Abstract
Different types of cell behavior including growth, motility, and navigation require actin proteins to assemble into filaments. Here, we describe a biochemical process that was able to disassemble actin filaments and limit their reassembly. Actin was a specific substrate of the multi-domain oxidation-reduction (Redox) enzyme, Mical, a poorly-understood actin disassembly factor that directly responds to Semaphorin/Plexin extracellular repulsive cues. Actin filament subunits were directly modified by Mical on their conserved pointed-end that is critical for filament assembly. Mical post-translationally oxidized the methionine 44 residue within the D-loop of actin, simultaneously severing filaments and decreasing polymerization. This mechanism underlying actin cytoskeletal collapse may have broad physiological and pathological ramifications.
The actin cytoskeleton underlies a diverse array of cellular behaviors (1) but its regulatory mechanisms are incompletely understood. Recently, an actin regulator, the multidomain cytosolic protein Mical, was shown to directly bind and disassemble individual and bundled actin filaments (F-actin) (2). Although still poorly understood, Mical-mediated actin remodeling alters cell morphology and navigation in response to one of the largest families of extracellular guidance cues, the semaphorins and their plexin receptors (2–5). Drosophila Mical and its vertebrate family members are known as the MICAL family of proteins and belong to a class of flavoprotein monooxygenase/hydroxylase enzymes that bind flavin adenine dinucleotide (FAD) and use the co-enzyme nicotinamide adenine dinucleotide phosphate (NADPH) in oxidation-reduction (Redox) reactions (3, 5). Although MICALs have no known substrate/s, they employ their Redox region to bind F-actin and disassemble filaments in an NADPH-dependent manner (Fig. 1A; (2)).
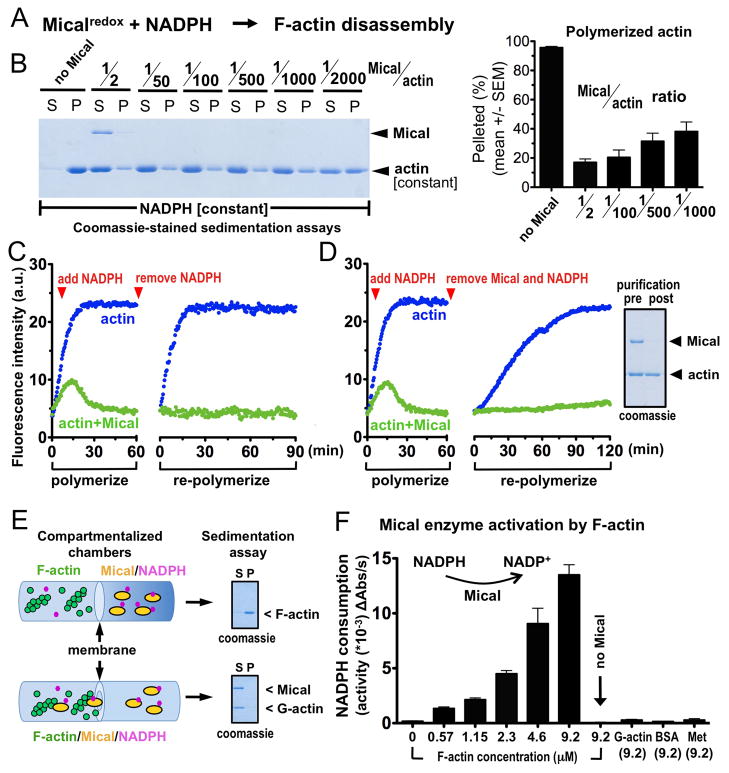
Fig. 1. Mical directly modifies F-actin.
(A) The Mical Redox domain alone with NADPH disassembles F-actin. (B) Low amounts of Mical, in comparison to actin, disassembles F-actin. S, soluble (G-actin); P, pellet (F-actin). n≥2/condition. [Actin]=2μM; [Mical]=1μM-1nM; [NADPH]=400μM. (C–D) Mical/NADPH-treated actin (green) does not re-polymerize after removal of NADPH (C) or Mical and NADPH (D). Modified actin (D, right) migrates normally and is not degraded. (E) Compartmentalized chambers with a membrane allowing small molecules, but not Mical, access to F-actin abolished disassembly (top). Compare to chambers with a punctured membrane (bottom). (F) Mical’s enzymatic activity (as determined by conversion of NADPH to NADP+), is substantially increased by F-actin, but not G-actin or other proteins (BSA), or free methionine (Met).
In in vitro actin biochemical assays only very low, substochiometric levels of Mical were required for F-actin disassembly (Fig. 1B), supporting the idea that a catalytic/post-translational mechanism underlies Mical-mediated F-actin disassembly. This Mical-treated actin failed to re-polymerize even after removal of Mical/NADPH (Figs. 1C-D), indicating that Mical stably modifies actin to alter polymerization. We next wondered if Mical, as an oxidoreductase enzyme, simply released diffusible oxidants to non-specifically alter polymerization. However, Mical does not alter polymerization of other proteins like tubulin (2) and preventing Mical-actin interactions abolished Mical’s effects on actin (Figs. 1E, S1–2). Indeed, unlike oxidases, which generate diffusible oxidants, most Mical-class monooxygenases/hydroxylases directly bind and are activated by their substrates (6). Likewise, Mical selectively binds F-actin (2) and increased its enzymatic activity by >100-fold in an F-actin-dependent manner (Fig. 1F). Thus, F-actin specifically activates the Mical enzyme and exhibits the characteristics of a direct Mical substrate.
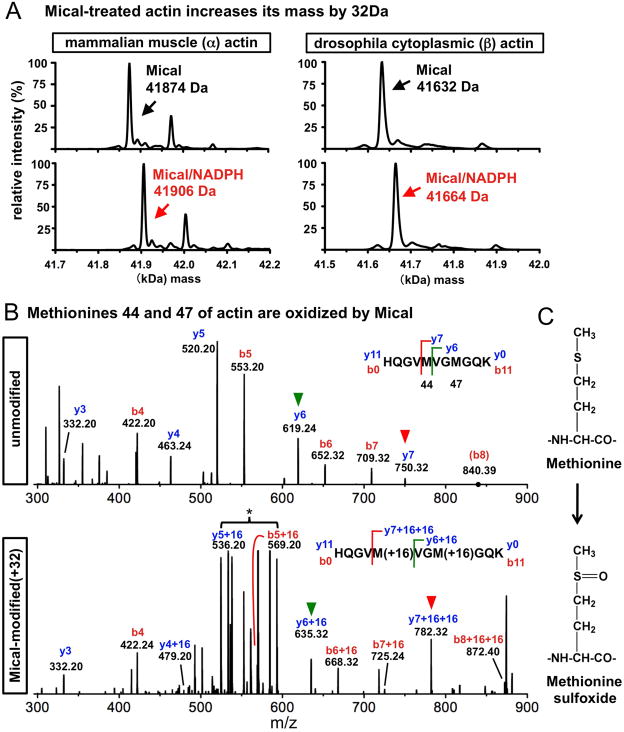
To determine if F-actin is a Mical substrate, we purified Mical/NADPH-treated actin (Fig. S3) and performed intact protein mass measurements. Mical/NADPH-treated actin increased its whole mass by 32 Daltons (Da) (Fig. 2A), which could represent the addition of two oxygens (16Da each) to actin. Further mass analysis revealed a substantial difference in the Mical/NADPH-treated actin peptide 40HQGVMVGMGQK50 (Figs. S3–4), and that actin’s methionine (M) 44 and M47 amino acid residues each had a mass increase of 16Da (Figs. 2B, S5). Moreover, Mical selectively modified only these two methionines and not any of actin’s 14 other methionines (Figs. S3–4, S6). Free methionine was also not a Mical substrate (Fig. 1F). M44 and M47 are poorly accessible to diffusible solvents including oxidants when actin is present in filaments (7–10), further indicating that these amino acid modifications are unlikely to be non-specific. Thus, Mical selectively adds 16Da (the equivalent of one oxygen) to both actin M44 and M47.
Fig. 2. Mical oxidizes actin M44 and M47.
(A) Mical/NADPH-treatment induces a 32Da shift in the whole mass of both α-actin and β-actin (Actin5C). (B) Spectra comparisons of the unmodified and modified (+32) peptide (Figs. S3–4) reveals 16Da increases on both M44 and M47. Individual amino acid bonds were broken from both ends of the peptide chain (y0-y11, b0-b11) and the mass determined for generated fragments. For example, comparing y6 (green arrowhead) and y7 (red arrowhead) fragments between and within samples, shows a +16 mass increase on the y7 ion (M44). Comparing b ions (breaking the peptide in the opposite direction) also reveals similar M44 and M47 mass increases. This Mical-modified spectrum (e.g., asterisk) is characteristic of methionine oxidation (see Fig. S5). (C) Methionine and its +16 form (methionine sulfoxide).
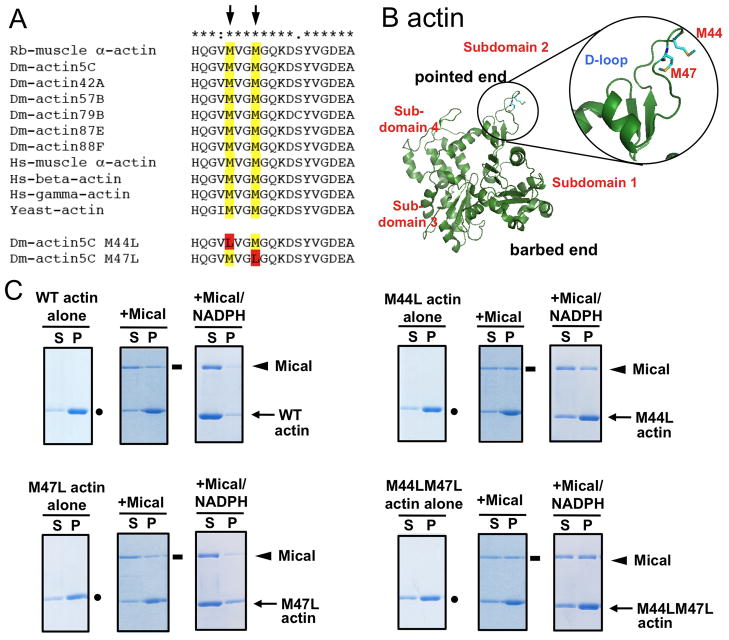
The M44 and M47 residues of actin are phylogenetically conserved and invariant among cardiac, muscle, and cytoplasmic actins (Fig. 3A), and lie within the D-loop (residues 39–51) of the subdomain 2 portion of actin (Fig. 3B), a region that mediates actin-actin contacts and polymerization (11, 12). To confirm that M44 and M47 are the functionally relevant sites on Mical-modified actin, we mutated each site, substituting chemically related leucine for methionine residues (Fig. 3A). We then expressed and purified wild-type, M44L, M47L, and the double mutant M44LM47L actin proteins (Figs. S7–8). In contrast to wild-type actin, which showed a 32Da difference after Mical treatment (Fig. 2A), M44LM47L actin was resistant to Mical-modification and exhibited a whole mass similar to control actin (Fig. S9). Thus, M44 and M47 are the only altered residues on Mical-treated actin and are required for Mical to post-translationally modify actin. We thus used these purified mutant actins to determine if modification of M44 and/or M47 induced Mical-mediated F-actin disassembly. All three mutant actins polymerized like wild-type actin and bound Mical (Fig. 3C). In contrast, F-actin generated by either wild-type or M47L depolymerized in the presence of Mical/NADPH, but filaments formed by M44L actin were resistant to Mical/NADPH and did not depolymerize (Fig. 3C). F-actin generated by M44LM47L was also resistant to Mical (Fig. 3C), revealing that Mical modifies actin M44 to induce F-actin disassembly.
Fig. 3. Mical disassembles F-actin by oxidizing M44.
(A) M44 and M47 (residue numbers from Rabbit) are conserved from yeast to humans. Rb, Rabbit; Dm, Drosophila; Hs, human. (B) The structure of monomeric actin including M44 and M47 in the D-loop of the pointed-end of the actin monomer. PDB ID is 2ZWH (13). (C) Co-sedimentation reveals that WT, M47L, M44L, and M44LM47L actins polymerize (dots, P fraction) and bind Mical (rectangles, P fraction). In contrast, Mical/NADPH disassembles WT and M47L but not M44L and M44LM47L actins (arrows, P fraction).
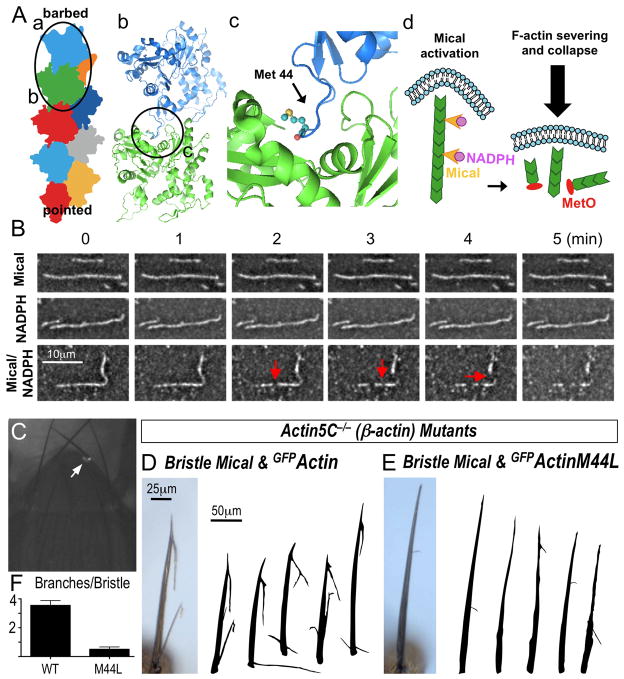
Methionine (2-amino-4-thiomethylbutanoic acid) is susceptible to the addition of an oxygen group on its sulfur atom, which generates methionine sulfoxide (MetO) (Fig. 2C). Our results, including the identification of MetO containing peptide fragments (Fig. S5), suggest that Mical directly converts M44 to MetO44, which disassembles F-actin and alters actin polymerization. The D-loop region containing M44 on the pointed-end of one actin monomer mediates the contact with the barbed-end of the adjacent actin monomer (Fig. 4A; (11, 13–16)). The side chain of methionine is normally flexible and uncharged, but the side chain of MetO with its oxygen atom is stiff and charged (17, 18). Thus, M44 oxidation would be predicted to affect the interaction between the pointed- and barbed-ends of individual actin subunits (12) and perhaps lead to F-actin disassembly. To examine if such a mechanism underlies Mical’s effects, we performed additional actin biochemical assays and visualized individual actin filaments directly using both real-time Total Internal Reflection Fluorescence (TIRF) and electron (EM) microscopy. Indeed, Mical cut actin filaments into multiple smaller pieces (Figs. 4B, S10–11; Movies S1–4), indicating that Mical-mediated M44 oxidation disrupts the association between individual actin monomers and thereby disassembles F-actin and alters re-polymerization.
Fig. 4. Mical-mediated M44 oxidation severs F-actin and triggers remodeling.
(A) Actin filament (a) formation involves amino acids within the D-loop at the pointed-end of one actin monomer associating with residues at the barbed-end of another actin monomer (b). The M44 residue is situated in this D-loop at a critical interface between adjacent actin monomers (c). Mical (d, yellow arrowheads) activation by NADPH oxidizes M44 (MetO), disrupting actin-actin interactions to cut filaments. (B) Time-lapse TIRF microscopy images reveal that individual actin filaments are cut (e.g., arrows) by Mical/NADPH but not by either Mical or NADPH only (and Movies S1–S4). (C–F) GFPActin5C M44L-marked single cell clone (C, arrow) mutant for actin5C. (D–F) Increasing Mical in single bristle cells generates F-actin reorganization and branching (D), which is suppressed by actin M44L expression (E-F). Mean±S.E.M., n≥10/genotype.
MICALs control the organization of actin in neurons, muscles, and bristles in vivo and mammalian cells in vitro (Fig. S12; (5)). A dominant mutation in the M44 residue (M44T) of skeletal muscle actin underlies a human musculoskeletal disease associated with actin accumulation and aggregation (nemaline myopathy (19)), and we sought to determine if M44 was necessary for Mical-mediated F-actin remodeling in vivo. Mical-mediated F-actin alterations, including that Mical is both activated and required for Semaphorin/Plexin-induced F-actin disassembly and remodeling, have been well-characterized using model Drosophila bristle processes (2), so we generated mutant bristle cells in which we replaced wild-type actin with M44L actin (Figs. 4C, S13). As in vitro (Fig. 3C), actin M44L incorporated into filaments in vivo (Fig. S13). In contrast, replacing wild-type actin with actin M44L suppressed the branching and shortening of bristles characteristic of elevated Mical activity (Figs. 4D–F), and generated Mical loss-of-function-like straight and tip-altered bristles (Fig. S13; (2)). Thus, Mical-mediated F-actin alterations in vivo, as in vitro, require the M44 residue of actin.
F-actin is thus a direct and specific substrate for Mical. This biochemical reaction alters actin at a specific amino acid residue disrupting actin-actin associations and fragmenting filaments. Also, this post-translationally-modified actin no longer polymerizes normally, differentiating Mical’s effects from other F-actin disassembly factors like cofilin which physically disassembles F-actin, recycles actin monomers, and promotes actin assembly (20). Furthermore, Mical modifies the pointed-end of actin proteins, and not the fast-growing, membrane-proximal barbed-end (Fig. 4A), providing a logic by which actin reassembly and branching (5) follows Semaphorin/Plexin/Mical-mediated F-actin collapse (Fig. S14). These results together present a specific oxidation-dependent mechanism (Figs. 4A, S15) that selectively regulates actin dynamics and cellular behavior.
Supplementary Material
Acknowledgments
We thank T. Brown, C. Cowan, H. Kramer, M. Rosen, and Terman lab members for assistance. Mass Spectrometry was performed by Y. Li (UT Southwestern Protein Chemistry Technology Center). We also thank Z. Chen, Y.-C. Kuo, K. Luby-Phelps, J. Merriam, H. Oda, V. Verkhusha, S. Weintraub, and Bloomington and Japanese Stock Centers. Supported by Cancer Prevention Research Institute of Texas (CPRIT) predoctoral (R.-J.H), NIH (DK091074) postdoctoral (C.W.P.), and NIH (NS073968) and Welch Foundation (I-1749) (J.R.T) grants. The data are presented in the figures and SOM.
Footnotes
References and Notes
- 1.Pollard TD, Cooper JA. Actin, a central player in cell shape and movement. Science. 2009;326:1208. doi: 10.1126/science.1175862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hung RJ, et al. Mical links semaphorins to F-actin disassembly. Nature. 2010;463:823. doi: 10.1038/nature08724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Terman JR, Mao T, Pasterkamp RJ, Yu HH, Kolodkin AL. MICALs, a family of conserved flavoprotein oxidoreductases, function in plexin-mediated axonal repulsion. Cell. 2002;109:887. doi: 10.1016/s0092-8674(02)00794-8. [DOI] [PubMed] [Google Scholar]
- 4.Tran TS, Kolodkin AL, Bharadwaj R. Semaphorin regulation of cellular morphology. Annu Rev Cell Dev Biol. 2007;23:263. doi: 10.1146/annurev.cellbio.22.010605.093554. [DOI] [PubMed] [Google Scholar]
- 5.Hung RJ, Terman JR. Extracellular inhibitors, repellents, and Semaphorin/Plexin/MICAL-mediated actin filament disassembly. Cytoskeleton. 2011;68:415. doi: 10.1002/cm.20527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Berkel WJ, Benen JA, Eppink MH, Fraaije MW. Flavoprotein kinetics. Methods Mol Biol. 1999;131:61. doi: 10.1385/1-59259-266-X:61. [DOI] [PubMed] [Google Scholar]
- 7.Dalle-Donne I, et al. Methionine oxidation as a major cause of the functional impairment of oxidized actin. Free Radic Biol Med. 2002;32:927. doi: 10.1016/s0891-5849(02)00799-2. [DOI] [PubMed] [Google Scholar]
- 8.Guan JQ, Almo SC, Reisler E, Chance MR. Structural reorganization of proteins revealed by radiolysis and mass spectrometry: G-actin solution structure is divalent cation dependent. Biochemistry. 2003;42:11992. doi: 10.1021/bi034914k. [DOI] [PubMed] [Google Scholar]
- 9.Guan JQ, Takamoto K, Almo SC, Reisler E, Chance MR. Structure and dynamics of the actin filament. Biochemistry. 2005;44:3166. doi: 10.1021/bi048021j. [DOI] [PubMed] [Google Scholar]
- 10.Takamoto K, Kamal JK, Chance MR. Biochemical implications of a three-dimensional model of monomeric actin bound to magnesium-chelated ATP. Structure. 2007;15:39. doi: 10.1016/j.str.2006.11.005. [DOI] [PubMed] [Google Scholar]
- 11.Sheterline PJ, Clayton, John C, Sparrow . Actin. 4. Oxford; 1998. [Google Scholar]
- 12.Dominguez R, Holmes KC. Actin structure and function. Annu Rev Biophys. 2011;40:169. doi: 10.1146/annurev-biophys-042910-155359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oda T, Iwasa M, Aihara T, Maeda Y, Narita A. The nature of the globular- to fibrous-actin transition. Nature. 2009;457:441. doi: 10.1038/nature07685. [DOI] [PubMed] [Google Scholar]
- 14.Murakami K, et al. Structural basis for actin assembly, activation of ATP hydrolysis, and delayed phosphate release. Cell. 2010;143:275. doi: 10.1016/j.cell.2010.09.034. [DOI] [PubMed] [Google Scholar]
- 15.Galkin VE, Orlova A, Schroder GF, Egelman EH. Structural polymorphism in F-actin. Nat Struct Mol Biol. 2010;17:1318. doi: 10.1038/nsmb.1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fujii T, Iwane AH, Yanagida T, Namba K. Direct visualization of secondary structures of F-actin by electron cryomicroscopy. Nature. 2010;467:724. doi: 10.1038/nature09372. [DOI] [PubMed] [Google Scholar]
- 17.Hoshi T, Heinemann S. Regulation of cell function by methionine oxidation and reduction. J Physiol. 2001;531:1. doi: 10.1111/j.1469-7793.2001.0001j.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Black SD, Mould DR. Development of hydrophobicity parameters to analyze proteins which bear post- or cotranslational modifications. Anal Biochem. 1991;193:72. doi: 10.1016/0003-2697(91)90045-u. [DOI] [PubMed] [Google Scholar]
- 19.Laing NG, et al. Mutations and polymorphisms of the skeletal muscle alpha-actin gene (ACTA1) Hum Mutat. 2009;30:1267. doi: 10.1002/humu.21059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ono S. Mechanism of depolymerization and severing of actin filaments and its significance in cytoskeletal dynamics. Int Rev Cytol. 2007;258:1. doi: 10.1016/S0074-7696(07)58001-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.