Abstract
The four serotypes of endemic dengue viruses (DENV) circulate between humans and peridomestic Aedes mosquitoes. At present endemic DENV infect 100 million people per year, and a third of the global population is at risk. In contrast, sylvatic DENV strains are maintained in a transmission cycle between nonhuman primates and sylvatic Aedes species, and are evolutionarily and ecologically distinct from endemic DENV strains. Phylogenetic analyses place sylvatic strains basal to each of the endemic serotypes, supporting the hypothesis that each of the endemic DENV serotypes emerged independently from sylvatic ancestors. We utilized complete genome analyses of both sylvatic and endemic DENV serotype 2 (DENV-2) to expand our understanding of their genetic relationships. A high degree of conservation was observed in both the 5′- and 3′-untranslated genome regions, whereas considerable differences at the nucleotide and amino acid levels were observed within the open reading frame. Additionally, replication of the two genotypes was compared in cultured cells, where endemic DENV strains produced a significantly higher output of progeny in human liver cells, but not in monkey kidney or mosquito cells. Understanding the genetic relationships and phenotypic differences between endemic and sylvatic DENV genotypes may provide valuable insight into DENV emergence and guide monitoring of future outbreaks.
Keywords: Dengue virus (DENV), Sylvatic DENV, Endemic DENV, Phylogenetic and phenotypic analysis
Introduction
The natural history of dengue viruses (DENV, genus Flavivirus, family Flaviviridae) encompasses two ecologically and evolutionarily distinct transmission cycles: sylvatic (forest) enzootic maintenance cycles probably involve transmission among nonhuman primates by arboreal mosquitoes of the genus Aedes, whereas endemic/epidemic cycles (henceforth called endemic) involve human reservoir and amplification hosts and peridomestic Aedes spp. vectors, primarily Ae. aegypti and Ae. albopictus (Gubler, 1998). The four serotypes of endemic DENV (DENV-1–4) are the etiologic agents of several forms of dengue disease, a cause of severe morbidity and mortality among urban and peri-urban populations in the tropics and neotropics worldwide. By current estimates, 100 million infections of dengue fever (DF) occur annually, including up to 500,000 cases of the more severe form of disease called dengue hemorrhagic fever (DHF) with a case fatality rate of up to 5% (Halstead, 1997). While the biology of endemic DENV has been studied extensively (Halstead et al., 1973a,b,c; 1977; Halstead and O'Rourke, 1977; Halstead and Palumbo,1973; Kuberski et al., 1977; Marchette et al., 1973; Rosen, 1958; Rosen and Gubler, 1974; Sabin, 1952; Scherer et al., 1978), much less is known about the sylvatic strains.
DENV sylvatic cycles have been demonstrated in Asia, where serologic evidence as well as virus isolation indicate transmission of sylvatic strains of DENV-1, -2, and -4 among Macaca and Presbytis spp. monkeys, vectored by Ae. niveus mosquitoes (Peiris et al., 1993; Rudnick and Lim, 1986). In West Africa, DENV-2 is the only sylvatic serotype known to circulate, between Erythrocebus patas and possibly other monkeys and various sylvatic Aedes sp., including Ae. taylori, Ae. furcifer, Ae. vitattus, and Ae. luteocephalus, in a sylvatic focus in the vicinity of Kedougou, Senegal (Diallo et al., 2003, 2005; Rodhain, 1991; Saluzzo et al., 1986b) as well as in other nearby nations.
Phylogenetic analyses of DENV based on envelope protein (E) gene nucleotide (nt) sequences representing strains from diverse localities around the world consistently place each sylvatic DENV serotype (DENV-1, -2 and -4) basal in relation to the corresponding endemic DENV serotype (Chang et al., 1994; Rico-Hesse, 1990; Twiddy et al., 2002a; Wang et al., 2000), consistent with an ancestral status of the sylvatic lineages and independent emergence of the endemic lineages from sylvatic ancestors. Furthermore, DENV-2 are grouped into five genotypes (Shurtleff et al., 2001; Twiddy et al., 2002a) based on the phylogenetic relationships of the E gene nucleotide sequences (Chang et al., 1994; Rico-Hesse, 1990; Wang et al., 2000). Varying degrees of adaptation of DENV-2 to distinct geographic and host–vector niches is reflected by the differences in incidence and virulence exhibited by these DENV-2 genotypes (Shurtleff et al., 2001). While initial evidence suggested that emergence of sylvatic DENV-2 into the endemic cycle was likely facilitated by adaptation to replicate more efficiently in the peridomestic mosquito vectors Ae. aegypti and Ae. albopictus (Moncayo et al., 2004), more extensive studies have indicated that this adaptation pertains only to the Asian genotype of endemic DENV-2 (Cologna et al., 2005). Moreover, comparison of the replication of sylvatic and endemic DENV-2 in models for human infection indicate that emergence of endemic DENV-2 from sylvatic progenitors may not have required adaptation to replicate more efficiently in humans (Vasilakis et al., 2007b).
Although limited spillover transmission of sylvatic DENV-2 to humans has been documented in West Africa (Saluzzo et al., 1986a; Zeller et al., 1992), there is no evidence that these sylvatic DENV are involved in secondary amplifications in humans, perhaps due to their confinement to forest habitats and lack of contact with the peridomestic DENV vectors (Diallo et al., 2003). However, recent reports have shown that the gallery forest-dwelling mosquito Ae. furcifer is highly susceptible to sylvatic DENV infection (Diallo et al., 2005) and shows a pattern of movement into peridomestic habitats (villages) in eastern Senegal (Diallo et al., 2003), suggesting that this species may act as a bridge vector for exchange between forest and peridomestic habitats. Furthermore, recent phylogenetic analyses based on complete DENV-2 genomes from human isolates associated with a 1966 outbreak of dengue fever in Nigeria suggest limited spillover outbreaks of sylvatic DENV (defined as the enzootic transmission into a small, localized group of people often confined to a village or a small area due to favorable ecological conditions, such as increased vector densities) (Carey et al., 1971; Vasilakis et al., 2008). Collectively, these data imply a high potential for sylvatic DENV-2 emergence into the endemic cycle.
Previous analyses of endemic DENV-2 based on full genomic sequences (Leitmeyer et al., 1999; Twiddy et al., 2002b) or of endemic and sylvatic DENV-2 based on envelope gene sequences (Wang et al., 2000) or complete sequences (Vasilakis et al., 2007a) identified various mutations that were hypothesized to play a role in DENV Darwinian evolution and pathogenesis. We sought to expand our understanding of the phylogenetic relationships between sylvatic and endemic DENV-2 by performing analyses of full genomic sequences, focusing mainly on isolates of mostly low passage histories. Moreover, we analyzed the distribution of specific mutations that are likely to contribute to phenotypic differences. We included in this study 25 endemic DENV-2 complete genomic sequences from GenBank that represent the full genotypic diversity of this serotype. We also included the complete genome sequences of 15 West African and one Southeast Asian DENV-2 sylvatic isolates (Table 1) utilized in our recent studies (Vasilakis et al., in press, a; 2008). To better link genetic and phenotypic variation, we also measured the replication of a subset of endemic and sylvatic isolates, including sylvatic strains isolated from humans, in both mammalian and mosquito cells in culture.
Table 1.
Passage history of DENV-2 strains
| Virus strain | Epidemiological typea | Host | Passage historyb | Location | Year | GenBank accession no. |
|---|---|---|---|---|---|---|
| 16681c | Endemic | Human | BSC-1 –X, MK2 – 6, Rh. Macaque – 1, Tx.amboinensis – 2, | Thailand | 1964 | U87411 |
| C6/36 – 4, MK2 – 1, C6/36 – 1 | ||||||
| 1349c | Endemic | Human | SM – 2, C6/36 – 2 | Burkina Faso | 1982 | EU056810 |
| IQT-1950 | Endemic | Human | C6/36 – 2 | Peru | 1995 | EU056811 |
| 1328 | Endemic | Human | Mosq - 2, C6/36 – 1 | Puerto Rico | 1977 | EU056812 |
| TVP1915c | Endemic | Ae. aegypti | SM – 4, C6/36 – 1 | Burkina Faso | 1986 | N/A |
| PM33974 | Sylvatic | Ae. africanus | Tx.amboinensis 1, C6/36 – 2 | Guinea | 1981 | EF105378 |
| DakAr 578 | Sylvatic | Ae. taylori | SM – 8, C6/36 – 1 | Cote d'Ivoire | 1980 | EF105380 |
| DakAr 510c | Sylvatic | Ae. taylori | SM – 4, C6/36 – 1 | Cote d'Ivoire | 1980 | EF105381 |
| DakAr 2039 | Sylvatic | Ae. luteocephalus | SM – 6, C6/36 – 1 | Burkina Faso | 1980 | EF105382 |
| DakAr A1247 | Sylvatic | Ae. taylori | SM – 5, C6/36 – 1 | Cote d'Ivoire | 1980 | EF105383 |
| DakAr HD10674c | Sylvatic | Human | SM – 25, mosq – 1 | Senegal | 1970 | EF105384 |
| DakAr D20761 | Sylvatic | Ae. luteocephalus | SM – 8, C6/36 – 1 | Senegal | 1974 | EF105385 |
| DakAr A2022 | Sylvatic | Ae. africanus | SM – 6, C6/36 – 1 | Burkina Faso | 1980 | EF105386 |
| DakAr D75505c | Sylvatic | Ae. luteocephalus | AP61 – 5, C6/36 – 1 | Senegal | 1991 | EF457904 |
| DakAr 141069 | Sylvatic | Ae. luteocephalus | AP61 – 1, C6/36 – 1 | Senegal | 1999 | EF105389 |
| DakAr 141070 | Sylvatic | Ae. luteocephalus | AP61 – 5, C6/36 – 1 | Senegal | 1999 | EF105390 |
| IBH11208c | Sylvatic | Human | SM – 30, C6/36 – 1 | Nigeria | 1966 | EF105387 |
| IBH11234c | Sylvatic | Human | SM – 16, C6/36 – 1 | Nigeria | 1966 | EU003591 |
| IBH11664 | Sylvatic | Human | SM – 5, C6/36 – 1 | Nigeria | 1966 | EF105388 |
| P8-1407c | Sylvatic | Sentinel Monkey | SM – 3, C6/36 – 2 | Malaysia | 1970 | EF105379 |
DENV isolates obtained from the UTMB World Reference Center for Emerging Viruses and Arboviruses and amplified once on C6/36 mosquito cells to achieve sufficiently high titers for further evaluation.
Endemic denotes human or Ae. aegypti isolates or strains that are associated with peridomestic transmission.
SM — suckling mouse; C6/36 — Ae. albopictus cell line; MK2 — Rhesus monkey kidney cells; BSC-1 — African green monkey kidney cells; AP61 — Aedes pseudoscutellaris cell line; N/A — Not available.
Virus strains selected for phenotypic characterization.
Results and discussion
Phylogenetic analyses
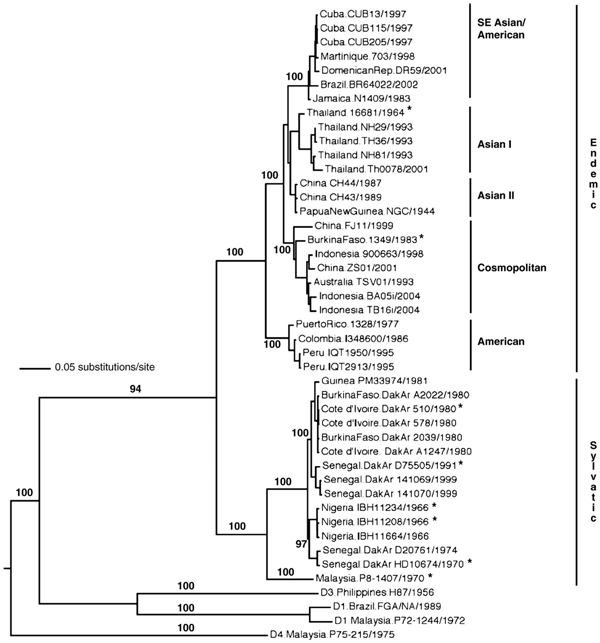
Phylogenetic analyses of 40 DENV-2 isolates (25 endemic and 15 sylvatic representing strains from diverse localities throughout the tropics and neotropics were performed based on complete genome sequences. The isolates selected for this study are representative members of all previously-defined major genotypes (Holmes and Twiddy, 2003; Twiddy et al., 2002a; Wang et al., 2000), and as expected they clustered into six distinct clades in this study as well (Fig. 1). Consensus trees based on Maximum-likelihood (ML) or Bayesian analyses exhibited identical strain topologies and only minor differences in bootstrap values and thus we present only Bayesian phylogenies. Sylvatic DENV-1 and -4, as well as endemic DENV-1 and -3 were used as outgroups to root the tree (Fig. 1). Considerable genetic diversity was apparent within each of the major DENV-2 genotypes, reflecting a continual process of divergence within each lineage. All sylvatic DENV-2 strains were distinct from and basal to the endemic clade, and the Malaysian and West African sylvatic DENV-2 strains were genetically distinct. All analyses also delineated a chronological divide among isolates within the sylvatic African DENV-2 clade; viruses isolated pre-1980 formed a group distinct from all post-1980 isolates. This observation supports recent evidence of rapid sylvatic DENV turnover (rapid generation of viral diversity) due to their high nucleotide substitution rates (Vasilakis et al., 2007a).
Fig. 1.
Phylogenetic analysis of sylvatic DENV-2 isolates. Phylogenetic tree derived from complete genome nucleotide sequences of sylvatic and representative endemic DENV-2 strains using Bayesian analysis. Genomic sequences from sylvatic dengue serotypes 1 (strain P72-1244) and 4 (strain P75-215), as well as endemic serotype 1 (strain FGA) and 3 (strain H87) were used as outgroups to root the DENV-2 tree. Numbers indicate bootstrap values for monophyletic groups to the right. Asterisks indicate DENV strains used to evaluate growth kinetics.
The endemic DENV-2 phylogeny also showed a subdivision between the American and Asian genotypes. Within the Asian genotype, two further sub-lineages represented the cosmopolitan and the Asian clades, with bootstrap support of 100%. The former included isolates that are geographically dispersed throughout the tropics (Twiddy et al., 2002a), whereas the latter included isolates found exclusively in Asia (Asian) as well as recent isolates that have been implicated with the appearance of DHF and displacement of American genotypes in the Americas (Asian/American) Cologna et al., 2005) (Fig. 1).
Genetic analysis
The DENV genome is a single stranded RNA approximately 10.7 kilobases (kb) long, capped and of positive polarity, comprising a single open reading frame (ORF) flanked by non-coding regions (NCR) at both the 5′- and 3′-ends. The ORF encodes three structural proteins: capsid (C), premembrane/membrane (prM) and envelope (E), as well as seven nonstructural proteins, NS1, NS2A, NS2B, NS3, NS4A, NS4B and NS5. The 5′-NCR is about 100 nucleotides long, whereas the 3′-NCR is approximately 400 nt long.
5′- and 3′-NCRs
As demonstrated in previous studies, both 5′- and 3′-NCRs were highly conserved (Hahn et al., 1987; Proutski et al., 1997; Rauscher et al., 1997; Thurner et al., 2004). Both endemic and sylvatic strains showed high similarity within and between groups in both the 5′- and 3′-NCR sequences (Table 2 and Table 3, Supplemental Table S1), as well as predicted secondary structures (data not shown). The flavivirus 3′-NCR can be subdivided into three subregions: the Variable Region (VR), the core, and the 3′ terminus (Markoff 2003). A number of highly conserved sequence elements have been identified in the core and 3′ terminus, including: (i) the last two plus-strand 3′-terminal nucleotides (UC-3′), which are complimentary to the 5′-terminal nucleotides (5′AG), and play a role in the cyclization of the genome during the early stages of replication (Markoff, 2003; Rice et al., 1985; Wengler, 1981); (ii) the pentanucleotide sequence 5′-CACAG, between nucleotides 43 and 47, which form a closed loop located within the conserved stem and loop structure (3′-SL) of the 3′-NCR terminal 95 nucleotides (Brinton et al., 1986; Hahn et al., 1987; Irie et al., 1989), and are thought to play a significant role during viral replication by facilitating the binding of cellular and/or viral factors to the 3′-SL (Blackwell and Brinton, 1995, 1997; Ta and Vrati, 2000). However, recent evidence suggests that conservation of the pentanucleotide sequence may not be required for replication (Silva et al., 2007); (iii) the cyclization sequence (CYC) 5′-ATATTGAC, a fully conserved sequence among DENV genotypes between nucleotides 97 and 102, located within the longer semi-conserved motif CS1 (Khromykh et al., 2001a; Markoff, 2003); and (iv) the tandem repeat sequences CS2, RCS2, TL1 (5′-GAAGCUGUACG) and TL2 (5′-GAAGCUGUA), the former two located approximately 20 and 90 nucleotides upstream of CS1, whereas the latter two are located upstream of RCS2 and between RCS2 and CS2, respectively. Several investigators have suggested that the conserved sequences (indicated above in bold) within TL1 and TL2 could interact through hydrogen bonding with the conserved sequences 5′-ACAGC and 5′-GCUGC (located downstream of CS2 and between RCS2 and CS2 respectively) to form pseudoknots (Olsthoorn and Bol, 2001; Proutski et al., 1997; Rauscher et al., 1997), possibly to facilitate the proper folding of the 3′-NCR, thus contributing to its proper function in flavivirus replication. However, a recent study suggested that substantial distortion of the pseudoknot secondary structure by mutations may be tolerated without abolishing viral replication (Romero et al., 2006).
Table 2.
Genetic sequence of selected dengue-2 sylvatic strains
| Virusa | Epidemiological type | Location | Genome length | % nt identityb | % aa identityb | ORF (nt) | 5′-NCR (nt) | 3′-NCR (nt) |
|---|---|---|---|---|---|---|---|---|
| PM33974 | Sylvatic | Guinea | 10722 | – | – | 10176 | 96 | 450 |
| P8-1407 | Sylvatic | Malaysia | 10719 | 87.7% | 98.6% | 10176 | 96 | 447 |
| DakAr510 | Sylvatic | Côte d' Ivoire | 10722 | 98.8% | 99.8% | 10176 | 96 | 450 |
| DakAr2039 | Sylvatic | Burkina Faso | 10722 | 98.8% | 99.7% | 10176 | 96 | 450 |
| DakAr A1247 | Sylvatic | Côte d' Ivoire | 10700 | 98.6% | 99.8% | 10176 | 96 | 428 |
| DakAr578 | Sylvatic | Côte d' Ivoire | 10722 | 98.7% | 99.8% | 10176 | 96 | 450 |
| DakAr HD10674 | Sylvatic | Senegal | 10724 | 96.7% | 99.4% | 10176 | 96 | 452 |
| DakAr A2022 | Sylvatic | Burkina Faso | 10721 | 98.8% | 99.9% | 10176 | 95 | 450 |
| DakArD20761 | Sylvatic | Senegal | 10724 | 96.6% | 99.3% | 10176 | 96 | 452 |
| DakArD77505 | Sylvatic | Senegal | 10724 | 97.4% | 99.6% | 10176 | 96 | 452 |
| DakAr141069 | Sylvatic | Senegal | 10723 | 97.3% | 99.5% | 10176 | 96 | 450 |
| DakAr141070 | Sylvatic | Senegal | 10717 | 97.2% | 99.5% | 10170 | 96 | 442 |
| IBH 11208 | Sylvatic | Nigeria | 10709 | 96.6% | 99.2% | 10176 | 96 | 437 |
| IBH 11234 | Sylvatic | Nigeria | 10515c | 94.8%d | 99.1% | 10176 | 96 | 243c |
| IBH 11664 | Sylvatic | Nigeria | 10711 | 96.6% | 99.2% | 10176 | 96 | 437 |
Low passage DENV isolates were obtained from the UTMB World Reference Center for Emerging Viruses and Arboviruses and amplified once on C6/36 mosquito cells to achieve sufficiently high titers for further evaluation.
Denotes % identity compared to West African PM33974 sylvatic strain.
Partial sequence.
% nt identity was based on partial sequence comparison.
Table 3.
Nucleotide identity (%) between the consensus sequence for the 5′-UTR (above the diagonal) and 3′-UTR (below the diagonal) of the Asian, American endemic, and sylvatic DENV-2
| 5′-UTR | Asian | American | Sylvatic |
|---|---|---|---|
| Asian | 100.0 | 96.9 | 92.8 |
| American | 92.7 | 100.0 | 91.8 |
| Sylvatic | 91.2 | 89.4 | 100.0 |
All of these sequence features were conserved in both the endemic and sylvatic DENV-2 strains. However, in the VR, the Asian and American endemic genotypes exhibited 2 and 10 nt deletions, respectively (Supplemental Table S2). It has been suggested that these deletions were introduced during their evolution from sylvatic progenitors (Leitmeyer et al., 1999; Shurtleff et al., 2001). Moreover sylvatic genotypes IBH11208 and IBH11664 contained a 14 nt deletion in this region. Among other flaviviruses, this region may act as a spacer to protect the well-conserved and structurally important distal 3′ terminus, which exhibits similar variability (Proutski et al., 1999).
Open Reading Frame (ORF)
Multiple sequence alignment of the ORFs of the endemic and sylvatic genotypes, and comparison of the sylvatic sequences to the prototype Southeast Asian endemic 16681 sequence, revealed a large number of nucleotide differences, many of which were nonsynonymous (Table 4).
Table 4.
Summary of consistent amino acid substitutions between sylvatic and endemic Southeast Asian, American DENV-2 genotypes
| Gene | Sitea | Typeb | Amino acid substitution | Comment |
|---|---|---|---|---|
| Capsid | 26 | E | I→V | |
| 79 | E | S→N | ||
| 104 | E | F→M/V/I | P8-1407 F→L | |
| 105 | E | L→I | ||
| 108 | E | V→L/M | P8-1407 V→I | |
| prM | 16 | E | K→R/I | |
| 17 | E | N→Q | P8-1407 N→H | |
| 28 | E-Am | K→E | ||
| 29 | E | N→D | P8-1407 N→D | |
| 31 | E-Am | I→T | ||
| 31 | E-As | I→V | P8-1407 I→M | |
| 49 | E | V→I | P8-1407, IBH11208 V→I | |
| 55 | E-As | F→L | ||
| 57 | E-As | K→R | ||
| 81 | E | S→T | ||
| 82 | E | S→T/A | P8-1407 S→T | |
| M (mature) | 28 | E | Q→H | |
| 44 | E | L→I/M | ||
| 48 | E | V→I | P8-1407 V→I | |
| 61 | E-Am | A→V/A | ||
| 70 | E-Am | V→I | ||
| E | 59 | E | F→Y | |
| 71 | E-Am | E→D | ||
| 81 | E-Am | S→T | ||
| 83 | E | V→N/K | ||
| 93 | E | R→K | P8-1407 R→K | |
| 122 | E | L→K | ||
| 124 | E | K→N | P8-1407 K→N | |
| 129 | E-Am | V→I | ||
| 139 | E-Am | I→V | ||
| 162 | E-As | V→I | ||
| 171 | E | A→T | P8-1407 A→T | |
| 181 | E | I→V | ||
| 236 | E | M→T | ||
| 247 | E | R→K | P8-1407 R→K | |
| 276 | E | I→L | P8-1407 I→L | |
| E | 279 | E | M→T | P8-1407 M→T |
| 330 | E | D→G | P8-1407 D→G | |
| 345 | E | K→R | ||
| 365 | E | I→V | ||
| 379 | E | V→I | P8-1407, IBH11208 V→1 | |
| 390 | E-Am | N→D | ||
| 425 | E | I→L | P8-1407 I→L | |
| 432 | E | V→I | ||
| 462 | E | V→I | P8-1407 V→I | |
| 478 | E | T→S | ||
| NS1 | 37 | E | D→E | |
| 73 | E-Am | P→S | ||
| 92 | E | E→D | P8-1407 E→D | |
| 99 | E-Am | A→V | ||
| 101 | E | R→K | P8-1407 R→K | |
| 105 | E | K→R | ||
| 112 | E-As | R→K | P8-1407 R→K | |
| 123 | E | V→M | ||
| 129 | E | Q→H | ||
| 141 | E | T→A | ||
| 146 | E | S→T | ||
| 162 | E | I→V | A2022 I→F | |
| 170 | E-Am | R→K | ||
| 177 | E-As | M/T→V/A | ||
| 178 | E | I/V→F | P8-1407 I/V→F | |
| 188 | E | V→I | ||
| 191 | E | D→N | P8-1407 D→N | |
| 205 | E | R→A | ||
| 212 | E | M→I/M | ||
| 224 | E | Y→H | ||
| 227 | E | R→K | ||
| 242 | E-Am | I→V | ||
| 256 | E-Am | Y→N | ||
| 261 | E | Y→H | ||
| 278 | E | E→D | ||
| 347 | E-Am | N→S | ||
| NS2A | 12 | E-As | I→V | |
| 18 | E | L→F | P8-1407 L→F | |
| 33 | E | L→I | ||
| NS2A | 38 | E | I→V | |
| 51 | E | K→R | ||
| 57 | E | I→M/V | ||
| 58 | E | I→V | P8-1407 I→V | |
| 63 | E | A→T | ||
| 67 | E | E→D | ||
| 68 | E | M→I | ||
| 82 | E | R→K | P8-1407 R→K | |
| 89 | E | V→A | P8-1407 V→A | |
| 102 | E | L→M | P8-1407 L→M | |
| 104 | E | A→T/A | P8-1407 A→T | |
| 108 | E | V→I | ||
| 115 | E-Am | N→T | ||
| 115 | E-As | N→S | ||
| 118 | E | G→E | P8-1407 G→E | |
| 120 | E-Am | V→I | ||
| 127 | E | I→L/W | ||
| 131 | E | I→M | ||
| 139 | E | S→N/K | ||
| 149 | E | V→I | P8-1407 V→I | |
| 152 | E | M→I | ||
| 155 | E | I→V | ||
| 158 | E | V→A | P8-1407 V→A | |
| 159 | E | M→V | P8-1407 T→V | |
| 163 | E | H→N | ||
| 168 | E | G→S | P8-1407 G→S | |
| 174 | E | A→V/A | IBH11208 A→V | |
| 189 | E | T→A | ||
| 195 | E | V→A | P8-1407, 20761, 10674 V→A | |
| 214 | E | P→T | ||
| 215 | E | N→S | ||
| 217 | E | T→K | ||
| NS2B | 55 | E | K→R | |
| 57 | E-Am | A→T | ||
| 58 | E | E→D | ||
| 62 | E-Am | E→D | ||
| 76 | E | V→I | ||
| 126 | E | T→V | ||
| NS3 | 13 | E | M→V | |
| 28 | E | R→K | ||
| 55 | E | V→M | P8-1407 V→M | |
| 117 | E-Am | K→R | ||
| 140 | E | I→V | ||
| 160 | E | T→A | ||
| 164 | E | S→A | ||
| 172 | E | V→I | P8-1407 V→M | |
| 186 | E | K→R | ||
| 213 | E | R→K/N | ||
| 248 | E | K→R | ||
| 273 | E-As | I→V | P8-1407 I→V | |
| 335 | E | V→M/I | P8-1407 V→M | |
| 351 | E | I→V | ||
| 353 | E | N→D | ||
| 366 | E | R→K | ||
| 367 | E-Am | A→T | ||
| 381 | E-As | R→K | ||
| 392 | E | T→S | P8-1407 T→S | |
| 395 | E | I→V | P8-1407 I→V | |
| 442 | E | V→I | P8-1407 V→I | |
| 466 | E-Am | K→R | ||
| 538 | E | K→R | P8-1407 K→R | |
| 552 | E | S→A | ||
| 561 | E | K→R | P8-1407 K→R | |
| 568 | E-Am | I→T | ||
| 569 | E-As | R→K/Q | ||
| 577 | E | M→V | ||
| 581 | E | V→I | ||
| 600 | E | T→I | ||
| NS4A | 5 | E | S→N | |
| 21 | E | T→A | ||
| 33 | E | S→T | P8-1407 S→T | |
| 36 | E | M→A/T | P8-1407 M→T | |
| 39 | E-Am | R→K | ||
| 59 | E | G→T | ||
| 78 | E | V→I | ||
| 93 | E | G→I/S | ||
| NS4A | 138 | E-Am | V→I | P8-1407 V→I |
| 144 | E | L→V | ||
| NS4B | 17 | E-As | G→S | |
| 17 | E-Am | G→H/N | ||
| 24 | E | I/T→S | P8-1407 I→A | |
| 155 | E | E→D | ||
| 201 | E-Am | I→V | ||
| 211 | E | K→R | ||
| 241 | E | R→K | P8-1407 R→K | |
| 245 | E-As | S→N | ||
| 246 | E | A→T | ||
| NS5 | 3 | E | T→N | |
| 21 | E | R→K | ||
| 71c | E | A→T | ||
| 134c | E-Am | T→V | ||
| 138c | E | R→K | ||
| 172 | E | G→N | ||
| 173 | E | S→N | ||
| 179 | E-As | V→I | ||
| 192 | E-As | R→K | ||
| 200 | E | F→Y | ||
| 268 | E | C→S | ||
| 273 | E | M→L | ||
| 374d | E | R→K | ||
| 382d | E | G→K | P8-1407 G→N | |
| 386d | E | R→K | ||
| 387d | E | R→K | P8-1407 R→K | |
| 389d | E | K→T | ||
| 392d | E | M→I | ||
| 396d | E | A→E | ||
| 99d | E | C→T | ||
| 400d | E-Am | N→K | ||
| 400d | E-As | N→R | ||
| 411 | E | V→I | ||
| 428 | E-Am | E→N/S | ||
| 428 | E-As | E→S/G | ||
| 435 | E | E→D | ||
| 443 | E | D→E | P8-1407 D→E | |
| 521e | E | I→V | ||
| 523e | E | R→K | IBH11208 R→K | |
| 525e | E | A→E | P8-1407 A→E | |
| NS5 | 543e | E-Am | S→I | |
| 543e | E-As | S→L | ||
| 552e | E-As | I→V | ||
| 554e | E | D→N | P8-1407 D→N | |
| 555e | E | Q→H | ||
| 557e | E-Am | E→A | ||
| 559e | E | R→E | P8-1407 R→K | |
| 627e | E | L→I/V | ||
| 632e | E | L→Q | P8-1407 L→Q | |
| 636e | E | P→A/V | P8-1407 P→S | |
| 637e | E-Am | Q→S | ||
| 637e | E-As | Q→T | ||
| 643e | E | H→K/Q | ||
| 644e | E-Am | A→D | ||
| 644e | E-As | A→N | ||
| 647e | E-As | V→A | ||
| 649e | E | E→V | ||
| 655e | E | A/T→S | ||
| 675e | E-Am | R→K | P8-1407 R→K | |
| 675e | E-As | R→S | ||
| 697e | E | K→R | ||
| 700e | E | S→N | P8-1407 S→N | |
| 722e | E-Am | K→T | ||
| 722e | E-As | K→V | ||
| 799e | E-As | S→K/T | P8-1407 S→T | |
| 818e | E-Am | Q→L | ||
| 819e | E | D→E | ||
| 834e | E | D→E | ||
| 871e | E | A→S | ||
| 875e | E | E→N/S | ||
| 890e | E | S→R | P8-1407 S→R | |
| 895e | E | S→A |
Amino acid to the left represents amino acid on sylvatic viruses and to the right on the endemic viruses. Arrows represent directionality of mutation.
Indicates amino acid location from the start of the gene.
E-Am: endemic American genotype; E-As: endemic Asian genotype; E: endemic genotype.
Indicates amino acid located within the S-adenosulmethionine-utilizing methyltransferase (SAM) domain.
Indicates amino acid located within the importin-β-binding domain and the adjacent nuclear localization sequence (NLS) domain.
Indicates amino acid located within the RNA-dependent RNA polymerase (RdRp) domain.
Structural genes
The capsid protein (C) an 11 kDa homodimer protein with an unusually high net charge (Trent, 1977) essential for the specific RNA genome encapsidation (Chang et al., 2001), contained 5 nonsynonymous (I26V, S79N, F104M/V/I, L105I and V108L/M) differences between all endemic versus sylvatic genotypes (Table 4). The a-helical rich content of the capsid structure (Jones et al., 2003; Kuhn et al., 2002; Ma et al., 2004) was fully conserved (V26-L35, K45-T58, A63-W69 and K74-N96) within all genotypes, except for endemic-specific differences at positions 26 (I26V) and 79 (S79N) located within the first and fourth a-helical domain. Similarly conserved was the hydrophobic region 46L-66L. Whether the observed residue changes have any effect in the assembly or processing of the CAP during the late stages of DENV infection is not known.
The membrane protein (prM), a 27–31 kDa N-glycosylated protein cleaved in the trans-Golgi by a resident furin-like protease during late stage virus assembly, contained 14 nonsynonymous mutations of which 4 [K28E, I31T (pr) and A61V and V70I (M)] were specific to American genotypes, and one (I31V) shared among all Asian-endemic genotypes (Table 4). Amino acid 68, the only available site for N-linked glycosylation, the six cysteine residues linked by disulfide bridges (Nowak and Wengler, 1987), as well as the consensus sequence 87R-XK- R90 proximal to the cleavage site were fully conserved.
The envelope (E) protein is a 53 kDa class II N-glycosylated dimeric membrane fusion protein that mediates virus binding and fusion to host cell membrane (Johnson et al., 1994; Modis et al., 2004; Rey et al., 1995), as well as confers protective immune responses by eliciting neutralizing, antifusion, and replication-enhancing antibodies (Chen et al., 1996; Roehrig et al., 1998). Each of the monomer subunits of the E protein is composed of three distinct domains: the centrally located domain I, a β-barrel structure oriented parallel to the viral membrane; domain II, a finger-like structure composed of a pair of discontinuous loops one of which is highly conserved among all flaviviruses functioning as an internal fusion peptide and is stabilized by three disulfide bridges; and the C-terminal domain III (aa 303–395), containing a single disulfide bond and located in the outer lateral surface of the dimer. Domain III has been suggested to contain residues that are responsible for the determination of host range, tropism and virulence among flaviviruses (Rey et al., 1995). Twelve cysteine residues that form the six disulfide bridges, as well as the two N-linked glycosylation sites (N67 and N153) (Johnson et al., 1994) were conserved among all DENV-2 strains. There were 25 evenly distributed, endemic-specific nonsynonymous mutations of which five (D330G, K345R, I365V, V379I and N390D) were located within the critical domain III (Table 4). One of these mutations (N390D), which is shared only by the American genotypes, is located within the putative glycosaminoglycan binding motif (386L-411M) responsible for the binding of DENV onto the host cell membrane via a non-Fc receptor (Chen et al., 1996). N390D has also been implicated as a potential virulence determinant in American DENV genotypes (Leitmeyer et al., 1999), and has been shown to alter virulence in mice (Sanchez and Ruiz, 1996). An amino acid substitution E71D that is also shared by American genotype strains has been implicated in controlling virulence in mice (Bray et al., 1998). Furthermore, residues 381–384, which facilitate the formation of a loop responsible for the serotype-specific attachment of domain III to mosquito cells, were conserved among all DENV-2 genotypes (Hung et al., 2004). Maximum-likelihood analysis indicated that amino acid positions L122K and D330G were subject to weak (possibly immune) positive selective pressure (Twiddy et al., 2002b). Among the DENV-2 genotypes, no nonsynonymous mutations resulted in a net change in charge in the E protein, an important observation since charged residues are important in the interaction of antigenic sites with antibodies (Hasegawa et al., 1992; Leitmeyer et al., 1999; Mandl et al., 1989). Many of the aforementioned amino acid substitutions have been mapped to epitopes that are recognized by elements of the cellular and humoral immune response, and some may play a role in cellular tropism via modification of the receptor binding sites. Disrupting the immune response's ability to detect and/or respond to DENV infection could have a dramatic effect on pathogenesis and transmission. However additional studies in nonhuman primates are needed to clearly define the roles of these mutations in vivo. Moreover, mosquito infections with viruses harboring these mutations have not been studied, effectively ignoring a critical part of the transmission cycle in nature that is essential to understanding how these mutations truly function. Thus, generation of sylvatic infectious clones utilizing reverse genetics may allow the interpretation of the functional importance of these mutations in cell tropism differences in sylvatic and endemic dengue viruses.
Nonstructural genes
Several nonsynonymous mutations were present throughout the nonstructural protein genes. In NS1, a 46 kDa dimeric N-glycosylated glycosyl-phosphatidylinositol (GPI) anchored protein that exists in both intra- and extracellular forms (Jacobs et al., 2000; Winkler et al., 1989), 26 endemic-specific nonsynonymous mutations were present. Of these, seven (P73S, A99V, R170K, M/T177A, I242V, Y256N and M347S) were shared among American genotypes, whereas 2 (R112K and M/T177V) were shared among all Asian genotypes (Table 4). However, the 12 cysteine residues that form disulfide bonds, as well as the two N-linked glycosylation sites (N131 and N218) were fully conserved.
In the NS2A, a 22 kDa protein involved in the coordination of change between RNA packaging and replication (Khromykh et al., 2001b) and possibly antagonism of interferon (IFN) (Jones et al., 2005; Munoz-Jordan et al., 2003), 34 endemic-specific nonsynonymous mutations were present, of which two pairs (I12V and N115V) and (N115T and V120I), were shared only among Asian and American genotypes respectively (Table 4). The lysine residue at position 190, an important determinant for infectious flavivirus production, was fully conserved (Kummerer and Rice, 2002). In NS2B, a membrane-associated 14 kDa protein that serves as a cofactor in the structural activation of the DENV serine protease of NS3 (Erbel et al., 2006; Leung et al., 2001), 6 endemic-specific nonsynonymous mutations were present of which two (A57T and E62D) were shared American genotype strains (Table 4). One of these mutations (V77I) is located within a 12-amino acid hydrophobic region (70GSSPILSITISE81), Φx3Φ, that associates directly with NS3 and results in reduced autoproteolytic efficiency (Brinkworth et al., 1999; Niyomrattanakit et al., 2004).
NS3, a 70 kDa multifunctional protein with a series of enzyme activities such as trypsin-like serine protease, helicase, and RNA triphosphatase (RTPase) (Gorbalenya et al., 1989; Li et al., 1999) involved in the processing of the polyprotein and RNA replication, had 29 endemic-specific, nonsynonymous mutations. Four (K117R, A367T, K466R and I568T) of these mutations were shared among the American genotype strains, whereas 3 (I273V, R381K and R569K/Q) were confined to the Asian genotype (Table 4). The mutation at amino acid position 186 (K186R) is located within a stretch of basic residues (184RKRR187) that is essential for the RTPase activity of NS3 (Li et al., 1999).
NS4A and NS4B, small hydrophobic proteins of 16 and 27 kDa, respectively, with the latter functioning as an interferon (IFN)-signaling inhibitor (Jones et al., 2005; Munoz-Jordan et al., 2003), showed 10 and 8 nonsynonymous mutations respectively. In the NS4A, two (R39K and V138I) mutations shared among the American genotype strains, whereas in the NS4B, two pairs (G17H, I201V) and (G17S, S245N) were shared among the American and Asian genotype strains, respectively (Table 4). None of the NS4B mutations occured at the positively selected sites identified in a recent study of sylvatic DENV-2 evolution (Vasilakis et al., 2007a). However, the G17H/S mutation and 10 out of 13 positively selected sites identified in our previous study (Vasilakis et al., 2007a) fell within a region previously identified as critical for IFN suppression (Munoz-Jordan et al., 2005).
NS5 is a large multifunctional, well-conserved protein of 103 kDa with RNA cap-processing, RNA-dependent RNA polymerase (Ackermann and Padmanabhan, 2001; Egloff et al., 2002), interleukin-8 (IL-8) (Medin et al., 2005) induction and nuclear localization (Pryor et al., 2007; Uchil et al., 2006) activities. It exhibited 56 nonsynonymous mutations, of which 10 and 12 were shared among the American and Asian genotype strains, respectively (Table 4). Three of the nonsynonymous mutations were located within the S-adenosyl-methionine-utilizing methyltransferase (SAM) domain, 10 within the importin-β-binding and nuclear localization sequence (NLS) domains and 43 are within the RNA-dependent RNA polymerase (RdRp). Interestingly, two of the 3 lysine residues (386K–387K) of the distal cluster located within the NLS, and shown to be indispensable for efficient nuclear import of NS5 (Pryor et al., 2007), were substituted with arginine (Table 4). Although these substitutions do not alter the net charge of the cluster, previous studies have demonstrated that net charge changes lead to virus attenuation (Hanley et al., 2002; Pryor et al., 2007), probably due to IFN-antagonist properties of IL-8 production (Khabar et al., 1997; Medin et al., 2005).
Although the functional importance of the nonstructural gene mutations is not yet known, reverse genetic techniques may elucidate their role, for example, in the emergence of endemic strains from sylvatic progenitors. Of particular interest is the concentration of putative positive selection in NS4B (Vasilakis et al., 2007a), a gene that functions as an interferon (IFN)-signaling inhibitor, and their possible role in distinguishing endemic and sylvatic DENV genotypes.
Phenotypic analyses
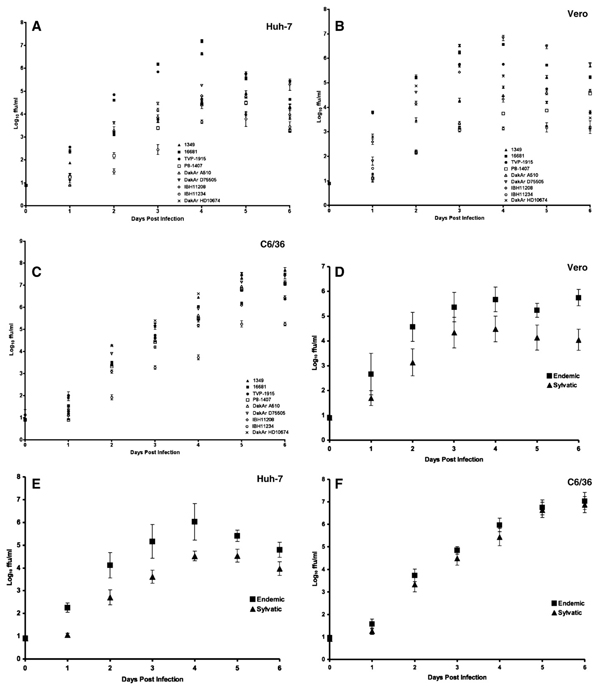
To assess replication in 2 primate cell lines as well as those of a mosquito vector, replication kinetics were compared between endemic and sylvatic DENV-2 genotypes. We included in our analysis 3 endemic strains (West African 1349 and TVP1915, and Southeast Asian 16681) and 6 sylvatic strains, including some isolated from humans in West Africa (Table 1). Titers of most viruses of both endemic and sylvatic genotypes increased until day 4 p.i. in both Huh-7 liver carcinoma cells and Vero (African green monkey kidney) (Figs. 2A and B respectively) and day 5 p.i. in C6/36 mosquito cells (Fig. 2C), after which titers plateaued or declined in all cell types. Huh-7 cells exhibited pronounced cytopathic effects (CPE) by day 4 p.i., which became severe by 6 p.i. due to infection from all strains (data not shown). Although Vero cells exhibited CPE due to infection from all strains, it was less severe than the CPE observed in Huh-7 cells; C6/36 cells exhibited no signs of CPE at all. Overall there was no noticeable difference in CPE in response to sylvatic versus endemic DENV infection in either Huh-7 or Vero cells.
Fig. 2.
Comparative growth curves of DENV-2 strains. (A–C) Virus output of sylvatic (West African DakAr A510, DakAr D75505, IBH11208, IBH11234, DakAr HD10674 and Asian P8-1407) and select endemic (Asian 16681, andWest African 1349 and TVP-1915) DENV-2 genotypes was evaluated on vertebrate cells lines Vero and Huh-7, and mosquito C6/36 cell line up to 6 days p.i. (A–C, respectively). (D–F) Comparison of mean virus output of endemic and sylvatic DENV-2 genotypes on vertebrate cells lines Vero and Huh-7, and mosquito C6/36 cell line up to 6 days p.i. and mosquito cell lines (D–F, respectively). Mean virus output for endemic and sylvatic genotypes are represented by square and triangle bars respectively. All infections were initiated at an MOI of 0.01.
Significant inter-genotypic variation in mean virus output was observed among strains of both endemic and sylvatic DENV-2 genotypes in all cell types (Figs. 2A–C). In Vero cells, repeated measure ANOVA revealed that both isolate (F0.05, 8, 32=554.0, P<0.001) and sampling day (F0.05, 4, 32=6,759.3, P<0.001) affected mean titers, and that there was a significant interaction between the two (F0.05, 32, 72=116.4, P<0.001), indicating that the shape of the replication curve differed among isolates. Moreover, the replication kinetics of every isolate differed from every other in Vero cells (Tukey–Kramer post-hoc test, P<0.05) with only two exceptions: sylvatic strains DakAr HD10674 and DakAr D75505 did not differ significantly nor did sylvatic strains DakAr A510 and P8-1407. In this cell line, sampling days 5 and 6 were excluded from analysis because titers declined after day 4. In Huh-7 cells, both isolate (F0.05, 8, 40=750.7, P<0.001) and sampling day (F0.05, 5, 40=3,3634.0, P<0.001) had a significant impact on titer and there was a significant interaction between the two (F0.05, 40,90=33.3, P<0.001). In this cell line, sampling day 6 was excluded from analysis. Replication dynamics of every isolate differed from every other in Huh-7 cells with five exceptions: sylvatic strain DakAr HD10674 did not differ from sylvatic strains IBH11208 or DakAr A510, sylvatic strain DakAr A510 did not differ from endemic strain 1349 or sylvatic strain IBH11208, and endemic strains 16681 and TVP1915 did not differ. Finally in C6/36 cells both isolate (F0.05, 8, 48=433.6, P<0.001) and sampling day (F0.05, 6, 48=6,678.4, P<0.001) had a significant impact on titer and there was a significant interaction between the two (F0.05, 48, 102=22.0, P<0.001). In C6/36 cells, all but five possible pairwise comparisons were significantly different; the exceptions were endemic strain 1349 and sylvatic strains DakAr D75505 and DakAr HD10674, respectively, sylvatic strain P8-1407 and endemic strain TVP 1915, and sylvatic strain DakAr A510 with both endemic strains 16681 and TVP1915.
The average replication dynamics and maximum titers of endemic and sylvatic strains in each of the three cell types are shown in Figs. 2D–F. In Huh-7 cells, the endemic strains reached significantly higher maximum titers than the sylvatic strains (t=2.6, df=7, P=0.04) and replication dynamicswere significantly different between the two groups (F0.05, 1, 5=8.5, P=0.022). This significant difference between endemic and sylvatic DENV-2 strains in both rapidity of replication and peak titer in Huh-7 cells in culture (Fig. 2A), differs from previous findings in vivo SCID mice engrafted with Huh-7 cells (SCID-Huh-7 mice) (Vasilakis et al., 2007b). It may be inappropriate to compare the two studies directly because different sets of isolates were used to represent the sylvatic and endemic ecotypes. Of particular importance, isolates from the American genotype of endemic DENV-2 were used in the in vivo study; to date several studies, including Vasilakis et al. (2007b) have reported that the American DENV-2 genotype achieves significantly lower titers in a variety of mammalian and mosquito models relative to the Asian I genotype (Armstrong and Rico-Hesse, 2003; Cologna et al., 2005; Cologna and Rico-Hesse, 2003; Vasilakis et al., 2007b). In Vero and C6/36 cells, neither maximum titer nor replication dynamics differed between the two genotypes (P>0.20 for all comparisons). These data support the previous conclusion that extant, sylvatic DENV strains could re-emerge into the human population (Vasilakis et al., 2007b).
Conclusions
Our phylogenetic analyses based on complete genome sequences demonstrate that sylvatic DENV-2 isolates are evolutionarily distinct from endemic DENV-2 isolates, supporting previous classification of DENV-2 into two discrete ecotypes. Although our genetic analyses revealed considerable differences at both the nucleotide and protein levels, several key elements (i.e. NCR secondary structures, disulfide bridges, glycosylation sites) were fully conserved.
Several studies have suggested the importance of the cyclization sequences in the replication of flaviviral genomes (Khromykh et al., 2001a; Khromykh and Westaway, 1997; Men et al., 1996; You and Padmanabhan, 1999). It is thought that both 5′- and 3′-NCR play an indispensable role throughout viral infection; in early stages facilitating the initiation of negative-strand synthesis and translation, whereas in late stage replication allowing for strand-switching thus facilitating the synthesis of plus-strand progeny RNAs and possibly their packaging in progeny virus particles. Thus, nucleotide changes in the 5′- and/or 3′-NCRs, as well as in the ORF, could alter the virus phenotype and virulence (Hanley et al., 2004; Romero et al., 2006; Sirigulpanit et al., 2007; Tajima et al., 2007; Whitehead et al., 2007). Furthermore, computer-generated (Mathews et al., 1999; Zuker, 2003) secondary structure predictions predict the involvement of these cis interactions in either the 5′- or 3′-NCRs. Such structures may also interact in trans, leading to the formation of double stranded panhandle structures that share similar shape and thermal energy (Markoff, 2003). Overall, in our analyses no differences of obvious importance in the 5′- and/or 3′-NCR were detected.
The roles of the amino acid substitutions revealed by our analyses in the emergence of endemic viruses from sylvatic progenitors, as well as on the transmissibility of sylvatic DENV-2 in the peridomestic mosquito vectors Ae. aegypti and Ae. albopictus, are not known and merit further study. Our replication analyses in mammalian and mosquito cell lines of select endemic and sylvatic genotypes, representing diverse localities throughout the tropics and neotropics, demonstrated no significant differences in peak virus outputs, implying the potential of sylvatic emergence into the endemic transmission cycle. Similarly, previous studies examining the fitness of the ancestral sylvatic DENV-2 compared to currently circulating endemic/epidemic strains, using surrogates for human infection (Vasilakis et al., 2007b) and experimental infections of vectors (Diallo et al., 2008, 2005) (K. Hanley, N. Vasilakis unpublished data), suggest that little or no adaptation was required for the initial emergence of endemic DENV-2 strains. This implies that the sylvatic cycles in Asia and West Africa will remain a potential source of re-emergence. Although several promising DENV vaccines under development may lead to the eradication of endemic DENV strains in the future (Whitehead et al., 2007), the sylvatic strains are not amenable to control and most likely remain a potential reservoir for re-emergence of endemic strains. Nonetheless, human herd immunity from prior exposure to endemic DENV strains as well as sterilizing immunity generated by live attenuated vaccine candidates is capable of sylvatic strain neutralization (Vasilakis et al., in press), indicating that sustained vaccination could prevent re-emergence. The above-described studies, albeit with imperfect models of human infection and testing of limited DENV strains, demonstrate that these viruses exhibit a high degree of phenotypic strain variation with substantial phenotypic overlap between ecotypes.
Utilization of reverse genetics to engineer specific mutations identified in this study into a sylvatic DENV-2 genome may provide valuable insights into the role of particular genes or amino acids in the emergence of endemic viruses from sylvatic progenitors. Although, evaluation of every observed mutation by reverse genetics would not be realistic or feasible, generation and evaluation of mutant viruses based on mutations located within ‘hot spots’ such as, in the NS4B, or E could provide clues for the genetic etiology of DENV emergence, as well as whether or not these mutations distinguish endemic and sylvatic DENV phenotypes. Moreover, many ecological traits of sylvatic DENV, including the identity of vertebrate hosts in the maintenance and amplification of sylvatic DENV and the degree and mechanism of contact between humans and the sylvatic DENV cycle, have received little attention for the last few decades. Recent evidence demonstrated the ability of sylvatic DENV-2 to cause limited spillover epidemics in urban settings in West Africa (Vasilakis et al., 2008). Therefore comprehensive ecological and epidemiological studies of sylvatic DENV in West Africa and Southeast Asia, where these strains circulate in proximity to human populations, are needed in order to evaluate the human infection potential and disease burden. Such studies will allow characterization of the disease caused by sylvatic DENV strains and quantification of viremia titers to determine the potential for infected people to initiate interhuman transmission if suitable vectors are present. Ecologic studies should also identify potential ‘bridge’ vectors that could introduce sylvatic DENV strains into a human transmission cycle. Although a recent genetic analysis of sylvatic DENV demonstrated that the rate of evolutionary change and pattern of natural selection are similar to those of endemic DENV (Vasilakis et al., 2007a), little is known about the processes that shape DENV evolution in this sylvatic cycle. Understanding the genetic relationships between endemic and sylvatic genotypes may enhance our understanding of dengue emergence, as well as allow us to explore the biological properties of sylvatic genotypes.
Materials and methods
Viruses and sequencing
Mostly low passage DENV isolates (Table 1) were obtained from the UTMB World Reference Center for Emerging Viruses and Arboviruses and amplified once in vitro in C6/36 mosquito cells to achieve sufficiently high titers. The reason for choosing a viral isolate of defined early passage history is mainly due to the error-prone nature of RNA polymerases leading to rapidly accumulating mutations due to selection for replication in cell cultures (Domingo and Holland, 1994; Drake and Holland, 1999; Duarte et al., 1994).
Genomic viral RNA (vRNA) was isolated with the Qiagen viral RNA isolation kit (Qiagen, Valencia). The 5′- and 3′-terminus sequences of the specific genomes were determined by their ligation and sequencing as demonstrated by Mandl et al. (1991). Initial overlapping cDNA fragments and amplicons of these viruses were generated using primer pairs and primers specific to DENV 16681 by utilizing the onestep reverse transcriptase polymerase chain reaction (RT-PCR) (Roche Diagnostics, Indianapolis), in eight overlapping cDNA fragments containing genome nucleotide regions 1–1396, 1333–2688, 2619–4049, 3986–5358, 5305–6736, 6667–8041, 7990–9382, and 9310–10723 (Kinney et al., 1997) (Supplemental Table 3). First strand synthesis was performed at 50 °C for 30 min, whereas amplification underwent for a total of 35 cycles with annealing temperature set at 5 °C below the lowest melting temperature (Tm) of the used PCR primer pairs and extension set at 1 min per 500 nucleotides. Amplified sequences were gel purified and automated sequencing with specific sequencing primers for both strands provided consensus sequences.
Nucleotide sequence accession numbers
The GenBank accession numbers for the genome sequences of DENV viruses used in the phylogenetic analyses are as follows: for DENV-2 strain 900663, accession number AB189122; TB16i, AY858036; BA05i, AY858035; TSV01, AY037116; FJ11, AF359579; CH43, AF204178; NGC, AF038403; CH44, AF204177; 16681, U87411; NH29, AF169678; TH36, AF169679; NH81, AF169688; CUB205, AY702038; CUB115, AY702036; CUB13, AY702034; 703, AF208496; DR59, AB122022; 64022, AF489932; IQT2913, AF100468; 348600, AY702040; and N.1409, M20558. DENV-1 and -3 strains, accession numbers AF226686 and NC_001475 respectively. The following DENV-2 genome sequences were determined in this study: P8-1407, EF105379; PM33974, EF105378; 1349, EU56810; IQT1950, EU56811; 1328, EU56812; DakAr 510, EF105381; DakAr A1247, EF105383; DakAr 2039, EF105382; DakAr A2022, EF105386; DakAr HD10674, EF105384; DakAr D20761, EF105385; DakAr 578, EF105380; DakAr 141069, EF105389; DakAr141070, EF105390; DakAr D75505, EF457904; IBH11208, EF105387; IBH11664, EF105388; and IBH11234, EU003591. DENV-1 and -4 sylvatic strains P72-1244, and P75-215, accession numbers EF457905 and EF457906 respectively. TVP-1915 is an endemic DENV-2 strain isolated from Ae. aegypti in 1986 in Burkina Faso and its sequence is not available.
Phylogenetic analyses
The obtained full genome sequences and representative genome sequences from the GenBank library were aligned using the ClustalW multiple sequence alignment function of MacVector version 8.0 (Accelrys) with default gap penalties. Phylogenetic analyses of the aligned genomic sequenceswere estimated with maximum likelihood and distance/neighbor joining using the PAUP* program version 4.10 (D.L. Swofford, Illinois Natural History Survey, Champaign) under the general time-reversible model of nucleotide substitution. The parameter values used for the substitution type, optimal base composition, proportion of invariable sites, as well as the shape parameter of the Γ distribution of rate variation among sites were estimated from the data and are available upon request. We also utilized Bayesian analysis (MrBayes v3.1.0) where 4 MCMC tree searches of one million generations each were run simultaneously sampling 1 in 100 trees and computing a 50% majority-rule consensus tree out of the last 9800 sampled trees. Although we observed identical strain topologies with either method, minor differences were observed among the bootstrap values and thus we present only Bayesian phylogenies. Homologous genome sequences from DENV-1, -3, and -4 were used as outgroups to root the DENV tree. Bootstrapping obtained by the Bayesian analysis was used to place confidence values on grouping within the consensus tree (Felsenstein, 1985). Character evolution was traced using Maclade 4.0 (Sinauer Associates, Sunderland, MA).
Viral kinetics
Comparative replication curves of select sylvatic (West African DakAr D77505, DakAr HD10674, IBH11234, IBH11208, DakAr 510 and Asian P8-1407) and endemic (Asian 16681, West African 1349 and TVP-1915) DENV-2 were generated in triplicate on simian kidney (Vero), human liver (Huh-7) and mosquito epithelial (C6/36) cells. Vero, Huh-7 and C6/36 cells, at 2.5×105, 2.5×105, and 5.0×105 cells per well respectively (~80% confluency), were plated in 12-well plates and infected with a multiplicity of 0.01 focus forming units per cell (ffu/cell), in triplicate. Three 12-well plates containing 80% confluent Vero, mosquito C6/36 or Huh-7 cell monolayers were also prepared and cell number was accurately determined. Human hepatoma Huh-7 cells (clone JTC-39) were obtained from the Japanese Health Sciences Foundation, Osaka. Viruses were diluted in minimal essential medium (MEM) supplemented with 5% heat inactivated fetal bovine serum (FBS), 2 mM l-Glutamine, 1% nonessential amino acids (NEAA), and 50 mg/ml penicillin/streptomycin (Invitrogen, Carlsbad), at an MOI of 0.01 ffu/cell are infected simultaneously in triplicate. Infected dishes were incubated for one hour with periodic gentle rocking to facilitate virus adsorption at 37 °C. Viral inocula were removed and cell monolayers were washed thrice with PBS to remove un-adsorbed virus. Two milliliters of complete cell media (MEM supplemented with 5% heat inactivated FBS, 2 mM l-Glutamine, 1% nonessential amino acids (NEAA), and 50 mg/ml penicillin/streptomycin) was then added and dishes were incubated at 29 °C or 37 °C for the mosquito or mammalian cell lines respectively. At various times (1, 2, 3, 4, 5 and 6 days) post infection, virus from individual wells were harvested, and purified by low speed centrifugation. Virus samples representing the various time points were assayed immediately in C6/36 cells to determine virus titer by focus forming immunoassay (FFA). Virus yield at each timepoint was recorded as ffu/cell, represented as the ratio of the total amount of virus present in the sample by the number of cells originally infected.
Focus forming assays and immunostaining
Ten-fold serial dilutions of virus in minimal essential medium (MEM) supplemented with 2% heat inactivated FBS and antibiotics (Invitrogen, Carlsbad), were added to confluent C6/36 cell monolayers attached to 12-well Costar plates, and incubated for one hour with periodic gentle rocking to facilitate virus adsorption at 37 °C. Subsequent to hour-long incubation wells were overlayed with 1 ml solution of 0.8% methylcellulose (Sigma-Aldrich, St. Louis) diluted in warm Optimem (Invitrogen, Carlsbad) supplemented with 2% heat inactivated FBS, antibiotics and 1% (w/v) l-glutamine and incubated undisturbed for 4 days at 28 °C. Methylcellulose overlay was aspirated and cell monolayer rinsed once with phosphate buffered saline (PBS), pH 7.4 (Invitrogen, Carlsbad) followed by fixation with a mixture of ice-cold acetone and methanol (1:1) solution and allowed to incubate for 30 min at room temperature (RT). Fixation solution was aspirated and plates were allowed to air dry. Plates were washed thrice with PBS supplemented with 3% FBS, followed by hour-long incubation with a dengue-specific mouse ascitic fluid. Plates were washed thrice followed by hour-long incubation with a secondary antibody conjugated to horseradish peroxidase (HRP) (KPL, Gaithersburg). Detection proceeded with the addition of aminoethylcarbazole (AEC) substrate (ENZO Diagnostics, Farmingdale) prepared according to vendor instructions. Antigen stained cells were counted against a white background and viral titers were recorded as the reciprocal of the highest dilution where adequate plaques (greater than 20 but lower than 90) and expressed as focus forming units per ml (FFU/ml).
Statistical analyses
The replication dynamics of each of the 9 isolates was compared using a repeated-measures ANOVA on the triplicate titer values of each isolate at each timepoint, followed by a Tukey–Kramer post-hoc test to compare individual pairs of isolates. Similarly, the replication curve of endemic and sylvatic isolates was compared using a repeated-measures ANOVA of the average titer value for each of the 6 sylvatic and 3 endemic isolates at each timepoint. Maximum titer values of endemic and sylvatic isolates were compared using a student's t-test. All analyses were conducted using Statview software (SAS Institutes, Inc., Cary, NC).
Acknowledgments
We thank Robert Tesh and Hilda Guzman for kindly providing DENV strains and antisera, Xiaodong Liang for expert technical assistance, Lark Coffey, Paige Adams and Naomi Forrester for their helpful comments on the manuscript, and The Japanese Health Sciences Foundation, Osaka, for providing the human hepatoma Huh-7 cells (clone JTC-39). NV was supported by the Centers for Disease Control and Prevention Fellowship Training Program in Vector-Borne Infectious Diseases, T01/CCT622892. KAH was supported by an NIH-NM-INBRE (P20 RR016480-05) grant. This research was supported in part by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, NIH.
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.virol.2008.04.044.
References
- Ackermann M, Padmanabhan R. De novo synthesis of RNA by the dengue virus RNA-dependent RNA polymerase exhibits temperature dependence at the initiation but not elongation phase. J. Biol. Chem. 2001;276(43):39926–39937. doi: 10.1074/jbc.M104248200. [DOI] [PubMed] [Google Scholar]
- Armstrong PM, Rico-Hesse R. Efficiency of dengue serotype 2 virus strains to infect and disseminate in Aedes aegypti. Am. J. Trop. Med. Hyg. 2003;68(5):539–544. doi: 10.4269/ajtmh.2003.68.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell JL, Brinton MA. BHK cell proteins that bind to the 3′ stem-loop structure of the West Nile virus genome RNA. J. Virol. 1995;69(9):5650–5658. doi: 10.1128/jvi.69.9.5650-5658.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell JL, Brinton MA. Translation elongation factor-1 alpha interacts with the 3′ stem-loop region of West Nile virus genomic RNA. J. Virol. 1997;71(9):6433–6444. doi: 10.1128/jvi.71.9.6433-6444.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray M, Men R, Tokimatsu I, Lai CJ. Genetic determinants responsible for acquisition of dengue type 2 virus mouse neurovirulence. J. Virol. 1998;72(2):1647–1651. doi: 10.1128/jvi.72.2.1647-1651.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkworth RI, Fairlie DP, Leung D, Young PR. Homology model of the dengue 2 virus NS3 protease: putative interactions with both substrate and NS2B cofactor. J. Gen. Virol. 1999;80(Pt 5):1167–1177. doi: 10.1099/0022-1317-80-5-1167. [DOI] [PubMed] [Google Scholar]
- Brinton MA, Fernandez AV, Dispoto JH. The 3′-nucleotides of flavivirus genomic RNA form a conserved secondary structure. Virology. 1986;153(1):113–121. doi: 10.1016/0042-6822(86)90012-7. [DOI] [PubMed] [Google Scholar]
- Carey DE, Causey OR, Reddy S, Cooke AR. Dengue viruses from febrile patients in Nigeria, 1964-68. Lancet 1. 1971;1(7690):105–106. doi: 10.1016/s0140-6736(71)90840-3. [DOI] [PubMed] [Google Scholar]
- Chang GJ, Trent DW, Vorndam AV, Vergne E, Kinney RM, Mitchell CJ. An integrated target sequence and signal amplification assay, reverse transcriptase-PCR-enzyme-linked immunosorbent assay, to detect and characterize flaviviruses. J. Clin. Microbiol. 1994;32(2):477–483. doi: 10.1128/jcm.32.2.477-483.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CJ, Luh HW, Wang SH, Lin HJ, Lee SC, Hu ST. The heterogeneous nuclear ribonucleoprotein K (hnRNP K) interacts with dengue virus core protein. DNA Cell Biol. 2001;20(9):569–577. doi: 10.1089/104454901317094981. [DOI] [PubMed] [Google Scholar]
- Chen Y, Maguire T, Marks RM. Demonstration of binding of dengue virus envelope protein to target cells. J. Virol. 1996;70(12):8765–8772. doi: 10.1128/jvi.70.12.8765-8772.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cologna R, Rico-Hesse R. American genotype structures decrease dengue virus output from human monocytes and dendritic cells. J. Virol. 2003;77(7):3929–3938. doi: 10.1128/JVI.77.7.3929-3938.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cologna R, Armstrong PM, Rico-Hesse R. Selection for virulent dengue viruses occurs in humans and mosquitoes. J. Virol. 2005;79(2):853–859. doi: 10.1128/JVI.79.2.853-859.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diallo M, Ba Y, Sall AA, Diop OM, Ndione JA, Mondo M, Girault L, Mathiot C. Amplification of the sylvatic cycle of dengue virus type 2, Senegal, 1999–2000: entomologic findings and epidemiologic considerations. Emerg. Infect. Dis. 2003;9(3):362–367. doi: 10.3201/eid0903.020219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diallo M, Sall AA, Moncayo AC, Ba Y, Fernandez Z, Ortiz D, Coffey LL, Mathiot C, Tesh RB, Weaver SC. Potential role of sylvatic and domestic African mosquito species in dengue emergence. Am. J. Trop. Med. Hyg. 2005;73(2):445–449. [PubMed] [Google Scholar]
- Diallo M, Ba Y, Faye O, Soumare ML, Dia I, Sall AA. Vector competence of Aedes aegypti populations from Senegal for sylvatic and epidemic dengue 2 virus isolated in West Africa. Trans. R. Soc. Trop. Med. Hyg. 2008;102(5):493–498. doi: 10.1016/j.trstmh.2008.02.010. [DOI] [PubMed] [Google Scholar]
- Domingo E, Holland JJ. Mutation rates and rapid evolution of RNA viruses. In: Morse SS, editor. Evolutionary Biology of Viruses. New York: Raven Press; 1994. pp. 161–184. [Google Scholar]
- Drake JW, Holland JJ. Mutation rates among RNA viruses. Proc. Natl. Acad. Sci. U. S. A. 1999;96(24):13910–13913. doi: 10.1073/pnas.96.24.13910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte EA, Novella IS, Weaver SC, Domingo E, Wain-Hobson S, Clarke DK, Moya A, Elena SF, de la Torre JC, Holland JJ. RNA virus quasispecies: significance for viral disease and epidemiology. Infect. Agents Dis. 1994;3(4):201–214. [PubMed] [Google Scholar]
- Egloff MP, Benarroch D, Selisko B, Romette JL, Canard B. An RNA cap (nucleoside-2′-O-)-methyltransferase in the flavivirus RNA polymerase NS5: crystal structure and functional characterization. EMBO J. 2002;21(11):2757–2768. doi: 10.1093/emboj/21.11.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erbel P, Schiering N, D'Arcy A, Renatus M, Kroemer M, Lim SP, Yin Z, Keller TH, Vasudevan SG, Hommel U. Structural basis for the activation of flaviviral NS3 proteases from dengue and West Nile virus. Nat. Struct. Mol. Biol. 2006;13(4):372–373. doi: 10.1038/nsmb1073. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39(4):783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Gorbalenya AE, Donchenko AP, Koonin EV, Blinov VM. N-terminal domains of putative helicases of flavi- and pestiviruses may be serine proteases. Nucleic Acids Res. 1989;17(10):3889–3897. doi: 10.1093/nar/17.10.3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler DJ. Dengue and dengue hemorrhagic fever. Clin. Microbiol. Rev. 1998;11(3):480–496. doi: 10.1128/cmr.11.3.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn CS, Hahn YS, Rice CM, Lee E, Dalgarno L, Strauss EG, Strauss JH. Conserved elements in the 3′ untranslated region of flavivirus RNAs and potential cyclization sequences. J. Mol. Biol. 1987;198(1):33–41. doi: 10.1016/0022-2836(87)90455-4. [DOI] [PubMed] [Google Scholar]
- Halstead SB. Epidemiology of dengue and dengue hemorrhagic fever. In: Gubler Kuno DJ, Kuno G, editors. Dengue and Dengue Hemorrhagic Fever. Oxon, UK: CBA international; 1997. [Google Scholar]
- Halstead SB, O'Rourke EJ. Dengue viruses and mononuclear phagocytes. I. Infection enhancement by non-neutralizing antibody. J. Exp. Med. 1977;146(1):201–217. doi: 10.1084/jem.146.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halstead SB, Palumbo NE. Studies on the immunization of monkeys against dengue. II. Protection following inoculation of combinations of viruses. Am. J. Trop. Med. Hyg. 1973;22(3):375–381. doi: 10.4269/ajtmh.1973.22.375. [DOI] [PubMed] [Google Scholar]
- Halstead SB, Casals J, Shotwell H, Palumbo N. Studies on the immunization of monkeys against dengue. I. Protection derived from single and sequential virus infections. Am. J. Trop. Med. Hyg. 1973a;22(3):365–374. doi: 10.4269/ajtmh.1973.22.365. [DOI] [PubMed] [Google Scholar]
- Halstead SB, Shotwell H, Casals J. Studies on the pathogenesis of dengue infection in monkeys. I. Clinical laboratory responses to primary infection. J. Infect. Dis. 1973b;128(1):7–14. doi: 10.1093/infdis/128.1.7. [DOI] [PubMed] [Google Scholar]
- Halstead SB, Shotwell H, Casals J. Studies on the pathogenesis of dengue infection in monkeys. II. Clinical laboratory responses to heterologous infection. J. Infect. Dis. 1973c;128(1):15–22. doi: 10.1093/infdis/128.1.15. [DOI] [PubMed] [Google Scholar]
- Halstead SB, O'Rourke EJ, Allison AC. Dengue viruses and mononuclear phagocytes. II. Identity of blood and tissue leukocytes supporting in vitro infection. J. Exp. Med. 1977;146(1):218–229. doi: 10.1084/jem.146.1.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanley KA, Lee JJ, Blaney JE, Jr, Murphy BR, Whitehead SS. Paired charge-to-alanine mutagenesis of dengue virus type 4 NS5 generates mutants with temperature-sensitive, host range, and mouse attenuation phenotypes. J. Virol. 2002;76(2):525–531. doi: 10.1128/JVI.76.2.525-531.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanley KA, Manlucu LR, Manipon GG, Hanson CT, Whitehead SS, Murphy BR, Blaney JE., Jr Introduction of mutations into the non-structural genes or 3′ untranslated region of an attenuated dengue virus type 4 vaccine candidate further decreases replication in rhesus monkeys while retaining protective immunity. Vaccine. 2004;22(25–26):3440–3448. doi: 10.1016/j.vaccine.2004.02.031. [DOI] [PubMed] [Google Scholar]
- Hasegawa H, Yoshida M, Shiosaka T, Fujita S, Kobayashi Y. Mutations in the envelope protein of Japanese encephalitis virus affect entry into cultured cells and virulence in mice. Virology. 1992;191(1):158–165. doi: 10.1016/0042-6822(92)90177-q. [DOI] [PubMed] [Google Scholar]
- Holmes EC, Twiddy SS. The origin, emergence and evolutionary genetics of dengue virus. Infect. Genet. Evol. 2003;3(1):19–28. doi: 10.1016/s1567-1348(03)00004-2. [DOI] [PubMed] [Google Scholar]
- Hung JJ, Hsieh MT, Young MJ, Kao CL, King CC, Chang W. An external loop region of domain III of dengue virus type 2 envelope protein is involved in serotype-specific binding to mosquito but not mammalian cells. J. Virol. 2004;78(1):378–388. doi: 10.1128/JVI.78.1.378-388.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irie K, Mohan PM, Sasaguri Y, Putnak R, Padmanabhan R. Sequence analysis of cloned dengue virus type 2 genome (New Guinea-C strain) Gene. 1989;75(2):197–211. doi: 10.1016/0378-1119(89)90266-7. [DOI] [PubMed] [Google Scholar]
- Jacobs MG, Robinson PJ, Bletchly C, Mackenzie JM, Young PR. Dengue virus nonstructural protein 1 is expressed in a glycosyl-phosphatidylinositol-linked form that is capable of signal transduction. FASEB J. 2000;14(11):1603–1610. doi: 10.1096/fj.14.11.1603. [DOI] [PubMed] [Google Scholar]
- Johnson AJ, Guirakhoo F, Roehrig JT. The envelope glycoproteins of dengue 1 and dengue 2 viruses grown in mosquito cells differ in their utilization of potential glycosylation sites. Virology. 1994;203(2):241–249. doi: 10.1006/viro.1994.1481. [DOI] [PubMed] [Google Scholar]
- Jones CT, Ma L, Burgner JW, Groesch TD, Post CB, Kuhn RJ. Flavivirus capsid is a dimeric alpha-helical protein. J. Virol. 2003;77(12):7143–7149. doi: 10.1128/JVI.77.12.7143-7149.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M, Davidson A, Hibbert L, Gruenwald P, Schlaak J, Ball S, Foster GR, Jacobs M. Dengue virus inhibits alpha interferon signaling by reducing STAT2 expression. J. Virol. 2005;79(9):5414–5420. doi: 10.1128/JVI.79.9.5414-5420.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khabar KS, Al-Zoghaibi F, Al-Ahdal MN, Murayama T, Dhalla M, Mukaida N, Taha M, Al-Sedairy ST, Siddiqui Y, Kessie G, Matsushima K. The alpha chemokine, interleukin 8, inhibits the antiviral action of interferon alpha. J. Exp. Med. 1997;186(7):1077–1085. doi: 10.1084/jem.186.7.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khromykh AA, Meka H, Guyatt KJ, Westaway EG. Essential role of cyclization sequences in flavivirus RNA replication. J. Virol. 2001a;75(14):6719–6728. doi: 10.1128/JVI.75.14.6719-6728.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khromykh AA, Varnavski AN, Sedlak PL, Westaway EG. Coupling between replication and packaging of flavivirus RNA: evidence derived from the use of DNA-based full-length cDNA clones of Kunjin virus. J. Virol. 2001b;75(10):4633–4640. doi: 10.1128/JVI.75.10.4633-4640.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khromykh AA, Westaway EG. Subgenomic replicons of the flavivirus Kunjin: construction and applications. J. Virol. 1997;71(2):1497–1505. doi: 10.1128/jvi.71.2.1497-1505.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinney RM, Butrapet S, Chang GJ, Tsuchiya KR, Roehrig JT, Bhamarapravati N, Gubler DJ. Construction of infectious cDNA clones for dengue 2 virus: strain 16681 and its attenuated vaccine derivative, strain PDK-53. Virology. 1997;230(2):300–308. doi: 10.1006/viro.1997.8500. [DOI] [PubMed] [Google Scholar]
- Kuberski T, Rosen L, Reed D, Mataika J. Clinical and laboratory observations on patients with primary and secondary dengue type 1 infections with hemorrhagic manifestations in Fiji. Am. J. Trop. Med. Hyg. 1977;26(4):775–783. doi: 10.4269/ajtmh.1977.26.775. [DOI] [PubMed] [Google Scholar]
- Kuhn RJ, Zhang W, Rossmann MG, Pletnev SV, Corver J, Lenches E, Jones CT, Mukhopadhyay S, Chipman PR, Strauss EG, Baker TS, Strauss JH. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell. 2002;108(5):717–725. doi: 10.1016/s0092-8674(02)00660-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kummerer BM, Rice CM. Mutations in the yellow fever virus nonstructural protein NS2A selectively block production of infectious particles. J. Virol. 2002;76(10):4773–4784. doi: 10.1128/JVI.76.10.4773-4784.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitmeyer KC, Vaughn DW, Watts DM, Salas R, Villalobos I, de C, Ramos C, Rico-Hesse R. Dengue virus structural differences that correlate with pathogenesis. J. Virol. 1999;73(6):4738–4747. doi: 10.1128/jvi.73.6.4738-4747.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung D, Schroder K, White H, Fang NX, Stoermer MJ, Abbenante G, Martin JL, Young PR, Fairlie DP. Activity of recombinant dengue 2 virus NS3 protease in the presence of a truncated NS2B co-factor, small peptide substrates, and inhibitors. J. Biol. Chem. 2001;276(49):45762–45771. doi: 10.1074/jbc.M107360200. [DOI] [PubMed] [Google Scholar]
- Li H, Clum S, You S, Ebner KE, Padmanabhan R. The serine protease and RNA-stimulated nucleoside triphosphatase and RNA helicase functional domains of dengue virus type 2 NS3 converge within a region of 20 amino acids. J. Virol. 1999;73(4):3108–3116. doi: 10.1128/jvi.73.4.3108-3116.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Jones CT, Groesch TD, Kuhn RJ, Post CB. Solution structure of dengue virus capsid protein reveals another fold. Proc. Natl. Acad. Sci. U. S. A. 2004;101(10):3414–3419. doi: 10.1073/pnas.0305892101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandl CW, Guirakhoo F, Holzmann H, Heinz FX, Kunz C. Antigenic structure of the flavivirus envelope protein E at the molecular level, using tick-borne encephalitis virus as a model. J. Virol. 1989;63(2):564–571. doi: 10.1128/jvi.63.2.564-571.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandl CW, Heinz FX, Puchhammer-Stockl E, Kunz C. Sequencing the termini of capped viral RNA by 5′-3′ ligation and PCR. Biotechniques. 1991;10(4):484–486. [PubMed] [Google Scholar]
- Marchette NJ, Halstead SB, Falkler WA, Jr, Stenhouse A, Nash D. Studies on the pathogenesis of dengue infection in monkeys. 3. Sequential distribution of virus in primary and heterologous infections. J. Infect. Dis. 1973;128(1):23–30. doi: 10.1093/infdis/128.1.23. [DOI] [PubMed] [Google Scholar]
- Markoff L. 5′- and 3′-noncoding regions in flavivirus RNA. In: Monath TP, Chambers TJ, editors. The Flaviviruses: Structure, Replication and Evolution. Advances in Virus Research. vol. 59. San Diego: Elsevier Academic Press; 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 1999;288(5):911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- Medin CL, Fitzgerald KA, Rothman AL. Dengue virus nonstructural protein NS5 induces interleukin-8 transcription and secretion. J. Virol. 2005;79(17):11053–11061. doi: 10.1128/JVI.79.17.11053-11061.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Men R, Bray M, Clark D, Chanock RM, Lai CJ. Dengue type 4 virus mutants containing deletions in the 3′ noncoding region of the RNA genome: analysis of growth restriction in cell culture and altered viremia pattern and immunogenicity in rhesus monkeys. J. Virol. 1996;70(6):3930–3937. doi: 10.1128/jvi.70.6.3930-3937.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modis Y, Ogata S, Clements D, Harrison SC. Structure of the dengue virus envelope protein after membrane fusion. Nature. 2004;427(6972):313–319. doi: 10.1038/nature02165. [DOI] [PubMed] [Google Scholar]
- Moncayo AC, Fernandez Z, Ortiz D, Diallo M, Sall A, Hartman S, Davis CT, Coffey L, Mathiot CC, Tesh RB, Weaver SC. Dengue emergence and adaptation to peridomestic mosquitoes. Emerg. Infect. Dis. 2004;10(10):1790–1796. doi: 10.3201/eid1010.030846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Jordan JL, Sanchez-Burgos GG, Laurent-Rolle M, Garcia-Sastre A. Inhibition of interferon signaling by dengue virus. Proc. Natl. Acad. Sci. U. S. A. 2003;100(24):14333–14338. doi: 10.1073/pnas.2335168100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Jordan JL, Laurent-Rolle M, Ashour J, Martinez-Sobrido L, Ashok M, Lipkin WI, Garcia-Sastre A. Inhibition of alpha/beta interferon signaling by the NS4B protein of flaviviruses. J. Virol. 2005;79(13):8004–8013. doi: 10.1128/JVI.79.13.8004-8013.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niyomrattanakit P, Winoyanuwattikun P, Chanprapaph S, Angsuthanasombat C, Panyim S, Katzenmeier G. Identification of residues in the dengue virus type 2 NS2B cofactor that are critical for NS3 protease activation. J. Virol. 2004;78(24):13708–13716. doi: 10.1128/JVI.78.24.13708-13716.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak T, Wengler G. Analysis of disulfides present in the membrane proteins of the West Nile flavivirus. Virology. 1987;156(1):127–137. doi: 10.1016/0042-6822(87)90443-0. [DOI] [PubMed] [Google Scholar]
- Olsthoorn RC, Bol JF. Sequence comparison and secondary structure analysis of the 3′ noncoding region of flavivirus genomes reveals multiple pseudoknots. Rna. 2001;7(10):1370–1377. [PMC free article] [PubMed] [Google Scholar]
- Peiris JS, Dittus WP, Ratnayake CB. Seroepidemiology of dengue and other arboviruses in a natural population of toque macaques (Macaca sinica) at Polonnaruwa, Sri Lanka. J. Med. Primatol. 1993;22(4):240–245. [PubMed] [Google Scholar]
- Proutski V, Gould EA, Holmes EC. Secondary structure of the 3′ untranslated region of flaviviruses: similarities and differences. Nucleic Acids Res. 1997;25(6):1194–1202. doi: 10.1093/nar/25.6.1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proutski V, Gritsun TS, Gould EA, Holmes EC. Biological consequences of deletions within the 3′-untranslated region of flaviviruses may be due to rearrangements of RNA secondary structure. Virus Res. 1999;64(2):107–123. doi: 10.1016/s0168-1702(99)00079-9. [DOI] [PubMed] [Google Scholar]
- Pryor MJ, Rawlinson SM, Butcher RE, Barton CL, Waterhouse TA, Vasudevan SG, Bardin PG, Wright PJ, Jans DA, Davidson AD. Nuclear localization of dengue virus nonstructural protein 5 through its importin alpha/beta-recognized nuclear localization sequences is integral to viral infection. Traffic. 2007;8(7):795–807. doi: 10.1111/j.1600-0854.2007.00579.x. [DOI] [PubMed] [Google Scholar]
- Rauscher S, Flamm C, Mandl CW, Heinz FX, Stadler PF. Secondary structure of the 3′-noncoding region of flavivirus genomes: comparative analysis of base pairing probabilities. Rna. 1997;3(7):779–791. [PMC free article] [PubMed] [Google Scholar]
- Rey FA, Heinz FX, Mandl C, Kunz C, Harrison SC. The envelope glycoprotein from tick-borne encephalitis virus at 2 A resolution. Nature. 1995;375(6529):291–298. doi: 10.1038/375291a0. [see comments] [DOI] [PubMed] [Google Scholar]
- Rice CM, Lenches EM, Eddy SR, Shin SJ, Sheets RL, Strauss JH. Nucleotide sequence of yellow fever virus: implications for flavivirus gene expression and evolution. Science. 1985;229:729–733. doi: 10.1126/science.4023707. [DOI] [PubMed] [Google Scholar]
- Rico-Hesse R. Molecular evolution and distribution of dengue viruses type 1 and 2 in nature. Virology. 1990;174(2):479–493. doi: 10.1016/0042-6822(90)90102-w. [DOI] [PubMed] [Google Scholar]
- Rodhain F. The role of monkeys in the biology of dengue and yellow fever. Comp. Immunol. Microbiol. Infect. Dis. 1991;14(1):9–19. doi: 10.1016/0147-9571(91)90036-d. [DOI] [PubMed] [Google Scholar]
- Roehrig JT, Bolin RA, Kelly RG. Monoclonal antibody mapping of the envelope glycoprotein of the dengue 2 virus, Jamaica. Virology. 1998;246(2):317–328. doi: 10.1006/viro.1998.9200. [DOI] [PubMed] [Google Scholar]
- Romero TA, Tumban E, Jun J, Lott WB, Hanley KA. Secondary structure of dengue virus type 4 3′ untranslated region: impact of deletion and substitution mutations. J. Gen. Virol. 2006;87(Pt 11):3291–3296. doi: 10.1099/vir.0.82182-0. [DOI] [PubMed] [Google Scholar]
- Rosen L. Experimental infection of New World monkeys with dengue and yellow fever viruses. Am. J. Trop. Med. Hyg. 1958;7(4):406–410. doi: 10.4269/ajtmh.1958.7.406. [DOI] [PubMed] [Google Scholar]
- Rosen L, Gubler D. The use of mosquitoes to detect and propagate dengue viruses. Am. J. Trop. Med. Hyg. 1974;23(6):1153–1160. doi: 10.4269/ajtmh.1974.23.1153. [DOI] [PubMed] [Google Scholar]
- Rudnick A, Lim TW. Dengue fever studies in Malaysia. Inst. Med. Res. Malays. Bull. 1986;23:51–152. [Google Scholar]
- Sabin AB. Research on dengue during World War II. Am. J. Trop. Med. Hyg. 1952;1(1):30–50. doi: 10.4269/ajtmh.1952.1.30. [DOI] [PubMed] [Google Scholar]
- Saluzzo JF, Cornet M, Adam C, Eyraud M, Digoutte JP. Dengue 2 in eastern Senegal: serologic survey in simian and human populations. 1974 – 85. Bull. Soc. Pathol. Exot. Filiales. 1986a;79(3):313–322. [PubMed] [Google Scholar]
- Saluzzo JF, Cornet M, Castagnet P, Rey C, Digoutte JP. Isolation of dengue 2 and dengue 4 viruses from patients in Senegal. Trans. R. Soc. Trop. Med. Hyg. 1986b;80(1):5. doi: 10.1016/0035-9203(86)90182-3. [DOI] [PubMed] [Google Scholar]
- Sanchez IJ, Ruiz BH. A single nucleotide change in the E protein gene of dengue virus 2 Mexican strain affects neurovirulence in mice. J. Gen. Virol. 1996;77(Pt 10):2541–2545. doi: 10.1099/0022-1317-77-10-2541. [DOI] [PubMed] [Google Scholar]
- Scherer WF, Russell PK, Rosen L, Casals J, Dickerman RW. Experimental infection of chimpanzees with dengue viruses. Am. J. Trop. Med. Hyg. 1978;27(3):590–599. doi: 10.4269/ajtmh.1978.27.590. [DOI] [PubMed] [Google Scholar]
- Shurtleff AC, Beasley DW, Chen JJ, Ni H, Suderman MT, Wang H, Xu R, Wang E, Weaver SC, Watts DM, Russell KL, Barrett AD. Genetic variation in the 3′ non-coding region of dengue viruses. Virology. 2001;281(1):75–87. doi: 10.1006/viro.2000.0748. [DOI] [PubMed] [Google Scholar]
- Silva PA, Molenkamp R, Dalebout TJ, Charlier N, Neyts JH, Spaan WJ, Bredenbeek PJ. Conservation of the pentanucleotide motif at the top of the yellow fever virus 17D 3′stem-loop structure is not required for replication. J. Gen. Virol. 2007;88(Pt 6):1738–1747. doi: 10.1099/vir.0.82811-0. [DOI] [PubMed] [Google Scholar]
- Sirigulpanit W, Kinney RM, Leardkamolkarn V. Substitution or deletion mutations between nt 54 and 70 in the 5′ non-coding region of dengue type 2 virus produce variable effects on virus viability. J. Gen. Virol. 2007;88(Pt 6):1748–1752. doi: 10.1099/vir.0.82455-0. [DOI] [PubMed] [Google Scholar]
- Ta M, Vrati S. Mov34 protein from mouse brain interacts with the 3′ noncoding region of Japanese encephalitis virus. J. Virol. 2000;74(11):5108–5115. doi: 10.1128/jvi.74.11.5108-5115.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima S, Nukui Y, Takasaki T, Kurane I. Characterization of the variable region in the 3′ non-translated region of dengue type 1 virus. J. Gen. Virol. 2007;88(Pt 8):2214–2222. doi: 10.1099/vir.0.82661-0. [DOI] [PubMed] [Google Scholar]
- Thurner C, Witwer C, Hofacker IL, Stadler PF. Conserved RNA secondary structures in Flaviviridae genomes. J. Gen. Virol. 2004;85(Pt 5):1113–1124. doi: 10.1099/vir.0.19462-0. [DOI] [PubMed] [Google Scholar]
- Trent DW. Antigenic characterization of flavivirus structural proteins separated by isoelectric focusing. J. Virol. 1977;22(3):608–618. doi: 10.1128/jvi.22.3.608-618.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twiddy SS, Farrar JJ, Vinh Chau N, Wills B, Gould EA, Gritsun T, Lloyd G, Holmes EC. Phylogenetic relationships and differential selection pressures among genotypes of dengue-2 virus. Virology. 2002a;298(1):63–72. doi: 10.1006/viro.2002.1447. [DOI] [PubMed] [Google Scholar]
- Twiddy SS, Woelk CH, Holmes EC. Phylogenetic evidence for adaptive evolution of dengue viruses in nature. J. Gen. Virol. 2002b;83(Pt 7):1679–1689. doi: 10.1099/0022-1317-83-7-1679. [DOI] [PubMed] [Google Scholar]
- Uchil PD, Kumar AV, Satchidanandam V. Nuclear localization of flavivirus RNA synthesis in infected cells. J. Virol. 2006;80(11):5451–5464. doi: 10.1128/JVI.01982-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasilakis N, Holmes EC, Fokuyam EB, Faye O, Diallo M, Sall AA, Weaver SC. Evolutionary processes among sylvatic Dengue-2 viruses. J. Virol. 2007a;81:9591–9595. doi: 10.1128/JVI.02776-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasilakis N, Shell EJ, Fokam EB, Mason PW, Hanley KA, Estes DM, Weaver SC. Potential of ancestral sylvatic dengue-2 viruses to re-emerge. Virology. 2007b;358(2):402–412. doi: 10.1016/j.virol.2006.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasilakis N, Tesh RB, Weaver SC. Sylvatic dengue virus type 2 activity in humans, Nigeria, 1966. Emerg. Infect. Dis. 2008;14(3):502–504. doi: 10.3201/eid1403.070843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasilakis N, Durbin A, Travassos da Rosa APA, Munoz-Jordan JL, Tesh RB, Weaver SC. Antigenic relationships between sylvatic and endemic dengue viruses. Am J. Trop. Med. Hygs. in press. [PubMed] [Google Scholar]
- Wang E, Ni H, Xu R, Barrett AD, Watowich SJ, Gubler DJ, Weaver SC. Evolutionary relationships of endemic/epidemic and sylvatic dengue viruses. J. Virol. 2000;74(7):3227–3234. doi: 10.1128/jvi.74.7.3227-3234.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wengler G. Terminal sequences of the genome and replicative-form RNA of the flavivirus West Nile virus: absence of poly(A) and possible role in RNA replication. Virology. 1981;113(2):544–555. doi: 10.1016/0042-6822(81)90182-3. [DOI] [PubMed] [Google Scholar]
- Whitehead SS, Blaney JE, Durbin AP, Murphy BR. Prospects for a dengue virus vaccine. Nat. Rev., Microbiol. 2007;5(7):518–528. doi: 10.1038/nrmicro1690. [DOI] [PubMed] [Google Scholar]
- Winkler G, Maxwell SE, Ruemmler C, Stollar V. Newly synthesized dengue-2 virus nonstructural protein NS1 is a soluble protein but becomes partially hydrophobic and membrane-associated after dimerization. Virology. 1989;171(1):302–305. doi: 10.1016/0042-6822(89)90544-8. [DOI] [PubMed] [Google Scholar]
- You S, Padmanabhan R. A novel in vitro replication system for Dengue virus. Initiation of RNA synthesis at the 3′-end of exogenous viral RNA templates requires 5′- and 3′-terminal complementary sequence motifs of the viral RNA. J. Biol. Chem. 1999;274(47):33714–33722. doi: 10.1074/jbc.274.47.33714. [DOI] [PubMed] [Google Scholar]
- Zeller HG, Traore-Lamizana M, Monlun E, Hervy JP, Mondo M, Digoutte JP. Dengue-2 virus isolation from humans during an epizootic in southeastern Senegal in November, 1990. Res. Virol. 1990;143(2):101–102. doi: 10.1016/s0923-2516(06)80088-9. [DOI] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31(13):3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]