Abstract
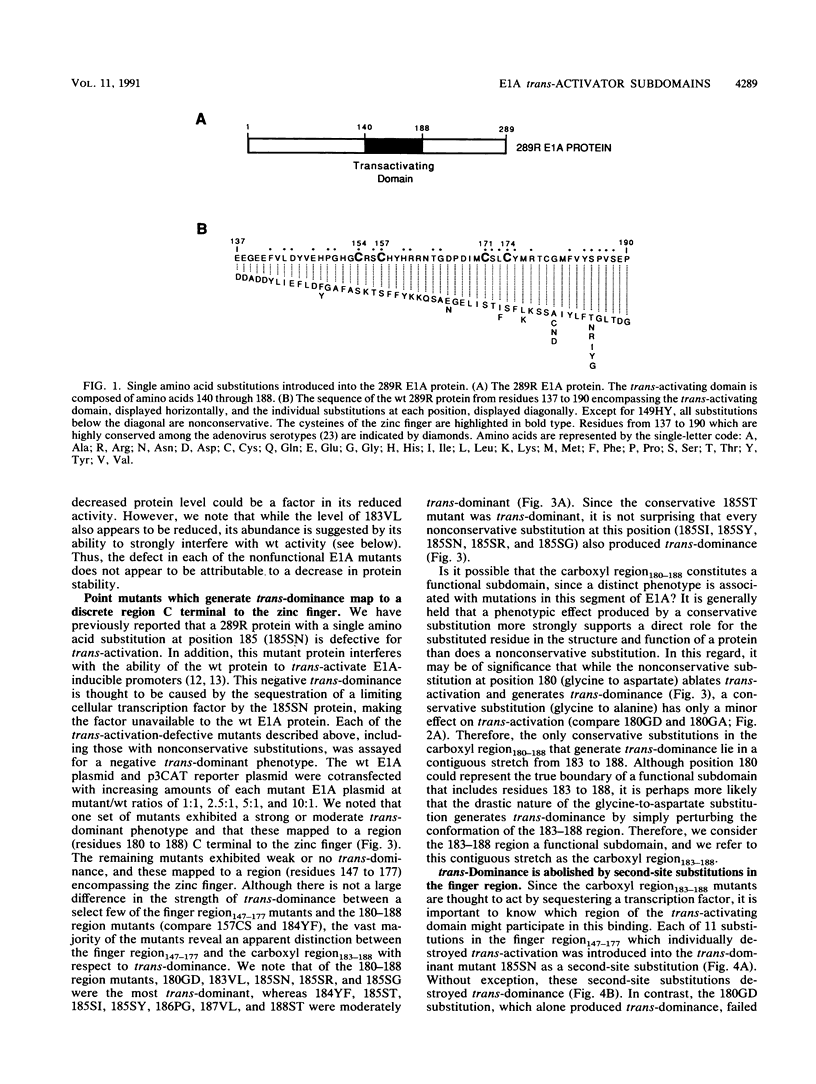
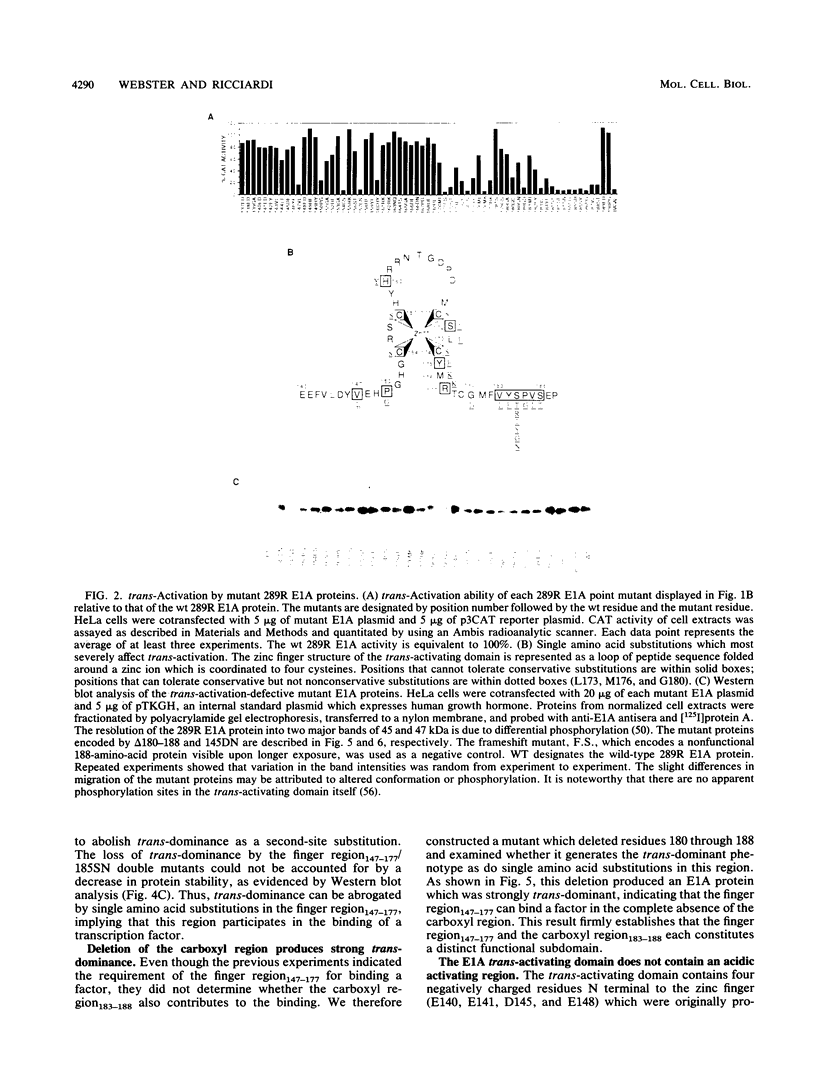
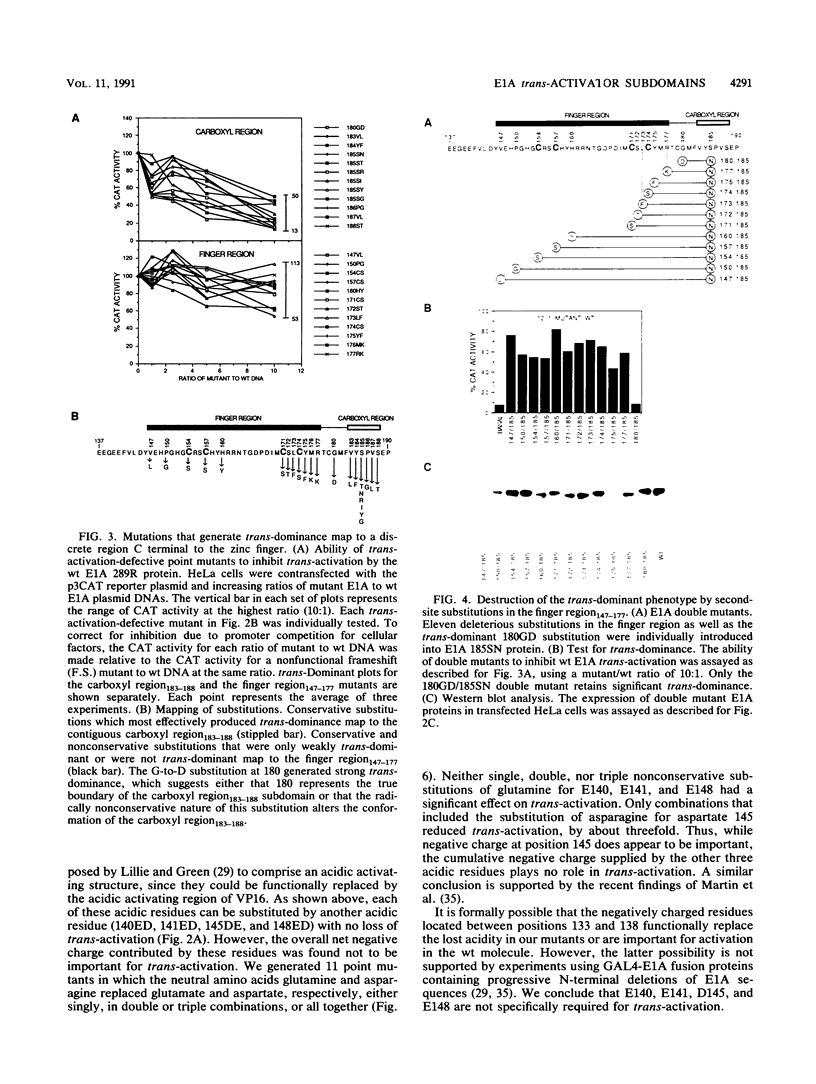
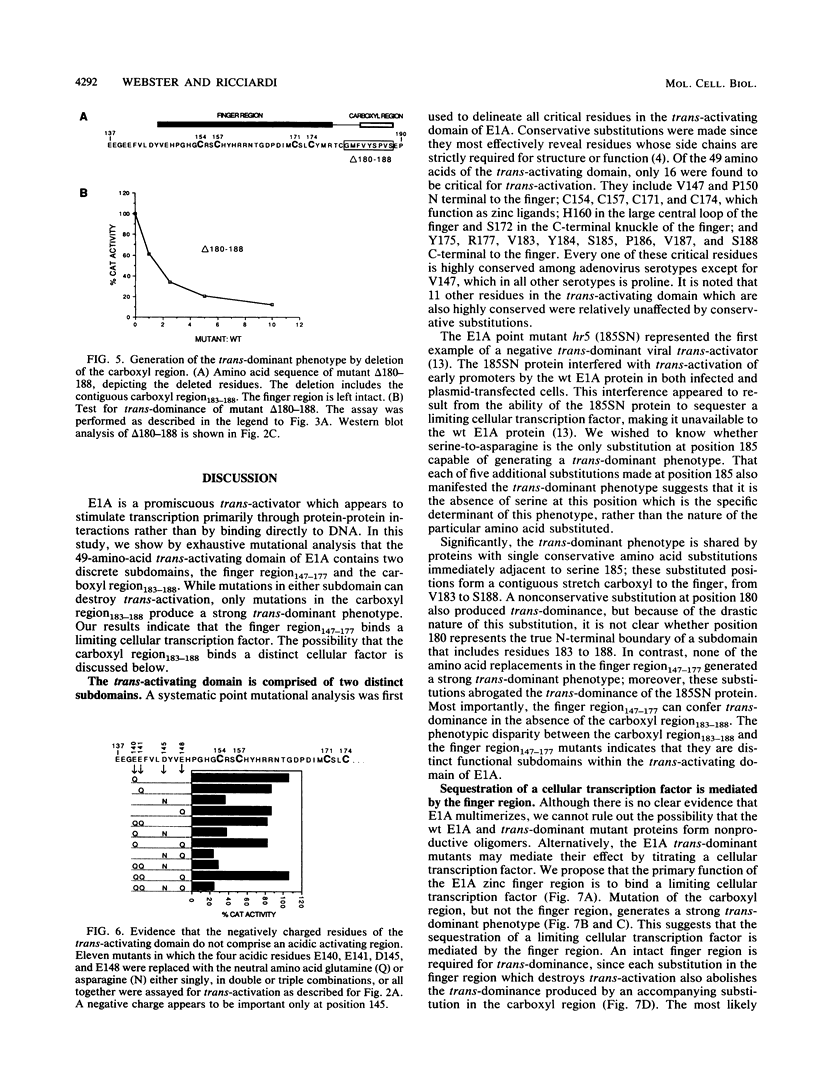
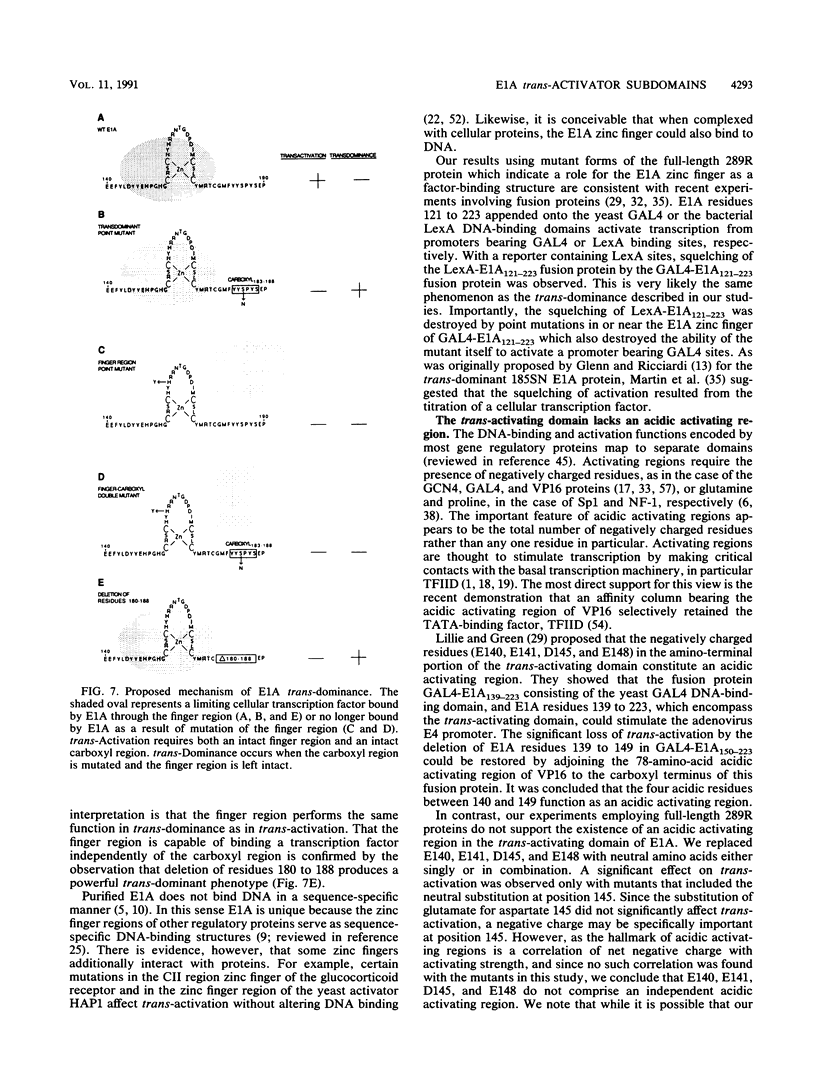
The 289R E1A protein of adenovirus stimulates transcription of early viral and certain cellular genes. trans-Activation requires residues 140 to 188, which encompass a zinc finger. Several studies have indicated that trans-activation by E1A is mediated through cellular transcription factors. In particular, the ability of the trans-dominant E1A point mutant hr5 (Ser-185 to Asn) to inhibit wild-type E1A trans-activation was proposed to result from the sequestration of a cellular factor. Using site-directed mutagenesis, we individually replaced every residue within and flanking the trans-activating domain with a conservative amino acid, revealing 16 critical residues. Six of the individual substitutions lying in a contiguous stretch C terminal to the zinc finger (carboxyl region183-188) imparted a trans-dominant phenotype. trans-Dominance was even produced by deletion of the entire carboxyl region183-188. Conversely, an intact finger region147-177 was absolutely required for trans-dominance, since second-site substitution of every critical residue in this region abrogated the trans-dominant phenotype of the hr5 protein. These data indicate that the finger region147-177 bind a limiting cellular transcription factor and that the carboxyl region183-188 provides a separate and essential function. In addition, we show that four negatively charged residues within the trans-activating domain do not comprise a distinct acidic activating region. We present a model in which the trans-activating domain of E1A binds to two different cellular protein targets through the finger and carboxyl regions.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abmayr S. M., Workman J. L., Roeder R. G. The pseudorabies immediate early protein stimulates in vitro transcription by facilitating TFIID: promoter interactions. Genes Dev. 1988 May;2(5):542–553. doi: 10.1101/gad.2.5.542. [DOI] [PubMed] [Google Scholar]
- Bagchi S., Raychaudhuri P., Nevins J. R. Phosphorylation-dependent activation of the adenovirus-inducible E2F transcription factor in a cell-free system. Proc Natl Acad Sci U S A. 1989 Jun;86(12):4352–4356. doi: 10.1073/pnas.86.12.4352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berk A. J. Adenovirus promoters and E1A transactivation. Annu Rev Genet. 1986;20:45–79. doi: 10.1146/annurev.ge.20.120186.000401. [DOI] [PubMed] [Google Scholar]
- Bowie J. U., Reidhaar-Olson J. F., Lim W. A., Sauer R. T. Deciphering the message in protein sequences: tolerance to amino acid substitutions. Science. 1990 Mar 16;247(4948):1306–1310. doi: 10.1126/science.2315699. [DOI] [PubMed] [Google Scholar]
- Chatterjee P. K., Bruner M., Flint S. J., Harter M. L. DNA-binding properties of an adenovirus 289R E1A protein. EMBO J. 1988 Mar;7(3):835–841. doi: 10.1002/j.1460-2075.1988.tb02882.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courey A. J., Tjian R. Analysis of Sp1 in vivo reveals multiple transcriptional domains, including a novel glutamine-rich activation motif. Cell. 1988 Dec 2;55(5):887–898. doi: 10.1016/0092-8674(88)90144-4. [DOI] [PubMed] [Google Scholar]
- Culp J. S., Webster L. C., Friedman D. J., Smith C. L., Huang W. J., Wu F. Y., Rosenberg M., Ricciardi R. P. The 289-amino acid E1A protein of adenovirus binds zinc in a region that is important for trans-activation. Proc Natl Acad Sci U S A. 1988 Sep;85(17):6450–6454. doi: 10.1073/pnas.85.17.6450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Sal G., Manfioletti G., Schneider C. The CTAB-DNA precipitation method: a common mini-scale preparation of template DNA from phagemids, phages or plasmids suitable for sequencing. Biotechniques. 1989 May;7(5):514–520. [PubMed] [Google Scholar]
- Evans R. M., Hollenberg S. M. Zinc fingers: gilt by association. Cell. 1988 Jan 15;52(1):1–3. doi: 10.1016/0092-8674(88)90522-3. [DOI] [PubMed] [Google Scholar]
- Ferguson B., Krippl B., Andrisani O., Jones N., Westphal H., Rosenberg M. E1A 13S and 12S mRNA products made in Escherichia coli both function as nucleus-localized transcription activators but do not directly bind DNA. Mol Cell Biol. 1985 Oct;5(10):2653–2661. doi: 10.1128/mcb.5.10.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman L. P., Luisi B. F., Korszun Z. R., Basavappa R., Sigler P. B., Yamamoto K. R. The function and structure of the metal coordination sites within the glucocorticoid receptor DNA binding domain. Nature. 1988 Aug 11;334(6182):543–546. doi: 10.1038/334543a0. [DOI] [PubMed] [Google Scholar]
- Glenn G. M., Ricciardi R. P. Adenovirus 5 early region 1A host range mutants hr3, hr4, and hr5 contain point mutations which generate single amino acid substitutions. J Virol. 1985 Oct;56(1):66–74. doi: 10.1128/jvi.56.1.66-74.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glenn G. M., Ricciardi R. P. An adenovirus type 5 E1A protein with a single amino acid substitution blocks wild-type E1A transactivation. Mol Cell Biol. 1987 Mar;7(3):1004–1011. doi: 10.1128/mcb.7.3.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman C. M., Moffat L. F., Howard B. H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982 Sep;2(9):1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham F. L., van der Eb A. J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973 Apr;52(2):456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- Hanas J. S., Hazuda D. J., Bogenhagen D. F., Wu F. Y., Wu C. W. Xenopus transcription factor A requires zinc for binding to the 5 S RNA gene. J Biol Chem. 1983 Dec 10;258(23):14120–14125. [PubMed] [Google Scholar]
- Hope I. A., Struhl K. Functional dissection of a eukaryotic transcriptional activator protein, GCN4 of yeast. Cell. 1986 Sep 12;46(6):885–894. doi: 10.1016/0092-8674(86)90070-x. [DOI] [PubMed] [Google Scholar]
- Horikoshi M., Carey M. F., Kakidani H., Roeder R. G. Mechanism of action of a yeast activator: direct effect of GAL4 derivatives on mammalian TFIID-promoter interactions. Cell. 1988 Aug 26;54(5):665–669. doi: 10.1016/s0092-8674(88)80011-4. [DOI] [PubMed] [Google Scholar]
- Horikoshi M., Hai T., Lin Y. S., Green M. R., Roeder R. G. Transcription factor ATF interacts with the TATA factor to facilitate establishment of a preinitiation complex. Cell. 1988 Sep 23;54(7):1033–1042. doi: 10.1016/0092-8674(88)90118-3. [DOI] [PubMed] [Google Scholar]
- Jones N. C., Richter J. D., Weeks D. L., Smith L. D. Regulation of adenovirus transcription by an E1a gene in microinjected Xenopus laevis oocytes. Mol Cell Biol. 1983 Dec;3(12):2131–2142. doi: 10.1128/mcb.3.12.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadonaga J. T., Carner K. R., Masiarz F. R., Tjian R. Isolation of cDNA encoding transcription factor Sp1 and functional analysis of the DNA binding domain. Cell. 1987 Dec 24;51(6):1079–1090. doi: 10.1016/0092-8674(87)90594-0. [DOI] [PubMed] [Google Scholar]
- Kim K. S., Guarente L. Mutations that alter transcriptional activation but not DNA binding in the zinc finger of yeast activator HAPI. Nature. 1989 Nov 9;342(6246):200–203. doi: 10.1038/342200a0. [DOI] [PubMed] [Google Scholar]
- Kimelman D., Miller J. S., Porter D., Roberts B. E. E1a regions of the human adenoviruses and of the highly oncogenic simian adenovirus 7 are closely related. J Virol. 1985 Feb;53(2):399–409. doi: 10.1128/jvi.53.2.399-409.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingston R. E., Baldwin A. S., Sharp P. A. Transcription control by oncogenes. Cell. 1985 May;41(1):3–5. doi: 10.1016/0092-8674(85)90049-2. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Leong K., Brunet L., Berk A. J. Factors responsible for the higher transcriptional activity of extracts of adenovirus-infected cells fractionate with the TATA box transcription factor. Mol Cell Biol. 1988 Apr;8(4):1765–1774. doi: 10.1128/mcb.8.4.1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewin B. Commitment and activation at pol II promoters: a tail of protein-protein interactions. Cell. 1990 Jun 29;61(7):1161–1164. doi: 10.1016/0092-8674(90)90675-5. [DOI] [PubMed] [Google Scholar]
- Lillie J. W., Green M. R. Transcription activation by the adenovirus E1a protein. Nature. 1989 Mar 2;338(6210):39–44. doi: 10.1038/338039a0. [DOI] [PubMed] [Google Scholar]
- Lillie J. W., Green M., Green M. R. An adenovirus E1a protein region required for transformation and transcriptional repression. Cell. 1986 Sep 26;46(7):1043–1051. doi: 10.1016/0092-8674(86)90704-x. [DOI] [PubMed] [Google Scholar]
- Lillie J. W., Loewenstein P. M., Green M. R., Green M. Functional domains of adenovirus type 5 E1a proteins. Cell. 1987 Sep 25;50(7):1091–1100. doi: 10.1016/0092-8674(87)90175-9. [DOI] [PubMed] [Google Scholar]
- Liu F., Green M. R. A specific member of the ATF transcription factor family can mediate transcription activation by the adenovirus E1a protein. Cell. 1990 Jun 29;61(7):1217–1224. doi: 10.1016/0092-8674(90)90686-9. [DOI] [PubMed] [Google Scholar]
- Ma J., Ptashne M. Deletion analysis of GAL4 defines two transcriptional activating segments. Cell. 1987 Mar 13;48(5):847–853. doi: 10.1016/0092-8674(87)90081-x. [DOI] [PubMed] [Google Scholar]
- Malim M. H., Böhnlein S., Hauber J., Cullen B. R. Functional dissection of the HIV-1 Rev trans-activator--derivation of a trans-dominant repressor of Rev function. Cell. 1989 Jul 14;58(1):205–214. doi: 10.1016/0092-8674(89)90416-9. [DOI] [PubMed] [Google Scholar]
- Martin K. J., Lillie J. W., Green M. R. Evidence for interaction of different eukaryotic transcriptional activators with distinct cellular targets. Nature. 1990 Jul 12;346(6280):147–152. doi: 10.1038/346147a0. [DOI] [PubMed] [Google Scholar]
- Merino A., Buckbinder L., Mermelstein F. H., Reinberg D. Phosphorylation of cellular proteins regulates their binding to the cAMP response element. J Biol Chem. 1989 Dec 15;264(35):21266–21276. [PubMed] [Google Scholar]
- Mermer B., Felber B. K., Campbell M., Pavlakis G. N. Identification of trans-dominant HIV-1 rev protein mutants by direct transfer of bacterially produced proteins into human cells. Nucleic Acids Res. 1990 Apr 25;18(8):2037–2044. doi: 10.1093/nar/18.8.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mermod N., O'Neill E. A., Kelly T. J., Tjian R. The proline-rich transcriptional activator of CTF/NF-I is distinct from the replication and DNA binding domain. Cell. 1989 Aug 25;58(4):741–753. doi: 10.1016/0092-8674(89)90108-6. [DOI] [PubMed] [Google Scholar]
- Miller J., McLachlan A. D., Klug A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 1985 Jun;4(6):1609–1614. doi: 10.1002/j.1460-2075.1985.tb03825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montell C., Fisher E. F., Caruthers M. H., Berk A. J. Resolving the functions of overlapping viral genes by site-specific mutagenesis at a mRNA splice site. Nature. 1982 Feb 4;295(5848):380–384. doi: 10.1038/295380a0. [DOI] [PubMed] [Google Scholar]
- Moran E., Mathews M. B. Multiple functional domains in the adenovirus E1A gene. Cell. 1987 Jan 30;48(2):177–178. doi: 10.1016/0092-8674(87)90418-1. [DOI] [PubMed] [Google Scholar]
- Moran E., Zerler B., Harrison T. M., Mathews M. B. Identification of separate domains in the adenovirus E1A gene for immortalization activity and the activation of virus early genes. Mol Cell Biol. 1986 Oct;6(10):3470–3480. doi: 10.1128/mcb.6.10.3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson L., Garcia J., Wu F., Modesti N., Nelson J., Gaynor R. A transdominant tat mutant that inhibits tat-induced gene expression from the human immunodeficiency virus long terminal repeat. Proc Natl Acad Sci U S A. 1990 Jul;87(13):5079–5083. doi: 10.1073/pnas.87.13.5079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei R., Berk A. J. Multiple transcription factor binding sites mediate adenovirus E1A transactivation. J Virol. 1989 Aug;63(8):3499–3506. doi: 10.1128/jvi.63.8.3499-3506.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptashne M., Gann A. A. Activators and targets. Nature. 1990 Jul 26;346(6282):329–331. doi: 10.1038/346329a0. [DOI] [PubMed] [Google Scholar]
- Ptashne M. How eukaryotic transcriptional activators work. Nature. 1988 Oct 20;335(6192):683–689. doi: 10.1038/335683a0. [DOI] [PubMed] [Google Scholar]
- Raychaudhuri P., Bagchi S., Nevins J. R. DNA-binding activity of the adenovirus-induced E4F transcription factor is regulated by phosphorylation. Genes Dev. 1989 May;3(5):620–627. doi: 10.1101/gad.3.5.620. [DOI] [PubMed] [Google Scholar]
- Raychaudhuri P., Rooney R., Nevins J. R. Identification of an E1A-inducible cellular factor that interacts with regulatory sequences within the adenovirus E4 promoter. EMBO J. 1987 Dec 20;6(13):4073–4081. doi: 10.1002/j.1460-2075.1987.tb02753.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricciardi R. P., Jones R. L., Cepko C. L., Sharp P. A., Roberts B. E. Expression of early adenovirus genes requires a viral encoded acidic polypeptide. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6121–6125. doi: 10.1073/pnas.78.10.6121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter J. D., Slavicek J. M., Schneider J. F., Jones N. C. Heterogeneity of adenovirus type 5 E1A proteins: multiple serine phosphorylations induce slow-migrating electrophoretic variants but do not affect E1A-induced transcriptional activation or transformation. J Virol. 1988 Jun;62(6):1948–1955. doi: 10.1128/jvi.62.6.1948-1955.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schena M., Freedman L. P., Yamamoto K. R. Mutations in the glucocorticoid receptor zinc finger region that distinguish interdigitated DNA binding and transcriptional enhancement activities. Genes Dev. 1989 Oct;3(10):1590–1601. doi: 10.1101/gad.3.10.1590. [DOI] [PubMed] [Google Scholar]
- Simon M. C., Fisch T. M., Benecke B. J., Nevins J. R., Heintz N. Definition of multiple, functionally distinct TATA elements, one of which is a target in the hsp70 promoter for E1A regulation. Cell. 1988 Mar 11;52(5):723–729. doi: 10.1016/0092-8674(88)90410-2. [DOI] [PubMed] [Google Scholar]
- Stringer K. F., Ingles C. J., Greenblatt J. Direct and selective binding of an acidic transcriptional activation domain to the TATA-box factor TFIID. Nature. 1990 Jun 28;345(6278):783–786. doi: 10.1038/345783a0. [DOI] [PubMed] [Google Scholar]
- Taylor I. C., Kingston R. E. E1a transactivation of human HSP70 gene promoter substitution mutants is independent of the composition of upstream and TATA elements. Mol Cell Biol. 1990 Jan;10(1):176–183. doi: 10.1128/mcb.10.1.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay M. L., Dumont D. J., Branton P. E. Analysis of phosphorylation sites in the exon 1 region of E1A proteins of human adenovirus type 5. Virology. 1989 Apr;169(2):397–407. doi: 10.1016/0042-6822(89)90165-7. [DOI] [PubMed] [Google Scholar]
- Triezenberg S. J., Kingsbury R. C., McKnight S. L. Functional dissection of VP16, the trans-activator of herpes simplex virus immediate early gene expression. Genes Dev. 1988 Jun;2(6):718–729. doi: 10.1101/gad.2.6.718. [DOI] [PubMed] [Google Scholar]
- Wachsman W., Cann A. J., Williams J. L., Slamon D. J., Souza L., Shah N. P., Chen I. S. HTLV x gene mutants exhibit novel transcriptional regulatory phenotypes. Science. 1987 Feb 6;235(4789):674–677. doi: 10.1126/science.3027894. [DOI] [PubMed] [Google Scholar]
- Weeks D. L., Jones N. C. E1A control of gene expression is mediated by sequences 5' to the transcriptional starts of the early viral genes. Mol Cell Biol. 1983 Jul;3(7):1222–1234. doi: 10.1128/mcb.3.7.1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte P., Williamson N. M., Harlow E. Cellular targets for transformation by the adenovirus E1A proteins. Cell. 1989 Jan 13;56(1):67–75. doi: 10.1016/0092-8674(89)90984-7. [DOI] [PubMed] [Google Scholar]
- Williams G. T., McClanahan T. K., Morimoto R. I. E1a transactivation of the human HSP70 promoter is mediated through the basal transcriptional complex. Mol Cell Biol. 1989 Jun;9(6):2574–2587. doi: 10.1128/mcb.9.6.2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winberg G., Shenk T. Dissection of overlapping functions within the adenovirus type 5 E1A gene. EMBO J. 1984 Aug;3(8):1907–1912. doi: 10.1002/j.1460-2075.1984.tb02066.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Rosser D. S., Schmidt M. C., Berk A. A TATA box implicated in E1A transcriptional activation of a simple adenovirus 2 promoter. Nature. 1987 Apr 2;326(6112):512–515. doi: 10.1038/326512a0. [DOI] [PubMed] [Google Scholar]