Abstract
Oval cell activation occurs under conditions of severe liver injury when normal hepatocyte proliferation is blocked. Recent studies have shown that a subset of hepatocellular carcinomas expresses oval cell markers, suggesting that these cells are targets of hepatocarcinogens. However, the signaling pathways that control oval cell activation and proliferation are not well characterized. Based on the role of the nutrient signaling kinase complex, mTORC1, in liver development, we investigated the role of this pathway in oval cell activation. Oval cell proliferation was induced in male Fisher rats by a modification of the traditional choline deficient plus ethionine model (CDE) or by 2-acetylaminoflourene treatment followed by 2/3 partial hepatectomy with or without initiation by diethylnitrosamine. To assess the role of mTOR in the oval cell response and development of preneoplastic foci, the effect of the mTORC1 inhibitor, rapamycin, was studied in all models. Rapamycin induced a significant suppression of the oval cell response in both models, an effect that coincided with a decrease in oval cell proliferation. Rapamycin administration did not affect the abundance of neutrophils or natural killer cells in CDE-treated liver or the expression of key cytokines. Gene expression studies revealed the fetal hepatocyte marker MKP-4 to be expressed in oval cells. In an experimental model of hepatic carcinogenesis, rapamycin decreased the size of preneoplastic foci and the rate of cell proliferation within the foci. mTORC1 signaling plays a key role in the oval cell response and in the development of preneoplastic foci. This pathway may be a target for the chemoprevention of hepatocellular carcinoma.
Keywords: Hepatic progenitor cells, Liver regeneration, Liver injury, mTOR, Hepatocellular carcinoma
Introduction
The adult mammalian liver displays an immense capacity for regeneration in response to injury caused by chemicals or toxins, and to surgical reduction in mass. The regenerative response is a complex, coordinated process whereby pre-existing, mature hepatocytes, biliary cells, and stromal cells re-enter the cell cycle to replace functional liver mass while maintaining their differentiated phenotype (Taub, 2004). In cases where adult liver cell proliferation is impaired, such as in chronic liver injury or acute liver failure, a progenitor cell compartment is activated and gives rise to transit amplifying cells (Mangnall et al., 2003). These cells, termed oval cells by Farber because of their ovoid nucleus and large nuclear to cytoplasmic ratio, are now known to be bipotential progenitor cells capable of differentiating into hepatocytes and ductal cells (Dabeva and Shafritz, 1993; Farber, 1956; Hixson, 2003; Hixson et al., 2000).
Oval cells can be induced in the rat by feeding a choline-deficient diet in combination with ethionine or by treating with 2-acetylaminofluorene (2-AAF) followed by 2/3 partial hepatectomy (PHx) (Hixson, 2003). Hepatic oval cells, referred to as progenitor cells in humans, have been identified in a number of human liver diseases, including hepatitis, alcoholic and non-alcoholic steatohepatitis, and extra-hepatic biliary atresia. Their abundance is correlated with disease severity (Lowes et al., 1999; Roskams et al., 2003). Many human liver diseases in which progenitor cell expansion occurs are associated with the development of liver cancer. In patients with viral hepatitis, proliferating oval cells are found in close proximity to hepatocellular carcinoma (HCC). Furthermore, a portion of the HCCs from these patients express oval cell markers, suggesting that persistent oval cells may acquire mutations that contribute to the development of HCC (Hixson, 2003; Lowes et al., 1999). This notion is supported by recent studies that have shown that a subset of HCC expresses progenitor cell markers, suggesting that these tumors arise from oval cells. This has special significance in light of the data supporting the existence of cancer stem cells and their role in tumor recurrence, metastasis, and resistance to chemotherapeutics (Alison et al., 2011; Bomken et al., 2010). In agreement with the cancer stem cell hypothesis, hepatic tumors expressing progenitor cell antigens have been correlated with a high rate of recurrence and poor patient survival (Yamashita et al., 2008; Yang et al., 2010).
Although the circumstances that result in oval cell activation have been well studied, the signaling pathways regulating their activation, proliferation, and differentiation remain unclear. To date, a TNF family member, TWEAK, has been the only mitogen identified to be selective for oval cells. However, inhibition of TWEAK by blockade or deletion of its receptor, Fn14, does not prevent oval cell activation or proliferation (Jakubowski et al., 2005). The Wnt/β-catenin pathway has also been shown to be activated in proliferating oval cells but, as with TWEAK, deletion of β-catenin only partially inhibits the oval cell response in mice placed on the 2-AAF/partial hepatectomy protocol (Apte et al., 2008).
Previous studies in our laboratory have focused on the signaling mechanisms that regulate hepatocyte growth and proliferation during development. We have found that the pathways regulating adult hepatocyte proliferation after 2/3 PHx are not active in late gestation fetal liver. In addition, fetal hepatocyte proliferation is insensitive to rapamycin, a specific inhibitor of the nutrient- and energy-sensing kinase mTOR (Boylan et al., 2001). Rapamycin is a macrolide antibiotic with immunosuppressive and antitumor activities (Eng et al., 1984; Thomson et al., 2009; Vezina et al., 1975). In mammalian cells, mTOR exists in two complexes, mTOR complex 1 (mTORC1) and mTOR complex 2 (mTORC2) (Abraham, 2002). mTORC1 is rapamycin sensitive while mTORC2 is rapamycin resistant and is primarily involved in the control of the actin cytoskeleton (Guertin and Sabatini, 2009).
Rapamycin inhibits mTOR's serine/threonine kinase activity, resulting in changes in the phosphorylation state and activity of downstream effectors of mTOR, including S6 kinase (S6K) and eukaryotic translation initiation factor 4E binding protein 1 (Fingar and Blenis, 2004; Hay and Sonenberg, 2004). Rapamycin's anti-cancer properties reflect the drug's ability to inhibit the normal biological functions of mTOR, including ribosome biogenesis and protein translation, promotion of cellular proliferation, and angiogenesis (Baldo et al., 2008; Gingras et al., 2001; Wiederrecht et al., 1995). Our previous work on the mTORC1/S6K pathway has shown that signaling via the pathway is sensitive to rapamycin during late gestation development in a number of tissues including liver, intestine, and kidney. However, cellular proliferation and normal tissue remodeling that occur as part of normal growth in the late gestation and early postnatal periods in the rat are rapamycin resistant (Sanders et al., 2008). In addition, we found that sensitivity to the anti-proliferative effects of rapamycin varied across a panel of hepatic cell lines and that sensitivity did not correlate with the degree of transformation (Jimenez et al., 2009). On the basis of these observations on the differential role of mTOR in fetal and adult hepatocyte proliferation, we examined the role of mTOR signaling in oval cell activation and proliferation.
Materials and methods
Animals
Timed pregnant female and 6–7 week old male Fisher F344 rats were obtained from Charles River Laboratories (Wilmington, MA). Animals were housed under standard conditions with access to food and water ad libitum. All animal experiments were performed in accordance with the guidelines of the National Institutes of Health and the Rhode Island Hospital Institutional Animal Care and Use Committee. Cesarean sections were performed on timed-pregnant rats under iso-flurane anesthesia on embryonic day 19 (E19).
For the CDE protocol, male Fisher rats were divided into three groups of 4 animals each. All groups were fed a choline-deficient diet (Dyets Inc., Bethlehem, PA) in combination with daily injections of ethionine (12 mg via i.p. injection; Fisher Scientific, Houston, TX) for up to 15 days. Rapamycin (2.5 μg/g; LC Laboratories, Woburn, MA) was administered by intraperitoneal injection on days 7, 10, and 13 of the CDE protocol. Controls included uninjected animals and animals receiving an equivalent amount of DMSO vehicle. BrdU was administered to all groups 24 h prior to euthanasia (60 μg/g; Sigma, St. Louis, MO).
The AAF/PHx protocol was performed on male rats, as previously described (Faris et al., 1991). Rapamycin (2.5 μg/g) or DMSO vehicle was administered 1 h before 2/3 partial hepatectomy and on days 3 and 5 post-hepatectomy. Rats received an i.p. injection of EdU 24 h prior to euthanasia (50 μg/g; Invitrogen, Carlsbad, CA). Rats were euthanized 7 days post-hepatectomy.
Preneoplastic foci were induced according to the protocol described by Solt and Farber (Solt et al., 1977). Briefly, male rats were administered a single intraperitoneal injection of diethylnitrosamine (DENA, Sigma, MO) at a dose of 200 mg/kg. After a 2-week recovery, rats were subjected to the AAF/PHx protocol with or without rapamycin treatment as described above.
Immunohistochemical analysis
Paraffin-embedded liver was analyzed by hematoxylin and eosin staining and immunohistochemistry for BrdU (Vector labs, Burlingame, CA) and Ki-67 (Dako, Carpinteria, CA) as previously described (Sanders et al., 2008). For CK19 staining, sections were incubated in 0.1% trypsin for 15 min, followed by a 1 h incubation in anti-CK19 antibody (Leica Microsystems, Bannockburn, IL). For GST-P staining, sections were incubated in anti-GST-P antibody (MBL International, Woburn, MA) for 1 h. For phospho-ribosomal protein S6 staining, sections were incubated in anti-phospho S6235/236 (Cell Signaling Technology, Beverly, MA) overnight at 4 °C. Slides were scanned using the Aperio ScanScope system (Aperio Technologies Inc., Vista, CA) and quantitative analyses performed on 10 fields acquired at a 20× zoom using NIH ImageJ Software. All sections were analyzed in a blinded fashion.
Immunofluorescence and image analysis
Immunofluorescent staining for each antigen was performed on multiple acetone fixed liver cryosections (5 μm). Primary monoclonal antibodies included the following: BD.2 (Faris et al., 1994) and OC.10 (Hixson et al., 2000), oval cell/cholangiocyte markers; RP-3, a neutrophil-specific marker (Sekiya et al., 1989); CD161a, a natural killer cell marker (BD Biosciences, San Diego, CA); and H.4, a hepatocyte specific marker (Faris et al., 1994; Hixson et al., 1997). For dual staining, the use of BD.2 versus OC.10 was determined by the isotype of the other primary antibody used. For double labeling with EdU, sections were incubated in either mAb BD.2, mAb H.4, or GST-P with AlexaFluor594 IgG followed by EdU staining as previously described by Salic and Mitchison (Salic and Mitchison, 2008). Slides were counterstained with 4′,6-diamidino-2-phenylinade (Vector Laboratories, Burlingame, CA). Gray-scale images (8 bit) were acquired with a Nikon E800 microscope (Nikon Inc., Mellville, NY) using a 10× or 20× PlanFluor objective and a spotII digital camera (Diagnostic Instruments, Sterling Heights, MI). The cameras built-in green filter was used to increase image contrast.
Western immunoblotting
Liver homogenates were prepared as previously described (Sanders et al., 2008). Samples were separated by SDS-PAGE, transferred to polyvinylidene membrane, and subjected to immunoblot analysis with antibodies directed toward ribosomal protein S6 or phosphorylated S6Ser235/236 (Cell Signaling Technology, Beverly, MA). Detection employed enhanced chemiluminescence. Equal amounts of protein were loaded per lane based on the protein concentration. Quantification of bands was performed by densitometry in LabWorks 4.5 software (UVP, Upland, CA).
RNA isolation and RT-PCR
Total RNA was isolated from frozen liver as previously described (Braun et al., 1990). For semi-quantitative RT-PCR, cDNA was synthesized using random hexamers and the Taqman Reverse Transcrption kit (Applied Biosystems, Foster City, CA) according to the manufacturer's instructions. Primer sequences used for amplification of fetal marker genes are as follows: (5′→3′): MKP-4, GCATCCGCTACATCCTCAAT and GACCAGGTCATAGGCATCGT; CRD-BP, AATGGAACAGTGTCCTTGCC and GGCATGGTTCTCCAGTTGAT; cyclin D1, GCGTACCCTGACACCAATCT and GCTCCAGAGACAAGAAACGG; cyclin D2, GGTAGCACACAGAGCGATGA and GCCAAAGGAAGGAGGTAAGG; cyclin D3, CGAGGCTCCTACTTCCAGTG and CTGAGCATGCTTTTTGACCA; and 18s, AGTTGGATCTTGGGAGCG and ATTAAGCCGCAGGCTCCAC. grb-10 primers were designed as described previously (Gruppuso et al., 2000). The optimal PCR cycle number for exponential amplification was determined in preliminary experiments.
Statistical analysis
Statistical analyses were performed using GraphPad Prism (San Diego, CA). Results were analyzed using a Student's t-test for CDE versus CDE rapamycin comparisons and one-way ANOVA with a post hoc Tukey test for comparing the relative abundance of the fetal markers. Chi-square analysis was used for comparison of AAF/PHx vehicle versus AAF/PHx rapamycin.
Results
A modified CDE protocol for oval cell activation
To induce oval cells, rats were placed on a modified CDE protocol. For this protocol, rats were fed a choline deficient diet and received daily intraperitoneal injections of ethionine (12 mg), instead of the traditional method where ethionine is administered orally. Liver was harvested on day 11 or day 15 of the protocol. Oval cell expansion was assessed by indirect immunofluorescence for the oval cell marker OC.10 and a marker of mitosis, phospho-histone H3. After 11 days on the CDE protocol, OC.10-positive oval cells were most prominent in the portal areas (Fig. 1A). On day 15, there were marked increases in the number of OC.10 positive cells and in the oval cell mitotic index. The oval cells were distributed throughout the liver and had formed duct-like structures (Fig. 1A). In accordance with our immunofluorescence data, hematoxylin and eosin-stained liver sections contained oval cells with a characteristic shape and high nucleus to cytoplasmic ratio at day 15 (Fig. 1B). The architecture of the liver lobule was disrupted by the oval cells, resulting in the isolation of clusters of hepatocytes. These sections also revealed inflammatory cells in close proximity to the oval cells.
Fig. 1.
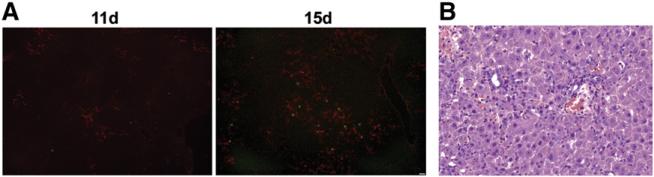
Oval cell activation in rats fed a choline deficient diet in combination with ethionine injection (CDE). (A) Liver cryosections from rats placed on the modified CDE protocol for 11 or 15 days underwent immunofluorescent staining for OC.10 (red) and phospho-histone H3 (green). Images were acquired at 10×. Scale bar: 50 μm. (B) Hematoxylin and eosin staining of 15 day CDE liver sections reveals the presence of oval cells and inflammatory cells. Image was acquired at 20×. Scale bar: 50 μm.
To assess the effect of the modified CDE protocol on adult hepatocyte proliferation, rats were subjected to 2/3 partial hepatectomy after 7 days on the protocol. A marked decrease in phospho-histone H3 staining was observed in treated rats compared to animals fed standard chow, confirming that the modified protocol was mito-inhibitory to adult hepatocytes (data not shown).
Rapamycin decreases oval cell abundance
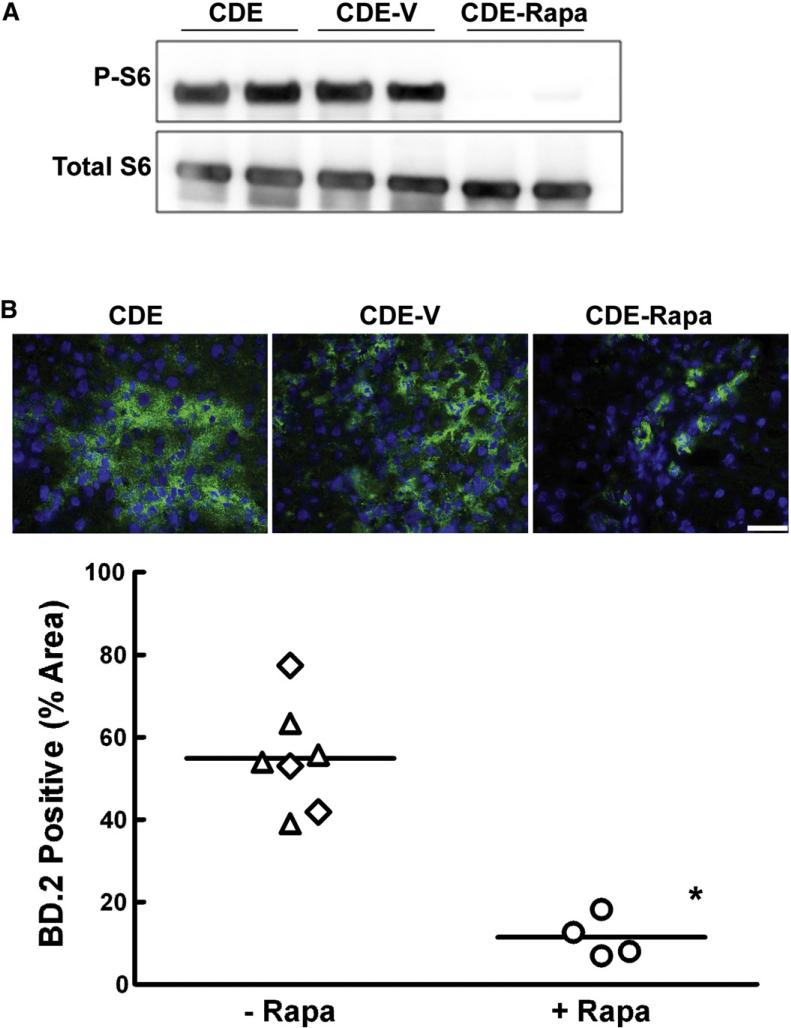
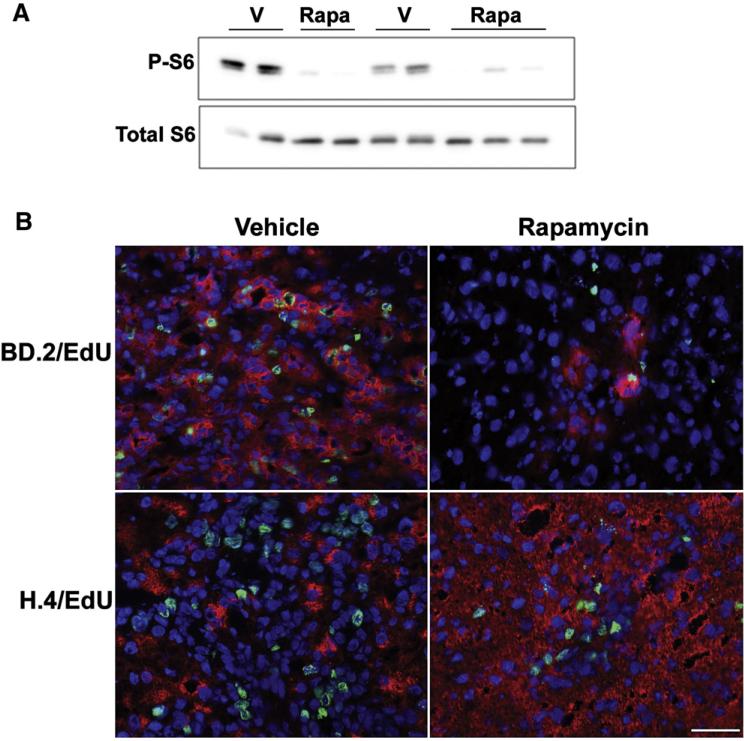
In order to assess the role of the mTOR/S6 kinase pathway in oval cell expansion, rats were placed on the modified CDE protocol and treated with vehicle or rapamycin on days 7, 10, and 13. To confirm the efficacy of rapamycin, Western immunoblotting for phosphorylated (P-S6Ser235/236) and total ribosomal protein S6 was performed. Rapamycin treatment resulted in a profound inhibition of S6 phosphorylation at these two phosphorylation sites (Fig. 2A). Hepatic oval cell abundance determined by the percent area of BD.2 positive cells was significantly decreased in animals that received rapamycin (Fig. 2B). Whereas oval cells in the CDE-treated control rats represented 55% of the total liver area on day 15, rapamycin administration reduced the area occupied by oval cells to 10%. This decrease was confirmed by histology, immunohistochemical staining for CK19, and immunofluorescence for OC.10 (data not shown).
Fig. 2.
The effect of rapamycin treatment on oval cell abundance in rats placed on the modified CDE protocol. (A) Western immunoblotting for phospho-S6Ser235/236 and total S6 was performed on total liver homogenates from rats placed on the CDE protocol, CDE plus vehicle (CDE-V), or CDE in combination with rapamycin treatment (CDE-Rapa). (B) Liver cryosections underwent immunofluorescent staining for an oval cell marker BD.2 and were counterstained with DAPI. Images were acquired at 40×. Scale bar: 50 μm. The graph shows the percent area of BD.2-positive cells in ten 20× fields. Results are shown as individual data points and the mean. ◇, CDE; △, CDE-V; ○, CDE–Rapa. *; p=0.003.
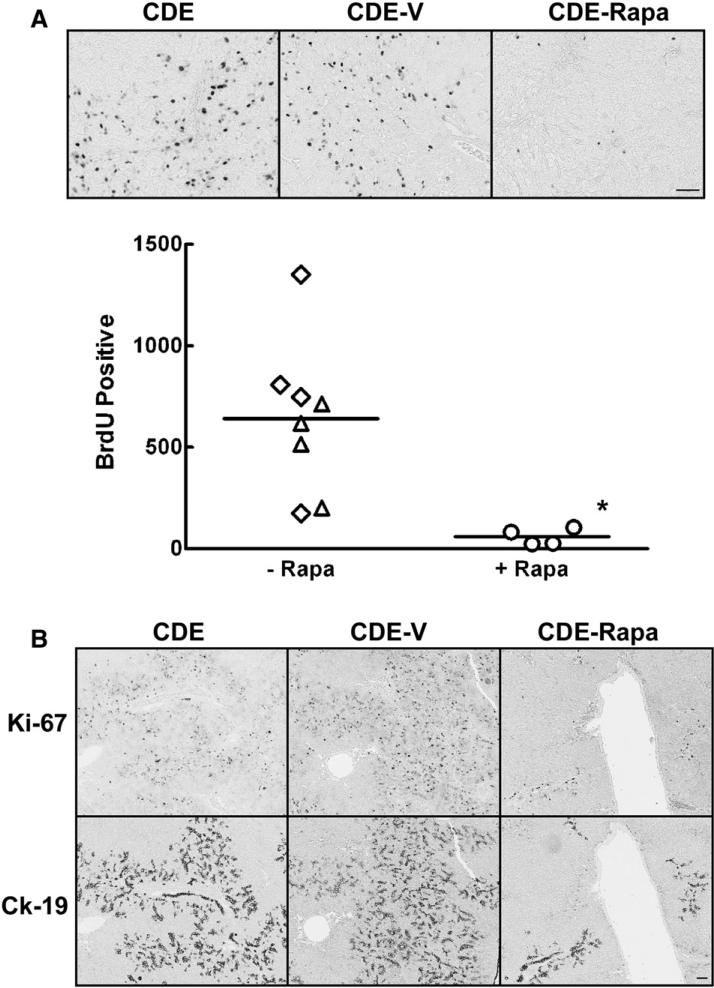
Having determined that rapamycin inhibition of the mTOR/S6K pathway was sufficient to cause a marked inhibition of oval cell expansion in this model, we went on to investigate whether rapamycin affected oval cell proliferation, apoptosis or both. DNA synthesis was measured by quantifying BrdU-positive nuclei in uninjected control, vehicle-treated and rapamycin-treated rats that had been placed on the modified CDE protocol (Fig. 3A). The number of BrdU-positive nuclei in the rapamycin-injected animals was significantly reduced compared to the two control groups (Fig. 3A). In order to further investigate the effect of rapamycin on oval cell proliferation, immunohistochemical staining for CK19 and Ki-67 was performed on serial sections. A subset of the oval cells present in the rapamycin treated animals was positive for Ki-67 (Fig. 3B). Similar results were obtained from serial sections stained with CK19 and BrdU (data not shown). From these results we concluded that rapamycin markedly reduced oval cell proliferation, but that a subpopulation of oval cells was resistant to the growth inhibitory effect of rapamycin.
Fig. 3.
The effect of rapamycin on oval cell DNA synthesis in CDE treated animals. (A) Immunohistochemical staining for BrdU was performed on sections from animals placed on the CDE protocol, CDE plus vehicle (CDE-V), or CDE in combination with rapamycin treatment (CDE-Rapa). Images were acquired at 20×. Scale bar: 50 μm. The graph depicts the number of BrdU-positive nuclei in ten 20× fields. Data are shown as individual points with the mean. ◇, CDE; △, CDE-V; ○, CDE-Rapa. *; p<0.02. (B) Immunohistochemical staining was performed on serial sections for the proliferation marker Ki-67 and an oval cell marker CK19. Images were acquired at 5×. Scale bar: 50 μm.
We next examined apoptosis by quantifying apoptotic bodies on hematoxylin and eosin-stained sections from CDE-treated control, vehicle-injected, and rapamycin-injected animals. Comparable, low numbers of apoptotic cells in the portal areas were observed in control and vehicle-treated rats. Rapamycin administration in the CDE-treated animals resulted in a 2-fold decrease in apoptotic bodies (data not shown).
Effect of rapamycin on inflammatory cell infiltration
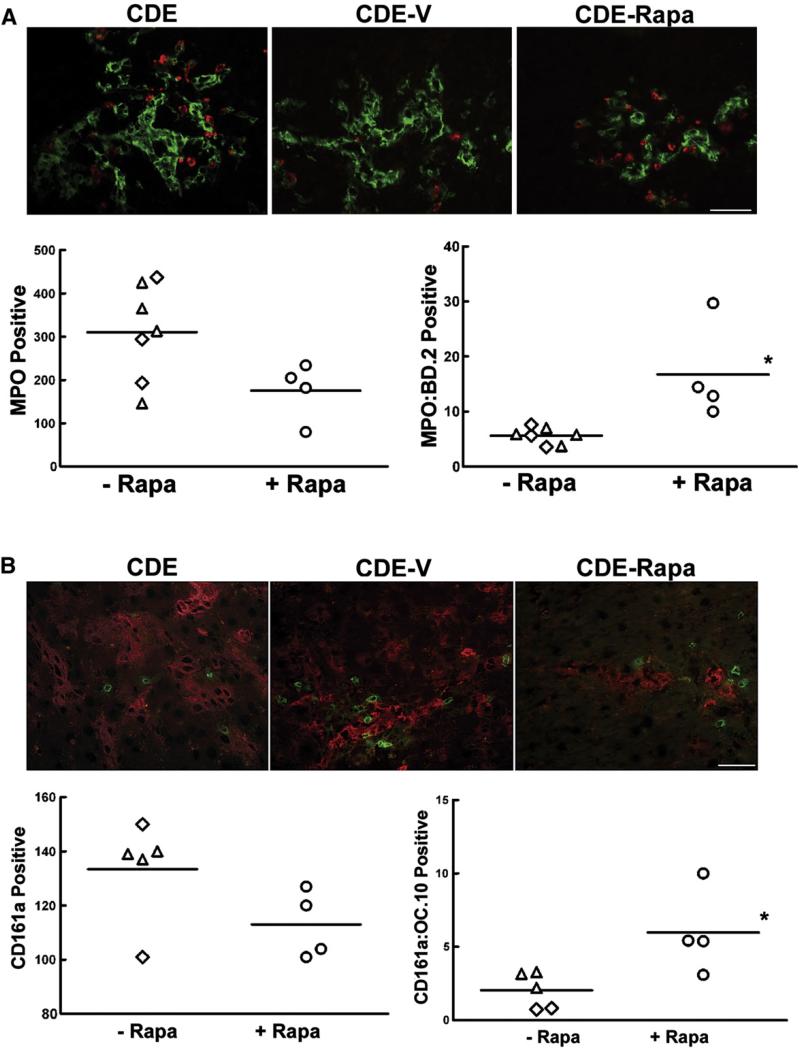
Crosstalk between the immune system and hepatic oval cells has been shown to promote the activation and expansion of oval cells in mice (Strick-Marchand et al., 2008). With the modified CDE protocol, we found that oval cell proliferation was accompanied by an inflammatory response. We hypothesized that the decrease in oval cell abundance was related to the immunosuppressant activity of rapamycin. To test this, sections from CDE-treated control, vehicle-injected and rapamycin-injected rats were immunostained using oval cell and neutrophil-specific or NK-specific antibodies. Neutrophils, recognized by their reactivity with the rat neutrophil specific antibody RP-3, were found to be interspersed between or in close contact with oval cells in all three groups (Fig. 4A). Rapamycin treatment had no effect on the total number of neutrophils in the liver. However, rapamycin induced a significant increase in the number of neutrophils in close proximity to oval cells. Numerous NK cells (CD161a+) were observed in the liver parenchyma. Clusters of CD161a+ cells were found to accumulate near oval cell colonies, while single NK cells could be found throughout the liver parenchyma. As with neutrophils, rapamycin did not affect the total number of NK cells in the liver, but did increase the number of NK cells in close association with oval cells (Fig. 4B). We also examined the expression of IL-1 alpha and beta, TWEAK, TNF-α, IL-6, and LT-beta as these cytokines have been proposed to play a role in the oval cell response. In accordance with previous studies, we found the expression of IL-1 beta, IL-6, and LT-beta to be significantly upregulated in rats placed on our CDE protocol (Knight et al., 2005). However, there was no significant effect of rapamycin on the expression of any of the cytokines analyzed (data not shown), suggesting that the effect of rapamycin on oval cells is direct.
Fig. 4.
Effect of rapamycin treatment on the number of neutrophils or NK cells in the liver of CDE treated rats. (A) Immunofluorescent staining for the oval cell marker BD.2 and the neutrophil marker RP-3. The graph depicts the number of RP-3-positive cells in ten 20× fields. (B) Immunofluorescent staining for the oval cell marker OC.10 and NK cell marker CD161a. The graph depicts the number of CD161a-positive cells in ten 20× fields. Data are shown as individual points with the mean. ◇, CDE; △, CDE-V; ○, CDE-Rapa. Images were acquired at 20×. Scale bar: 50 μm. Image analysis was undertaken as described in the Supplemental Information.
The expression of fetal markers in oval cells
To further characterize the phenotype of oval cells in the modified CDE model, we performed RT-PCR for candidate growth-regulating genes that we had previously identified as differentially expressed in fetal and adult liver (Fig. 5). Although these genes were found to be overexpressed in late gestation fetal liver and in primary cultures of fetal hepatocytes, they were not induced during liver regeneration following 2/3 partial hepatectomy (Boylan and Gruppuso, 2005; Gruppuso et al., 2000; Leeds et al., 1997).
Fig. 5.
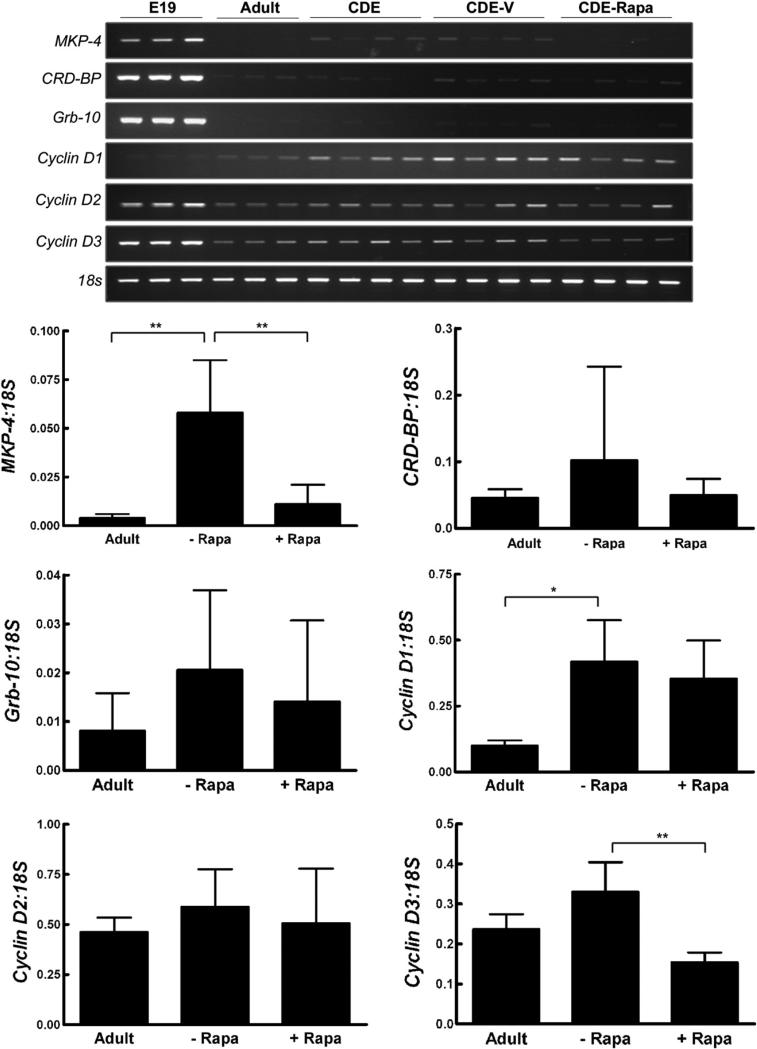
Expression profile of a panel of fetal marker genes in liver from CDE treated rats. RT-PCR for genes previously identified as markers of fetal hepatocytes was performed on RNA isolated from E19 fetal and adult rats or from rats placed on the CDE protocol, CDE plus vehicle (CDE-V), and CDE in combination with rapamycin treatment (CDE-Rapa). Expression levels of the various genes were quantified and expressed as the ratio to 18 s. Data are expressed as means+1SD. *; p<0.05. **; p<0.01.
Total RNA was isolated from triplicate E19 fetal and adult rat livers, and from rats placed on the modified CDE protocol with and without rapamycin administration. The fetal marker MKP-4, and cyclin D1, the predominant d-type cyclin upregulated during normal liver regeneration, were significantly induced in CDE-treated liver. Rapamycin administration in CDE-treated rats led to a reduction in MKP-4 to levels comparable to normal adult liver, an observation consistent with MKP-4 being expressed in oval cells. There was no effect on cyclin D1. Cyclin D3 was not induced by CDE treatment but was significantly decreased by rapamycin treatment in the modified CDE model. The expression of the other fetal markers, Grb-10, CRD-BP, and cyclin D2, was not affected by CDE treatment or CDE in combination with rapamycin administration.
Rapamycin decreases oval cell activation in a second model of oval cell expansion
We extended our findings on the role of the mTOR/S6K pathway in the oval cell response to the well characterized model employing 2-AAF plus 2/3 partial hepatectomy (Tatematsu et al., 1984). Rats implanted with a time-release 2-AAF pellet were administered rapamycin or DMSO vehicle 1 h prior to partial hepatectomy and on days 3 and 5 post-hepatectomy. To assess the efficacy of rapamycin (Fig. 6A) liver harvested 7 days post-hepatectomy was analyzed by immunoblotting for phosphorylated (P-S6Ser235/236) and total ribosomal protein S6.
Fig. 6.
Effect of rapamycin treatment on oval cell abundance and proliferation in rats treated with 2-AAF followed by 2/3 partial hepatectomy (PHx). (A) Western immunoblotting for phospho-S6Ser235/236 and total S6 was performed on total liver homogenates from vehicle (V) or rapamycin (Rapa) treated rats placed on the 2-AAF PHx protocol and harvested 7 days post-hepatectomy. (B) Cryosections from these animals underwent immunofluorescent staining for an oval cell marker BD.2 and a marker of DNA synthesis EdU or (C) a hepatocyte marker H.4 and EdU. Slides were counterstained with DAPI. Images were acquired at 40×. Scale bar: 50 μm. Image analysis was undertaken as described in the Supplemental Information.
Similar to the modified CDE model, rapamycin administration resulted in profound inhibition of S6 phosphorylation and suppressed oval cell activation and proliferation assessed by immunostaining for BD.2 and by quantifying the incorporation of EdU into DNA (Fig. 6B). The percent of the liver occupied by oval cells was reduced from 43% in vehicle-injected animals to 12.5% in rapamycin-treated animals. The percentage of oval cells that had traversed S-phase and was EdU positive was also significantly decreased by rapamycin (8.7% vs 0.9%, vehicle vs. rapamycin). Immunofluorescent staining for the hepatic marker H.4 confirmed the effect of rapamycin on oval cell expansion (Fig. 6C). The decrease in oval cells resulted in a dramatic increase in the number of H.4 labeled cells per field in rapamycin-injected rats compared to control. In accordance with the mito-inhibitory effect of 2-AAF on hepatocytes, we did not observe H.4+/EDU+ cells. As in the CDE model, rapamycin did not affect the expression of IL-1 alpha or beta, TWEAK, TNF-α, IL-6, or LT-beta (data not shown).
Rapamycin decreases the size of preneoplastic foci
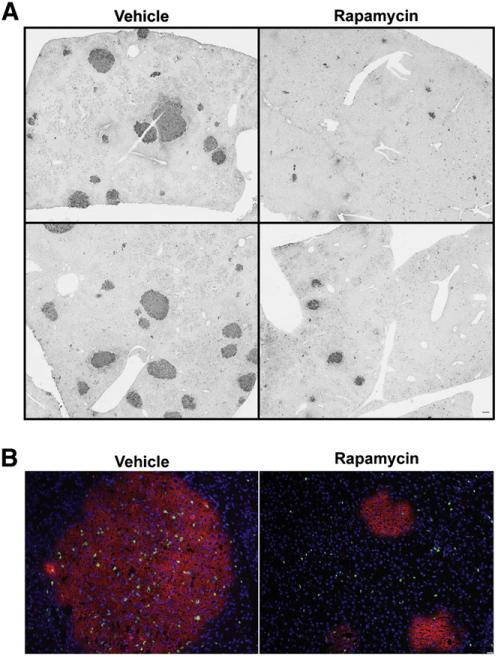
Given the effect of rapamycin on oval cell abundance and the contribution of progenitor cells to HCC, we performed a preliminary series of experiments to analyze the effect of rapamycin on the development of preneoplastic foci in a model of liver carcinogenesis. Rats were administered a single injection of DENA and subjected to the AAF/PHx protocol described above. Rapamycin inhibition of mTORC1 was confirmed by analysis of S6 phosphorylation as described in our other experiments (data not shown). One week post-hepatectomy, preneoplastic foci were clearly distinguishable from the surrounding normal parenchyma by staining for a marker of neoplastic and preneoplastic cells, GST-P (Satoh et al., 1991). Rapamycin treatment resulted in a striking decrease in the size of GST-P+ foci (Fig. 7A). To investigate the mechanism by which rapamycin inhibited the development of preneoplastic foci, we performed dual immunofluorescence for GST-P and EdU. Rapamycin treatment resulted in a decrease in the incorporation of EdU into DNA in preneoplastic foci (Fig. 7B). These data suggest that rapamycin inhibition of cellular proliferation markedly reduces the size of preneoplastic foci in this particular model.
Fig. 7.
Effect of rapamycin treatment on the abundance and size of preneoplastic foci in rats placed on the Solt–Farber protocol. Rats subjected to the Solt–Farber protocol were treated with rapamycin or DMSO vehicle and liver harvested 7 days post-hepatectomy. (A) Sections from these animals were stained with anti-GST-P. Images from duplicate animals were acquired at 2×. Scale bar: 50 μm. (B) Cryosections from these animals underwent immunofluorescent staining for GST-P and a marker of DNA synthesis EdU. Slides were counterstained with DAPI. Images were acquired at 10×. Scale bar: 50 μm. Image analysis was undertaken as described in the Supplemental Information.
Discussion
The signaling mechanisms that mediate the hepatic oval cell response are poorly understood. Given that previous studies have shown that oval cell differentiation recapitulates hepatoblast differentiation during development, and that oval cells express cell-surface markers that are also found on fetal hepatocytes (Farber, 1956), we set out to test the hypothesis that the signaling phenotype of oval cells would resemble that of late gestation fetal hepatocytes in vivo.
One of the most striking differences our laboratory has identified between fetal and adult hepatocyte signaling is the role of the mTORC1 pathway. Activation of this pathway is required for adult hepatocyte proliferation that occurs during normal liver regeneration following 2/3 partial hepatectomy, but is dispensable for hepatocyte proliferation during late gestation (Boylan et al., 2001). In the present study, we found that the oval cell response in rats fed a choline deficient diet in combination with ethionine, or subjected to the AAF/PHx model, was inhibited by rapamycin. Coincident with the decreased abundance of hepatic oval cells, oval cell proliferation in both models was significantly decreased by rapamycin administration. These results indicate that similar to adult hepatocytes, the mTORC1 pathway is a critical mediator of hepatic oval cell proliferation. In the CDE model, we observed a subset of oval cells that were resistant to the anti-proliferative effects of rapamycin. Based on these data and the expression of fetal hepatic markers in oval cells, there may be a subpopulation of oval cells that recapitulate the fetal hepatocyte phenotype.
In the context of these studies, we developed a novel modification to the standard CDE protocol where ethionine is administered orally (Shinozuka et al., 1978). In our new model, ethionine was given by intraperitoneal injection for up to 15 days. By day 7, single periductal cells had begun to proliferate. Over the next several days, a significant increase in the number of proliferating cells positive for oval cell markers was observed in the periportal area. These observations indicate that our modified protocol provides a rapid and reproducible experimental system for studying the earliest events in the oval cell response.
An inflammatory response accompanied the appearance of oval cells in the CDE-treated animals. As noted above, previous studies have suggested that immune cross-talk can promote the expansion of oval cells (Strick-Marchand et al., 2008). Since rapamycin has immunosuppressant activity, largely through inhibition of the expansion of specific immune cells (Thomson et al., 2009; Wai et al., 2008), we questioned whether there was a reduction in the number of immune cells present in the parenchyma in the rapamycin-treated animals. Neutrophil and NK cell number were unaffected by rapamycin. In fact, we observed an increase in the proportion of immune cells in close proximity to oval cells. We also examined the expression of cytokines proposed to play a role in the oval cell response. While the expression of IL-1 beta, IL-6, and LT-beta was induced by placing rats on the CDE protocol, rapamycin did not affect the expression of any of the cytokines studied. The most straightforward interpretation of our data is that mTORC1 signaling contributes to the oval cell response through a direct effect.
We examined the expression of candidate growth regulating genes that our laboratory had previously identified as being differentially expressed in late gestation fetal compared to adult liver (Boylan and Gruppuso, 2005; Gruppuso et al., 2000; Leeds et al., 1997). The expression of one fetal marker in this panel, MKP-4 (mitogen-activated protein kinase phosphatase 4), mirrored the abundance of oval cells. Based on the temporal pattern of its expression, we conclude that MKP-4 is an oval cell marker. MKP-4 is a member of a class of dual spec-ificity phosphatases with specific activity against threonine and tyrosine residues. MKP-4 preferentially recognizes and inactivates the classical ERK1/2 pathway (Christie et al., 2005). Immunohistochemical staining of CDE-treated liver revealed that ERK1/2 was phosphorylated in oval cells (data not shown). Although further studies are necessary to investigate the functional role of MKP-4 in oval cells, its induction in the CDE model suggests a recapitulation of the fetal hepatocyte phenotype.
The mTOR pathway is activated in a number of human tumors, including HCC. Approximately 50% of human HCCs have abnormal mTOR signaling with the majority of cases accounted for by activation of the IGF-1 signaling pathway, EGF upregulation, or mutations in PTEN (Villanueva et al., 1983). In vitro and in vivo studies have shown that rapamycin reduces HCC growth and enhances sensitivity to other chemotherapeutic agents (Ong et al., 2009; Piguet et al., 2008; Shi and August, 2008). Clinical trials using rapamycin as a therapeutic agent for HCC are in the earliest stages. In one pilot study, rapamycin was well tolerated but minimally effective in patients with advanced HCC (Schoniger-Hekele and Muller, 2010). In our preliminary experiments, we showed that rapamycin decreased the size of preneoplastic foci by inhibiting the proliferation of GST-P positive cells within the foci.
In conclusion, our findings reveal a direct role for mTOR in the oval cell response as treatment with rapamycin significantly decreases the abundance of oval cells in the liver without affecting the infiltration of immune cells or the expression of cytokines. Treatment with rapamycin also led to a reduction in the size of preneoplastic foci. Given the role of oval cells in chronic liver disease and in the development of HCC, our data raise the possibility that rapamycin and other mTOR inhibitors may be useful as chemopreventative agents in conditions that predispose to HCC.
Supplementary Material
Acknowledgments
We thank Virginia Hovanesian for assistance with image acquisition and Joan Boylan for critical reading of this manuscript.
Grant support
These studies were supported by National Institutes of Health grants P20 RR017695 (J.A.S.), R01 HD24455 (P.A.G.) and R01 CA93840 (D.C.H.) and by the Department of Pediatrics.
Footnotes
Conflict of interest statement
The authors declare that there are no conflicts of interest.
References
- Abraham RT. Identification of TOR signaling complexes: more TORC for the cell growth engine. Cell. 2002;111:9–12. doi: 10.1016/s0092-8674(02)01009-7. [DOI] [PubMed] [Google Scholar]
- Alison MR, Lim SM, Nicholson LJ. Cancer stem cells: problems for therapy? The Journal of Pathology. 2011;223:147–161. doi: 10.1002/path.2793. [DOI] [PubMed] [Google Scholar]
- Apte U, Thompson MD, Cui S, Liu B, Cieply B, Monga SP. Wnt/beta-catenin signaling mediates oval cell response in rodents. Hepatology. 2008;47:288–295. doi: 10.1002/hep.21973. [DOI] [PubMed] [Google Scholar]
- Baldo P, Cecco S, Giacomin E, Lazzarini R, Ros B, Marastoni S. mTOR pathway and mTOR inhibitors as agents for cancer therapy. Current Cancer Drug Targets. 2008;8:647–665. doi: 10.2174/156800908786733513. [DOI] [PubMed] [Google Scholar]
- Bomken S, Fiser K, Heidenreich O, Vormoor J. Understanding the cancer stem cell. British Journal of Cancer. 2010;103:439–445. doi: 10.1038/sj.bjc.6605821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boylan JM, Gruppuso PA. d-type cyclins and G1 progression during liver development in the rat. Biochemical and Biophysical Research Communications. 2005;330:722–730. doi: 10.1016/j.bbrc.2005.03.042. [DOI] [PubMed] [Google Scholar]
- Boylan JM, Anand P, Gruppuso PA. Ribosomal protein S6 phosphorylation and function during late gestation liver development in the rat. Journal of Biological Chemistry. 2001;276:44457–44463. doi: 10.1074/jbc.M103457200. [DOI] [PubMed] [Google Scholar]
- Braun L, Gruppuso P, Mikumo R, Fausto N. Transforming growth factor beta 1 in liver carcinogenesis: messenger RNA expression and growth effects. Cell Growth & Differentiation. 1990;1:103–111. [PubMed] [Google Scholar]
- Christie GR, Williams DJ, Macisaac F, Dickinson RJ, Rosewell I, Keyse SM. The dual-specificity protein phosphatase DUSP9/MKP-4 is essential for placental function but is not required for normal embryonic development. Molecular and Cellular Biology. 2005;25:8323–8333. doi: 10.1128/MCB.25.18.8323-8333.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabeva MD, Shafritz DA. Activation, proliferation, and differentiation of progenitor cells into hepatocytes in the d-galactosamine model of liver regeneration. American Journal of Pathology. 1993;143:1606–1620. [PMC free article] [PubMed] [Google Scholar]
- Eng CP, Sehgal SN, Vezina C. Activity of rapamycin (AY-22,989) against transplanted tumors. Journal of Antibiotics (Tokyo) 1984;37:1231–1237. doi: 10.7164/antibiotics.37.1231. [DOI] [PubMed] [Google Scholar]
- Farber E. Similarities in the sequence of early histological changes induced in the liver of the rat by ethionine, 2-acetylamino-fluorene, and 3′-methyl-4-dimethylaminoazobenzene. Cancer Research. 1956;16:142–148. [PubMed] [Google Scholar]
- Faris RA, Monfils BA, Dunsford HA, Hixson DC. Antigenic relationship between oval cells and a subpopulation of hepatic foci, nodules, and carcinomas induced by the “resistant hepatocyte” model system. Cancer Research. 1991;51:1308–1317. [PubMed] [Google Scholar]
- Faris RA, McBride A, Yang L, Affigne S, Walker C, Cha CJ. Isolation, propagation, and characterization of rat liver serosal mesothelial cells. American Journal of Pathology. 1994;145:1432–1443. [PMC free article] [PubMed] [Google Scholar]
- Fingar DC, Blenis J. Target of rapamycin (TOR): an integrator of nutrient and growth factor signals and coordinator of cell growth and cell cycle progression. Oncogene. 2004;23:3151–3171. doi: 10.1038/sj.onc.1207542. [DOI] [PubMed] [Google Scholar]
- Gingras AC, Raught B, Sonenberg N. Regulation of translation initiation by FRAP/mTOR. Genes & Development. 2001;15:807–826. doi: 10.1101/gad.887201. [DOI] [PubMed] [Google Scholar]
- Gruppuso PA, Boylan JM, Vaslet CA. Identification of candidate growth-regulating genes that are overexpressed in late gestation fetal liver in the rat. Biochimica et Biophysica Acta. 2000;1494:242–247. doi: 10.1016/s0167-4781(00)00244-x. [DOI] [PubMed] [Google Scholar]
- Guertin DA, Sabatini DM. The pharmacology of mTOR inhibition. Science Signaling. 2009;2:e24. doi: 10.1126/scisignal.267pe24. [DOI] [PubMed] [Google Scholar]
- Hay N, Sonenberg N. Upstream and downstream of mTOR. Genes & Development. 2004;18:1926–1945. doi: 10.1101/gad.1212704. [DOI] [PubMed] [Google Scholar]
- Hixson DC. Animal Models for Assessing the Contribution of Stem Cells to Liver Development. In: Sell S, editor. Stem Cells Handbook. Humana Press; Towanda: 2003. [Google Scholar]
- Hixson DC, Chapman L, McBride A, Faris R, Yang L. Antigenic phenotypes common to rat oval cells, primary hepatocellular carcinomas and developing bile ducts. Carcinogenesis. 1997;18:1169–1175. doi: 10.1093/carcin/18.6.1169. [DOI] [PubMed] [Google Scholar]
- Hixson DC, Brown J, McBride AC, Affigne S. Differentiation status of rat ductal cells and ethionine-induced hepatic carcinomas defined with surface-reactive monoclonal antibodies. Experimental and Molecular Pathology. 2000;68:152–169. doi: 10.1006/exmp.2000.2302. [DOI] [PubMed] [Google Scholar]
- Jakubowski A, Ambrose C, Parr M, Lincecum JM, Wang MZ, Zheng TS, Browning B, Michaelson JS, Baetscher M, Wang B, Bissell DM, Burkly LC. TWEAK induces liver progenitor cell proliferation. The Journal of Clinical Investigation. 2005;115:2330–2340. doi: 10.1172/JCI23486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez RH, Boylan JM, Lee JS, Francesconi M, Castellani G, Sanders JA, Gruppuso PA. Rapamycin response in tumorigenic and non-tumorigenic hepatic cell lines. PLoS One. 2009;4:e7373. doi: 10.1371/journal.pone.0007373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight B, Matthews VB, Akhurst B, Croager EJ, Klinken E, Abraham LJ, Olynyk JK, Yeoh G. Liver inflammation and cytokine production, but not acute phase protein synthesis, accompany the adult liver progenitor (oval) cell response to chronic liver injury. Immunology and Cell Biology. 2005;83:364–374. doi: 10.1111/j.1440-1711.2005.01346.x. [DOI] [PubMed] [Google Scholar]
- Leeds P, Kren BT, Boylan JM, Betz NA, Steer CJ, Gruppuso PA, Ross J. Developmental regulation of CRD-BP, an RNA-binding protein that stabilizes c-myc mRNA in vitro. Oncogene. 1997;14:1279–1286. doi: 10.1038/sj.onc.1201093. [DOI] [PubMed] [Google Scholar]
- Lowes KN, Brennan BA, Yeoh GC, Olynyk JK. Oval cell numbers in human chronic liver diseases are directly related to disease severity. American Journal of Pathology. 1999;154:537–541. doi: 10.1016/S0002-9440(10)65299-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangnall D, Bird NC, Majeed AW. The molecular physiology of liver regeneration following partial hepatectomy. Liver International. 2003;23:124–138. doi: 10.1034/j.1600-0676.2003.00812.x. [DOI] [PubMed] [Google Scholar]
- Ong LC, Song IC, Jin Y, Kee IH, Siew E, Yu S, Thng CH, Huynh H, Chow PK. Effective inhibition of xenografts of hepatocellular carcinoma (HepG2) by rapamycin and bevacizumab in an intrahepatic model. Molecular Imaging and Biology. 2009;11:334–342. doi: 10.1007/s11307-009-0213-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piguet AC, Semela D, Keogh A, Wilkens L, Stroka D, Stoupis C, St Pierre MV, Dufour JF. Inhibition of mTOR in combination with doxorubicin in an experimental model of hepatocellular carcinoma. Journal of Hepatology. 2008;49:78–87. doi: 10.1016/j.jhep.2008.03.024. [DOI] [PubMed] [Google Scholar]
- Roskams TA, Libbrecht L, Desmet VJ. Progenitor cells in diseased human liver. Seminars in Liver Disease. 2003;23:385–396. doi: 10.1055/s-2004-815564. [DOI] [PubMed] [Google Scholar]
- Salic A, Mitchison TJ. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:2415–2420. doi: 10.1073/pnas.0712168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders JA, Lakhani A, Phornphutkul C, Wu KY, Gruppuso PA. The effect of rapamycin on DNA synthesis in multiple tissues from late gestation fetal and post-natal rats. American Journal of Physiology. Cell Physiology. 2008;295:C406–C413. doi: 10.1152/ajpcell.00450.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh K, Hatayama I, Tsuchida S, Sato K. Biochemical characteristics of a preneoplastic marker enzyme glutathione S-transferase P-form (7–7). Archives of Biochemistry and Biophysics. 1991;285:312–316. doi: 10.1016/0003-9861(91)90365-p. [DOI] [PubMed] [Google Scholar]
- Schoniger-Hekele M, Muller C. Pilot study: rapamycin in advanced hepatocellular carcinoma. Alimentary Pharmacology & Therapeutics. 2010;32:763–768. doi: 10.1111/j.1365-2036.2010.04404.x. [DOI] [PubMed] [Google Scholar]
- Sekiya S, Gotoh S, Yamashita T, Watanabe T, Saitoh S, Sendo F. Selective depletion of rat neutrophils by in vivo administration of a monoclonal antibody. Journal of Leukocyte Biology. 1989;46:96–102. doi: 10.1002/jlb.46.2.96. [DOI] [PubMed] [Google Scholar]
- Shi Y, August DA. A new trick for an old drug: mTOR inhibitor rapamycin augments the effect of fluorouracil on hepatocellular carcinoma by inducing cell senescence. Cancer Biology & Therapy. 2008;7:397–398. doi: 10.4161/cbt.7.3.5871. [DOI] [PubMed] [Google Scholar]
- Shinozuka H, Lombardi B, Sell S, Iammarino RM. Enhancement of DL-ethionine-induced liver carcinogenesis in rats fed a choline-devoid diet. Journal of the National Cancer Institute. 1978;61:813–817. [PubMed] [Google Scholar]
- Solt DB, Medline A, Farber E. Rapid emergence of carcinogen-induced hyperplastic lesions in a new model for the sequential analysis of liver carcinogenesis. American Journal of Pathology. 1977;88:595–618. [PMC free article] [PubMed] [Google Scholar]
- Strick-Marchand H, Masse GX, Weiss MC, Di Santo JP. Lymphocytes support oval cell-dependent liver regeneration. Journal of Immunology. 2008;181:2764–2771. doi: 10.4049/jimmunol.181.4.2764. [DOI] [PubMed] [Google Scholar]
- Tatematsu M, Ho RH, Kaku T, Ekem JK, Farber E. Studies on the proliferation and fate of oval cells in the liver of rats treated with 2-acetylaminofluorene and partial hepatectomy. American Journal of Pathology. 1984;114:418–430. [PMC free article] [PubMed] [Google Scholar]
- Taub R. Liver regeneration: from myth to mechanism. Nature Reviews. Molecular Cell Biology. 2004;5:836–847. doi: 10.1038/nrm1489. [DOI] [PubMed] [Google Scholar]
- Thomson AW, Turnquist HR, Raimondi G. Immunoregulatory functions of mTOR inhibition. Nature Reviews. Immunology. 2009;9:324–337. doi: 10.1038/nri2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vezina C, Kudelski A, Sehgal SN. Rapamycin (AY-22,989), a new antifungal antibiotic. I. Taxonomy of the producing streptomycete and isolation of the active principle. Journal of Antibiotics (Tokyo) 1975;28:721–726. doi: 10.7164/antibiotics.28.721. [DOI] [PubMed] [Google Scholar]
- Villanueva A, Chiang DY, Newell P, Peix J, Thung S, Alsinet C, Tovar V, Roayaie S, Minguez B, Sole M, Battiston C, Van Laarhoven S, Fiel MI, Di Feo A, Hoshida Y, Yea S, Toffanin S, Ramos A, Martignetti JA, Mazzaferro V, Bruix J, Waxman S, Schwartz M, Meyerson M, Friedman SL, Llovet JM. Pivotal role of mTOR signaling in hepatocellular carcinoma. Gastroenterology. 1983;135:1972–1983. doi: 10.1053/j.gastro.2008.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wai LE, Fujiki M, Takeda S, Martinez OM, Krams SM. Rapamycin, but not cyclosporine or FK506, alters natural killer cell function. Transplantation. 2008;85:145–149. doi: 10.1097/01.tp.0000296817.28053.7b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiederrecht GJ, Sabers CJ, Brunn GJ, Martin MM, Dumont FJ, Abraham RT. Mechanism of action of rapamycin: new insights into the regulation of G1-phase progression in eukaryotic cells. Progress in Cell Cycle Research. 1995;1:53–71. doi: 10.1007/978-1-4615-1809-9_5. [DOI] [PubMed] [Google Scholar]
- Yamashita T, Forgues M, Wang W, Kim JW, Ye Q, Jia H, Budhu A, Zanetti KA, Chen Y, Qin LX, Tang ZY, Wang XW. EpCAM and alpha-fetoprotein expression defines novel prognostic subtypes of hepatocellular carcinoma. Cancer Research. 2008;68:1451–1461. doi: 10.1158/0008-5472.CAN-07-6013. [DOI] [PubMed] [Google Scholar]
- Yang XR, Xu Y, Yu B, Zhou J, Qiu SJ, Shi GM, Zhang BH, Wu WZ, Shi YH, Wu B, Yang GH, Ji Y, Fan J. High expression levels of putative hepatic stem/progenitor cell biomarkers related to tumour angiogenesis and poor prognosis of hepatocellular carcinoma. Gut. 2010;59:953–962. doi: 10.1136/gut.2008.176271. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.