNPs associated with blood lipid levels on the following outcomes: (i) the odds of being identified as a ‘high-risk’ individual as determined by a Framingham 10-year CVD risk >20%, which is the qualifying threshold used to identify such individuals in Britain (and many other countries), and is a reference against which many other methods of risk prediction are routinely assessed; (ii) the odds of actually receiving lipid-lowering treatment, as guidelines encourage primary therapeutic intervention for these high-risk individuals; and (iii) the risk of coronary disease events. Analysis was carried out in the Whitehall II Study (WHII) and the British Women's Health and Heart Study (BWHHS), in which prescribing decisions were made without knowledge of participants' genotype. For comparison, we looked at the association of the Framingham 10-year CVD risk score, which is based on phenotypic rather than genetic measurements, with the odds of receiving lipid medication and CHD outcome.
Methods
Study populations and non-genetic measures
WHII Study: The study recruited 10 308 participants (70% men), aged 35–55 years, between 1985 and 1989 from 20 London-based Civil service departments.11 Blood samples were collected from 6156 participants in 2002–2004 (see Supplementary material). Clinical and questionnaire data collected between 1991 and 1993 provided the first comprehensive phenotypic dataset for WHII and was considered the baseline phase for the purpose of the current analysis. Follow-up clinical and questionnaire data were also collected between 2003 and 2004 (10 years later). Information on CHD events and lipid medication from the follow-up phase was used for this analysis. The study was approved by the UCL Research Ethics Committee and participants gave informed consent to each aspect of the study
BWHHS: This is a prospective cohort study of 4286 British women between the ages of 60 and 79 years at baseline (1999–2001). Participants were randomly selected from general practice registers in 23 British towns.12 Baseline demographic, anthropometric, and biological data were collected between 1999 and 2001 and a DNA repository was made from 3884 participants (see Supplementary material). Only questionnaire data were collected during follow-up in 2007 (8 years later). Information on CHD events and lipid medication from this follow-up phase was used for this analysis. Ethical committee approval was obtained for the study.
Detailed information on lipid measurements and record of coronary events in both studies are described in the Supplementary material.
Use of lipid-lowering drug therapy
In WHII, participants were asked to name any medication taken in the 14 days prior to the survey at each phase of data collection. For BWHHS, lipid-lowering drug use was determined by face-to-face interview at baseline. Participants were asked to bring to the assessment their repeat medication slips or their actual medications. For subsequent phases, information on medication was obtained from self-administered postal questionnaires. For questions on medication use, participants were encouraged to write medication details direct from their repeat prescription sheet and/or mail a copy of the prescription sheet back with the questionnaire. Detailed information on medication data collection is provided in Supplementary material.
Genotyping
The Illumina HumanCVD BeadChip,13 a gene-centric array containing ∼50 000 SNPs covering ∼2000 CVD candidate loci (see Supplementary material), was used to genotype 5592 subjects from the WHII study10 and 3445 women from BWHHS12 with sufficient DNA preparation (see Supplementary material). Genotypes were generated using Illumina Beadstudio software. After quality control, information from 5059 WHII and 3414 BWHHS Caucasian samples were used in the analysis. The genomic inflation factors14 were close to 1, indicating negligible influence from population structure or genotyping errors. The APOE ɛ2/ɛ3/ɛ4 haplotype, made up of two SNPs (rs429358 and rs7412) and resulting in different isoforms of the ApoE protein, is an important genetic determinant of LDL-C and TC levels. The two APOE SNPs had been separately typed in both studies and APOE haplotypes determined as previously described.15,16
Derivation of lipid genetic scores
Typically many SNPs in a region harbouring one or more causal variant(s) demonstrate univariate associations with the traits of interest, but the majority of these associations are indirect and operate through linkage disequilibrium (LD) with the causal site(s). Previously reported SNPs that exhibited an association with each of baseline lipid measurements in WHII10 were therefore passed through a stepwise variable selection scheme with the Akaike’s Information Criterion (AIC)17 implemented separately for each chromosome (as we expect independence between SNPs on different chromosomes) to remove redundant SNPs (due to correlation) and select the best genetic predictors for each lipid trait.10 The derived SNPs used to construct genetic scores included 19 SNPs plus the APOE haplotype for TC, 21 SNPs plus the APOE haplotype for LDL-C, 12 SNPs for HDL-C, and 16 SNPs for TG (Supplementary material online, Tables S1–S4).
For each lipid trait, genetic scores for each participant were calculated by summing the number of risk alleles (0, 1, or 2 for each SNP). On the basis of prior knowledge of the relationship between the APOE haplotypes and LDL-C levels,18 the APOE risk count was calculated as follows: ɛ2ɛ2/ɛ2ɛ3/ɛ2ɛ4 = 0, ɛ3ɛ3 = 1 and ɛ3ɛ4/ɛ4ɛ4 = 2. Weighting each allele by the corresponding beta coefficient provided very similar estimates (Supplementary material online, Table S5) and the simpler model is presented here for ease of interpretation. Individuals with missing genotypes were excluded. The genetic scores were calculated in the same manner for BWHHS participants.
Estimation of the 10-year absolute risk of cardiovascular disease
Using the equation from the Framingham Heart Study,5 the 10-year risk of developing CVD was estimated using baseline measures in both studies in individuals with complete phenotype data. The equation incorporates information on gender, age, diabetes status, smoking habit, systolic blood pressure, TC, and HDL-C. As the equation incorporates TC levels and is designed for estimating risk in individuals without heart disease, participants on lipid-lowering medication or with CHD at baseline were excluded. The risk is presented as the percentage chance of developing CVD (CHD or stroke) within 10 years.
Association of genetic scores with lipid levels
We expressed the effect of genetic scores on baseline lipid concentration per additional allele (equivalent to a unit change in the score) and as the difference in lipid value between participants in the highest and lowest quintile of the genetic score distribution. Individuals on lipid medication at baseline were excluded from this analysis. Linear regression analysis was performed unadjusted and adjusted for gender (only in WHII) and age. HDL-C and TG variables were log-transformed prior to analysis.
Association of genetic scores with ‘high-risk’ status
We used the Framingham 10-year risk of CVD >20% as an assessment of high-risk status. Using logistic regression, the odds ratio (OR) for having CVD risk >20% were calculated for individuals in the top quintile of the lipid score distribution with reference to individuals in the lowest quintile, unadjusted and adjusted for the respective lipid fraction.
Association of genetic scores with actual use of lipid-lowering medication
Using logistic regression, the ORs for lipid medication use for primary prevention were calculated for individuals in the top quintile of the genetic score distribution with reference to individuals in the lowest quintile, unadjusted and adjusted for the respective lipid fraction. Baseline lipid-medication users were excluded. To ensure that analysis was restricted to subjects receiving lipid-lowering treatment for primary rather than secondary prevention, we excluded those individuals who had a CHD event prior to receiving lipid medication. For comparison, we calculated the unadjusted OR of lipid drug use for individuals with baseline Framingham 10-year CVD risk >20% compared with those with lower risk.
Association of genetic scores with coronary events
We used logistic regression to calculate the OR for developing CHD for those in the top fifth of the lipid score distribution compared with those in the bottom fifth, both unadjusted and adjusted for the relevant lipid level. As genotype precedes outcome, all individuals with CHD at the follow-up phase (including those with CHD at baseline) were included in the analysis. For comparison, we calculated the unadjusted OR of developing CHD for individuals with high baseline risk (>20%) compared with those with lower risk.
Utility of genetic scores for prediction of lipid-lowering drug use and coronary heart disease events
To evaluate the potential value of the lipid genetic scores for discrimination, we calculated the area under the receiver operating characteristic (AUROC) for all four lipid genetic scores for distinguishing individuals that would qualify (i.e. high-risk individuals) and actually use lipid medication as well as for CHD outcome. To determine whether the genetic scores improve discrimination above non-genetic risk factors, we also calculated AUROC for the combined Framingham risk and lipid genetic scores.
The data points used for each analysis in both studies are summarized in Supplementary material online, Table S6.
Results
Participant characteristics
The baseline characteristics of the participants from the two studies are shown in Table 1. Median and ranges for HDL and triglycerides, which had skewed distributions, are provided in the Supplementary material. In WHII, of those individuals that did not have CHD and were not on lipid medication at baseline, 8% had an estimated 10-year CVD risk >20%. On follow-up (∼10 years later) 32% of these ‘high-risk’ individuals were on lipid medication, whereas only 7% of individuals with baseline CVD risk ≤20% were on lipid medication at follow-up. In BWHHS, 49% had high CVD risk (>20%) at baseline, of which 34% were on medication at follow-up (∼8 years later). In comparison, only 8% of BWHHS individuals with baseline CVD risk ≤20% were on lipid medication at follow-up.
Table 1.
Study characteristics
| Whitehall II |
BWHHS | ||
|---|---|---|---|
| Men (N = 3721) | Women (N = 1338) | Women (N = 3443) | |
| Baseline | |||
| Age (years) | 49.1 (5.9) | 49.6 (6.1) | 68.8 (5.5) |
| BMI (kg/m2) | 25.0 (3.1) | 25.3 4.7) | 27.6 (4.9) |
| % Smokers (current) | 11 | 15 | 11 |
| % Smokers (ex/current) | 51 | 46 | 44 |
| Diastolic BP (mmHg) | 80.7 (8.9) | 76.1 (9.3) | 79.4 (11.7) |
| Systolic BP (mmHg) | 121.5 (12.8) | 116.6 (13.7) | 146.9 (25.3) |
| Framingham 10-year risk (%) | 10.6 (6.9) | 5.6 (4.9) | 22.1 (11.7) |
| Total cholesterol (mmol/l) | 6.5 (1.1) | 6.4 (1.2) | 6.6 (1.2) |
| LDL-C (mmol/l) | 4.4 (1.0) | 4.2 (1.1) | 4.2 (1.1) |
| HDL-C (mmol/l) | 1.3 (0.4) | 1.7 (0.4) | 1.7 (0.5) |
| Triglyceride (mmol/l) | 1.6 (1.2) | 1.1 (0.7) | 1.9 (1.2) |
| Baseline | |||
| Lipid drug users | 33 (0.9%) | 10 (0.7%) | 204 (5.9%) |
| CHD cases | 96 (2.6%) | 25 (1.9%) | 460 (13.4%) |
| Follow-up phase | |||
| Duration from baseline | ∼10 years | ∼8 years | |
| Lipid drug users | 426 (11.4%) | 121 (9.0%) | 692 (20.1%) |
| CHD cases | 334 (9.0%) | 87 (6.5) | 802 (23.3%) |
Association of genetic scores with blood lipids
The genic locations of SNPs used in genetic score are shown in Supplementary material online, Tables S1–S4. As expected, though per allele effects on lipid values are small, there is substantial difference in mean lipid levels between individuals in the highest and lowest quintile of the genetic score distribution (Table 2).
Table 2.
Association of genetic scores with lipid levels
| Genetic score | Outcome | Study | Per allele effect |
Top vs. bottom quintile |
||
|---|---|---|---|---|---|---|
| Beta-coefficient (95% CI)a | P value | Beta-coefficient (95%CI)a | P value | |||
| TC | Total cholesterol | WHII | 0.10 (0.09, 0.11) | <1.0 × 10−50 | 0.96 (0.85, 1.07) | <1.0 × 10−50 |
| BWHHS | 0.08 (0.07, 0.09) | 1.2 × 10−26 | 0.62 (0.46, 0.78) | 8.7 × 10−14 | ||
| LDL | LDL-C | WHII | 0.09 (0.08, 0.10) | <1.0 × 10−50 | 0.85 (0.76, 0.94) | <1.0 × 10−50 |
| BWHHS | 0.07 (0.06, 0.08) | 6.7 × 10−32 | 0.63 (0.50, 0.76) | 3.5 × 10−21 | ||
| HDL | Log (HDL-C) | WHII | −0.03 (−0.03, −0.03) | <1.0 × 10−50 | −0.20 (−0.23, −0.18) | <1.0 × 10−50 |
| BWHHS | −0.02 (−0.02, −0.01) | 1.9 × 10−16 | −0.15 (−0.19, 0.12) | 8.6 × 10−16 | ||
| TG | Log (triglyceride) | WHII | 0.04 (0.04, 0.05) | <1.0 × 10−50 | 0.38 (0.33, 0.43) | <1.0 × 10−50 |
| BWHHS | 0.04 (0.04, 0.05) | 2.1 × 10−27 | 0.26 (0.21, 0.31) | 1.5 × 10−21 | ||
aFor untransformed outcomes the beta coefficient represents the mmol/l change in lipid levels. For log-transformed outcomes the beta coefficient represents the percentage change in lipid levels. In each case the beta coefficients are adjusted for gender (only in WHII) and age.
Association of genetic scores with ‘high-risk’ status
Individuals in the top quintile of the distributions of each of the four lipid genetic scores tended to have a higher odds of being identified as ‘high risk’, as determined by the Framingham 10-year CVD risk >20% (Table 3). Adjusting for the respective baseline lipid levels completely attenuated the association of all genetics scores (Table 3). The TG genetic score showed the strongest association, with individuals in the top quintile of the TG score distribution having a 1.99 (1.39–2.85)- and 1.56 (1.22–2.00)-fold higher odds of having CVD risk >20% compared with those in the bottom quintile in WHII and BWHHS, respectively. None of the lipid genetic scores were associated with risk factors incorporated in the Framingham risk equation other than blood lipids (shown for WHII in Supplementary material online, Table S7).
Table 3.
Association of genetic scores with “high-risk” status (Framingham 10-year CVD risk >20%)
| Exposure | Study | Top vs. bottom quintile unadjusted analysis |
Top vs. bottom quintile adjusted for lipid fraction |
||||
|---|---|---|---|---|---|---|---|
| OR (95%CI) | P-value | N | OR (95%CI) | P value | N | ||
| TC score | WHII | 1.53 (1.03–2.26) | 0.034 | 1401 | 0.90 (0.58–1.39) | 0.63 | 1401 |
| BWHHS | 1.30 (0.95–1.77) | 0.097 | 702 | 1.05 (0.76–1.45) | 0.78 | 702 | |
| LDL score | WHII | 1.36 (0.93–1.98) | 0.11 | 1654 | 0.80 (0.53–1.20) | 0.28 | 1654 |
| BWHHS | 1.49 (1.14–1.94) | 0.003 | 885 | 1.04 (0.78–1.39) | 0.79 | 865 | |
| HDL score | WHII | 1.94 (1.37–2.74) | 0.00017 | 1837 | 0.93 (0.64–1.37) | 0.72 | 1837 |
| BWHHS | 1.23 (0.91–1.66) | 0.17 | 747 | 0.86 (0.62–1.19) | 0.35 | 747 | |
| TG score | WHII | 1.99 (1.39–2.85) | 0.00017 | 1576 | 0.95 (0.63–1.44) | 0.81 | 1576 |
| BWHHS | 1.56 (1.22–2.00) | 0.00037 | 1118 | 1.03 (0.79–1.35) | 0.81 | 1118 | |
Association of genetic scores with actual lipid medication use
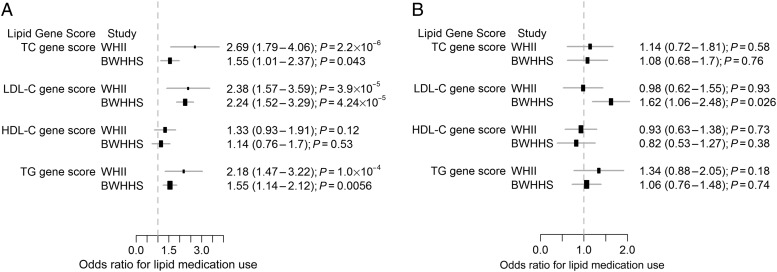
Individuals in the top quintile of the LDL genetic score had a 2.38 (1.57–3.59)- and 2.24 (1.52–3.29)-fold higher odds of receiving lipid medication than those in the lowest quintile (Figure 1A) in WHII and BWHHS, respectively. However, adjustment for LDL-C concentration completely attenuated this association in WHII. In BWHHS, though the association was substantially reduced, it remained significant (Figure 1B). Individuals in the top quintile of the TC and TG genetic scores were more likely to use lipid medication and these associations were attenuated to the null after adjusting for TC and TG levels (Table 3 and Figure 1). The HDL genetic score was not associated with actual use of lipid medication (Figure 1). As expected, individuals with an estimated CVD risk >20% had a higher likelihood [WHII = 4.15 (3.04–5.67); BWHHS = 2.98 (2.32–3.83)] of receiving lipid medication.
Figure 1.
Association of lipid genetic scores with actual lipid medication use. Odds ratio of using lipid-modifying drugs in top vs. bottom quintiles of the genetic score distribution. (A) Unadjusted odds ratios and (B) adjusted for the respective lipid fraction.
Association of genetic scores with coronary heart disease events
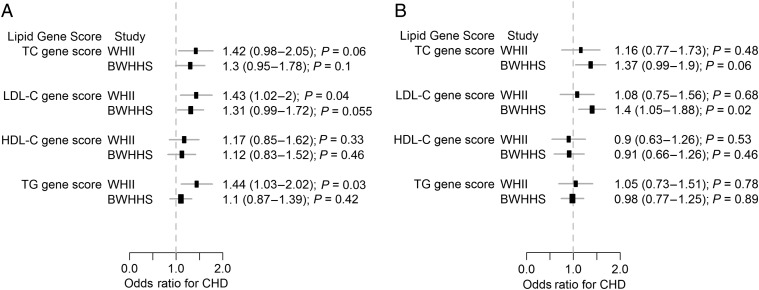
Individuals in the top quintile (compared with bottom quintile) of the LDL-C genetic score distribution had a higher risk of CHD [WHII: OR=1.43 (1.02–2.00) and BWHHS: OR=1.31 (0.99–1.72)] (Figure 2). After adjusting for LDL-C levels, this association was completely attenuated in WHII but not in BWHHS (Figure 2). Similar associations were seen in both studies for the TC genetic score (Figure 2). The TG score showed association with higher risk of CHD in WHII but not in BWHHS (Figure 2). The HDL genetic score was not associated with CHD. By comparison, individuals with 10-year CVD risk >20% had a 4.21 (3.08–5.75)- and 2.49 (1.80–3.44)-fold higher odds of CHD in WHII and BWHHS, respectively.
Figure 2.
Association of lipid genetic scores with coronary heart disease. Odds ratio of coronary heart disease for individuals in the top quintile of the lipid genetic score distribution compared with individuals in the bottom quintile. Odds ratios and P values are shown for (A) unadjusted analyses and (B) adjusted for the respective lipid fraction.
Comparison of genotype-based and phenotype-based discrimination of coronary heart disease events
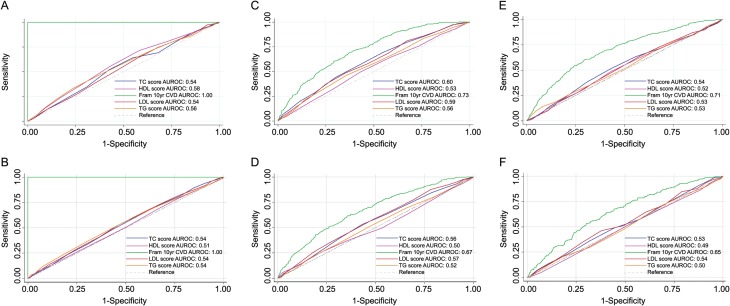
The performance of lipid genetic scores for discriminating high-risk individuals, those who become users of lipid-lowering medication or develop CHD is shown in Figure 3. None of the genetic scores exhibited an AUROC greater than 0.60 whereas the Framingham risk equation exhibited much better discrimination for CHD events (Figure 3). The addition of the individual genetic scores to the Framingham risk score did not improve discrimination of CHD events (Table 4). We also calculated the net reclassification index (NRI) for risk models incorporating Framingham risk and lipid genetic scores. All NRIs were <0.009 (Supplementary material online, Table S8).
Figure 3.
Prediction of lipid drug usage and coronary heart disease events in Whitehall II using lipid genetic scores area under the receiver operating characteristic for discriminating between high-risk individuals based on a Framingham risk >20% in (A) the Whitehall II study and (B) the British Women's Heart and Health Study, actual use of lipid medication in (C) the Whitehall II and (D) the British Women's Heart and Health Study, and coronary heart disease in (E) the Whitehall II and (F) the British Women's Heart and Health Study.
Table 4.
Area under the receiver operating curve for combined Framingham 10-year cardiovascular disease risk score and lipid genetic scores
| Exposure | Study | AUROC for actual drug use | AUROC for CHD |
|---|---|---|---|
| Framingham 10-year CVD risk | WHII | 0.73 (0.70–0.76) | 0.71 (0.67–0.74) |
| BWHHS | 0.67 (0.64–0.70) | 0.65 (0.61–0.69) | |
| Framingham + TC genetic score | WHII | 0.74 (0.71–0.77) | 0.68 (0.65–0.72) |
| BWHHS | 0.68 (0.65–0.71) | 0.65 (0.61–0.69) | |
| Framingham + LDL genetic score | WHII | 0.74 (0.71–0.77) | 0.68 (0.64–0.71) |
| BWHHS | 0.68 (0.65–0.71) | 0.65 (0.61–0.69) | |
| Framingham + HDL genetic score | WHII | 0.71 (0.67–0.74) | 0.70 (0.66–0.73) |
| BWHHS | 0.67 (0.64–0.70) | 0.64 (0.60–0.68) | |
| Framingham + TG genetic score | WHII | 0.72 (0.69–0.76) | 0.70 (0.66–0.73) |
| BWHHS | 0.67 (0.64–0.70) | 0.64 (0.60–0.68) |
Using PLINK to impute lipid genetic scores in individuals with missing data did not substantially alter our results (data available on request).
Discussion
Statement of principal findings
Individuals in the top quintile of the LDL and TC genetic score distributions, calculated using 22 LDL-associated and 20 TC-associated genetic variants, respectively, tended to have greater odds of having high CVD-risk status, receiving lipid-lowering medication and having a CHD event than individuals in the bottom quintile, in two UK studies of middle-aged men and women. Despite predisposing to lifelong differences in levels of blood lipids, the strength of the genetic associations was insufficiently large to usefully discriminate individuals likely to require lipid-lowering treatment or develop CHD. The Framingham risk equation which incorporates a single mid-life measurement of TC and HDL-C as well as other non-genetic risk factors, performed better than genetic scores for the prediction of CHD events, and the addition of the individual genetic scores to the Framingham risk calculation did not improve discrimination.
Comparison with previous studies
Murray et al.19 found that LDL-C and TG genetic scores based on 7 and 11 SNPs, respectively, were associated with the likelihood of exceeding the lipid thresholds for intervention as advocated by the US guidelines (http://www.nhlbi.nih.gov/guidelines/cholesterol/index.htm), in an Italian sample of 1155 individuals over 65 years, but that a score based on nine HDL-C-associated variants was not. However, this study did not examine associations with estimates of absolute CVD risk, nor the number of individuals actually treated with lipid-modifying drugs. In our study the genetic scores were based on SNPs included in the Illumina HumanCVD Beadarray, which has denser SNP coverage of many of the loci16 associated with blood lipid fractions, but lesser genome coverage than whole-genome SNP arrays. SNPs previously studied by Murray et al.19 were either present or had proxy SNPs (based on HapMap CEU LD estimates; R2 > 0.8) present on the array used in our study. Although the loci marked by our lipid genetic scores exhibited substantial overlap with those studied by Murray et al.,19 the denser SNP coverage at each locus, enabled variable selection methods to identify the best genetic predictors of lipid levels from the large number of significant associations observed in each region.
Ripatti et al.20 assessed the performance of a genetic score based on 13 SNPs previously associated with CHD itself rather than blood lipids. Although individuals in the top quintile of the score had a higher relative hazard of CHD compared with individuals in the bottom quintile, this CHD genetic score also did not improve the discrimination of CHD events over non-genetic risk factors and family history. A study by Kathiresan et al.21 examined the utility of LDL and HDL genetic scores for the discrimination of CVD events. They selected a much smaller subset of seven SNPs for LDL-C and four for HDL-C for the score calculation, based on published studies. Although the genetic scores were associated with incident CVD events even after adjustment for lipid levels they did not improve the discrimination of CVD events.21 However, this study did not look at other clinically relevant outcomes.
Implications for clinicians and policymakers
For nearly 2 decades, UK and European guidelines have recommended the use of an absolute risk estimate, based on information on multiple non-genetic risk factors (such as lipid levels, blood pressure, and lifestyle factors) to identify individuals best suited to receive lipid-lowering medication for the primary prevention of CVD. Current guidance for England and Wales from NICE (http://www.nice.org.uk/CG67) offers clinicians the option to use Framingham risk or QRISK. However, measurement error and biological variability could in theory reduce the predictive performance of a single blood lipid measurement because this may not accurately reflect an individual's life time exposure to altered lipid levels. By contrast, genotypes are invariant over time, are measured with minimal error, and can influence lipid levels over a lifetime.21 For this reason, it might be supposed that genetic predictors of healthcare outcomes might outperform cross-sectional phenotypic measurements. However, consistent evidence from this and other studies now indicates that prediction based on phenotype outdoes prediction based on common genotypic variation. Despite this, at the group or population level, genotype might have utility as an index of prescribing because the prevailing average threshold for prescribing is inducible from the relative proportions of genotypes receiving medication.22
Limitations of the study
We studied a large number of SNPs associated with the major blood lipid fractions, but these variants collectively explain only a small proportion of the variance in blood lipid levels and only a fraction of the heritability.10 As the current work was completed, the Global Lipids Genetic Consortium conducted a meta-analysis of genome-wide association studies involving >100 000 participants which has increased the list of loci influencing the major blood lipid fractions to almost 100.23 Scores based on a larger number of lipid-related SNPs will likely explain a larger proportion of the variance in blood lipids and have larger average differences in lipid concentrations in individuals at opposite extremes of the score distribution.23 However, the ability of genetic scores incorporating these additional SNPs to identify individuals with high-risk status or CHD events may not be correspondingly large because the effect sizes of additional loci identified in very large meta-analysis tend to be extremely small. Additionally, new SNPs are also distributed across different chromosomes and inherited independently, so that only a small proportion of the population carries a large burden of lipid-raising alleles. Further analysis based on all known lipid-related loci will be needed to determine whether the interpretations of our findings, based on the HumanCVD array-derived genetic scores, on the utility of lipid-related SNPs for predicting important healthcare outcomes will substantially alter.
Although the content of the array we used for genotyping was based on haplotype tagging SNPs, we may not have captured all the genetic variation at the loci we studied. Ongoing efforts to fine map causal variants at these loci may increase the number of eligible SNPs and improve the performance of lipid-related genetic scores. The effects of gene–gene and gene–environment interactions were not modelled and may also contribute to the missing phenotype variance explained. We also used an approach of simply counting the number of trait-raising alleles, which assumes that the SNPs act independently and additively and with equal effect. However, given that the effect sizes for most SNPs are very small, using a weighted genetics score is unlikely to make a significant difference. Current efforts to deeply resequence for rare variants at the relevant genomic regions may also identify highly penetrant (albeit rare) alleles with a larger effect24 on blood lipid levels than those studied here.
The associations observed in WHII are likely to be overestimated as the same data were used both for SNP discovery and evaluation of the performance of the allele scores. However, the associations and performance estimates were broadly similar in BWHHS. As genotype precedes phenotype, individuals with baseline CHD could be included for the analysis of genetic score with CHD outcome. However, as the Framingham equation is used for the calculation of risk in healthy individuals, those with baseline CHD were excluded for the analysis of Framingham risk score with CHD outcome. Exclusion of higher-risk CHD patients from the latter analysis may have blunted the true association.
A final limitation is that data regarding an individual's use of lipid-lowering medications were obtained by self-recall at interview and from a questionnaire mailed to participants, as access to the participant's medical records was restricted. However, as participants are unaware of their genetic score any reporting error would be non-differential with respect to this, and the statistical expectation would be an underestimation of the reported association.
Conclusion
At the population level, the common SNPs examined here that are associated with TC and LDL-C contribute to variation in blood lipid levels, high cardiovascular risk, use of lipid-lowering medications, and the risk of CHD events. However, the effects are too small to be useful for discriminating the need for lipid-lowering medication for primary prevention, or the future risk of CHD in an individual.
Supplementary material
Supplementary material is available at European Heart Journal online.
Funding
This work on WHII was supported by the British Heart Foundation (BHF) (PG/07/133/24260, RG/08/008, SP/07/007/23671) and a Senior Fellowship to A.D.H. (FS/2005/125). S.E.H. is a BHF Chairholder. M.K. and M.K.'s time on this manuscript was partially supported by the National Heart Lung and Blood Institute (NHLBI:HL36310). The WHII study has been supported by grants from the Medical Research Council; British Heart Foundation; Health and Safety Executive; Department of Health; National Institute on Aging (AG13196), US, National Institute of Health; Agency for Health Care Policy Research (HS06516); and the John D and Catherine T MacArthur Foundation Research Networks on Successful Midlife Development and Socio-economic Status and Health. The British Women’s Heart and Health Study has been supported by funding from the BHF (PG/07/131/24254) and the UK Department of Health Policy Research Programme. The BWHHS Illumina HumanCVD BeadChip work is funded by the BHF (PG/07/131/24254, PI Tom Gaunt).
Conflict of interest: P.J.T., S.E.H., D.A.L., A.D.H., I.N.M.D., and M.K. have support from MRC, BHF, Department of Health (England), NHLBI for the submitted work. J.C.W. is an employee of GlaxoSmithKline. All authors declare no other relationships or activities that could appear to have influenced the submitted work.
Supplementary Material
Acknowledgements
The authors thank all of the participants and the general practitioners, research nurses, and data management staff who supported data collection and preparation. A.H., J.P.C., M.K., and S.H. were responsible for the study concept and design. J.P. and M.K. were involved in the acquisition of the data. The BWHHS is co-ordinated by S.E. (PI), D.A.L., and J.P.C. S.S. and T.G. did the analysis of the data. S.S., J.P.C., A.H., and S.H. drafted the manuscript and all authors critically revised the manuscript for important intellectual content.
References
- 1.Baigent C, Blackwell L, Emberson J, Holland L, Reith C, Bhala N, Peto R, Barnes EH, Keech A, Simes J, Collins R. Efficacy and safety of more intensive lowering of LDL cholesterol: a meta-analysis of data from 170 000 participants in 26 randomised trials. Lancet. 2010;376:1670–1681. doi: 10.1016/S0140-6736(10)61350-5. doi:10.1016/S0140-6736(10)61350-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Emberson JR, Whincup PH, Morris RW, Walker M. Re-assessing the contribution of serum total cholesterol, blood pressure and cigarette smoking to the aetiology of coronary heart disease: impact of regression dilution bias. Eur Heart J. 2003;24:1719–1726. doi: 10.1016/s0195-668x(03)00471-8. doi:10.1016/S0195-668X(03)00471-8. [DOI] [PubMed] [Google Scholar]
- 3.Law MR, Wald NJ. Risk factor thresholds: their existence under scrutiny. BMJ. 2002;324:1570–1576. doi: 10.1136/bmj.324.7353.1570. doi:10.1136/bmj.324.7353.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Di Angelantonio E, Sarwar N, Perry P, Kaptoge S, Ray KK, Thompson A, Wood AM, Lewington S, Sattar N, Packard CJ, Collins R, Thompson SG, Danesh J. Major lipids, apolipoproteins, and risk of vascular disease. JAMA. 2009;302:1993–2000. doi: 10.1001/jama.2009.1619. doi:10.1001/jama.2009.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anderson KM, Odell PM, Wilson PW, Kannel WB. Cardiovascular disease risk profiles. Am Heart J. 1991;121:293–298. doi: 10.1016/0002-8703(91)90861-b. doi:10.1016/0002-8703(91)90861-B. [DOI] [PubMed] [Google Scholar]
- 6.Hippisley-Cox J, Coupland C, Vinogradova Y, Robson J, May M, Brindle P. Derivation and validation of QRISK, a new cardiovascular disease risk score for the United Kingdom: prospective open cohort study. BMJ. 2007;335:136. doi: 10.1136/bmj.39261.471806.55. doi:10.1136/bmj.39261.471806.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nashef SA, Roques F, Michel P, Gauducheau E, Lemeshow S, Salamon R. European system for cardiac operative risk evaluation (EuroSCORE) Eur J Cardiothorac Surg. 1999;16:9–13. doi: 10.1016/s1010-7940(99)00134-7. doi:10.1016/S1010-7940(99)00134-7. [DOI] [PubMed] [Google Scholar]
- 8.Assmann G, Cullen P, Schulte H. Simple scoring scheme for calculating the risk of acute coronary events based on the 10-year follow-up of the prospective cardiovascular Münster (PROCAM) study. Circulation. 2002;105:310–315. doi: 10.1161/hc0302.102575. doi:10.1161/hc0302.102575. [DOI] [PubMed] [Google Scholar]
- 9.Krauss RM. What can the genome tell us about LDL cholesterol? Lancet. 2008;371:450–452. doi: 10.1016/S0140-6736(08)60213-5. doi:10.1016/S0140-6736(08)60213-5. [DOI] [PubMed] [Google Scholar]
- 10.Talmud PJ, Drenos F, Shah S, Shah T, Palmen J, Verzilli C, Gaunt TR, Pallas J, Lovering R, Li K, Casas JP, Sofat R, Kumari M, Rodriguez S, Johnson T, Newhouse SJ, Dominiczak A, Samani NJ, Caulfield M, Sever P, Stanton A, Shields DC, Padmanabhan S, Melander O, Hastie C, Delles C, Ebrahim S, Marmot MG, Smith GD, Lawlor DA, Munroe PB, Day IN, Kivimaki M, Whittaker J, Humphries SE, Hingorani AD. Gene-centric association signals for lipids and apolipoproteins identified via the HumanCVD BeadChip. Am J Hum Gen. 2009;85:628–642. doi: 10.1016/j.ajhg.2009.10.014. doi:10.1016/j.ajhg.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marmot M, Brunner E. Cohort Profile: the Whitehall II study. Int Journal Epidemiol. 2005;34:251–256. doi: 10.1093/ije/dyh372. doi:10.1093/ije/dyh372. [DOI] [PubMed] [Google Scholar]
- 12.Lawlor DA, Bedford C, Taylor M, Ebrahim S. Geographical variation in cardiovascular disease, risk factors, and their control in older women: British Women's Heart and Health Study. J Epidemiol Community Health. 2003;57:134–140. doi: 10.1136/jech.57.2.134. doi:10.1136/jech.57.2.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keating BJ, Tischfield S, Murray SS, Bhangale T, Price TS. Concept, design and implementation of a cardiovascular gene-centric 50 k SNP array for large-scale genomic association studies. PloS One. 2008;3:e3583. doi: 10.1371/journal.pone.0003583. doi:10.1371/journal.pone.0003583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. doi:10.1111/j.0006-341X.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 15.Sabia S, Kivimaki M, Kumari M, Shipley MJ, Singh-Manoux A. Effect of Apolipoprotein E epsilon4 on the association between health behaviours and cognitive function in late midlife. Mol Neurodegener. 2010;1:23. doi: 10.1186/1750-1326-5-23. doi:10.1186/1750-1326-5-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abdollahi MR, Guthrie PAI, Smith GD, Lawlor DA, Ebrahim S, Day INM. Integrated single-label liquid-phase assay of APOE codons 112 and 158 and a lipoprotein study in British women. Clin Chem. 2006;52:1420–1423. doi: 10.1373/clinchem.2006.067082. doi:10.1373/clinchem.2006.067082. [DOI] [PubMed] [Google Scholar]
- 17.Akaike H. A new look at the statistical model identification. IEEE Trans Automatic Control. 1974;19:716–723. doi:10.1109/TAC.1974.1100705. [Google Scholar]
- 18.Bennet AM, Di Angelantonio E, Ye Z, Wensley F, Dahlin A, Ahlbom A, Keavney B, Collins R, Wiman B, de Faire U, Danesh J. Association of apolipoprotein E genotypes with lipid levels and coronary risk. JAMA. 2007;298:1300–1311. doi: 10.1001/jama.298.11.1300. doi:10.1001/jama.298.11.1300. [DOI] [PubMed] [Google Scholar]
- 19.Murray A, Cluett C, Bandinelli S, Corsi AM, Ferrucci L, Guralnik J, Singleton A, Frayling T, Melzer D. Common lipid-altering gene variants are associated with therapeutic intervention thresholds of lipid levels in older people. Eur Heart J. 2009;30:1711–1719. doi: 10.1093/eurheartj/ehp161. doi:10.1093/eurheartj/ehp161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ripatti S, Tikkanen E, Orho-Melander M, Havulinna AS, Silander K, Sharma A, Guiducci C, Perola M, Jula A, Sinisalo J, Lokki M, Nieminen MS, Melander O, Salomma V, Peltonen L, Kathiresan S. A multilocus genetic risk score for coronary heart disease: case-control and prospective cohort analyses. Lancet. 2010;376:1393–1400. doi: 10.1016/S0140-6736(10)61267-6. doi:10.1016/S0140-6736(10)61267-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kathiresan S, Melander O, Anevski D, Guiducci C, Burtt NP, Roos C, Hirschhorn JN, Berglund G, Hedblad B, Groop L, Altshuler DM, Newton-Cheh C, Orho-Melander M. Polymorphisms associated with cholesterol and risk of cardiovascular events. NEJM. 2008;358:1240–1249. doi: 10.1056/NEJMoa0706728. doi:10.1056/NEJMoa0706728. [DOI] [PubMed] [Google Scholar]
- 22.Davies NM, Windmeijer F, Martin RM, Abdollahi MR, Davey Smith G, Lawlor DA, Ebrahim S, Day IN. Use of genotype frequencies in medicated groups to investigate prescribing practice: APOE and statins as a proof of principle. Clin Chem. 2011;57:502–510. doi: 10.1373/clinchem.2010.156356. doi:10.1373/clinchem.2010.156356. [DOI] [PubMed] [Google Scholar]
- 23.Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. doi:10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, Cho JH, Guttmacher AR, Kong A, Kruglyak L, Mardis E, Rotimi CN, Slatkin M, Valle D, Whittemore A, Boehnke M, Clark AG, Eichler EE, Gibson G, Haines JL, Mackay TFC, McCarroll SA, Visscher PM. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. doi:10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.