Abstract
Although prostate cancer (PCa) is the second leading cause of cancer death among men worldwide, not all men diagnosed with PCa will die from the disease. A critical challenge, therefore, is to distinguish indolent PCa from more advanced forms to guide appropriate treatment decisions. We used Enhanced Reduced Representation Bisulfite Sequencing, a genome-wide high-coverage single-base resolution DNA methylation method to profile seven localized PCa samples, seven matched benign prostate tissues, and six aggressive castration-resistant prostate cancer (CRPC) samples. We integrated these data with RNA-seq and whole-genome DNA-seq data to comprehensively characterize the PCa methylome, detect changes associated with disease progression, and identify novel candidate prognostic biomarkers. Our analyses revealed the correlation of cytosine guanine dinucleotide island (CGI)-specific hypermethylation with disease severity and association of certain breakpoints (deletion, tandem duplications, and interchromosomal translocations) with DNA methylation. Furthermore, integrative analysis of methylation and single-nucleotide polymorphisms (SNPs) uncovered widespread allele-specific methylation (ASM) for the first time in PCa. We found that most DNA methylation changes occurred in the context of ASM, suggesting that variations in tumor epigenetic landscape of individuals are partly mediated by genetic differences, which may affect PCa disease progression. We further selected a panel of 13 CGIs demonstrating increased DNA methylation with disease progression and validated this panel in an independent cohort of 20 benign prostate tissues, 16 PCa, and 8 aggressive CRPCs. These results warrant clinical evaluation in larger cohorts to help distinguish indolent PCa from advanced disease.
Introduction
Prostate cancer (PCa) is the most common cancer in men worldwide [1]. Although 1 in 6 men will likely be diagnosed with PCa during his lifetime, only 1 in 36 will ultimately die from the disease, reflecting a 5-year survival rate close to 100% [2]. Overtreatment is a major concern in PCa, as many patients diagnosed with clinically localized PCa do not require definitive treatment [3,4]. Patients presenting with advanced forms, including metastatic and castration-resistant prostate cancer (CRPC), however, have much worse outcomes; therefore, distinguishing indolent from advanced PCa has been an imperative task in PCa research.
In this study, we sought to investigate how genome-wide DNA methylation patterns might help differentiate between indolent and advanced PCa. We also reasoned that single-nucleotide resolution analysis of DNA methylation might provide mechanistic insights into the evolution of PCa toward more advanced forms. DNA methylation primarily arises at cytosine guanine dinucleotide (CpG) sites and is associated with epigenetic regulation of gene expression [5]. Perturbed DNA methylation patterns have been shown to arise during PCa tumorigenesis and have been implicated in PCa etiology and disease progression [6]. DNA methylation profiling studies using microarrays, MethylPlex-next-generation sequencing, and MeDIP-Seq have indeed identified a large number of DNA methylation changes in PCa [7–11]. However, few studies have investigated DNA methylation changes in CRPC, in part due to limited availability of tissues. DNA hypomethylation has been found to occur during PCa progression by evaluating 5-methylcytosine content in genomic DNA, while hypermethylation at certain CpG island (CGI) promoter genes were found in both localized PCa and metastatic PCa [10,12–14]. An array-based DNA methylation profiling study of CRPC recently found that alterations in DNA methylation arose more frequently than mutations or copy number changes [15], further strengthening the rationale for interrogating metastatic PCa to identify biomarkers of DNA methylation related to PCa disease progression. To our knowledge, the present study is the first to perform broad coverage single-nucleotide resolution analysis of CRPC.
We previously used next-generation sequencing to characterize the whole genome and transcriptome of seven clinically localized PCa together with matched benign adjacent prostate tissues as well as of seven CRPC cases [16,17]. In an effort to understand how aberrant DNA methylation may contribute to PCa and to the metastatic phenotype, we profiled the global DNA methylation patterns of these same cases using Enhanced Reduced Representation Bisulfite Sequencing (ERRBS). ERRBS with single-base resolution gives a broader genome-wide coverage by extending to CGI shores, compared to Reduced Representation Bisulfite Sequencing [18]. Since bisulfite sequencing provides single-base resolution, it allowed us to investigate how frequently allele-specific methylation (ASM) occurs in genetically and epigenetically unstable PCa and CRPC samples. ASM is mainly studied in the context of genomic imprinting, whose role is to ensure and maintain parent-of-origin effects on gene expression in a small set of genes [19,20]; its occurrence in non-imprinted genomic regions of cancer cells is less clear. In this study, we integrated data of methylation and SNPs to delineate and understand ASM in clinical PCa cases.
We report novel insights into DNA methylation patterns in PCa progression, association between DNA methylation and breakpoints, ASM changes, and the identification of a panel of CGIs highly related to PCa progression. We propose that differential methylation changes could be used not only as early detection biomarkers but also as new prognostic biomarkers in assisting treatment management decisions.
Materials and Methods
Prostate Tissue Samples
All tissue samples (benign prostate tissues and PCa and CRPC samples) were collected as part of an Institutional Review Board-approved protocol at Weill Cornell Medical College (WCMC). PCa (hormone naïve PCa) and CRPC were obtained, and DNA extraction was performed as previously described [16,17]. None of the PCa samples were from patients who received preoperative hormonal therapy, chemotherapy, or radiotherapy. Hematoxylin and eosin (H&E) slides of each prostate tissue sample were evaluated by the study's pathologists (M.A.R., K.P., and J.M.M.) to determine areas of benign and cancer; tumors were selected for high-density cancer foci (>90% tumor tissue). Biopsy cores (1.5 mm) from frozen tissue blocks (ERRBS) or formalinfixed, paraffin-embedded (FFPE) blocks (validation) were prepared for DNA extraction. Table W1 shows Gleason grade, age, preoperative serum prostate-specific antigen levels, and staging of all of the patients from whom prostate tissues were obtained for this study. Seven separately collected benign prostatic tissue samples matched to the cancer tissues (obtained from the same patients) were used as normal controls. Samples for the validation cohort consisted of 20 benign prostate tissues and 16 PCa and 8 CPRC samples. The benign prostate tissues also came from the matched PCa and CRPC patients. All CRPC samples were from patients with variable degrees of neuroendocrine differentiation, and three of the eight cases were pure neuroendocrine histology and lacked protein androgen receptor (AR) expression. The PCa samples used in ERRBS were from patients without any recurrence following radical prostatectomy for 5 to 6 years and 3 to 7 years for the PCa patients in the validation cohort (surgeries performed between 2006 and 2010). Benign prostate tissues were benign glands from a cancer-free region of the matched PCa or CRPC patients, taken from a separate frozen or FFPE tissue block without a cancer region as determined by the study's pathologists. DNA from FFPE blocks was extracted using the DNeasy Blood and Tissue Kit (Qiagen, Germantown, MD) according to the manufacturer's instructions and then evaluated by the FFPE Quality Control Kit (Illumina, San Diego, CA). Samples that passed the quality control (QC) test were used in the validation. Clinical parameters of samples used in ERRBS and the validation study are listed in Tables W1 and W2.
Enhanced Reduced Representation Bisulfite Sequencing
Sample preparations were performed at the WCMC Epigenomics Core Facility as previously described [18]. In brief, the preparation steps included: 1) MspI enzyme digestion; 2) end repair of digested DNA; 3) adenylation; 4) adapter ligation, with pre-annealed 5-methylcytosine-containing Illumina adapters; 5) isolation of library fragments of 150 to 400 bp from a 1.5% agarose gel (using low-range ultra-agarose from Bio-Rad, Des Plaines, IL); 6) bisulfite conversion using the EZ DNA Methylation Kit (Zymo Research, Irvine, CA) with the following changes: i) CT conversion incubation in a thermocycler (Eppendorf, Hauppauge, NY) with the condition: 30 seconds at 95°C followed by 15 minutes at 50°C for 55 cycles, and ii) elution into nuclease-free water; 7) polymerase chain reaction (PCR) amplification; each library was prepared with FastStart High Fidelity DNA Polymerase (Roche, Indianapolis, IN) and Illumina PCR primers PE1.0 and 2.0. The thermocycler conditions were given as follows: 5 minutes at 94°C, 18 cycles of 20 seconds at 94°C, 30 seconds at 65°C, 1 minute at 72°C, followed by 3 minutes at 72°C. PCR products were isolated using Agencourt AMPure XP (Beckman Coulter, Brea, CA) beads per manufacturer's recommended protocol (Agencourt) All amplified libraries were evaluated using a Qubit 1.0 fluorometer and Quant-iT dsDNA HS Assay Kit (Invitrogen, Grand Island, NY) for quantitation and bioanalyzer visualization (Agilent 2100 Bioanalyzer; Agilent, Santa Clara, CA). The WCMC Computational Genomics Core Facility supported ERRBS data analysis.
Quantitative DNA Methylation Analysis by MassARRAY EpiTYPER
Measurement of DNA methylation levels was performed at the WCMC Epigenomics Core Facility with matrix-assisted laser desorption ionization/time-of-flight mass spectrometry using MassARRAY EpiTYPER assays (Sequenom, San Diego, CA) as previously described [21]. Genomic DNA was bisulfite-converted using the EZ DNA Methylation Kit according to the manufacturer's protocol (Zymo Research). The spectra's methylation ratios were then calculated using EpiTYPER software v1.0 (Sequenom). The EpiTYPER primers were designed through the Sequenom EpiDesigner web site at http://www.epidesigner.com/start3.html. The reverse primers were tagged with a T7 promoter sequence: cagtaatacgactcactatagggagaaggct. EpiTYPER primer sequences are listed in Table W4.
Computational Approaches
Bisulfite-treated read alignment and methylation calls were performed as previously described [18]. Briefly, bisulfite reads were aligned to the bisulfite-converted hg19 reference genome using Bismark [22]. All samples had bisulfite conversion rates >99.5%. Differentially methylated CpGs were identified using the Fisher exact test with correction for Benjamini-Hochberg multiple testing. Differentially methylated regions (DMRs) were defined as regions containing at least five differentially methylated CpGs [false discovery rate (FDR) = 10%] and whose total methylation difference was more than 10%. DMRs were annotated using ChIPseeqerAnnotate from the ChIPseeqer package [23]. Motif and pathway analyses were performed with default parameters using FIRE [24] and iPAGE [25], respectively. When comparing methylation at CGIs between groups of samples, we used the LIMMA approach on log-transformed methylation values. Methylation of a specific region was calculated by averaging the methylation levels of all covered CpGs in that region. Clonality rates were calculated by counting how many times rare non-converted cytosine among reads mapped at the same genomic location and orientation, as described in Gertz et al. [26]. Random Forests [27] and 10-fold cross-validation were used to build classification models based on the samples in the validation cohort. Both classification and the estimation of sensitivity, specificity, and area under the receiver operating characteristic (ROC) curve [area under curve (AUC)] were performed in R using the Caret package (at http://caret.r-forge.r-project.org/).
Data Deposition Statement
The ERRBS data have been deposited for public access in the National Center for Biotechnology Information's (NCBI) gene expression omnibus (GEO) database and is accessible through GEO Series accession number GSE41701. DNA-seq and RNA-seq data are available as described in Beltran et al. and Berger et al. [16,17].
Results
Profound and Widespread Changes in DNA Methylation in Prostate Adenocarcinoma
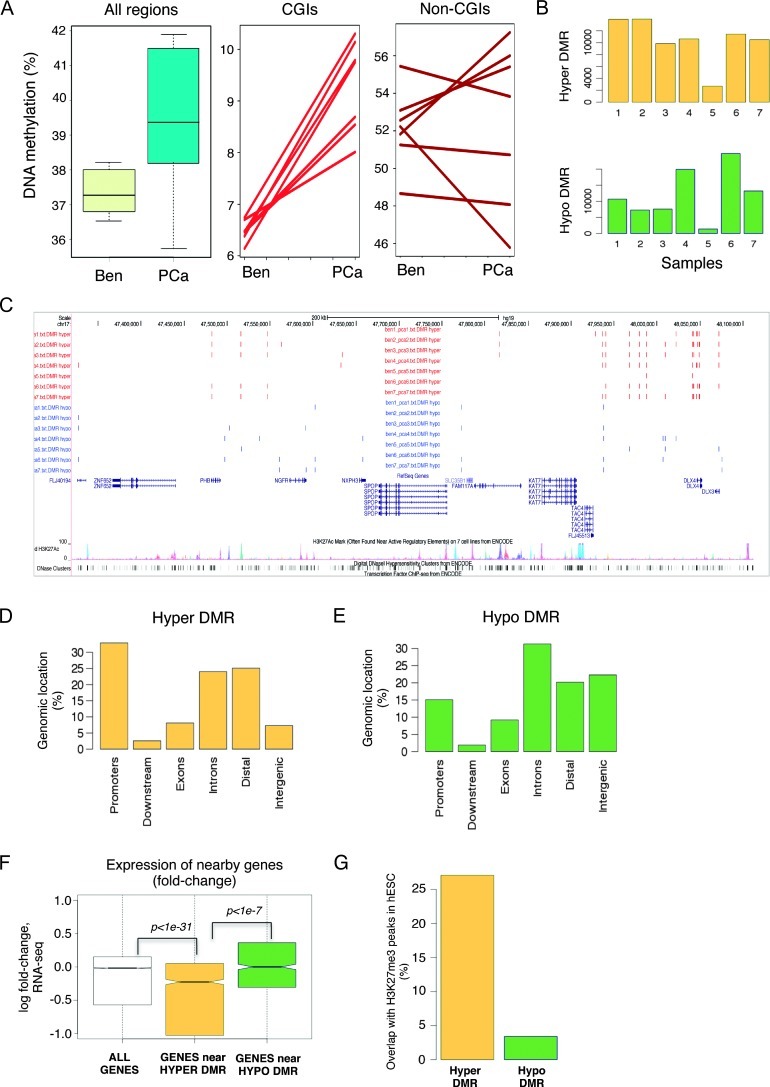
We first interrogated seven pairs of localized PCa and their matched benign prostate tissues. ERRBS provided more than 10-fold sequencing coverage on >2.5 million CpG sites genome-wide for each sample. By combining methylation levels from all interrogated CpG sites, we observed an overall increase in DNA methylation in PCa (Figure 1A), driven in large part by a sharp and systematic methylation increase at CGIs (P = .0002). The changes at non-CGI regions (defined as at least 10 kb away from known CGIs) were more diverse and less systematic among PCa patients (P = .85); there was a trend toward global hypomethylation in four of the seven cases. This analysis also indicated that global methylation levels were significantly more heterogeneous in PCa compared to benign prostate tissues. We confirmed this heterogeneity when we determined DMRs within each pair and detected differing numbers of hypermethylated and hypomethylated regions in each case (Figure 1B), with some cases showing more hypomethylated DMRs while others showed more hypermethylated DMRs. Despite this heterogeneity and the different genetic background of each patient, we found that many hypermethylated and hypomethylated DMRs resided at the same locations between individuals, suggesting that aberrant DNA methylation in PCa follows a predetermined path (Figure 1C). When we examined DNA methylation patterns across different genomic regions, we found that hypermethylated DMRs were mostly enriched at promoters [defined as ±2 kb windows centered on RefSeq transcription start sites (TSS); Figure 1D]. However, hypo-methylated DMRs were mostly located at introns and intergenic regions (Figure 1E).
Figure 1.
Overall increased DNA methylation in PCa, consistent across cases. (A) Percentages of DNA methylation in all regions, CGIs, and non-CGIs of matched benign prostate tissues (Ben) and PCa. (B) Numbers of hypermethylated or hypomethylated DMRs of individual PCa samples. (C) Example of hypermethylated and hypomethylated DMRs, many of which occur independently in different patients. (D) Percentage of hypermethylated regions occurring within indicated genomic locations. (E) Percentage of hypomethylated regions occurring within indicated genomic locations. (F) Expression fold changes of nearby genes in regard of hypermethylated or hypomethylated DMRs (data shown for case 1). (G) Percentage of hypermethylated or hypomethylated DMRs overlapped with H3K27me3 peaks in human embryonic stem cells (hESC).
We then sought to integrate RNA-seq data performed on the same samples to examine the relationship between aberrant methylation and gene expression changes between PCa and matched benign tissue from the same patient. We found that overall genes located near hyper-methylated DMRs had decreased expression (P <1e-31), while genes near hypomethylated DMRs had increased expression (Figure 1F; data shown for patient 1). This indicates that, within controlled isogenic conditions, the frequently assumed but broadly elusive negative correlation between DNA methylation and gene expression tends to hold. We also found that hypermethylated DMRs overlapped strongly with regions enriched for H3K27me3 in human embryonic stem cells [28] (Figure 1G), confirming previous reports of such concurrent epigenomic changes in PCa cell lines and other cancer types [29–32].
De novo DNA motif analysis was carried out to examine the sequences of DMRs. We focused the analysis on differentially methylated CGIs, since these regions behaved in a consistent way across cases (Figure 1A). We used non-differentially methylated CGIs as control, thus controlling for CpG content. This analysis identified three DNA motifs preferentially enriched with hypermethylated DMRs, including one that resembled the motif bound by the transcription factor early growth response gene 1 (EGR1; Figure W1A). When examining RNA-seq data, we found that EGR1 expression levels were lower in PCa compared to benign prostate tissues (Figure W1B). This down-regulation was confirmed by immunohistochemistry staining (Figure W1C). Although more extensive work is required to investigate the role of EGR1 in PCa, we speculate that EGR1 binding to CGIs may prevent DNA methylation, while loss of EGR1 expression may lead to increased DNA methylation.
Structural Variations in PCa Are Correlated with DNA Methylation Patterns in Benign Tissues
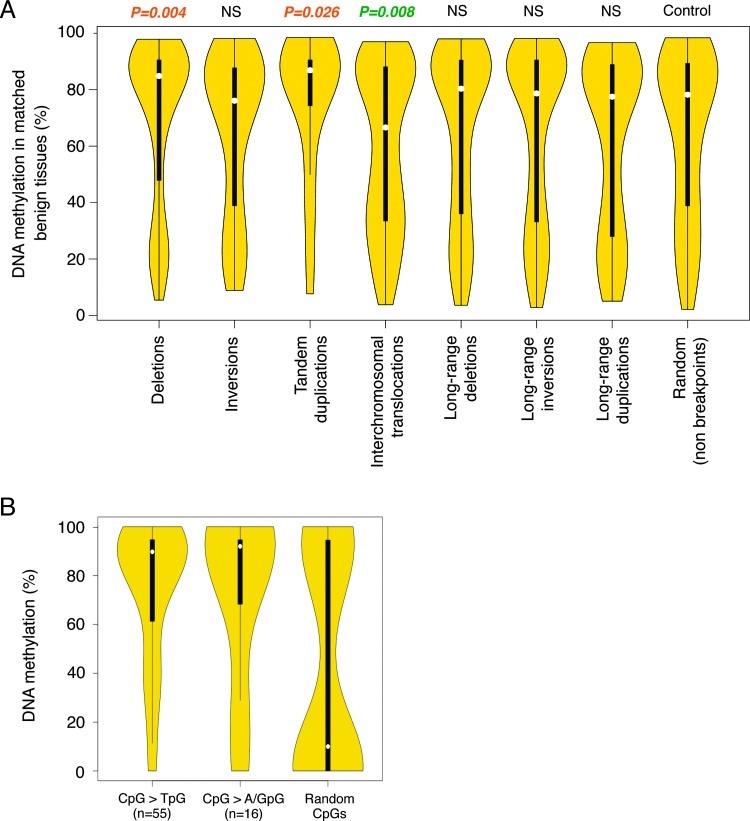
The previously described genomic characterization of our seven PCa samples provided a unique opportunity to integrate genomic and epigenomic information and better understand the events that lead to progression from benign lesion to PCa [17]. To evaluate whether DNA methylation patterns in normal tissues may influence the formation of genomic alterations that lead to PCa, we examined structural variation breakpoints that have occurred in PCa and determined their DNA methylation levels in matched benign prostate tissues. For this analysis, we examined DNA methylation in 10-kb windows centered on each breakpoint. We found that in benign tissues, deletion and tandem duplication breakpoints that occurred in PCa were less methylated than control random regions (P = .004 and P = .026, respectively); however, interchromosomal translocation breakpoints were more methylated than expected (P = .008; Figure 2A). Thus, certain structural variation breakpoints have non-random methylation levels in benign prostate tissues, hinting at a potential role of the epigenome in mediating these alterations. Moreover, Berger et al. found that CpG sites had significantly higher mutation rate (more than 10-fold) than all other genomic positions did [17]. When we examined CpGs that were mutated in PCa samples, we found that these CpGs were highly methylated in their matched benign tissues (Figure 2B). By comparison, a random selection of non-mutated CpGs showed much lower DNA methylation in the benign tissues. Most mutations occurring at CpGs were C-to-T transition mutations; however, we also found the less frequent C-to-A and C-to-G transversion mutations. The C-to-T transition may come from spontaneous deamination of methylated cytosine to thymidine. While many of these mutations may be passenger mutations that have been deaminated in the initial malignant clone, our results altogether suggest that many genetic alterations in PCa have a non-random methylation signature in normal prostate cells. Moreover, our results suggest that non-C-to-T mutations at methylated cytosines occur at a non-negligible rate and that highly methylated CpG sites are inherently fragile and mutation prone, while non-methylated CpGs are protected from alterations.
Figure 2.
DNA methylation patterns in benign prostate tissues are associated with certain types of mutations in PCa. (A) Percentage of DNA methylation in matched benign tissues for various breakpoints. The width of the violin plots correlates with the fraction of mutations with corresponding methylation levels (Y axis). NS, not significant. (B) Percentage of DNA methylation of CpG sites in benign prostate tissues, where C>T, C>A/G mutations occur.
Widespread Allele-Specific DNA Methylation in PCa
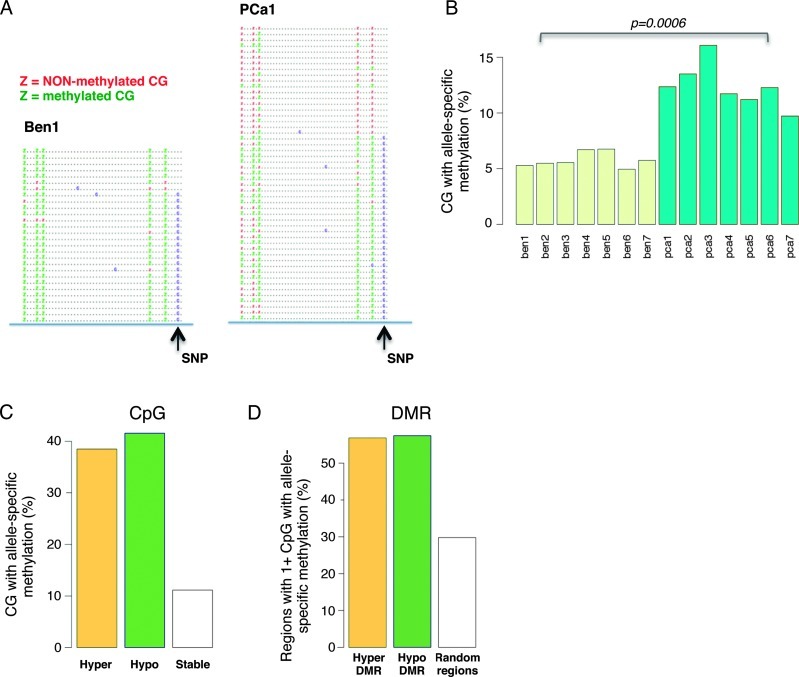
Taking advantage of single CpG resolution and high coverage provided by ERRBS, we sought to investigate whether certain regions of the methylome showed ASM and whether ASM levels differ between matched tumors and benign tissues. We therefore mined the ERRBS data and looked for statistical associations between SNPs (defined here as mismatches between the samples and the reference genome) and CpG methylation within the same ERRBS reads (Figure 3A). Comparing CpGs covered in both benign tissues and matched tumors, we identified between 60,000 and 150,000 CpGs that coincided with a heterozygous SNP within the same read and for which we could perform a statistical test of association (Fisher exact test) [17]. After correction for multiple testing (Benjamini-Hochberg, FDR = 20%), we identified several thousand CpGs with ASM in both benign tissues and PCa. When we compared the fraction of CpGs with ASM, we observed a significant increase in PCa compared to benign tissues (P = .0009, t test; Figure 3B). Importantly, we found that unequal read clonality (PCR duplicates) could not explain these differences, as clonality levels were approximately equal between PCa and matched benign tissues (Figure W2).
Figure 3.
Allele-specific DNA methylation is highly frequent in PCa. (A) Methylated and non-methylated CpGs correlate with the presence or absence of a nearby SNP in the first pair of benign prostate tissue and PCa. (B) Fraction of CpGs associated with ASM in benign prostate tissue and PCa. Only CpGs with a nearby SNP are considered in this analysis. (C) Fraction of CpGs with ASM in hypermethylated or hypomethylated CGIs. (D) Fraction of regions containing at least one CpG with ASM in hypermethylated or hypomethylated DMRs.
We speculated that allele-specific changes in methylation could be responsible for a significant fraction of all changes in DNA methylation (that is, one allele undergoes methylation changes, while the other does not). Accordingly, we found that ASM preferentially occurred in differentially methylated CpGs and DMRs, for both hypermethylated and hypomethylated (Figure 3, C and D). Altogether, these results suggest that a large fraction of DNA methylation changes occur on a single-allele basis. We also sought to determine whether ASM occurred in the context of allele-specific expression. While we found overlaps (i.e., genes whose promoter showed ASM and with different allelic usage between DNA-seq and RNA-seq; not shown), the coverage differences between ERRBS, DNA-seq, and RNA-seq data limited the analysis and precluded a definitive conclusion.
DNA Methylation at CGIs Increases with Disease Severity
To examine how DNA methylation patterns may have evolved in more advanced PCa, we profiled the DNA methylome of six CRPC samples from patients who failed endocrine therapy and/or developed a predominantly androgen-independent PCa associated with lack of AR expression and extensive neuroendocrine differentiation [16]. Examining DNA methylation levels at CGIs, we found that CRPC had higher levels compared to PCa (Figure 4A). Taken together, our analyses indicate that DNA methylation levels at CGIs increase with disease severity from benign tissue to PCa onto CRPC. There was also a less systematic and less pronounced average trend toward DNA methylation increase with disease severity in non-CGI regions (Figure 4A). Nevertheless, DNA methylation levels in CRPC were highly variable among cases. To further investigate the global DNA methylation increase, we focused on CGIs and performed gene ontology analysis of the genes near CGIs that were differentially methylated in PCa and CRPC using iPAGE [25] (LIMMA analysis, FDR = 5%; Figure W3A). Interestingly, the genes associated with hypermethylated CGIs had specific enrichments related to G-protein signaling, cAMPmediated signaling, and hormone activity. This suggests thatDNA methylation increases during the transition between indolent PCa and highly aggressive CRPC are at least partially functionally coherent. Wealso found that EGR1 mRNA levels are further decreased in CRPC compared to PCa, implicating the potential role of EGR1 down-regulation in enabling CGI hypermethylation (Figure W3B).
Figure 4.
DNA methylation increases with and may predict disease severity. (A) Percentage of DNA methylation in CGIs and non-CGIs of benign prostate tissues (Ben), PCa, and CRPC. (B) Venn diagram displays the methylation overlap observed between two sections: increased methylation from benign prostate tissues to PCa and increased methylation from PCa to CRPC; the inset number represents the coverage in each section. Two hundred twenty-eight CGIs appear in both sections with P <1e-104. (C) Left: Heat map of DNA methylation levels of the panel of 13 CGIs in the three groups, 20 benign prostate tissues and 16 PCa and 8 CRPC samples (value range, 0 to 1). Right: Comparison between the three groups with adjusted log P values (Benjamini-Hochberg correction FDR controlled at 0.05). (D) Boxplots of the average DNA methylation levels of the panel for individual samples. (E) Boxplots of the average DNA methylation levels of the panel for the three groups. The AUC, sensitivity, and specificity for the comparison of benign and PCa are 0.9375, 100%, and 75%, and those for the comparison of PCa and CRPC are 0.975, 95%, and 95%.
We then sought to identify a subset of these CGIs whose methylation increases with disease severity. We determined CGIs whose methylation levels increased systematically between benign prostate tissues and PCa (LIMMA analysis, FDR = 5%, n = 692). We also performed the same analysis for CGIs whose methylation levels increased consistently and reproducibly between PCa and CRPC (LIMMA analysis, FDR = 5%, n = 1229). We found that the overlap between two groups of hypermethylated CGIs was higher than expected (P <1e-104, hypergeometric P value; Figure 4B). This indicates that, to a large extent, the same genomic regions gain methylation in the transition from benign prostate tissue to PCa, and this trend continues onto CRPC. There were 228 CGIs in this overlap and the DNA methylation heat map of the 228 CGIs provided a visual demonstration of the positive association between PCa disease progression and increasing DNA methylation levels (Figure W3C). Clusteranalysisbased on CGI methylation not only further confirmed the strong DNA methylation signature uncovered here but also showed that some PCa samples clustered with CRPC samples, suggesting a propensity of these cases to progress toward a more advanced disease (Figure W3D). As a control, we performed the same analysis for hypomethylated CGIs; in that analysis, hypomethylated CGIs between benign tissues and PCa and between PCa and CRPC did not overlap more frequently than expected by chance (Figure W3E). These results indicate that unlike hypermethylation, hypomethylation may occur randomly during progression to advanced PCa. Likewise, when we analyzed ASM in CRPC, we found that ASM occurred at the same levels as benign prostate tissues (Figure W3F).
We then sought to validate these observations on an independent patient cohort. We integrated gene expression from the RNA-seq data with the 228 CGIs and selected a panel of 13 CGIs for validation (Table W3). The 13 CGIs showed persistently increasing hypermethylation from benign prostate tissues to PCa, and from PCa to CRPC, while genes associated with these CGIs also demonstrated differential expression (LIMMA, FDR = 5%, fold change > 1.5). Among the panel of 13 CGIs, three associated genes, general receptor for phosphoinositides (GRASP), glutathione S-transferase π 1 (GSTP1), and tropomyosin 4 (TPM4), have been reported as differentially methylated in PCa versus normal tissues in Mahapatra et al. [8], thus providing independent validation of our analysis. Tissues from FFPE blocks of an independent cohort of 20 benign prostate tissues and 16 PCa and 8 CRPC samples were used to examine the panel of 13 CGIs. We used MassARRAY EpiTYPER assay to directly interrogate methylation of the 13 CGIs, thus seeking not only biologic validation in an independent cohort but also technical validation of the methylation measurements provided by ERRBS. As shown in Figure 4, C and D, we observed the same increase in DNA methylation correlating with disease severity in FFPE samples using MassARRAY EpiTYPER assay as that revealed by ERRBS on frozen tissues. We found that all 13 CGIs had increased methylation levels between benign and primary PCa cases (FDR = 5%); 8 of the 13 CGIs had increased methylation levels between primary cases and CRPC. Intriguingly, four advanced cases (FF18, 55, 57, and 59) that had no previous treatment showed high levels of DNA methylation at these selected CGIs. Using Random Forests [27], we built two binary classification models to discriminate benign prostate tissues from PCa, as well as PCa from CRPC, based on the methylation levels of the 13 CGIs. In 10-fold cross-validation, the AUC scores for the predictive models were high, 0.9375 and 0.975, respectively (Figure 4E), indicating a high sensitivity and specificity. Taken together, our data suggest that both models provide excellent discriminatory power and the panel may be useful in predicting disease aggressiveness at diagnosis.
Discussion
In this study, we performed global DNA methylation profiling in primary PCa samples, matched benign prostate tissues, and CRPC and integrated DNA methylation patterns with whole-genome and transcriptome sequencing data. We used ERRBS to profile genome-wide DNA methylation status, thus interrogating >2.5 M single CpGs in each sample. ERRBS has recently been developed to capture more broadly genome-wide DNA methylation at single-base resolution, including CGI shores [18]. ERRBS yields quantitative comparisons of methylation levels and enhances methylation detection at various genomic regions. By providing deep coverage of regulatory regions, especially CGIs, and low coverage of repeat regions (e.g., LINE1 elements), ERRBS enables a range of new analyses, including ASM analyses.
The analyses described in this study led to a number of new findings. First, by examining global methylation patterns in benign prostate tissue versus PCa versus CRPC, we found increasing levels of DNA methylation at CGIs correlated with increasing severity of the disease. As most CRPCs still retain AR expression, the methylation patterns observed in this study may not be representative of most CRPCs. Our motif analysis indicates that EGR1 DNA motifs are overrepresented at hypermethylated CGIs in PCa and CRPC. Previous studies using genome-wide chromatin immunoprecipitation (ChIP) assays in other cell types have indeed shown that EGR1 binds heavily to CGIs [33]. Our analyses also showed that EGR1 mRNA and protein levels decreased with disease severity. These data suggest that down-regulation of EGR1 may enable DNA methylation to accumulate at CGIs, perhaps through the recruitment of CGI-binding protein complexes that include DNA methyltransferase; however, a broader evaluation of EGR1 expression and a further investigation of the role of EGR1 in PCa disease progression are warranted. Of the 228 CGIs that showed increasing methylation during disease progression, we selected a panel of 13 CGIs for further validation, as the expression of their associated genes also exhibited significant changes and they may play important roles in PCa progression, especially toward a late stage and lethal neuroendocrine phenotype. We propose examination of the methylation status of this panel of 13 CGIs as putative diagnostic and prognostic biomarkers, which may aid in directing treatment decisions.
According to RNA-seq data, the majority of genes on this panel were downregulated (70%, 9 of the 13 CGIs). GSTP1, a well-known DNA methylation biomarker for PCa, was among the associated genes shown to have a continuous decline in gene expression during PCa disease progression. Its silencing may render prostate cells more sensitive to environmental insults and consequently more susceptible to genomic damage [6]. Capping protein (actin filament), gelsolin-like (CAPG), a proposed tumor suppressor in certain cancers [34], has been shown to modulate cancer cell invasion in breast and PCa cell lines [35]. Retinoic acid receptor responder 2 (RARRES2), also known as chemerin or tazarotene-induced gene 2, was found to attract dendritic cells and macrophages to the site of inflammation [36]. As such, down-regulation of RARRES2 may affect the dynamics of the PCa microenvironment. TPM4 has been connected to human cancer, even though its role is less clear. Invasive squamous cervical cancer showed lower expression of TPM4, while lymph node metastasis in breast cancer has a significant association with TPM4 up-regulation [37,38]. Family with sequence similarity 107, member A (FAM107A), also known as DRR1 or TU3A, was previously reported to be downregulated and deleted in renal cell carcinoma, and the expression of FAM107A inhibited cell growth [39]. Filamin A interacting protein 1-like (FILIP1L) has also been shown downregulated in ovarian cancer, and its down-regulation is inversely correlated with promoter methylation and advanced phenotype [40].
Furthermore, the association of gene-body methylation with active gene expression has been found in human cells and other organisms [41,42]. Similarly, T-box containing transcription factor (TBX1) was found to have a hypermethylated CGI around its second exon and elevated gene expression in our cohort. A functional link has previously been established between TBX1 and retinoic acid (RA) signaling where overexpressed TBX1 inhibits RA signaling, in part, due to downregulation of ALDH1a2 and PCNA [43,44]. As down-regulation of ALDH1a2 was detected in human PCa and in the TRAMP mouse model of PCa [45,46], this study suggests a regulation of ALDH1a2 and RA signaling through up-regulation of TBX1 by DNA methylation during PCa progression. Likewise, the CGI that overlapped with the second exon of potassium voltage-gated channel subfamily C (KCNC2) was also hypermethylated in PCa, with concurrent up-regulation of the gene. KCNC2 encodes a membrane protein for voltage-gated potassium channel. Its function in PCa is unclear and may be related to the neuroendocrine phenotype of CRPC in this cohort. Intriguingly, both RARRES2 and TBX1 were identified in the panel and related to aberrant regulation of RA signaling during PCa progression. Even though we found that their hypermethylation resulted in opposite directions of gene expression (i.e., down-regulation in RARRES2 and up-regulation in TBX1), the overall outcome may down-regulate the RA signaling.
Another important finding in our study is that certain breakpoints, like deletion, tandem duplications, or interchromosomal translocations, are associated with distinct DNA methylation patterns in premalignant prostate tissue. We also found that highly methylated CpG sites are likely more fragile and more frequently targeted for somatic base mutations during PCa tumorigenesis compared to unmethylated CpGs. However, given that the benign tissues are benign glands taken from a cancer-free region of the matched PCa patient, we cannot rule out that epigenomic changes may have arisen in these morphologically normal tissues. Thus, the methylation patterns detected in benign tissues and associated with PCa breakpoints may not be present in the truly normal tissues (e.g., from autopsy) taken from other individuals. Altogether, our study points to an important role of the epigenome in mediating or facilitating a broad range of genomic alterations in PCa. Whether this role is more central in PCa than in other malignancies is currently unclear.
Our study revealed for the first time (to the best of our knowledge) frequent ASM changes in PCa. It also showed that a vast fraction of DNA methylation changes might occur on a single-allele basis. The mechanisms behind these single-allele methylation changes are unclear and may involve differential binding of transcription factors to each allele due to the presence of SNPs or different nuclear localization patterns. ASM was recently found at the H3K27me3-bound regions in a study of bisulfite sequencing of chromatin-immunoprecipitated DNA using PCa cell lines [30]. Thus, it is possible that a complex interplay exists between DNA methylation and H3K27me3, and this interplay precludes changes from occurring on certain alleles marked with either epigenetic mark.
Recent large-scale genome-wide DNA methylation profiling studies have discovered large numbers of DMRs. Our analysis on paired samples further identified a consistent trend in many DMRs located similarly across the entire genome, suggesting that DNA methylation changes may follow certain genetic cues during PCa pathogenesis. Moreover, we noted that within paired samples hypermethylation was significantly correlated with down-regulation of gene expression, while hypomethylation associates with up-regulation. Although this concept is broadly used, recent genome-wide studies have observed more complex regulations [18,42].
Both hypermethylation and hypomethylation take place in PCa as well as other types of cancer. On the basis of the analysis of 5-methylcytosine content of genomic DNA, previous studies have suggested that hypomethylation occurs later in course of metastatic PCa [13,14]. In contrast, through the use of sequencing-based DNA methylation analysis, the results from this study and that of Kim et al. did not find decreased CpG methylation levels of metastatic PCa [10]. However, our approach is biased toward regions of high guanine-cytosine (GC) content, and LINE-1 and other repetitive regions shown to be hypomethylated during PCa progression are underrepresented by ERRBS; it stands to reason that a significant amount of hypomethylation may occur later at low or intermediate GC content regions, which are not adequately represented by ERRBS or M-NGS.
Gene fusions between members of the ETS family and the androgen-regulated TMPRSS2 are frequently detected in PCa [47]. Differences in global methylation patterns or LINE-1 methylation were reported when categorizing PCa according to ETS gene fusion status [10,11]. Consistent with the study of Kim et al., we observed lower global methylation in TMPRSS2:ERG fusion-negative PCa (Figure W4), in contrast to the study of Borno et al., which showed increased levels of global methylation in the fusion-negative PCa. This discrepancy may be explained by the different approaches that were employed in these studies, as the levels of GC content coverage varied considerably: both ERRBS and M-NGS focus on regions of high GC content with minimal coverage of repeat elements, while MeDIP-seq has a broader coverage over CpGs in repetitive elements [48].
From a clinical point of view, the robust observation that DNA methylation increases with disease severity, especially at CGIs, provides new opportunities for detecting and diagnosing advanced PCa earlier than currently possible and for assisting treatment decisions. As a first step toward this goal, we provide a panel of 13 CGIs whose methylation reliably and reproducibly increases with disease severity, including association with a late stage and lethal neuroendocrine phenotype. Classification models based on the DNA methylation levels of these 13 CGIs and using Random Forest analysis deliver great discriminatory power, pointing toward a potential clinical use of the panel.
Larger studies with long-term follow-up, and inclusion of various stages of disease progression including CRPCs with retained AR expression, will be required to assess the true clinical value of these 13 CGIs and of other epigenetic alterations in the diagnosis, prognosis, and treatment of PCa.
Supplementary Material
Acknowledgments
We acknowledge the important contribution of the WCMC Epigenomics Core Facility and the WCMC Computational Genomics Core Facility and the technical assistance provided by Naoki Kitabayashi, Theresa McDonald, and Yifang Liu. We appreciate Ari M. Melnick and Daniel Di Bartolo for helpful suggestions.
Abbreviations
- ASM
allele-specific methylation
- CGI
cytosine guanine dinucleotide (CpG) island
- CRPC
castration-resistant prostate cancer
- ERRBS
Enhanced Reduced Representation Bisulfite Sequencing
- PCa
prostate cancer
Footnotes
This study was funded by the Early Detection Research Network grant U01 CA 11275-07 (M.A.R.), the Prostate Cancer Foundation Young Investigator Award(H.B.), and the National Science Foundation CAREER grant (O.E.).
This article refers to supplementary materials, which are designated by Tables W1 to W4 and Figures W1 to W4 and are available online at www.neoplasia.com.
References
- 1.Bray F, Jemal A, Grey N, Ferlay J, Forman D. Global cancer transitions according to the Human Development Index (2008–2030): a population-based study. Lancet Oncol. 2012;13:790–801. doi: 10.1016/S1470-2045(12)70211-5. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 3.Prensner JR, Rubin MA, Wei JT, Chinnaiyan AM. Beyond PSA: the next generation of prostate cancer biomarkers. Sci Transl Med. 2012;4:127rv123. doi: 10.1126/scitranslmed.3003180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilt TJ, Brawer MK, Jones KM, Barry MJ, Aronson WJ, Fox S, Gingrich JR, Wei JT, Gilhooly P, Grob BM, et al. Radical prostatectomy versus observation for localized prostate cancer. N Engl J Med. 2012;367:203–213. doi: 10.1056/NEJMoa1113162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 6.Nelson WG, De Marzo AM, Yegnasubramanian S. Epigenetic alterations in human prostate cancers. Endocrinology. 2009;150:3991–4002. doi: 10.1210/en.2009-0573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kobayashi Y, Absher DM, Gulzar ZG, Young SR, McKenney JK, Peehl DM, Brooks JD, Myers RM, Sherlock G. DNA methylation profiling reveals novel biomarkers and important roles for DNA methyltransferases in prostate cancer. Genome Res. 2011;21:1017–1027. doi: 10.1101/gr.119487.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mahapatra S, Klee EW, Young CY, Sun Z, Jimenez RE, Klee GG, Tindall DJ, Donkena KV. Global methylation profiling for risk prediction of prostate cancer. Clin Cancer Res. 2012;18:2882–2895. doi: 10.1158/1078-0432.CCR-11-2090. [DOI] [PubMed] [Google Scholar]
- 9.Kron K, Pethe V, Briollais L, Sadikovic B, Ozcelik H, Sunderji A, Venkateswaran V, Pinthus J, Fleshner N, van der Kwast T, et al. Discovery of novel hyper-methylated genes in prostate cancer using genomic CpG island microarrays. PLoS One. 2009;4:e4830. doi: 10.1371/journal.pone.0004830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim JH, Dhanasekaran SM, Prensner JR, Cao X, Robinson D, Kalyana-Sundaram S, Huang C, Shankar S, Jing X, Iyer M, et al. Deep sequencing reveals distinct patterns of DNA methylation in prostate cancer. Genome Res. 2011;21:1028–1041. doi: 10.1101/gr.119347.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Borno ST, Fischer A, Kerick M, Falth M, Laible M, Brase JC, Kuner R, Dahl A, Grimm C, Sayanjali B, et al. Genome-wide DNA methylation events in TMPRSS2:ERG fusion negative prostate cancers implicate an EZH2 dependent mechanism with miRNA-26a hypermethylation. Cancer Discov. 2012;2(11):1024–1035. doi: 10.1158/2159-8290.CD-12-0041. [DOI] [PubMed] [Google Scholar]
- 12.Yegnasubramanian S, Kowalski J, Gonzalgo ML, Zahurak M, Piantadosi S, Walsh PC, Bova GS, De Marzo AM, Isaacs WB, Nelson WG. Hypermethylation of CpG islands in primary and metastatic human prostate cancer. Cancer Res. 2004;64:1975–1986. doi: 10.1158/0008-5472.can-03-3972. [DOI] [PubMed] [Google Scholar]
- 13.Bedford MT, van Helden PD. Hypomethylation of DNA in pathological conditions of the human prostate. Cancer Res. 1987;47:5274–5276. [PubMed] [Google Scholar]
- 14.Yegnasubramanian S, Haffner MC, Zhang Y, Gurel B, Cornish TC, Wu Z, Irizarry RA, Morgan J, Hicks J, DeWeese TL, et al. DNA hypomethylation arises later in prostate cancer progression than CpG island hypermethylation and contributes to metastatic tumor heterogeneity. Cancer Res. 2008;68:8954–8967. doi: 10.1158/0008-5472.CAN-07-6088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friedlander TW, Roy R, Tomlins SA, Ngo VT, Kobayashi Y, Azameera A, Rubin MA, Pienta KJ, Chinnaiyan A, Ittmann MM, et al. Common structural and epigenetic changes in the genome of castration-resistant prostate cancer. Cancer Res. 2012;72:616–625. doi: 10.1158/0008-5472.CAN-11-2079. [DOI] [PubMed] [Google Scholar]
- 16.Beltran H, Rickman DS, Park K, Chae SS, Sboner A, MacDonald TY, Wang Y, Sheikh KL, Terry S, Tagawa ST, et al. Molecular characterization of neuroendocrine prostate cancer and identification of new drug targets. Cancer Discov. 2011;1:487–495. doi: 10.1158/2159-8290.CD-11-0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berger MF, Lawrence MS, Demichelis F, Drier Y, Cibulskis K, Sivachenko AY, Sboner A, Esgueva R, Pflueger D, Sougnez C, et al. The genomic complexity of primary human prostate cancer. Nature. 2011;470:214–220. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Akalin A, Garrett-Bakelman FE, Kormaksson M, Busuttil J, Zhang L, Khrebtukova I, Milne TA, Huang Y, Biswas D, Hess JL, et al. Base-pair resolution DNA methylation sequencing reveals profoundly divergent epigenetic landscapes in acute myeloid leukemia. PLoS Genet. 2012;8:e1002781. doi: 10.1371/journal.pgen.1002781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reik W, Walter J. Genomic imprinting: parental influence on the genome. Nat Rev Genet. 2001;2:21–32. doi: 10.1038/35047554. [DOI] [PubMed] [Google Scholar]
- 20.Kerkel K, Spadola A, Yuan E, Kosek J, Jiang L, Hod E, Li K, Murty VV, Schupf N, Vilain E, et al. Genomic surveys by methylation-sensitive SNP analysis identify sequence-dependent allele-specific DNA methylation. Nat Genet. 2008;40:904–908. doi: 10.1038/ng.174. [DOI] [PubMed] [Google Scholar]
- 21.Ehrich M, Nelson MR, Stanssens P, Zabeau M, Liloglou T, Xinarianos G, Cantor CR, Field JK, van den Boom D. Quantitative high-throughput analysis of DNA methylation patterns by base-specific cleavage and mass spectrometry. Proc Natl Acad Sci USA. 2005;102:15785–15790. doi: 10.1073/pnas.0507816102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krueger F, Andrews SR. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011;27:1571–1572. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giannopoulou EG, Elemento O. An integrated ChIP-seq analysis platform with customizable workflows. BMC Bioinformatics. 2011;12:277. doi: 10.1186/1471-2105-12-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Elemento O, Slonim N, Tavazoie S. A universal framework for regulatory element discovery across all genomes and data types. Mol Cell. 2007;28:337–350. doi: 10.1016/j.molcel.2007.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goodarzi H, Elemento O, Tavazoie S. Revealing global regulatory perturbations across human cancers. Mol Cell. 2009;36:900–911. doi: 10.1016/j.molcel.2009.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gertz J, Varley KE, Reddy TE, Bowling KM, Pauli F, Parker SL, Kucera KS, Willard HF, Myers RM. Analysis of DNA methylation in a three-generation family reveals widespread genetic influence on epigenetic regulation. PLoS Genet. 2011;7:e1002228. doi: 10.1371/journal.pgen.1002228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Breiman L. Random forests. Mach Learn. 2001;45:5–32. [Google Scholar]
- 28.Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, Presser A, Nusbaum C, Xie X, Chi AS, et al. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet. 2008;4:e1000242. doi: 10.1371/journal.pgen.1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gal-Yam EN, Egger G, Iniguez L, Holster H, Einarsson S, Zhang X, Lin JC, Liang G, Jones PA, Tanay A. Frequent switching of Polycomb repressive marks and DNA hypermethylation in the PC3 prostate cancer cell line. Proc Natl Acad Sci USA. 2008;105:12979–12984. doi: 10.1073/pnas.0806437105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Statham AL, Robinson MD, Song JZ, Coolen MW, Stirzaker C, Clark SJ. Bisulfite sequencing of chromatin immunoprecipitated DNA (BisChIP-seq) directly informs methylation status of histone-modified DNA. Genome Res. 2012;22:1120–1127. doi: 10.1101/gr.132076.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schlesinger Y, Straussman R, Keshet I, Farkash S, Hecht M, Zimmerman J, Eden E, Yakhini Z, Ben-Shushan E, Reubinoff BE, et al. Polycombmediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet. 2007;39:232–236. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
- 32.Velichutina I, Shaknovich R, Geng H, Johnson NA, Gascoyne RD, Melnick AM, Elemento O. EZH2-mediated epigenetic silencing in germinal center B cells contributes to proliferation and lymphomagenesis. Blood. 2010;116:5247–5255. doi: 10.1182/blood-2010-04-280149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kubosaki A, Tomaru Y, Tagami M, Arner E, Miura H, Suzuki T, Suzuki M, Suzuki H, Hayashizaki Y. Genome-wide investigation of in vivo EGR-1 binding sites in monocytic differentiation. Genome Biol. 2009;10:R41. doi: 10.1186/gb-2009-10-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Watari A, Takaki K, Higashiyama S, Li Y, Satomi Y, Takao T, Tanemura A, Yamaguchi Y, Katayama I, Shimakage M, et al. Suppression of tumorigenicity, but not anchorage independence, of human cancer cells by new candidate tumor suppressor gene CapG. Oncogene. 2006;25:7373–7380. doi: 10.1038/sj.onc.1209732. [DOI] [PubMed] [Google Scholar]
- 35.Van den Abbeele A, De Corte V, Van Impe K, Bruyneel E, Boucherie C, Bracke M, Vandekerckhove J, Gettemans J. Downregulation of gelsolin family proteins counteracts cancer cell invasion in vitro. Cancer Lett. 2007;255:57–70. doi: 10.1016/j.canlet.2007.03.023. [DOI] [PubMed] [Google Scholar]
- 36.Wittamer V, Franssen JD, Vulcano M, Mirjolet JF, Le Poul E, Migeotte I, Brezillon S, Tyldesley R, Blanpain C, Detheux M, et al. Specific recruitment of antigen-presenting cells by chemerin, a novel processed ligand from human inflammatory fluids. J Exp Med. 2003;198:977–985. doi: 10.1084/jem.20030382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li DQ, Wang L, Fei F, Hou YF, Luo JM, Zeng R, Wu J, Lu JS, Di GH, Ou ZL, et al. Identification of breast cancer metastasis-associated proteins in an isogenic tumor metastasis model using two-dimensional gel electrophoresis and liquid chromatography-ion trap-mass spectrometry. Proteomics. 2006;6:3352–3368. doi: 10.1002/pmic.200500617. [DOI] [PubMed] [Google Scholar]
- 38.Lomnytska MI, Becker S, Bodin I, Olsson A, Hellman K, Hellstrom AC, Mints M, Hellman U, Auer G, Andersson S. Differential expression of ANXA6, HSP27, PRDX2, NCF2, and TPM4 during uterine cervix carcinogenesis: diagnostic and prognostic value. Br J Cancer. 2011;104:110–119. doi: 10.1038/sj.bjc.6605992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang L, Darling J, Zhang JS, Liu W, Qian J, Bostwick D, Hartmann L, Jenkins R, Bardenhauer W, Schutte J, et al. Loss of expression of the DRR 1 gene at chromosomal segment 3p21.1 in renal cell carcinoma. Genes Chromosomes Cancer. 2000;27:1–10. doi: 10.1002/(sici)1098-2264(200001)27:1<1::aid-gcc1>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 40.Burton ER, Gaffar A, Lee SJ, Adeshuko F, Whitney KD, Chung JY, Hewitt SM, Huang GS, Goldberg GL, Libutti SK, et al. Downregulation of Filamin A interacting protein 1-like is associated with promoter methylation and induces an invasive phenotype in ovarian cancer. Mol Cancer Res. 2011;9:1126–1138. doi: 10.1158/1541-7786.MCR-11-0162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 42.Ball MP, Li JB, Gao Y, Lee JH, LeProust EM, Park IH, Xie B, Daley GQ, Church GM. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat Biotechnol. 2009;27:361–368. doi: 10.1038/nbt.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Caterino M, Ruoppolo M, Fulcoli G, Huynth T, Orru S, Baldini A, Salvatore F. Transcription factor TBX1 overexpression induces downregulation of proteins involved in retinoic acid metabolism: a comparative proteomic analysis. J Proteome Res. 2009;8:1515–1526. doi: 10.1021/pr800870d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ryckebusch L, Bertrand N, Mesbah K, Bajolle F, Niederreither K, Kelly RG, Zaffran S. Decreased levels of embryonic retinoic acid synthesis accelerate recovery from arterial growth delay in a mouse model of DiGeorge syndrome. Circ Res. 2010;106:686–694. doi: 10.1161/CIRCRESAHA.109.205732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Touma SE, Perner S, Rubin MA, Nanus DM, Gudas LJ. Retinoid metabolism and ALDH1A2 (RALDH2) expression are altered in the transgenic adenocarcinoma mouse prostate model. Biochem Pharmacol. 2009;78:1127–1138. doi: 10.1016/j.bcp.2009.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim H, Lapointe J, Kaygusuz G, Ong DE, Li C, van de Rijn M, Brooks JD, Pollack JR. The retinoic acid synthesis gene ALDH1a2 is a candidate tumor suppressor in prostate cancer. Cancer Res. 2005;65:8118–8124. doi: 10.1158/0008-5472.CAN-04-4562. [DOI] [PubMed] [Google Scholar]
- 47.Rubin MA, Maher CA, Chinnaiyan AM. Common gene rearrangements in prostate cancer. J Clin Oncol. 2011;29:3659–3668. doi: 10.1200/JCO.2011.35.1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Harris RA, Wang T, Coarfa C, Nagarajan RP, Hong C, Downey SL, Johnson BE, Fouse SD, Delaney A, Zhao Y, et al. Comparison of sequencing-based methods to profile DNA methylation and identification of monoallelic epigenetic modifications. Nat Biotechnol. 2010;28:1097–1105. doi: 10.1038/nbt.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.