Abstract
Insect cells, like other eucaryotic cells, modify many of their proteins by N-glycosylation. However, the endogenous insect cell N-glycan processing machinery generally does not produce complex, terminally sialylated N-glycans such as those found in mammalian systems. This difference in the N-glycan processing pathways of insect cells and higher eucaryotes imposes a significant limitation on their use as hosts for baculovirus-mediated recombinant glycoprotein production. To address this problem, we previously isolated two transgenic insect cell lines that have mammalian β1,4-galactosyltransferase or β1,4- galactosyltransferase and α2,6-sialyltransferase genes. Unlike the parental insect cell line, both transgenic cell lines expressed the mammalian glycosyltransferases and were able to produce terminally galactosylated or sialylated N-glycans. The purpose of the present study was to investigate the structures of the N-glycans produced by these transgenic insect cell lines in further detail. Direct structural analyses revealed that the most extensively processed N-glycans produced by the transgenic insect cell lines were novel, monoantennary structures with elongation of only the α1,3 branch. This led to the hypothesis that the transgenic insect cell lines lacked adequate endogenous N-acetylglucosaminyltransferase II activity for biantennary N-glycan production. To test this hypothesis and further extend the N-glycan processing pathway in Sf9 cells, we produced a new transgenic line designed to constitutively express a more complete array of mammalian glycosyltransferases, including N-acetylglucosaminyltransferase II. This new transgenic insect cell line, designated SfSWT-1, has higher levels of five glycosyltransferase activities than the parental cells and supports baculovirus replication at normal levels. In addition, direct structural analyses showed that SfSWT-1 cells could produce biantennary, terminally sialylated N-glycans. Thus, this study provides new insight on the glycobiology of insect cells and describes a new transgenic insect cell line that will be widely useful for the production of more authentic recombinant glycoproteins by baculovirus expression vectors.
Like other eucaryotic cells, insect cells covalently modify many of their proteins by N-glycosylation. The complexity of insect protein N-glycosylation pathways is intermediate between those of Saccharomyces cerevisiae and mammalian cells (reviewed in refs 1–6). Each begins with the cotranslational transfer of a dolichol-linked precursor oligosaccharide to a specific recognition sequence within newly synthesized glycoproteins. In all three systems, exoglycosidases in the endoplasmic reticulum remove terminal glucose and mannose residues from the precursor oligosaccharide to produce a common intermediate, Man8GlcNAc2-N-Asn. However, this intermediate is subsequently converted to distinct end products in these three different systems. In S. cerevisiae, mannosyltransferase adds peripheral mannose residues to the trimmed precursor to produce polymannosidic N-glycans. In insect and mammalian cells, exoglycosidases and a glycosyltransferase catalyze trimming and elongation reactions, which yield the common intermediate, GlcNAcMan3-GlcNAc2-N-Asn. In mammalian cells, terminal glycosyltransferases can elongate this common intermediate to produce hybrid and complex N-glycans with terminal sialic acids. In contrast, insect cells have only extremely low levels, if any, of these terminal glycosyltransferase activities and, in some cases, have a competing exoglycosidase that can remove the terminal N-acetylglucosamine residue from GlcNAcMan3GlcNAc2-N-Asn. Hence, the major processed N-glycan produced by insect cells is usually the paucimannosidic structure, Man3GlcNAc2-N-Asn.
It is important to understand insect protein N-glycosylation pathways because they occupy an important evolutionary niche among eucaryotic organisms. Additionally, insect cells occupy an important biotechnological niche as hosts for the production of recombinant glycoproteins by baculovirus expression vectors (2, 7, 8). In the context of this application, one must understand the differences between insect and mammalian N-glycan processing pathways because Nglycans are known to influence many different protein properties, such as enzymatic activity, conformation, immunogenicity, physical stability, and pharmacokinetics (9–13)American Chemical Due to the influence of N-glycans on glycoprotein structure and function and the general inability of insect cells to produce complex, terminally sialylated N-glycans, it is important to try to engineer insect cells to enable them to produce more extensively processed, mammalian-like Nglycans.
The first attempt to extend the N-glycan processing pathway in insect cells involved coinfecting a widely used insect cell line with two recombinant baculoviruses, one encoding human N-acetylglucosaminyltransferase I (GlcNAc-TI)1 and the other encoding a recombinant viral glycoprotein (14). This significantly increased GlcNAc-TI activity levels and enabled these cells to produce hybrid N-glycans with terminal N-acetylglucosamine residues instead of the usual paucimannose N-glycans. At about the same time, it was shown that a baculovirus encoding bovine β,4-galactosyltransferase (β4Gal-T) under the control of an immediate early promoter could produce this enzyme early in infection, which enabled insect cells to produce a viral glycoprotein with terminally β-galactosylated N-glycans later in infection (15). More recent efforts to engineer insect N-glycosylation pathways have focused on the creation of transgenic insect cell lines that constitutively express mammalian glycosyltransferase genes. In the first example of this approach, a Spodoptera frugiperda cell line (Sf9; 16) was transformed with a constitutively expressible bovine β4Gal-T gene (17). Unlike the parental Sf9 cells, these Sfβ4GalT cells were able to produce recombinant glycoproteins with terminally galactosylated N-glycans after baculovirus infection. This led to the isolation of transgenic Sf9 and Trichoplusia ni (BTI Tn 5B1-4; 18) derivatives that were doubly transformed with mammalian β4Gal-T and α2,6-sialyltransferase (ST6GalI) genes (19, 20). Lectin blotting analyses and limited glycan structural analyses showed that Sfβ4GalT/ST6 and Tnβ4GalT/ ST6 cells, unlike the parental cell lines, could produce recombinant glycoproteins with terminally sialylated Nglycans. Together, all of these studies have shown that N-glycan processing pathways in lepidopteran insect cells can be extended by the addition of mammalian genes encoding functions that are missing or limited, relative to mammalian cells.
The initial purpose of the current study was to examine the structures of the N-glycans produced by the transgenic insect cell lines isolated in our previous studies. The results of these more detailed structural analyses confirmed that both Sfβ4GalT and Sfβ4GalT/ST6 cells can produce more extensively processed N-glycans than their parental counterparts, as expected (17, 19). However, they also revealed that the most extensively processed N-glycans produced by these cells are monoantennary structures, in which only the mannose on the R1,3 branch is elongated. These results suggested that these insect cell lines have insufficient levels of N-acetylglucosaminyltransferase II (GlcNAc-TII) activity to produce conventional biantennary N-glycans. To examine this possibility, we constructed a new transgenic cell line, SfSWT-1, by transforming Sfβ4GalT cells with several mammalian glycosyltransferase genes, including GlcNAc-TII. Extensive analyses demonstrated that this new cell line can produce biantennary N-glycans terminating in sialic acid or galactose, which are structurally identical to complex N-glycans produced by mammalian cells. These results demonstrate that a limitation in GlcNAc-TII activity is one reason Sf9 cells typically fail to produce biantennary N-glycans. In addition, the new insect cell line described in this study will be useful as an improved host for the production of more authentic recombinant glycoproteins by baculovirus expression vectors.
EXPERIMENTAL PROCEDURES
Construction of Immediate Early Expression Plasmids
pIE1Hygro, pIE1GlcNAcTI, pIE1GlcNAcTII, pIE1ST6GalI, and pIE1ST3GalIV are immediate early expression plasmids encoding Escherichia coli hygromycin phosphotransferase (21), human GlcNAc-TI (22), human GlcNAc-TII (23), rat ST6GalI (24), and a mouse α2,3-sialyltransferase (ST3GalIV; 25), respectively, under the control of a baculovirus ie1 promoter (26). The ie1 promoter provides constitutive foreign gene expression in uninfected insect cells. Therefore, immediate early expression plasmids can be used to genetically transform established insect cell lines (27). The methods used to construct pIE1Hygro and pIE1ST6GalI have been previously described (19, 20). The method used to construct pIE1GlcNAcTI involved introducing a BamHI site upstream of the open reading frame in the human GlcNAc-TI cDNA clone (22), excising and gel purifying the 2.6 kb BamHI–XbaI fragment from the resulting plasmid, and subcloning this fragment into the unique BamHI site of pIE1600BamHR (28). pIE1ST3GalIV was constructed in similar fashion, by excising and gel purifying the 1.032 kb BamHI–AccI fragment encoding full-length ST3GalIV from pBSSKST3GalIV (25) and then subcloning this fragment into the BamHI-NruI sites of pIE1HR4 (28). In contrast, a PCR-based method was used to construct pIE1GlcNAcTII. Gene-specific forward (5′-TTACTAGTGGAGACCATGAGGTTCCGCATC) and reverse (5′-CCACTAGTGAAACCAGTTCCTAAACTC) primers were used to produce a 1.441 kb amplimer with SpeI sites (underlined in the primer sequences) flanking the open reading frame from a human GlcNAc-TII cDNA clone (pHG30; 23). This amplimer was inserted into pCR-2.1-TOPO (Invitrogen, Carlsbad, CA), multiple clones were sequenced, and a clone with a wild-type open reading frame was used as the source of the SpeI fragment, which was gel purified and subcloned into the SpeI site of pIE1HR4.
Cells and Viruses
Sf9 (16), Sfβ4GalT (17), Sfβ4GalT/ ST6 (19), and SfSWT-1 (this study) cells were routinely maintained as shake flask cultures in TNM-FH medium containing 10% fetal bovine serum (HyClone, Logan, UT) and 0.1% (w/v) pluronic F68, as previously described (7). This medium was designated complete TNM-FH. In addition, all four cell lines were maintained as shake flask cultures in HyQ Sfx-Insect protein-free medium (Hyclone). SfSWT-1 is a new transgenic derivative of Sf9 cells that was produced for this study using a modification of an established procedure (27). Briefly, Sfβ4GalT cells (17) were cotransfected with a mixture containing 5 µg each of pIE1Hygro, pIE1Glc-NAcTI, pIE1GlcNAcTII, pIE1ST6GalI, and pIE1ST3GalIV using a modified calcium phosphate method (7). The transfected cells were allowed to recover for 1 day and were treated with 1 mg of hygromycin (Sigma, St. Louis, MO)/ mL of complete TNM-FH medium, and hygromycin-resistant clones were isolated by limiting dilution, as previously described (19). After stepwise amplification into larger cultures, individual clones were assayed for expression of the unselected markers using RNA dot-blot analyses, as described below.
Autographa californica multicapsid nucleopolyhedrovirus (AcMNPV) strain E2 (29) was used as the wild-type baculovirus. AcGST-SfManI (30) is a recombinant baculovirus that was used to express a soluble, glutathione S-transferase tagged version of an Sf9 class I R-mannosidase (GST-SfManI; 31) under the control of the polyhedrin promoter (30). Ac-βTP is a recombinant baculovirus that was used to express the human β-trace protein under the control of the polyhedrin promoter (H. Conradt, unpublished).
RNA Dot Blots
Total RNA was prepared from each hygromycin-resistant clone using the Tri-Reagent (MRC, Cincinnati, OH) according to the manufacturer’s directions. Aliquots containing 1, 5, or 10 µg of RNA were adjusted to a total volume of 0.12 mL of TE (10 mM Tris-HCl, pH 8, and 1.0 mM EDTA) and supplemented with 0.072 mL of 20× SSC buffer (3.0 M NaCl and 0.3 M trisodium citrate) and 0.048 mL of 37% (w/v) formaldehyde. Each sample was then heated for 15 min at 60 °C and applied to nitrocellulose membranes under light vacuum in a minifold apparatus (Schleicher & Schuell Inc., Keene, NH). The membranes were baked for 90 min at 80 °C under vacuum and cut into replicate strips, and each strip was prehybridized for 1 h at 42 °C in 50% formamide and 1.5× SSPE (15 mM NaH2-PO4, pH 7.4, 225 mM NaCl, and 1.5 mM EDTA), 0.5% blotto [10% (w/v) nonfat dried milk and 0.2% (w/v) NaN3], 1% sodium dodecyl sulfate (SDS), and 150 µg/mL salmon sperm DNA. Each strip was then probed with a uniformly radiolabeled DNA probe (32) for 12 h at 42 °C in the same buffer. The DNA fragments used as probes in these experiments were a 0.831 kb PstI fragment of GlcNAc-TI, a 1.451 kb SpeI fragment of GlcNAc-TII, a 1.089 kb BsaI fragment of β4Gal-T (33), a 1.170 kb SacII/BamHI fragment of ST6GalI, and a 0.687 kb AflII fragment of ST3GalIV. After hybridization, the blots were washed once with 2× SSC containing 0.1% SDS, twice with 0.1× SSC containing 0.1% SDS, and once with 0.1× SSC containing 1% SDS, which was preheated to 50 °C. After washing, the membranes were blotted dry, and hybridization was visualized by autoradiography.
One-Step Growth Curves
Sf9 and SfSWT-1 cells were seeded at a density of 0.5 million cells/mL into 250 mL shake flasks containing a total of 100 mL of complete TNM-FH. The cells were allowed to reach middle log phase (1.25 million cells/mL), and then they were mock infected or infected with AcMNPV at a multiplicity of 5 plaque-forming units per cell. The virus and cells were incubated on a rocking platform for 1 h at 28 °C, and then the cells were pelleted and washed three times with complete TNM-FH. The cells were resuspended at a density of 1.0 million cells/mL in a total of 100 mL of complete TNM-FH and transferred to 250 mL shake flasks, and triplicate samples of the growth media and cells were harvested at various times after infection. Progeny virus production was measured by plaque assays on Sf9 cells as previously described (7), and the average titers were plotted against hours postinfection.
Glycosyltransferase Assays
Concurrent with the one-step growth curve experiment, samples of mock- or AcMNPV-infected Sf9 and SfSWT-1 cells were harvested at various times after infection and used for glycosyltransferase assays. For the β4Gal-T and ST6GalI assays, duplicate aliquots of cells were pelleted, washed in ice-cold Tris-buffered saline (50 mM Tris-HCl, pH 7.5. and 150 mM NaCl), resuspended in either β4Gal-T (10 mM HEPES, pH 7.4, 140 mM NaCl, 20 mM MnCl2, and 0.5% Nonidet P-40) or ST6GalI (50 mM Na2HPO4, pH 7.5, 100 mM NaCl, and 1.5% CF-54) assay buffer, and frozen at -85 °C. Prior to performing the assays, the extracts were thawed and clarified in a microcentrifuge, and protein concentrations were determined using a commercial bicinchoninic acid assay (Pierce, Rockford, IL) with BSA as the standard. The β4Gal-T and ST6GalI assays were performed as previously described (19), except we used the β4Gal-T and ST6GalI reaction buffers described above. For the GlcNAc-TII assays, extracts were prepared as described above, except the cells were washed with GlcNAc-TII buffer (100 mM MES, pH 6.1, and 100 mM NaCl) and frozen in this same buffer plus 1% (v/v) Triton X-100 (Sigma). Prior to the assay, the extracts were clarified by centrifugation, and protein concentrations were determined as described above. Replicate samples of each extract, containing 100 µg of total protein, were incubated at 37 °C for 1 h in a final reaction mixture containing 67 mM MES (pH 6.1), 67 mM NaCl, 15 mM MnCl2, 6.7 mM AMP, 133 mM N-acetylglucosamine, 0.0833 mM ManR1,6(GlcNAcβ1,2ManR1,3)- Manβ-octyl (Toronto Research Chemicals, Ontario, Canada), and 0.9 µCi of uridine diphosphate [6-3H]-N-acetylglucosamine (36 Ci/mmol; New England Nuclear, Boston, MA). The reactions were quenched with 0.5 mL of ice-cold water and frozen at −85 °C. The reactions were then thawed and passed through SepPak C18 cartridges (Millipore, Bedford, MA), and the unincorporated radiolabel was washed out with water. Finally, the acceptor was eluted into liquid scintillation vials by washing the SepPak cartridges with 3 mL of methanol, 5 mL of Scintisafe plus 50% (Fisher Scientific, New Lawn, N. J.) liquid scintillation cocktail was added, and incorporated radioactivity was measured with a model LS-6500 liquid scintillation spectrometer (Beckman-Coulter Instruments, Palo Alto, CA).
Purification of the Recombinant β-Trace Protein
Sf9, Sfβ4GalT, and Sfβ4GalT/ST6 cells were seeded at a density of 0.5 million cells/mL in a total volume of 50 mL of HyQ Sfx-Insect protein-free medium (HyClone) supplemented with bovine fetuin (1 mg/mL; Sigma). After the cells reached middle log phase, they were infected at a multiplicity of 5 plaque-forming units per cell with Ac-βTP, washed, and resuspended in the same media. At 72 h postinfection cell-free medium was prepared by low-speed centrifugation. The budded virus was removed by ultracentrifugation at 20000gin a Beckman model XL-100 ultracentrifuge. The supernatant was passed over a 1 mL immunoaffinity matrix packed in a 10 × 0.7 (l × i.d.) cm glass column. The matrix was washed with 10 column volumes of 10 mM Na2HPO4 (pH 6.8), and the bound material was eluted with 0.1 M glycine (pH 2.6) while 1.0 mL fractions were collected. Each fraction was immediately neutralized with 0.05 mL of 1.0 M Na2HPO4 (pH 8.0), samples were analyzed by SDS–PAGE, and peak fractions of the purified β-trace protein were pooled. This material was used to produce tryptic peptides for various analyses, as described below.
Purification of Recombinant GST-SfManI Protein and Preparation of N-Glycans
Sf9, Sfβ4GalT, Sfβ4GalT/ST6, and SfSWT-1 cells were infected with AcGST-SfManI at a multiplicity of 5 plaque-forming units per cell, washed, and resuspended in complete TNM-FH, as described above. At 72 h postinfection, the cultures were harvested, and cell-free media were prepared by low-speed centrifugation. The budded virus was removed by ultracentrifugation at 20000g in a Beckman Ti-45 rotor for 30 min at 4 °C in a Beckman model XL-100 ultracentrifuge, then the supernatant was harvested, and poly(ethylene glycol) 8000 was added to a final concentration of 15% (w/v). The sample was incubated for 6 h at 4 °C, and the resulting precipitate was harvested by centrifugation, dissolved in glutathione column binding buffer [25 mM Tris-HCl, pH 8.0, 250 mM NaCl, and 1.5% (v/v) Triton X-100], and dialyzed against the same buffer. The dialyzed supernatant was applied three times to an immobilized glutathione–agarose affinity column (Pierce), and then the column was washed once with excess column binding buffer and once with excess glycosidase buffer (5 mM NaH2PO4, pH 7.5). The bound GST-SfManI was eluted with glycosidase buffer supplemented with 10 mM glutathione, and the eluted material was dialyzed against glycosidase buffer with no glutathione. Total protein concentrations were determined using a commercial Bradford assay (Bio-Rad, Hercules, CA) with BSA as the standard. Five microgram samples of the purified proteins were analyzed by SDS–polyacrylamide gel electrophoresis (34) with Coomassie blue staining or by immunoblotting (35) with a commercial polyclonal antiserum against glutathione S-transferase (Sigma) as the probe. Samples containing from 150 to 300 µg of the purified GST-SfManI produced by the various cell lines were treated with 2000 milliunits/mL peptide N-glycosidase F for 12 h at 37 °C, and then the N-glycans were isolated on graphitized carbon cartridges (Carbograph; Alltech, Deerfield, IL) for high-pH anion-exchange chromatography with pulsed amperometric detection (HPAEC-PAD; 36), as described below.
Mass Spectrometry of the β-Trace
The purified β-trace preparations described above were digested with trypsin, and tryptic peptides were analyzed by high-performance liquid chromatography coupled with electrospray ionization mass spectrometry (HPLC/ESI-MS). Briefly, mixtures of the tryptic peptides obtained by digesting approximately 150 pmol of the β-trace protein purified from each cell line were analyzed on an Applied Biosystems 172A microbore HPLC system using a 1.0 mm × 100 mm Aquapore OD-300 C18 column. Peptides were eluted with a linear gradient running from 4% to 56% acetonitrile containing 0.06% trifluoroacetic acid at a flow rate of 40 µL/min. Elution was monitored by UV absorption at 214 nm and by MS on a TSQ 700 triple quadrupol instrument equipped with a Finnigan ES ion source connected on-line to the HPLC system. In addition, individual β-trace glycopeptides were isolated by preparative HPLC, using the same conditions described above, and analyzed by tandem mass spectrometry (ESI QTOF MS/MS). Briefly, 3 µL aliquots of the purified tryptic glycopeptides were placed into gold-coated nanospray glass capillaries, and the tips were placed orthogonally in front of the entrance hole of a QTOF 2 mass spectrometer (Micromass, Manchester, England) equipped with a nanospray ion source. A voltage of approximately 700–1000 V was applied. For collision-induced dissociation experiments, parent ions were selectively transmitted from the quadrupole mass analyzer into the collision cell. Argon was used as the collision gas, and the kinetic energy was set at around +25–30 eV for optimal fragmentation. The resulting daughter ions were then separated by an orthogonal time of flight mass analyzer.
HPAEC-PAD of N-Glycans from GST-SfManI
N-Glycans were isolated from the GST-SfManI produced by various insect cell lines and analyzed by HPAEC-PAD using a Dionex Bio LC system (Dionex, Sunnyvale, CA) equipped with a 0.4 × 25 cm CarboPac PA1 column (36, 37). Detector potentials (E) and pulse durations (T) were E1 +50 mV, T1 480 ms; E2 +500 mV, T2 120 ms; E3 −500 mV, T3 60 ms; the output range was 500–1500 nA. The CarboPac PA1 column was equilibrated with 100% solvent A (0.2 M NaOH) prior to injection of the N-glycans. The elution profile included an initial 5 min isocratic run with 100% solvent A, a 30 min linear gradient from 0% to 20% solvent B (0.2 M sodium hydroxide and 0.6 M sodium acetate), and a 2 min linear gradient to 100% solvent B, each at a flow rate of 1 mL/min. HPAEC-PAD also was used to prepare total N-glycans, which were pooled and desalted on graphitized carbon cartridges, for subsequent mass spectrometry, as described below.
Mass Spectrometry of N-Glycans from GST-SfManI
Total N-glycans from the GST-SfManI produced by various insect cell lines were analyzed by matrix-assisted laser desorption ionization time of flight (MALDI/TOF) mass spectrometry (MS). Briefly, N-glycans were analyzed by positive ion MALDI/TOF-MS using a Bruker REFLEX TOF instrument equipped with delayed extraction and reflectron systems and a N2 laser (337 nm) operating with 3 ns pulse width and 107–8 W/cm2 irradiance at the surface of 0.2 mm2 spots. One microliter of samples containing equal volumes of the N-glycan solution (~3–10 pmol/L) plus the ultraviolet-absorbing matrix (19 mg of α-cyano-4-hydroxycinnamic acid in 400 µL of acetonitrile plus 600 µL of 0.1% trifluoroacetic acid in water) was spotted onto a stainless steel target and dried at room temperature. Spectra were recorded at an acceleration voltage of +20 kV using the delayed extraction facility and the reflectron for enhanced resolution. In addition, N-glycans isolated from each form of GST-SfManI were reduced and permethylated, as previously described (38), and dissolved in an aqueous solution of 66% methanol (v/v) and 1 mM NaCl, and the resulting single or double charged sodium adducts of the oligosaccharide derivatives were characterized by ESI QTOF MS/MS.
Methylation Analysis of N-Glycans from GST-SfManI
Samples of the total N-glycans from the GST-SfManI produced by various insect cell lines were permethylated, hydrolyzed, reduced, and acetylated, as previously described (38). The resulting partially methylated alditol acetates were subsequently analyzed on a ThermoQuest CGQ ion trap GC/ MS system (MS, EI mode; GC, 30 m DB 5 column).
RESULTS AND DISCUSSION
Structures of the N-Glycans Produced by Existing Transgenic Insect Cell Lines
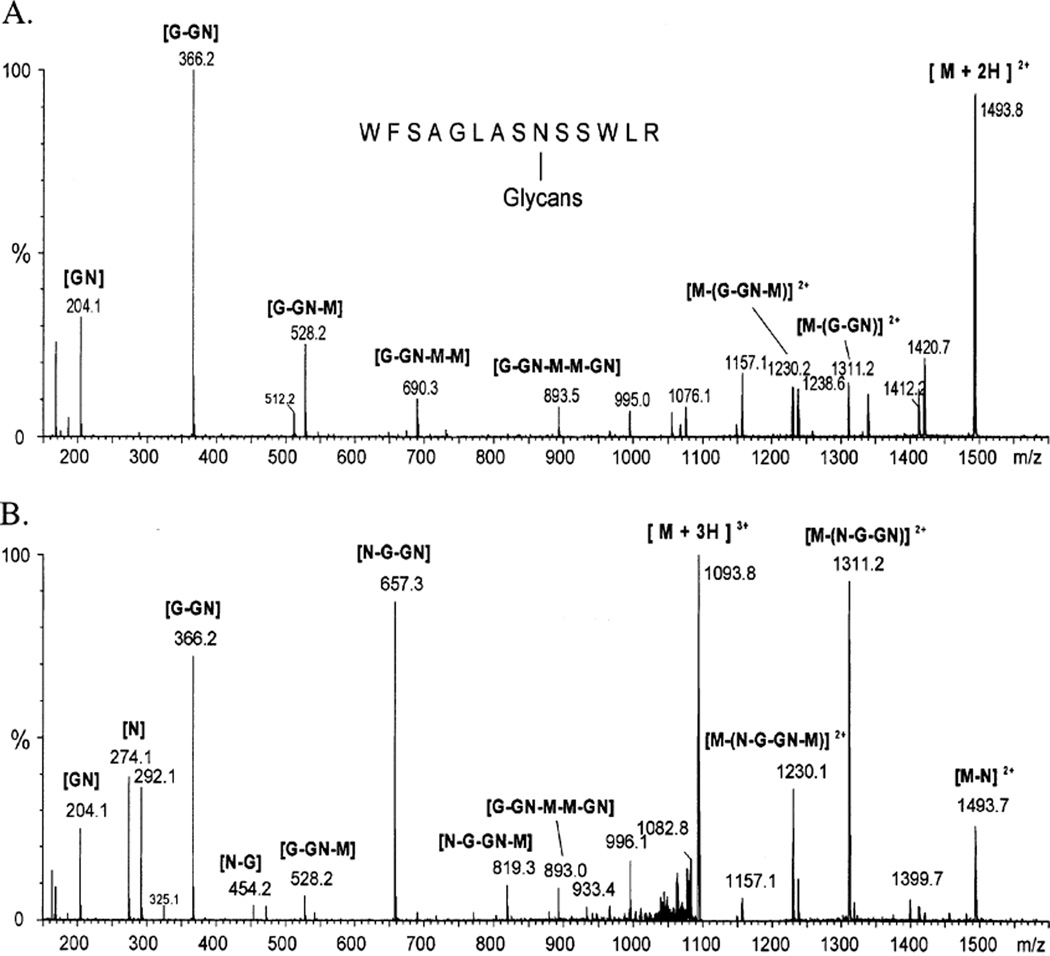
In a previous study, we isolated two transgenic insect cell lines, Sfβ4GalT and Sfβ4GalT/ST6, that constitutively express mammalian β4Gal-T or β4Gal-T and ST6GalI genes, respectively (17, 19). We also demonstrated that these new cell lines, unlike the parental Sf9 cells, could produce recombinant glycoproteins with terminally galactosylated or sialylated N-glycans, respectively. The initial purpose of the present study was to extend these observations by characterizing the structures of the N-glycans produced by these transgenic insect cell lines. The general approach was to infect the transgenic or parental cell lines with a recombinant baculovirus encoding a model glycoprotein and then to use the glycoprotein produced by each cell line as a source of N-glycans for the structural determinations. The model glycoprotein initially used for these studies was the human β-trace protein, a prostaglandin-D synthase (39). The β-trace protein produced during infection of each cell line was immunoaffinity purified from the extracellular growth medium, and tryptic glycopeptides were isolated and analyzed by HPLC/ESI-MS, as described in Experimental Procedures. The observed masses of the N-glycans found on the β-trace glycopeptide with the amino acid sequence WFSAGLASNSSWLR are shown in Table 1. This table also shows the proposed structures of these N-glycans, based on their masses, their theoretical masses, and their relative abundance. The predominant N-glycans the glycopeptide produced by Sf9 cells had molecular masses corresponding to fucosylated and nonfucosylated paucimannose structures [Man3GlcNAc2(±Fuc)]. This result was expected from our current understanding of the N-glycan processing pathway in this insect cell line (4, 5). Analysis of this same tryptic glycopeptide from Sfβ4GalT or Sfβ4GalT/ ST6 cells revealed that these cell lines, too, produced paucimannosidic N-glycans. However, the results revealed that these cells also produced several novel N-glycans that were not produced by Sf9 cells. Interestingly, none of these glycans had molecular masses corresponding to biantennary structures. Their masses corresponded, instead, to monoantennary structures terminating in galactose or sialic acid. This interpretation was confirmed by ESI QTOF MS/MS analysis of selected novel species, which were also observed by MALDI/TOF-MS (data not shown), as the resulting daughter ion spectra corresponded to monoantennary structures (Figure 1). Particularly, the fragment ion series at m/z 292 [NeuNAc + H]+, 454 [NeuNAc-Gal + H]+, and 657 [NeuNAc-Gal-GlcNAc + H]+ and the complementary ions generated by the elimination of these fragments from the molecular ion clearly show the presence of sialylated N-acetyllactosamine antennas in the protein produced in Sfβ4GalT/ST6 cells. Together, these data confirmed our original conclusion that both Sfβ4GalT and Sfβ4GalT/ST6 cells can produce more extensively processed N-glycans than Sf9 cells (17, 19, 40). In addition, these more detailed structural analyses showed that the most highly processed N-glycans produced by these cells have novel, monoantennary structures, in which the α1,6 branch is not elongated. These results are consistent with previous studies indicating that monoantennary N-glycans are the most highly processed structures produced when untransformed Sf9 or Tn-5B1-4 cells are infected with an immediate early baculovirus expression vector encoding bovine β4Gal-T (41, 42).
Table 1.
Major N-Glycans on the β-Trace Protein Produced by Various Insect Cell Lines
| Sf9a | Sfβ4GalT | Sfβ4GalT/ST |
|---|---|---|
| 2474.0 (2474.1): M3 GN2 (8) | 2457.4 (2458.0): M2 GN2 F (4) | 2457.6 (2458.0): M2 GN2 F (6) |
| 2619.7 (2620.1): M3 GN2 F (71) | 2473.8 (2474.1): M3 GN2 (5) | 2475.0 (2474.1): M3 GN2 (10) |
| 2797.8 (2798.2): M5 GN2 (4) | 2619.6 (2620.1): M3 GN2 (38) | 2619.8 (2620.1): M3 GN2 F (30) |
| 2823.0 (2823.2): M3 GN3 F (3) | 2839.6 (2839.2): G M3 GN3 F (4) | 2839.8 (2839.2): G M3 GN3 F (9) |
| 2960.4 (2960.2): M6 GN2 (7) | 2983.8 (2984.8): G M3 GN3 F (44) | 2984.0 (2984.8): G M3 GN3 F (38) |
| 3122.6 (3122.3): M7 GN2 (4) | 3163.0 (3163.3): G M5 GN3 (5) | 3129.4 (3130.0): NG M3 GN3 (1) |
| 3284.5 (3284.4): M8 GN2 (4) | 3162.8 (3163.3): G M5 GN3 (3) | |
| 3275.6 (3276.0): NG M3 GN3F (3) |
The data in the table show the masses of the major N-glycans linked to tryptic glycopeptides from the recombinant β-trace protein produced by Sf9, Sfβ4GalT, and Sfβ4GalT/ST6 cells observed by HPLC/ESI-MS [M + H]+. The masses are followed by the calculated masses of each glycan in parentheses, the proposed glycan structures, and the relative abundance (%) of each species. Monosaccharide residues attached due to the action of the additional transferases are printed in bold and underlined. The presence of monoantennary structures terminanting in galactose or sialic acid was unequivocally confirmed by ESI QTOF MS/MS (Figure 1).
Abbreviations: N, N-acetylneuraminic acid; G, galactose; M, mannose; GN, N-acetylglucosamine; F, fucose.
FIGURE 1.
ESI QTOF MS/MS analysis of novel glycopeptides from the β-trace protein produced by existing transgenic insect cell lines. A glycopeptide was isolated from the β-trace protein produced by Sfβ4GalT (A) or Sfβ4GalT/ST6 (B) cells and analyzed by ESI QTOF MS/MS, as described in Experimental Procedures. The amino acid sequence of the peptide is shown at the top, and the mass spectra are labeled with the glycan structures of key daughter ions. The monosaccharide abbreviations are defined in the legend of Table 1.
Isolation and Initial Characterization of a New Transgenic Insect Cell Line
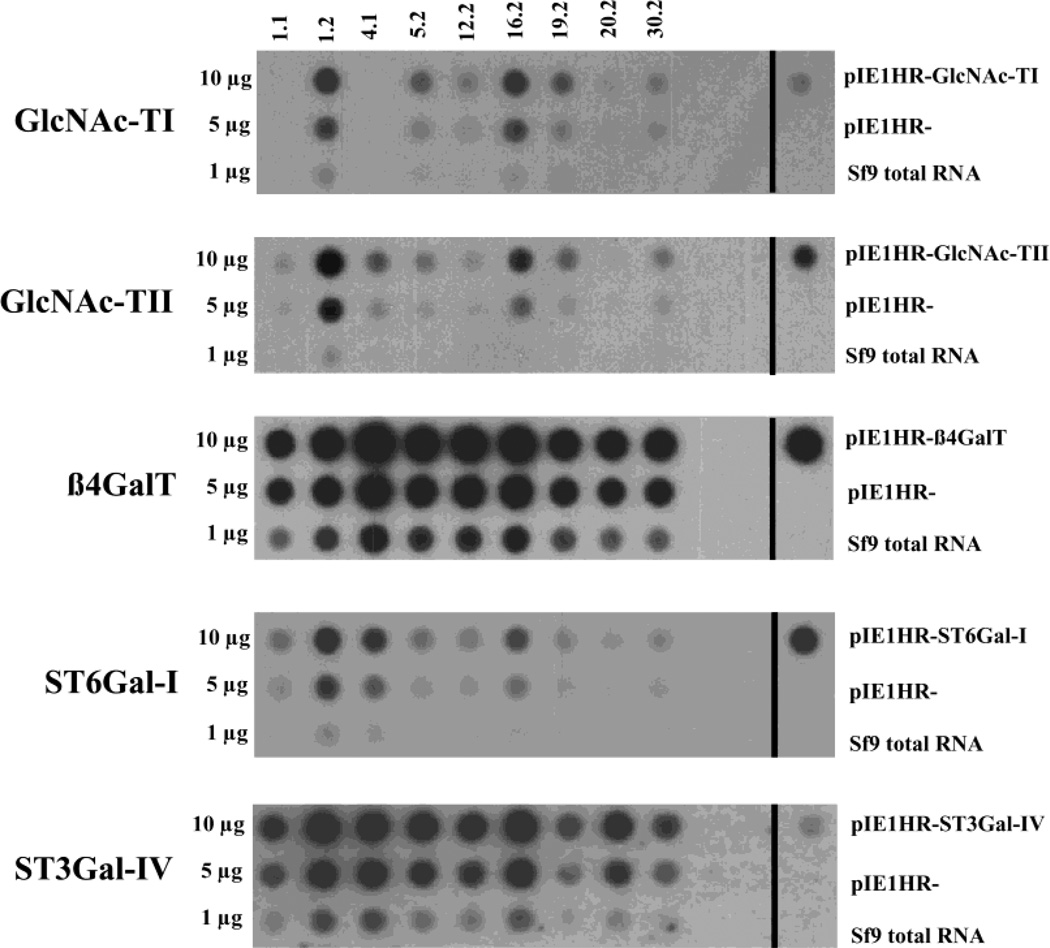
The monoantennary nature of the N-glycans produced by Sfβ4GalT and Sfβ4GalT/ST6 cells led us to hypothesize that these cells had insufficient levels of endogenous GlcNAc-TII activity to support the production of biantennary, complex N-glycans. This hypothesis was consistent with the previous demonstration that Sf9 cells contain only exceedingly low levels of GlcNAc-TII activity (43). To test this hypothesis, we isolated a new transgenic insect cell line designed to express a more complete array of mammalian glycosyltransferases, including GlcNAc-TII. Hygromycin-resistant clones were isolated by limiting dilution and screened for the ability to express each of the unselected transgenes by RNA dot-blot analysis, as described in Experimental Procedures. The DNA probes for each mammalian glycosyltransferase cDNA clearly hybridized to total RNA from some transformed clones but not to RNA from untransformed cells or to empty vector controls (Figure 2). All five glycosyltransferase probes hybridized at optimal or nearly optimal levels with RNA from clone 1.2. In addition, preliminary enzyme assays showed that this clone had significantly higher levels of GlcNAc-TII, β4Gal-T, and ST6GalI activities than Sf9 cells (data not shown). Thus, clone 1.2 was designated SfSWT-1 and used for the remainder of the experiments described in this report.
FIGURE 2.
Dot-blot analysis of transformed Sf9 cell clones. Various amounts of the total RNA isolated from nine independent transformed Sf9 cell clones were spotted onto replicate filters and hybridized with various glycosyltransferase probes, as described in Experimental Procedures. Samples of the plasmids used for transformation, the empty vectors (pIE1HR−), and total RNA from the parental Sf9 cells were spotted on the far right, as controls. The radiolabeled probes used for each filter are indicated on the left, the various transformed cell clones are indicated on the top, and the controls used for each filter are indicated on the right of the photographs.
Baculovirus Infection of SfSWT-1 Cells
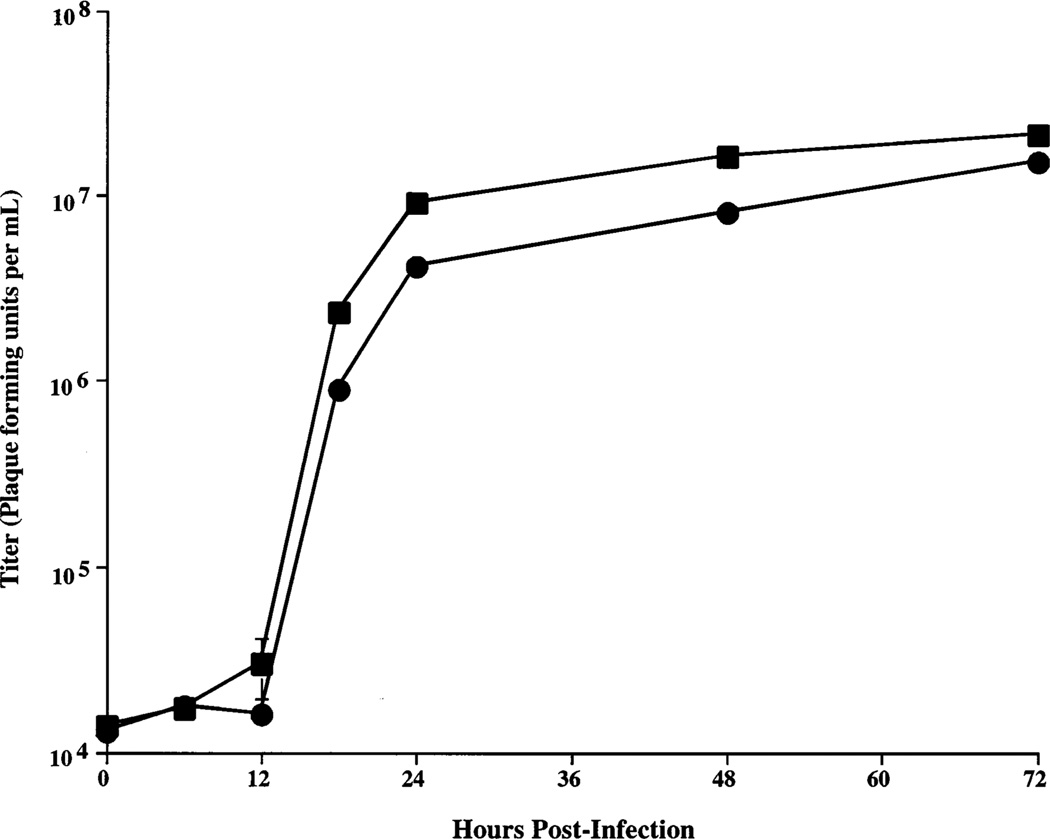
The baculovirus expression system is a binary system consisting of a recombinant baculovirus vector and its insect cell host. The recombinant baculovirus delivers the foreign cDNA and encodes the machinery for its high-level transcription, while the insect cells provide the machinery needed for protein translation and glycosylation. Thus, in order for SfSWT-1 cells to be useful for recombinant glycoprotein production, they had to be able to support normal levels of baculovirus infection. To examine this capability, a one-step baculovirus growth curve experiment was performed using Sf9 or SfSWT-1 cells as the hosts. Each cell line was infected with a wild-type baculovirus, AcMNPV, at a high multiplicity of infection, and then the amounts of infectious progeny-budded virus produced at various times after infection were measured by plaque assays, as described in Experimental Procedures. The results showed that SfSWT-1 cells supported baculovirus replication at least as well as Sf9 cells (Figure 3), indicating that they can be used as hosts for baculovirus-mediated foreign gene expression.
FIGURE 3.
One-step baculovirus growth curves. Sf9 (circles) or SfSWT-1 (squares) cells were infected with a wild-type baculovirus, triplicate samples were removed from the infected cultures at various times postinfection, and the viral titers were measured by plaque assays, as described in Experimental Procedures. Average titers were plotted against hours postinfection, and standard deviations are shown, but in most cases, the error bars are too small to show up on the plot.
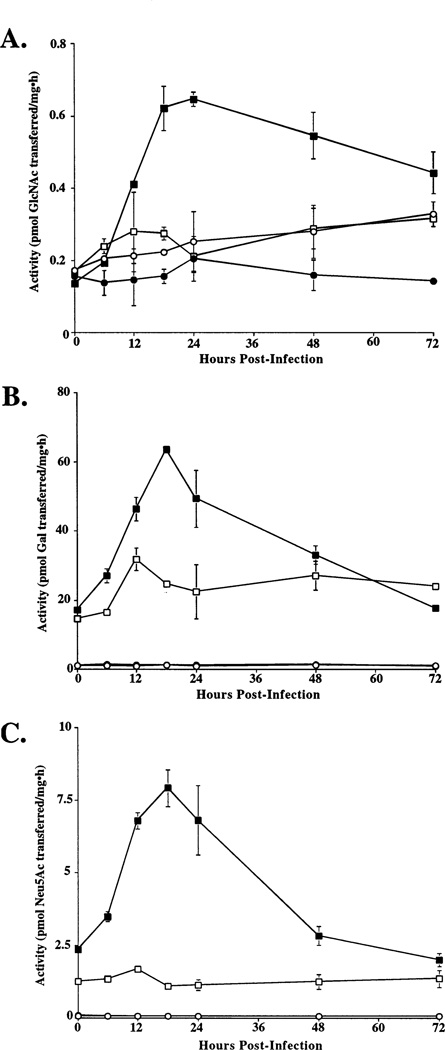
Generally, baculoviruses appear to shut down host gene expression at the transcriptional level (44). Thus, it also was important to examine the mammalian glycosyltransferase activity levels in SfSWT-1 cells during the course of a baculovirus infection. The results of this analysis showed that each mammalian glycosyltransferase activity examined was transiently induced at early times after baculovirus infection (Figure 4), probably due to transactivation of the ie1 promoter in the integrated transgenes by baculovirus-encoded transcription factors (45). Each activity decayed as the infection progressed but remained about as high as in the uninfected SfSWT-1 cell controls until at least 72 h postinfection. Importantly, SfSWT-1 cells had higher levels of each of the transgenic glycosyltransferase activities than the parental Sf9 cells throughout infection, indicating that they could function as improved hosts for baculovirusmediated glycoprotein production. It should be noted that GlcNAc-TI activity was not measured in this experiment because the product of this enzyme is the substrate for GlcNAc-TII, which is also expressed by SfSWT-1 cells. Hence, standard GlcNAc-TI assays could not be used to measure GlcNAc-TI activity levels in cell lysates because the GlcNAc-TII in these lysates would have transferred additional radioactivity from the donor substrate into secondary product and obscured the measurement. We did not undertake the effort necessary to develop an alternative assay because the dot-blot assays had indicated that SfSWT-1 cells expressed the GlcNAc-TI transgene and, in any event, the presence or absence of this activity was peripheral to this study.
FIGURE 4.
Influence of baculovirus infection on glycosyltransferase activities. Sf9 (circles) or SfSWT-1 (squares) cells were mock-infected (open symbols) or infected with a wild-type baculovirus (filled symbols), samples were removed at various times post-infection, and glycosyltransferase assays were performed in triplicate. The average GlcNAc-TII (A), β4Gal-T (B), and ST6GalI (C) activities were plotted against hours postinfection.
Expression and Purification of GST-SfManI from Various Insect Cell Lines
The ability of SfSWT-1 cells to express mammalian GlcNAc-TII, β4Gal-T, and ST6GalI indicated that these cells had the array of glycosyltransferase activities needed to produce biantennary, terminally sialylated Nglycans. To examine this hypothesis and compare the N-glycan processing capabilities of Sf9, Sfβ4GalT, Sfβ4GalT/ ST6, and SfSWT-1 cells, we determined the structures of the N-glycans isolated from a model glycoprotein produced by each cell line. The model glycoprotein used for this analysis was a GST-tagged version of the endogenous secretory pathway glycoprotein, SfManI (31). We used GST-SfManI as the model for these studies because it is expressed at high levels and it has only one N-glycan, which is efficiently processed to an endoglycosidase-H-resistant form (30, 46).
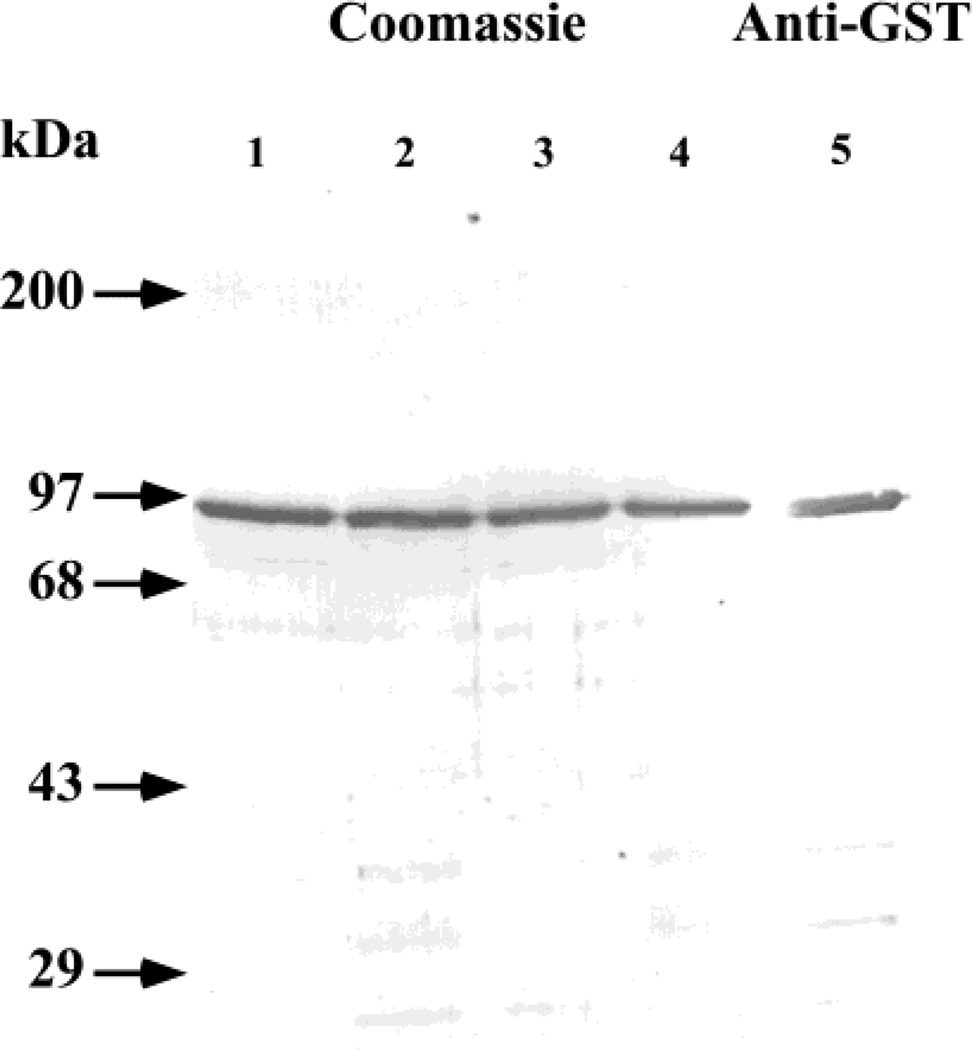
GST-SfManI was isolated by affinity chromatography from cell- and virus-free supernatants of Sf9, Sfβ4GalT, Sfβ4GalT/ST6, and SfSWT-1 cells infected with the recombinant baculovirus, AcGST-SfManI, as described in Experimental Procedures. Aliquots of the affinity-purified material from each cell line were examined by SDS–polyacrylamide gel electrophoresis with Coomassie blue staining. The results showed that GST-SfManI had been effectively purified from each cell line, though some degradation had occurred (Figure 5). These preparations were deglycosylated with peptide N-glycosidase F, and the N-glycans produced by each cell line were recovered and used for various structural determinations, as described in Experimental Procedures.
FIGURE 5.
Expression and purification of GST-SfManI. Sf9 (lane 1), Sfβ4GalT (lane 2), Sfβ4GalT/ST6 (lane 3), or SfSWT-1 (lane 4) cells were infected with AcGST-SfManI, and the recombinant protein was harvested and purified by affinity chromatography, as described in Experimental Procedures. Samples of each purified protein preparation were analyzed by SDS–polyacrylamide gel electrophoresis with Coomassie blue staining. The position of GST-SfManI was also determined by immunoblotting, using a duplicate sample of the GST-SfManI from SfSWT-1 cells (lane 5) and anti-GST as the probe. The positions of protein standards are marked on the left-hand side of the photograph.
HPAEC-PAD Analysis of N-Glycans Produced by Various Insect Cell Lines
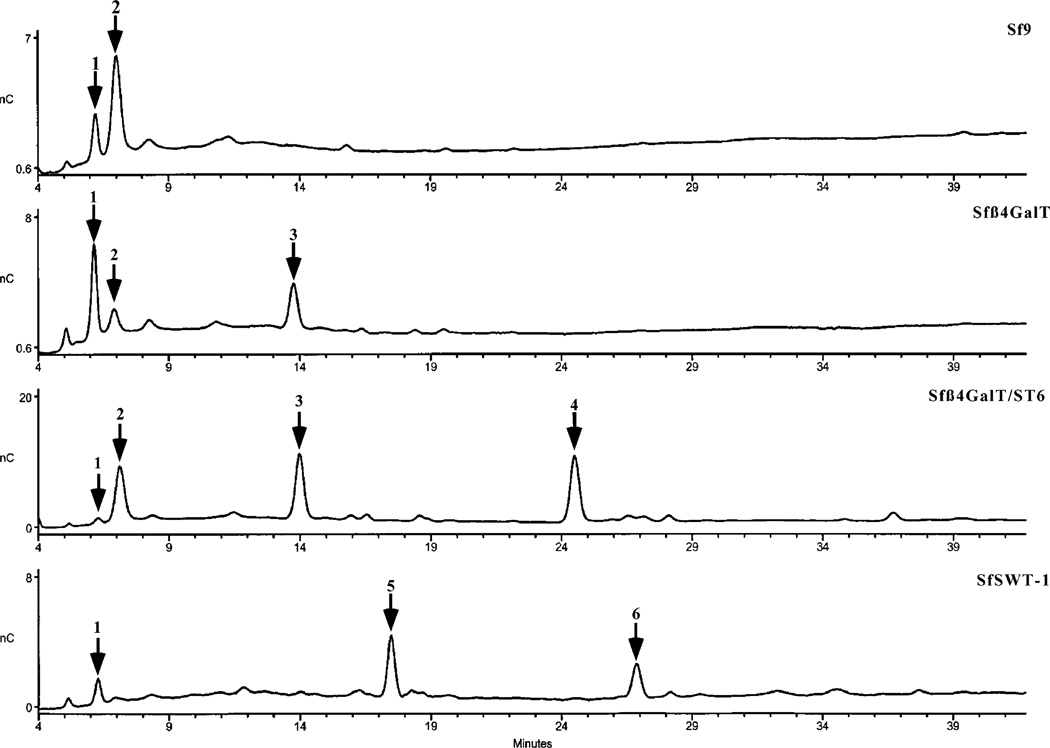
The N-glycans released from the GST-SfManI isolated from each insect cell line were initially analyzed by HPAEC-PAD (36) to obtain preliminary structural assignments by comparison of their elution times to those of N-glycan standards. The results of these analyses showed that the GST-SfManI produced by the parental Sf9 cells had two major N-glycans (peaks 1 and 2), which coeluted with paucimannosidic standards with (Man3GlcNAc2- Fuc) and without (Man3GlcNAc2) a core fucose residue, respectively (Figure 6). These two peaks were also observed upon analysis of the N-glycans from the GST-SfManI produced by Sfβ4GalT cells, though their relative abundance was reversed. Furthermore, this preparation contained an additional N-glycan (peak 3) that did not coelute with any standard. Similarly, the N-glycans from the GST-SfManI produced by Sfβ4GalT/ST6 cells consisted of four species, two of which coeluted with the paucimannosidic standards (peaks 1 and 2) and two of which failed to coelute with any N-glycan standard (peaks 3 and 4). Considering the results of our previous analyses of the N-glycans from the human β-trace protein produced by these same cell lines (Figures 1–2), we tentatively concluded that peaks 3 and 4 must represent monoantennary N-glycans terminating in galactose or sialic acid, respectively.
FIGURE 6.
HPAEC-PAD analysis of N-glycans from the GST-SfManI produced by various cell lines. N-Glycans were isolated from the GST-SfManI produced by Sf9, Sfβ4GalT, Sfβ4GalT/ST6, or SfSWT-1 cells and analyzed by HPAEC-PAD, as described in Experimental Procedures. The positions of major N-glycan peaks are marked with arrows and numbered.
Most interestingly, HPAEC-PAD analysis of the N-glycans isolated from the GST-SfManI from SfSWT-1 cells revealed only small amounts of the paucimannosidic species coeluting with Man3GlcNAc2 (peak 1), together with two new, major peaks (peaks 5 and 6) that coeluted with biantennary N-glycan standards terminating in galactose on both branches or in galactose on one branch and sialic acid on the other. The appearance of these peaks provided additional evidence that Sfβ4GalT and Sfβ4GalT/ST6 cells produced only monoantennary, terminally galactosylated or sialylated structures, respectively. Furthermore, the ability of SfSWT-1 cells to produce biantennary N-glycans supported the idea that Sfβ4GalT and Sfβ4GalT/ST6 cells cannot produce this type of N-glycan structure because they lack adequate levels of endogenous GlcNAc-TII activity.
MALDI/TOF-MS Analysis of N-Glycans Produced by Various Insect Cell Lines
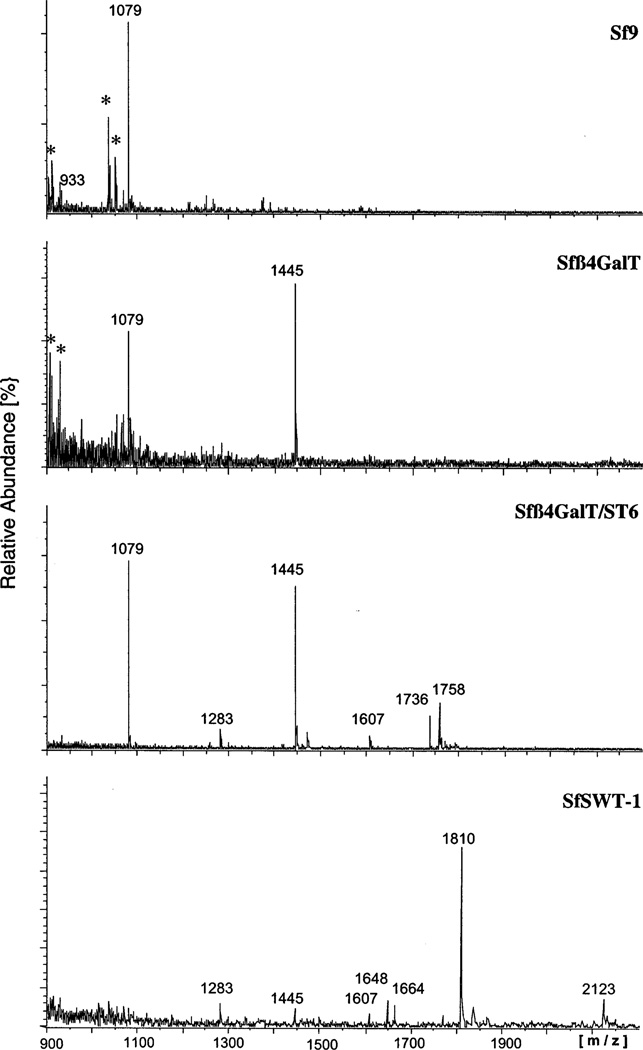
To obtain further structural information, MALDI/TOF-MS was used to determine the molecular masses of the N-glycans produced by the transgenic and parental insect cell lines. As above, peptide N-glycosidase F was used to release the N-glycans from the GST-SfManI isolated from each cell line, and then total N-glycans were separated from contaminants by preparative HPAEC-PAD, pooled, desalted, and analyzed by MALDI/TOF-MS, as described in Experimental Procedures. The results are shown in Figure 7. Structures were tentatively assigned to various N-glycans on the basis of their observed masses in comparison to the calculated masses of known N-glycans (Table 2). Briefly, the spectrum of N-glycans from the parental cell line included two molecular ions with masses of 933 and 1079 Da, which correspond to the calculated masses of the paucimannosidic N-glycans, Man3GlcNAc2 and Man3GlcNAc2Fuc. The spectrum of N-glycans from Sfβ4GalT cells included an additional molecular ion with a mass of 1445 Da, which corresponds to the mass of the monoantennary, terminally galactosylated N-glycan, GalGlcNAcMan3-GlcNAc2Fuc. The N-glycans from Sfβ4GalT/ST6 cells included two additional molecular ions with masses of 1758 and 1736 Da, which correspond to the calculated masses of the monosialylated, monoantennary N-glycan, Neu5AcGalGlc- NAcMan3GlcNAc2Fuc plus or minus an additional Na+. Finally, the spectrum of N-glycans from SfSWT-1 cells included two molecular ions with masses of 1810 and 2123 Da, which correspond to the calculated masses of the biantennary N-glycans, Gal2GlcNAc2Man3GlcNAc2Fuc and Neu5AcGal2GlcNAc2Man3GlcNAc2Fuc, respectively. Together, the data obtained from the HPAEC-PAD (Figure 6) and MALDI/TOF-MS (Figure 7 and Table 2) analyses demonstrated that the recombinant GST-SfManI produced by Sf9 cells had exclusively paucimannosidic N-glycans, whereas the same protein produced by the transgenic insect cell lines had N-glycans that are elongated on one or both branches. Furthermore, the difference between the structures of the N-glycans produced by Sfβ4GalT/ST6 and SfSWT-1 cells strongly supports the hypothesis that the endogenous level of GlcNAc-TII in Sf9 cells is too low to support the production of biantennary N-glycans, at least on recombinant β-trace and GST-SfManI proteins.
FIGURE 7.
MALDI/TOF MS analysis of N-glycans from the GST-SfManI produced by various cell lines. N-Glycans were isolated from the GST-SfManI produced by Sf9, Sfβ4GalT, Sfβ4GalT/ST6, or SfSWT-1 cells and analyzed by MALDI/TOF MS, as described in Experimental Procedures. The masses of key species are indicated above the peaks. Peaks resulting from noncarbohydrate contaminants are marked with asterisks.
Table 2.
MALDI/TOF-MS Analysis of the Major N-Glycans Produced by Various Insect Cell Lines
| Predicted glycan structure | HPAEC Peak # | Mass | |
|---|---|---|---|
| Predicted | Observed | ||
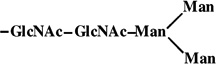 |
1 | 933 | 933 |
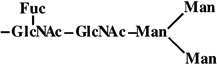 |
2 | 1079 | 1079 |
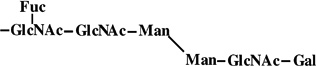 |
-a | 1283 | 1283 |
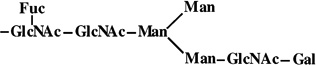 |
3 | 1445 | 1445 |
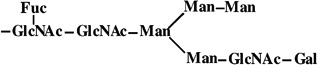 |
- | 1607 | 1606 |
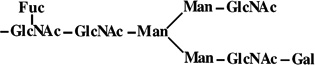 |
- | 1648 | 1648 |
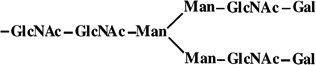 |
- | 1664 | 1664 |
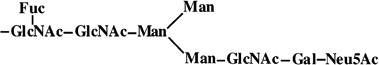 |
4 | 1736 | 1736 |
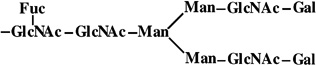 |
5 | 1810 | 1810 |
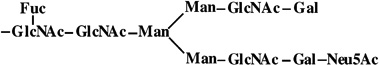 |
6 | 2123 | 2123 |
Minor N-glycan species detected by MALDI/TOF-MS but below the detection limit for HPAEC-PAD.
Tandem MS Analysis of Selected N-Glycans Produced by Various Insect Cell Lines
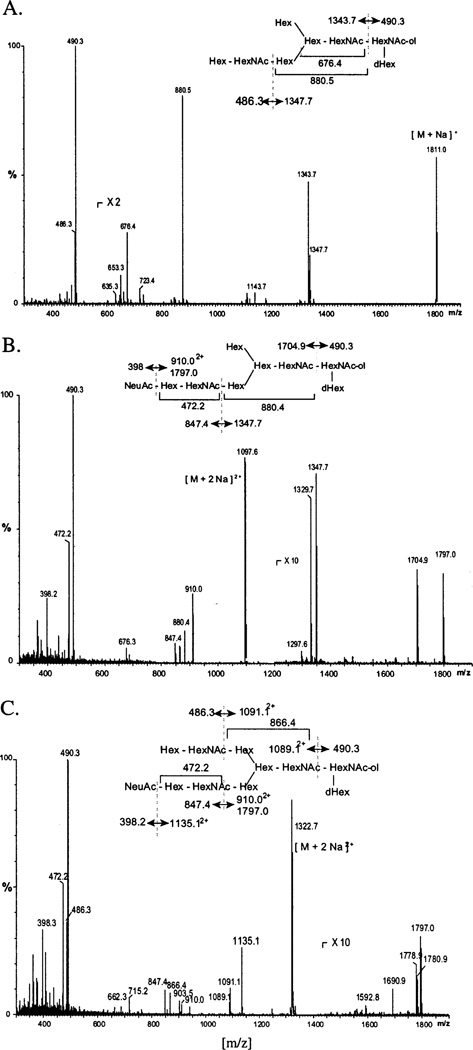
N-Glycans were reduced and permethylated as described in Experimental Procedures, and selected ions were characterized by ESI QTOF MS/MS with collision-induced dissociation. The resulting daughter ion spectra unequivocally demonstrated that the most highly processed N-glycans produced by Sfβ4GalT (Figure 8A) and Sfβ4GalT/ST6 (Figure 8B) cells are novel monoantennary structures with the α1,3 branch elongated by the addition of N-acetylglucosamine and galactose or N-acetylglucosamine, galactose, and sialic acid, respectively. Furthermore, the tandem MS results unequivocally demonstrated that the most highly processed N-glycans produced by SfSWT-1 cells (Figure 8C) are biantennary structures terminating with galactose on one branch and sialic acid on the other. On the basis of the previous finding that human ST6GalI acts preferentially on the α1,3 branch of N-glycans (47), we believe that the single sialic acid residue on the monosialylated N-glycans produced by SfSWT-1 cells is found predominantly on this branch, as indicated by the structures drawn above Figure 8B,C.
FIGURE 8.
ESI QTOF MS/MS analysis of selected N-glycans from the GST-SfManI produced by various cell lines. Selected N-glycans isolated from the GST-SfManI produced by Sfβ4GalT (A), Sfβ4GalT/ST6 (B), or SfSWT-1 (C) cells were reduced and permethylated and then analyzed by ESI QTOF MS/MS, as described in Experimental Procedures. The drawings above each spectrum indicate the structures and fragmentation schemes deduced from the daughter ion spectra.
Methylation Analysis of Selected N-Glycans Produced by Various Insect Cell Lines
Methylation analysis was used to extend the MALDI/TOF-MS and ESI QTOF MS/MS results by establishing the nature of the linkages between individual monosaccharides in the N-glycans produced by the parental and transgenic insect cell lines. This was of particular interest because SfSWT-1 cells encode both ST6GalI and ST3GalIV, and we wanted to determine which enzyme was responsible for terminal sialylation of the N-glycans on GST-SfManI. The results of the methylation analyses (Table 3) indicated that virtually all of the galactose residues were 6-substituted, indicating that the α2,6- and not the α2,3-sialyltransferase is responsible for terminal sialylation of the N-glycans on the GST-SfManI produced by SfSWT-1 cells. Significantly, the methylation data also showed that the N-glycans produced by SfSWT-1 cells have far less terminal mannose than those produced by any of the other cell lines, indicating that conversion of the N-glycans on GST-SfManI to complex, biantennary structures by SfSWT-1 cells is a highly efficient process.
Table 3.
Methylation Analysis of N-Glycans Produced by Various Insect Cell Lines
| peracetylated derivatives of |
substituted in position |
Sf9 | Sfβ4GalT | Sfβ4GalT/ST6N | SfSWT |
|---|---|---|---|---|---|
| fucitol | |||||
| 2,3,4-tri-O-methyl- | terminal | 0.9 | 0.9 | 0.9 | 0.9 |
| mannitol | |||||
| 2,3,4,6-tetra-O-methyl- | terminal | 2.2 | 1.4 | 1.9 | 0.2 |
| 3,4,6-tri-O-methyl- | 2 | 0.2 | 0.8 | 0.8 | 1.8 |
| 3,6-di-O-methyl- | 2,4 | ||||
| 3,4-di-O-methyl- | 2,6 | <0.1 | 0.2 | 0.1 | |
| 2,4-di-O-methyl- | 3,6 | 0.9 | 1.0 | 1.1 | 0.9 |
| galactitol | |||||
| 2,3,4,6-tetra-O-methyl- | terminal | 0.7 | 0.2 | 1.5 | |
| 2,4,6-tri-O-methyl- | 3 | <0.05 | <0.05 | ||
| 2,3,4-tri-O-methyl- | 6 | 0.2 | 0.5 | ||
| 2-N-methylacetamido-2-deoxyglucitol | |||||
| 1,3,4,6-tetra-O-methyl- | 4 | 0.1 | <0.1 | <0.1 | 0.1 |
| 3,4,6-tri-O-methyl- | terminal | <0.1 | |||
| 1,3,5-tri-O-methyl- | 4,6 | 0.9 | 0.9 | 0.9 | 0.9 |
| 3,6-di-O-methyl- | 4 | 1.0 | 1.7 | 1.4 | 2.9 |
CONCLUSIONS
This study provides the first comprehensive, direct structural analyses of the N-glycans produced by transgenic insect Engineering Protein N-Glycosylation in Insect cell lines previously described by our group (17, 19). The results clearly demonstrated that the most highly processed N-glycans produced by Sfβ4GalT and Sfβ4GalT/ST6 cells are novel, monoantennary structures terminating in galactose or galactose and sialic acid, respectively. These structural data are important for at least two reasons. First, they unequivocally support our previous conclusion that insect cell lines can be engineered for sialoglycoprotein production without engineering CMP-sialic acid production pathways (19, 20, 40). The source of the CMP-sialic acid donor substrate for the ST6GalI in these cells is unclear and under investigation. Nevertheless, the ability of these cells to produce sialoglycoproteins reveals unexpected and previously unrecognized infrastructure for glycoprotein biosynthesis in insect cells. Second, the structural data revealed that the transgenic insect cell lines previously created by our group could not produce biantennary N-glycans. This result directed the creation of a new transgenic line, which can produce biantennary structures identical to those produced by mammalian cells. This result confirmed the suspected underlying basis for the inability of our previous cell lines to produce biantennary N-glycans and taught us more about insect cell glycobiology. Finally, this study described the properties and capabilities of a new transgenic insect cell line that will be of great interest to researchers who want to use baculovirus expression vectors for recombinant glycoprotein production. By using SfSWT-1 cells as improved hosts, it should be possible to produce recombinant glycoproteins with more authentic biantennary, complex, terminally sialylated Nglycans, which are more likely to be efficacious, particularly for in vivo therapeutic applications.
ACKNOWLEDGMENT
We thank Drs. Harry Schachter, Pamela Stanley, Joel Shaper, Nancy Shaper, Jim Paulson, and Shuichi Tsuji for providing cDNAs for this work. We also thank Invitrogen for providing supplies and travel funds in support of this project. Finally, we thank HyClone for providing serum-free insect cell media for our studies.
Footnotes
This study was supported by grants from the National Institutes of Health (GM049734 to D.L.J.) and the National Science Foundation (BES-9818001 to D.L.J.) and by the Gesellschaft für Biotechnologische Forschung mbH (H.C.).
Abbreviations: β4Gal-T, β1,4-galactosyltranferase; ESI QTOF MS/ MS, electrospray ionization time of flight tandem mass spectrometry; GlcNAc-TI, N-acetylglucosaminyltransferase I; GlcNAc-TII, N-acetylglucosaminyltransferase II; GST-SfManI, glutathione S-transferase tagged Spodoptera frugiperda class I R-mannosidase; HPAEC-PAD, high-pH anion-exchange chromatography with pulsed amperometric detection; HPLC, high-performance liquid chromatography; HPLC/ESIMS, high-performance liquid chromatography coupled with electrospray ionization mass spectrometry; MALDI/TOF, matrix-assisted laser desorption ionization time of flight; MS, mass spectrometry; SDS, sodium dodecyl sulfate; ST6GalI, α2,6-sialyltransferase; ST3GalIV, α2,3-sialyltransferase.
References
- 1.Marz L, Altmann F, Staudacher E, Kubelka V. In: Glycoproteins. (Montreuil J, Vliegenthart JFG, Schachter H, editors. Amsterdam: Elsevier; 1995. pp. 543–563. [Google Scholar]
- 2.Jarvis DL. In: The BaculoViruses. (Miller LK, editor. New York: Plenum Press; 1997. pp. 389–431. [Google Scholar]
- 3.Varki A, Cummings R, Esko J, Freeze H, Hart G, Marth J. Essentials of Glycobiology. Cold Spring Harbor, NY: Cold Spring Harbor Press; 1999. [PubMed] [Google Scholar]
- 4.Altmann F, Staudacher E, Wilson IB, Marz L. Glycoconjugate J. 1999;16:109–123. doi: 10.1023/a:1026488408951. [DOI] [PubMed] [Google Scholar]
- 5.Marchal I, Jarvis DL, Cacan R, Verbert A. Biol. Chem. 2001;382:151–159. doi: 10.1515/BC.2001.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Montreuil J, Vliegenthart JFG, Schachter H. Glycoproteins. 29a. Amsterdam: Elsevier; 1995. [Google Scholar]
- 7.Summers MD, Smith GE. Tex. Agric. Exp. Stn.,[Bull.] 1987;(No. 1555) [Google Scholar]
- 8.O’Reilly DR, Miller LK, Luckow VA. BaculoVirus expression Vectors. New York: W. H. Freeman and Co; 1992. [Google Scholar]
- 9.Bhatia PK, Mukhopadhyay A. AdV. Biochem. Eng. Biotechnol. 1999;64:155–201. doi: 10.1007/3-540-49811-7_5. [DOI] [PubMed] [Google Scholar]
- 10.Jenkins N, Parekh RB, James DC. Nat. Biotechnol. 1996;14:975–981. doi: 10.1038/nbt0896-975. [DOI] [PubMed] [Google Scholar]
- 11.Jenkins N, Curling EMA. Enzymol. Microb. Technol. 1994;16:354–364. doi: 10.1016/0141-0229(94)90149-x. [DOI] [PubMed] [Google Scholar]
- 12.Weigel PH. BioEssays. 1994;16:519–524. doi: 10.1002/bies.950160713. [DOI] [PubMed] [Google Scholar]
- 13.Varki A. Glycobiology. 1993;3:97–130. doi: 10.1093/glycob/3.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wagner R, Liedtke S, Kretzschmar E, Geyer H, Geyer R, Klenk HD. Glycobiology. 1996;6:165–175. doi: 10.1093/glycob/6.2.165. [DOI] [PubMed] [Google Scholar]
- 15.Jarvis DL, Finn EE. Nat. Biotechnol. 1996;14:1288–1292. doi: 10.1038/nbt1096-1288. [DOI] [PubMed] [Google Scholar]
- 16.Vaughn JL, Goodwin RH, Thompkins GJ, McCawley P. In Vitro. 1977;13:213–217. doi: 10.1007/BF02615077. [DOI] [PubMed] [Google Scholar]
- 17.Hollister JR, Shaper JH, Jarvis DL. Glycobiology. 1998;8:473–480. doi: 10.1093/glycob/8.5.473. [DOI] [PubMed] [Google Scholar]
- 18.Wickham TJ, Davis T, Granados RR, Shuler ML, Wood HA. Biotechnol. Prog. 1992;8:391–396. doi: 10.1021/bp00017a003. [DOI] [PubMed] [Google Scholar]
- 19.Hollister J, Jarvis DL. Glycobiology. 2001;11:1–9. doi: 10.1093/glycob/11.1.1. [DOI] [PubMed] [Google Scholar]
- 20.Breitbach K, Jarvis DL. Biotechnol. Bioeng. 2001;74:230–239. doi: 10.1002/bit.1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yates JL, Warren N, Sugden B. Nature (London) 1985;313:812–815. doi: 10.1038/313812a0. [DOI] [PubMed] [Google Scholar]
- 22.Kumar R, Yang J, Larsen RD, Stanley P. Proc. Natl. Acad. Sci. U.S. A. 1990;87:9948–9952. doi: 10.1073/pnas.87.24.9948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tan J, D’Agostaro AF, Bendiak B, Reck F, Sarkar M, Squire JA, Leong P, Schachter H. Eur. J. Biochem. 1995;231:317–328. doi: 10.1111/j.1432-1033.1995.tb20703.x. [DOI] [PubMed] [Google Scholar]
- 24.Weinstein J, Lee EU, McEntee K, Lai P-H, Paulson JC. J. Biol. Chem. 1987;262:17735–17743. [PubMed] [Google Scholar]
- 25.Kono M, Ohyama Y, Lee YC, Hamamoto T, Kojima N, Tsuji S. Glycobiology. 1997;7:469–479. doi: 10.1093/glycob/7.4.469. [DOI] [PubMed] [Google Scholar]
- 26.Guarino LA, Summers MD. J. Virol. 1987;61:2091–2099. doi: 10.1128/jvi.61.7.2091-2099.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jarvis DL, Fleming JA, Kovacs GR, Summers MD, Guarino LA. Bio/Technology. 1990;8:950–955. doi: 10.1038/nbt1090-950. [DOI] [PubMed] [Google Scholar]
- 28.Jarvis DL, Weinkauf C, Guarino LA. Protein Expression Purif. 1996;8:191–203. doi: 10.1006/prep.1996.0092. [DOI] [PubMed] [Google Scholar]
- 29.Smith GE, Summers MD. Virology. 1978;89:517–527. doi: 10.1016/0042-6822(78)90193-9. [DOI] [PubMed] [Google Scholar]
- 30.Kawar Z, Romero PA, Herscovics A, Jarvis DL. Glycobiology. 2000;10:347–355. doi: 10.1093/glycob/10.4.347. [DOI] [PubMed] [Google Scholar]
- 31.Kawar Z, Herscovics A, Jarvis DL. Glycobiology. 1997;7:433–443. doi: 10.1093/glycob/7.3.433. [DOI] [PubMed] [Google Scholar]
- 32.Feinberg AP, Vogelstein B. Anal. Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 33.Shaper NL, Shaper JH, Meuth JL, Fox JL, Chang H, Kirsch IR, Hollis GF. Proc. Natl. Acad. Sci. U.S. A. 1986;83:1573–1577. doi: 10.1073/pnas.83.6.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laemmli UK. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 35.Towbin H, Staehelin T, Gordon J. Proc. Natl. Acad. Sci. U.S. A. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hardy MR, Townsend RR. Proc. Natl. Acad. Sci. U.S. A. 1988;85:3289–3293. doi: 10.1073/pnas.85.10.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hermentin P, Witzel R, Doenges R, Bauer R, Haupt H, Patel T, Parekh RB, Brazel D. Anal. Biochem. 1992;206:419–429. doi: 10.1016/0003-2697(92)90388-n. [DOI] [PubMed] [Google Scholar]
- 38.Anumula KR, Taylor PB. Anal. Biochem. 1992;203:101–108. doi: 10.1016/0003-2697(92)90048-c. [DOI] [PubMed] [Google Scholar]
- 39.Hoffmann A, Conradt HS, Gross G, Nimtz M, Lottspeich F, Wurster U. J. Neurochem. 1993;61:451–456. doi: 10.1111/j.1471-4159.1993.tb02145.x. [DOI] [PubMed] [Google Scholar]
- 40.Seo N-S, Hollister J, Jarvis DL. Protein Expression Purif. 2001;22:234–241. doi: 10.1006/prep.2001.1432. [DOI] [PubMed] [Google Scholar]
- 41.Wolff MW, Murhammer DW, Jarvis DL, Linhardt RJ. Glycoconjugate J. 1999;16:753–756. doi: 10.1023/a:1007131611378. [DOI] [PubMed] [Google Scholar]
- 42.Ailor E, Takahashi N, Tsukamoto Y, Masuda K, Rahman BA, Jarvis DL, Lee YC, Betenbaugh MJ. Glycobiology. 2000;10:837–847. doi: 10.1093/glycob/10.8.837. [DOI] [PubMed] [Google Scholar]
- 43.Altmann F, Kornfeld G, Dalik T, Staudacher E, Glossl J. Glycobiology. 1993;3:619–625. doi: 10.1093/glycob/3.6.619. [DOI] [PubMed] [Google Scholar]
- 44.Ooi BG, Miller LK. Virology. 1988;166:515–523. doi: 10.1016/0042-6822(88)90522-3. [DOI] [PubMed] [Google Scholar]
- 45.Jarvis DL. J. Virol. 1993;67:2583–2591. doi: 10.1128/jvi.67.5.2583-2591.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kawar ZS, Jarvis DL. Insect Biochem. Mol. Biol. 2001;31:289–297. doi: 10.1016/s0965-1748(00)00121-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grabenhorst E, Hoffmann A, Nimtz M, Zettlmeissl G, Conradt HS. Eur. J. Biochem. 1995;232:718–725. doi: 10.1111/j.1432-1033.1995.718zz.x. [DOI] [PubMed] [Google Scholar]