Abstract
Limited outcome data exist for patients with chronic lymphocytic leukemia (CLL) with a poor-risk cytogenetic profile and a good-risk (mutated) immunoglobulin heavy-chain variable (IGHV) sequence. In a retrospective, single-institution review, 1 in 4 patients with del(11q or 17p) CLL had a mutated IGVH sequence. We found that mutational status was a better predictor for treatment intervention and OS. IGHV has prognostic value in del(11q) and del(17p) patients with CLL.
Background
Most patients with CLL with a poor-risk cytogenetic profile have an unmutated IGHV sequence. Limited clinical information exists for patients with CLL who have a poor-risk cytogenetic profile and a mutated or good-risk IGHV status. We retrospectively screened all patients with CLL seen at our institution from 2006 onward who harbored a del(11q) or del(17p) CLL detected by fluorescence in situ hybridization (FISH) analysis for whom an IGHV analysis was requested. In 66 evaluable patients, 50 (76%) had an unmutated IGHV sequence. Thirty-nine patients (59%) had del(11q) and 27 patients (41%) had del(17p); no patient in this series had both del(11q) and del(17p). The patients’ initial clinical presentations were similar in both mutational groups. Patients with an unmutated IGHV sequence were more likely to receive treatment and to have a shorter survival, with an estimated 3-year overall survival (OS) of 81% compared with 100% in the group with a mutated IGHV sequence (log rank, P = .06). These data suggest that IGHV mutational status has prognostic relevance even in patients with CLL who are defined as poor risk by genomic FISH analysis.
Keywords: Clinical outcomes, CLL, del(17p), del(11q), IGHV
Introduction
The importance of interphase fluorescence in situ hybridization (FISH) DNA analysis as a predictive tool for survival in patients with chronic lymphocytic leukemia (CLL) is well established.1 However, even among cytogenetically defined groups, survival is variable.2-4 Importantly, in patients with del(13q) CLL, an unmutated immunoglobulin heavy-chain variable (IGHV) region mutational sequence predicts for (1) rapid Rai stage disease progression, (2) treatment initiation, and (3) poor overall survival (OS).5 In this article, the influence of IGHV mutational status on OS in patients with CLL harboring the poor prognostic FISH findings of del(11q) and del(17p) is analyzed.
Methods
Patients
After institutional review board approval, we retrospectively screened all patients with CLL evaluated at our institution between January 2006 and March 2011 for whom an IGHV mutational status was requested. This report includes all patients who had IGHV sequence homology analysis performed and also had del(11q) or del (17p) detected by FISH evaluation during the clinical course of the disease. At our center, standard laboratory practice for patients with CLL is to perform interphase FISH DNA analysis and IGHV mutational status analysis; however all evaluations are at physician discretion.
FISH and IGHV Mutation Analyses
All FISH analyses were performed at a single reference laboratory by interphase analysis using probes for 6 centromere (cen)/6q23.3 (CMYB), 11 cen/11q22.3 (ATM), 12 cen/12q15 (MDM2), 13q14 (D13S319)/13q34(LAMP1), 17 cen/17p13.1 (p53), 11q13 (CCND1-XT)/14q32 (IGH-XT). The 95% cutoff for homozygous del(13q) and dual del(13q) was 7.0% and 1.5%, respectively. All IGHV mutational analyses were performed at a single reference laboratory using cycle sequencing analysis. An IGHV nonhomologous sequence of < 2% was defined as unmutated (a low mutational status) and an IGHV nonhomologous sequence of ≥ 2% was defined as mutated (a high mutational status).6-10
Statistical Methods
Characteristics of patients by mutational status were compared using Wilcoxon rank sum tests for continuous measurements and the Fisher exact test for categorical measurements. OS was calculated as the time from (1) diagnosis and (2) initial poor-risk FISH documentation to death or last known follow-up. Treatment-free survival was calculated as the time from diagnosis to treatment or last known follow-up at which the patient had not received treatment. Event-time distributions were estimated by the Kaplan-Meier method and compared by the log-rank test. All analyses were completed using statistical software R, version 2.11.1 (SAS, Inc, Cary, NC) based on outcomes reported through May 19, 2011.
Results
Between January 2006 and March 2011, IGHV mutational studies were identified in 159 patients with CLL of whom 70 (44%) had documented del(11q) or del(17p) by interphase FISH during their clinical course. Four patients’ IGHV mutational status could not be analyzed and they were excluded from further analysis. Of the remaining 66 patients, 39 (59%) had del(11q) and 27 (41%) had del(17p); no patient in this cohort had both high-risk deletions and 50 patients (71%) had an unmutated IGHV sequence and 16 patients (29%) had a mutated IGHV sequence. Patient characteristics are shown in Table 1. The median time from diagnosis to last follow-up for patients with an unmutated IGHV sequence was 3.1 years (range, 0.1-14.6 years) and for the patients with a mutated IGHV sequence, it was 3.1 years (range, 0.6-19.4 years).
Table 1.
Characteristics of Patients With del(11q)/del(17p) CLL: Overall and by IGHV Mutation Percentagea
| Characteristic | All Patients N = 66 | IGHV < 2% n = 50 | IGHV ≥ 2% n = 16 | P Value |
|---|---|---|---|---|
| Age at Diagnosis (Years), Median (Range) | 57.6 (26.8, 81.2) | 57.6 (26.9, 81.2) | 56.5 (30.3, 78.4) | 0.90 |
| Stage at Diagnosis, No. (%) | ||||
| Rai 0 | 21 (33) | 15 (31) | 6 (38) | < 0.99 |
| Rai 1 | 28 (44) | 21 (44) | 7 (44) | |
| Rai 2 | 10 (16) | 8 (17) | 2 (12) | |
| Rai 3 | 3 (5) | 2 (4) | 1 (6) | |
| Rai 4 | 2 (3) | 2 (4) | 0 (0) | |
| Unknown | 2 | 2 | 0 | |
| Time from Diagnosis to Initial JHU Consultation (Years), Median (Range) | 0.5 (0, 16.3) | 0.6 (0, 13.0) | 0.4 (0, 16.3) | 0.539 |
| Stage at Initial JHU Consultation, No. (%) | ||||
| Rai 0 | 20 (31) | 12 (24) | 8 (50) | .062 |
| Rai 1 | 28 (43) | 21 (43) | 7 (44) | |
| Rai 2 | 5 (8) | 5 (10) | 0 (0) | |
| Rai 3 | 2 (3) | 1 (2) | 1 (6) | |
| Rai 4 | 10 (15) | 10 (20) | 0 (0) | |
| Richter transformation | 0 (0) | 0 (0) | 0 (0) | |
| Unknown | 1 | 1 | 0 | |
| Received Treatment Before JHU Consultation | ||||
| No | 48 (73) | 33 (66) | 15 (94) | .05 |
| Yes | 18 (22) | 17 (34) | 1 (6) | |
| Time from Diagnosis to FISH (Years) - Median (Range) | 1.2 (0, 18.9) | 1.9 (0, 13.2) | 0.7 (0, 18.9) | .26 |
| Documented del(11q) - No. (%) | ||||
| No | 27 (41) | 22 (44) | 5 (31) | < .001 |
| Yes | 39 (59) | 28 (56) | 11 (69) | |
| Documented del(17p) - No. (%) | ||||
| No | 39 (59) | 28 (56) | 11 (69) | .40 |
| Yes | 27 (41) | 22 (44) | 5 (31) |
Abbreviations: IGHV = immunoglobulin heavy-chain variable; JHU = Johns Hopkins University.
P values for Fisher exact test or Wilcoxon rank sum tests for differences in variables between mutation groups.
Patients With an Unmutated IGHV Sequence Were More Likely to Receive Treatment
When comparing patients with an unmutated IGHV sequence to patients with a mutated IGHV sequence, the median age of 57.6 vs. 51.6 years (P = .43) and the presenting Rai stage (P > .99) were similar. The median time to first CLL evaluation at Johns Hopkins was also similar in both groups: 0.6 years after diagnosis in group with an unmutated IGHV sequence and 0.4 years in the group with a mutated IGHV sequence (P = .43). Although the majority of patients were treatment naive at the time of consultation, patients with an unmutated IGHV sequence were more likely to have received treatment beforehand (34% vs. 6%; P = .05).
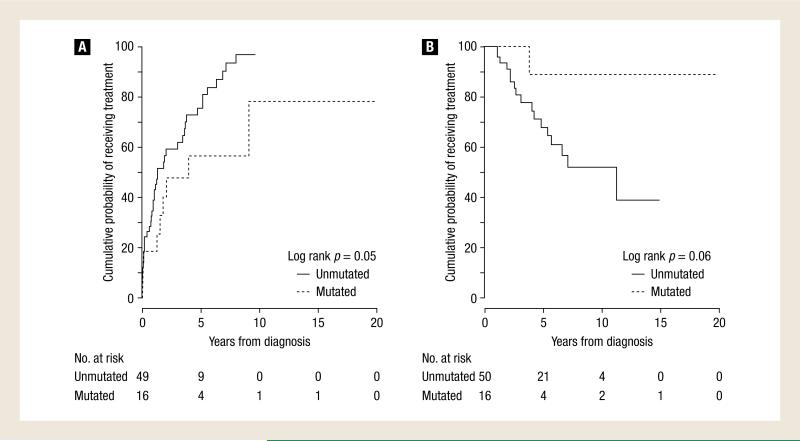
No patient died without first receiving treatment for CLL. The most common reasons for treatment initiation were either bulky adenopathy or progression to an advanced Rai stage. For the entire group, the median time to treatment initiation was 1.9 years (range, 0-19.4 years) and the cumulative probability of receiving treatment (CPT) at 3 years was 57% (95% confidence interval [CI], 42%-67%). The estimated 3-year CPT for the group with an unmutated IGHV sequence was 59% (95% CI, 42%-71%) compared with 48% (95% CI, 14%-68%) for the group with a mutated IGHV sequence (log rank, P = .05) (Figure 1A; Table 2). Of the 41 patients (82%) with unmutated disease who received treatment, a median of 2 (range, 1-6) regimens were used. Of the 9 patients (56%) with mutated disease who received treatment, a median of 2 regimens (range, 1-8) were used (Table 3).
Figure 1.
Estimated Overall Survival and Cumulative Probability of Receiving Treatment
Table 2.
Overall Survival (OS) from Diagnosis for the Entire Cohort and Cumulative Time to Treatment (CPT) for the Whole Cohort and by Mutational Analysis
| Variable | N | Median (95% CI) (Years) | 3 Years | Log Rank P | HR | 95% CI |
|---|---|---|---|---|---|---|
| OS: All Patients | 66 | 11.3 (6.6, 11.3+) | 85 (76, 95) | |||
| OS: by Mutation Status | ||||||
| Low IGVH Mutation (<2) | 50 | 11.3 (5.7, 19.9+) | 81 (69, 94) | 0.06 | 1.00 | - |
| High IGVH Mutation (2+) | 16 | NR | 100 (100, 100) | 0.21 | (0.03, 1.6) | |
| OS: del(11q) Patients | ||||||
| Low IGVH Mutation (< 2) | 28 | 11.3 (11.3, 19.9+) | 88 (75, 100) | 0.05 | 1.00 | - |
| High IGVH Mutation (2+) | 11 | NR | 100 (100, 100) | 0.47 | (0.05, 4.14) | |
| OS: del(17p) Patients | ||||||
| Low IGVH Mutation (<2) | 22 | 4.9 (3.1, 14.9+) | 72 (53, 96) | 0.04 | 1.00 | - |
| High IGVH Mutation (2+) | 5 | NR | 100 (100, 100) | b | - | |
| CPT: All Patients | 65 | 1.8 (1.3, 3.8) | 57 (42, 67) | |||
| CPT: By Mutation Status | ||||||
| Low IGVH Mutation (< 2) | 49 | 1.3 (0.96, 3.7) | 59 (42, 71) | 0.05 | 1.00 | - |
| High IGVH Mutation (2+) | 16 | 4.0 (1.5, 19.9+) | 48 (14, 68) | 0.50 | (0.24, 1.05) | |
| CPT: del(11q) Patients | ||||||
| Low IGVH Mutation (<2) | 28 | 3.7 (1.9, 6.9) | 38 (16, 55) | 0.87 | 1.00 | - |
| High IGVH Mutation (2+) | 11 | 1.5 (1.3, 19.9+) | 63 (12, 84) | 0.93 | (0.38, 2.25) | |
| CPT: del(17p) Patients | ||||||
| Low IGVH Mutation (< 2) | 21 | 0.96 (0.21, 1.33) | 86 (59, 95) | 0.001 | 1.00 | - |
| High IGVH Mutation (2+) | 5 | NR | 20 (0, 48) | 0.14 | (0.03, 0.63) |
Abbreviations: NR = Median survival not reached.
aValues for median survival are in years. Other values are survival probability (95% CI).
Hazard ratio could not be estimated due to no deaths in the mutated del(17p) patients.
Table 3.
Treatment Regimens Delivered
| Treatment | Total N = 66 | Unmutated IGHV Sequence N = 50 | Mutated IGHV Sequence N = 16 |
|---|---|---|---|
| Received Treatment | 50 (76%) | 41 (82%) | 9 (56%) |
| Treatment Base | |||
| Rituximab | 49 (74%) | 40 (80%) | 9 (56%) |
| Ofatumumab | 3 (5%) | 2 (4%) | 1 (6%) |
| Alemtuzumab | 11 (17%) | 10 (20%) | 1 (6%) |
| Anthracycline | 12 (18%) | 9 (18%) | 3 (19%) |
| Bendamustine | 18 (27%) | 16 (32%) | 2 (11%) |
| Pentostatin | 6 (9%) | 4 (8%) | 2 (11%) |
| Fludarabine | 14 (21%) | 11 (22%) | 3 (19%) |
| Alkylator | 8 (12%) | 6 (12%) | 2 (11%) |
| mTor inhibitors/IMIDs | 5 (8%) | 4 (8%) | 1 (6%) |
| Allogeneic BMT | 11 (17%) | 11 (22%) | 0 (0%) |
| Other | 3 (5%) | 2 (4%) | 1 (6%) |
Abbreviations: IGHV = immunoglobulin heavy-chain variable; IMIDs = immunomodulatory drugs.
Eleven of 66 patients (17%) underwent allogeneic bone marrow transplantation, at a median of 2.0 years (range, 0.9-11.4 years) from the date of diagnosis. All 11 transplantation procedures were performed in the unmutated IGHV population (11/50; 22%). Of these patients, 6 died (two of transplant- related causes) and 5 remained disease free at a median of 8 months (range, 3-32 months) after transplantation.
Patients With an Unmutated IGHV Sequence Were More Likely to Have Shorter Survivals
The estimated 3-year OS was 85% (95% CI, 76%-95%) for the entire group. Of the 18 deaths, only 1 occurred in the cohort of patients with a mutated IGHV sequence. The median OS for the patients with an unmutated IGHV sequence was 11.3 years with a 3-year OS probability of 81% (95% CI, 69%-94%). In the patients with a mutated IGHV sequence, the median OS was not reached and the 3-year OS probability was 100% (95% CI, 100%-100%) (log rank, P = .06) (Figure 1B; Table 2). Additionally, no difference in 3-year OS probability was found when comparing outcomes dichotomized by abnormal chromosomal disease burden (< 10% or < 20%) found at first abnormal FISH analysis (data not shown).
The median OS for our patients with del(11q) was not reached, and the 3-year OS probability was 88% (95% CI, 75%-100%). When examining patients with del(11q) by mutational status, we found the median OS for the patients without the mutation to be 11.26 years, whereas the median OS for the patients with the mutation was not reached. The 3-year OS probability for patients with del(11q) and an unmutated IGHV sequence compared with patients with del(11q) and a mutated IGHV sequence was 88% (95% CI, 75%-100%) vs. 100% (95% CI, 100%-100%), respectively (log rank, P = .46). The corresponding hazard ratio for patients with del(11q) and a mutated IGHV sequence vs. patients with del(11q) and an unmutated IGHV sequence was 0.47 (95% CI, 0.05-4.14). The median OS for our patients with del(17p) was 5.4 years with a 3-year OS probability of 77% (95% CI, 62%- 97%). When examining patients with del(17p) by mutational status, we found the median OS for the patients with an unmutated IGHV sequence to be 4.9 years, whereas the median survival for the patients with a mutated IGHV sequence was not reached. The 3-year OS probability for patients with del(17p) and an unmutated IGHV sequence compared with patients with a mutated IGHV sequence was 72% (95% CI, 53%- 96%) vs. 100% (no deaths in the group with a mutated IGHV sequence), respectively (log rank, P = .04).
Of the 18 deaths, only 1 was not a direct result of CLL but was caused by squamous cell lung carcinoma; this death occurred 4 years after the CLL diagnosis in a patient with del(17p) and an unmutated IGHV sequence. A Richter transformation occurred in 2 patients with an unmutated IGHV sequence and in 1 patient with a mutated IGHV sequence.
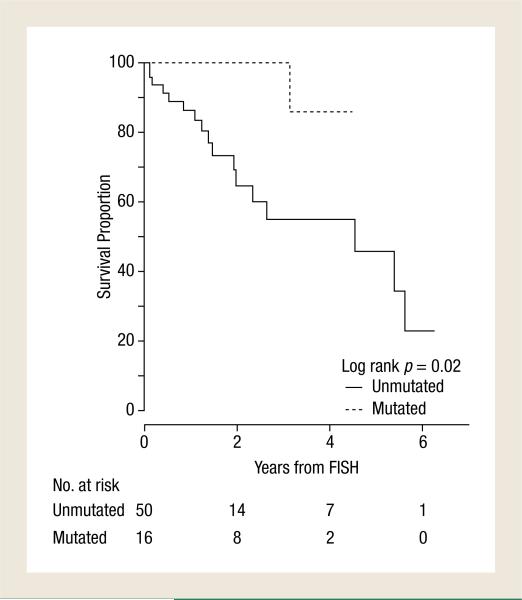
The estimated 3-year OS from time of first high-risk abnormal FISH analysis to last follow-up was 67% (95% CI, 53%-84%) for the entire group. The median OS from the first abnormal FISH analysis for the patients with an unmutated IGHV sequence was 4.5 years with a 3-year OS probability of 55% (95% CI, 39%-78%). In the patients with a mutated IGHV sequence, the median OS from first abnormal FISH analysis was not reached and the 3-year OS probability was 100% (95% CI, 100%-100%) (log rank, P = .02 (Figure 2, Table 4).
Figure 2.
Estimated Overall Survival From Time of First Poor-Risk Fluorescence in Situ Hybridization (FISH) Analysis
Table 4.
Overall Survival (OS) from Time of First High-Risk FISH Analysis for the Entire Cohort by Mutational Analysis
| N | Median | (95% CI) (Years) | 3 Years | Log Rank P | HR | 95% CI | |
|---|---|---|---|---|---|---|---|
| OS: All Patients | 66 | 5.4 (3.1,5.6+) | 67 (53, 84) | ||||
| OS, By Mutational Status | |||||||
| Low IGVH Mutation (< 2) | 50 | 4.5 (2.0, 6.2+) | 55 (39, 78) | .02 | 1.00 | - | |
| High IGVH Mutation (2+) | 16 | NR | 100 (100, 100) | .15 | (0.02, 1.2) | ||
| OS: Patients With del(11q) | |||||||
| Low IGVH Mutation (< 2) | 28 | 11.3 (11.3, 19.9+) | 70 (49, 98) | .31 | 1.00 | - | |
| High IGVH Mutation (2+) | 11 | NR | 100 (100, 100) | .37 | (0.04, 3.1) | ||
| OS: Patients With del(17p) | |||||||
| Low IGVH Mutation (< 2) | 22 | 1.9 (1.2, 5.6+) | 34 (15, 78) | .02 | 1.00 | - | |
| High IGVH Mutation (2+) | 5 | NR | 100 (100, 100) | a |
Values for median survival are in years. Other values are survival probability (95% CI).
Abbreviations: Hazard ratio could not be estimated because no deaths occurred in the patients with del(17p) mutated IGVH sequence.
Discussion
Both interphase FISH and IGHV mutational status analyses are used to predict clinical outcome in patients with CLL. A mutated IGHV sequence and del(13q) are generally considered markers for a favorable course. Conversely, an unmutated IGVH sequence and the presence of del(11q) and especially del(17p) are considered poor prognostic factors.10 Although favorable and unfavorable cytogenetic abnormalities generally track with the similarly prognostic IGHV mutational status, a significant fraction of patients with CLL exhibit cytogenetic profiles and IGHV mutations that are prognostically discordant. We recently reported that patients with del(13q) and CLL with an unmutated IGHV sequence tend to have aggressive clinical disease.5 In this study we similarly found the IGHV mutational status to predict clinical outcomes in patients with CLL harboring del(11q) and/or del(17p), with a mutated IGHV sequence predicting an indolent clinical course. In comparison to other studies, even our patients with an unmutated IGHV sequence had a favorable median OS; this can be explained by lead-time basis: our patients were followed from the time of diagnosis not from the time of treatment.11
Fortunately, only a single death was observed in the group with mutated IGHV sequence—a clinically important observation; however, the low incidence of terminal events, coupled with the size of the studied population, limits the ability to detect significant differences in survival in each genotypic group. Nonetheless, the clinical importance of a mutated status is suggested by the estimate of a 53% reduction in the risk of death in the patients with del(11q) and a mutated IGHV sequence and the pattern of the survival curves. Therefore although the small sample size limits the ability to demonstrate statistically significant differences in individual cytogenetically defined subgroups, the similar outcomes in patients with del(11q) and del(17p) justifies their analysis together and generates a strong clinical signal.
The discordance between cytogenetic and IGHV mutational risk assessments in almost one third of patients may offer an explanation for the variable clinical outcomes reported for high-risk cytogenetic groups. Tam et al also previously found that mutational status was prognostic in patients with del(17p).2 These observations, taken together with our previous findings in patients with del(13q),5 suggest CLL aberrations seen on FISH analysis largely predict clinical outcome because they track with a specific IGHV mutational status. Moreover, the outcomes in the patients with del(13q)/del(11q) and del(13q)/del(17p) are quite similar when dichotomized by IGVH mutational status.5 The signal produced by these analyses, although requiring confirmation from a larger study, is so provocative that at our institution we do not routinely offer allogeneic transplantation in first response12,13 for patients with CLL who have a high-risk cytogenetic profile and harbor a mutated IGHV sequence. Additional outcome research is indicated to best guide clinical care in patients with CLL who harbor a combination of high- and low-risk factors.
Clinical Practice Points.
Groups of patients with CLL cytogenetically defined by interphase FISH DNA analysis have variable outcomes.
An unmutated IGHV sequence predicts for an aggressive CLL phenotype in patients with del(13q) and CLL.
We found that 25% of our patients with CLL with the poor-risk cytogenetic finding of either del(11q) or del(17p) concurrently had a mutated or good-prognosis IGHV sequence.
IGHV is a better predictor of clinical outcomes than is FISH analysis even in patients with del(11q) and del(17p) and CLL.
Patients with a poor-risk cytogenetic profile and a mutated IGHV sequence may not require allogeneic bone marrow transplantation in first response.
Footnotes
Author Contributions
Douglas E. Gladstone: Main author for all sections.
Amanda Blackford: Statistician.
Eunpi Cho: Collected data and help write the result section.
Lode Swinnen: Collected data and helped write the result section.
Yvette Kasamon: Reviewed all data and helped write the discussion section.
Christopher D. Gocke: Collected and reviewed the IGHV data; helped write discussion.
Constance A. Griffin: Collected and reviewed the all the FISH data; helped write result section.
Javier Bolaños-Meade: Collected survival data; helped write result section.
Richard J. Jones: Reviewed all data; helped write the discussion section.
Disclosure
The authors have stated that they have no conflicts of interest.
References
- 1.Dohner H, Stilgenbauer S, Benner A, et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 2000;343:1910–6. doi: 10.1056/NEJM200012283432602. [DOI] [PubMed] [Google Scholar]
- 2.Tam CS, Shanafelt TD, Wierda WG, et al. De novo deletion 17p13.1 chronic lymphocytic leukemia shows significant clinical heterogeneity: the M. D. Anderson and Mayo Clinic experience. Blood. 2009;114:957–64. doi: 10.1182/blood-2009-03-210591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tomomatsu J, Isobe Y, Oshimi K, et al. Chronic lymphocytic leukemia in a Japanese population: varied immunophenotypic profile, distinctive usage of frequently mutated IGH gene, and indolent clinical behavior. Leuk Lymphoma. 2010;51:2230–9. doi: 10.3109/10428194.2010.527403. [DOI] [PubMed] [Google Scholar]
- 4.Best OG, Gardiner AC, Davis ZA, et al. A subset of Binet stage A CLL patients with TP53 abnormalities and mutated IGHV genes have stable disease. Leukemia. 2009;23:212–4. doi: 10.1038/leu.2008.260. [DOI] [PubMed] [Google Scholar]
- 5.Gladstone DE, Swinnen L, Kasamon Y, et al. Importance of immunoglobulin heavy chain variable region mutational status in del(13q) chronic lymphocytic leukemia. Leuk Lymphoma. 2011;52:1873–81. doi: 10.3109/10428194.2011.585529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Damle RN, Wasil T, Fais F, et al. Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood. 1999;94:1840–7. [PubMed] [Google Scholar]
- 7.Hamblin TJ, Davis Z, Gardiner A, et al. Unmutated Ig V(H) genes are associated with a more aggressive form of chronic lymphocytic leukemia. Blood. 1999;94:1848–54. [PubMed] [Google Scholar]
- 8.Hamblin TJ, Orchard JA, Ibbotson RE, et al. CD38 expression and immunoglobulin variable region mutations are independent prognostic variables in chronic lymphocytic leukemia, but CD38 expression may vary during the course of the disease. Blood. 2002;99:1023–9. doi: 10.1182/blood.v99.3.1023. [DOI] [PubMed] [Google Scholar]
- 9.Kharfan-Dabaja MA, Chavez JC, Khorfan KA, et al. Clinical and therapeutic implications of the mutational status of IgVH in patients with chronic lymphocytic leukemia. Cancer. 2008;113:897–906. doi: 10.1002/cncr.23671. [DOI] [PubMed] [Google Scholar]
- 10.Trojani A, Montillo M, Nichelatti M, et al. ZAP-70, IgVh, and cytogenetics for assessing prognosis in chronic lymphocytic leukemia. Cancer Biomark. 2010;6:1–9. doi: 10.3233/CBM-2009-0114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Catovsky D, Richards S, Matutes E, et al. Assessment of fludarabine plus cyclophosphamide for patients with chronic lymphocytic leukaemia (the LRF CLL4 trial): a randomised controlled trial. Lancet. 2007;370:230–9. doi: 10.1016/S0140-6736(07)61125-8. [DOI] [PubMed] [Google Scholar]
- 12.Dreger P, Dohner H, Ritgen M, et al. Allogeneic stem cell transplantation provides durable disease control in poor-risk chronic lymphocytic leukemia: long-term clinical and MRD results of the German CLL Study Group CLL3X trial. Blood. 2010;116:2438–47. doi: 10.1182/blood-2010-03-275420. [DOI] [PubMed] [Google Scholar]
- 13.Schetelig J, van Biezen A, Brand R, et al. Allogeneic hematopoietic stem-cell transplantation for chronic lymphocytic leukemia with 17p deletion: a retrospective European Group for Blood and Marrow Transplantation analysis. J Clin Oncol. 2008;26:5094–100. doi: 10.1200/JCO.2008.16.2982. [DOI] [PubMed] [Google Scholar]