Abstract
To rejuvenate tissues and/or repair wounds, stem cells must receive extrinsic signals from their surrounding environment and integrate them with their intrinsic abilities to self-renew and differentiate to make tissues. Increasing evidence suggests that the superfamily of transforming growth factor-βs (TGF-βs) constitute integral components in the intercellular crosstalk between stem cells and their microenvironment. In this review, we summarize recent advances in our understanding of TGF-β superfamily functions in embryonic and adult stem cells. We discuss how these pathways help to define the physiological environment where stem cells reside, and how perturbations in the signaling circuitry contribute to cancers.
Introduction
A single fertilized egg gives rise to all the cell types in the body. As individual tissues form, they set aside reservoirs (niches) of stem cells that become more restricted in their lineage options. These fascinating cells are long-lived and their purpose is to make and replenish the differentiated cells within their resident tissues that are lost through normal stress and injury. Stem cells also have the remarkable property to replenish themselves, a process known as self-renewal. Although not a universal feature, many stem cells are used sparingly, often giving rise to rapidly proliferating but transient cells that perform the lion’s share of tissue regeneration. Stem cells that spend much of their time in a quiescent state are protected from unnecessary cell divisions that can lead to cancer.
Stem cell niches must be dynamic to enable stem cells to respond promptly to tissue demands (Hsu and Fuchs, 2012). Stem cells that receive signals from their surrounding environment galvanize intracellular transduction pathways, which deliver information to the genome in the form of activated transcription factors. These factors recognize specific sequence motifs in the genome, where they associate with different transcription factors, co-activators and chromatin remodelers to exert their effects. This combinatorial action of intrinsic and extrinsic factors allows for the same signaling pathways to be used in multiple cellular environments and elicit a diverse array of responses (Massagué and Xi, 2012).
Signaling by the transforming growth factor β (TGF-β) superfamily features prominently in embryonic development, tissue homeostasis and regeneration, immune responses, tumor suppression and metastasis (Li and Flavell, 2008; Massagué, 2008; Derynck and Miyazono, 2008; Wu and Hill, 2009). Not surprisingly, these factors also govern the behaviors of many stem cell populations. TGF-β factors are secreted, and depending upon on cell type, context, ligand expression and dosage, they can exert pleiotropic and sometimes opposing cellular effects ranging from proliferation, differentiation, migration and death. In addition to three TGF-βs (TGF-β 1–3), the superfamily includes bone morphogenetic proteins (BMPs), growth and differentiation factors (GDFs), anti-Müllerian hormone (AMH), Activins and Nodal. Many family members and their downstream pathway components are well-conserved across metazoan evolution, and indeed, BMPs and Nodal-type ligands are found in both vertebrates and invertebrates. An exception could be the superfamily’s namesakes, vertebrate TGF-β 1–3, which do not appear to have counterparts in nematodes (C. elegans) or insects (Drosophila) (Lapraz et al., 2006).
The TGF-β superfamily members are intriguing in that their activities are controlled at multiple levels. All are translated as larger polypeptides whose amino-terminal prodomains are required for their proper folding and dimerization (Gray and Mason, 1990). Even though cleavage by a furin-like protease occurs intracellularly, noncovalent association between TGF-βs and their prodomains persists after secretion. For some TGF-β superfamily members, like TGF-β 1–3, BMP10 and GDF-8/myostatin, their prodomains (latency associated proteins or LAPs) render them inactive and orchestrate their association with other inhibitory, latent TGF binding proteins (LTBPs). In these cases, the prodomains are sufficient to confer latency, while LTBPs target the complex to extracellular matrix (ECM) components such as fibrillins (Munger and Sheppard, 2011). For BMP4, 5 and 7 and some other processed family members, however, prodomain association does not affect activity (Sengle et al., 2011).
Biochemical studies have often used TGF-β1 as a paradigm. Although TGF-β1 synthesis and expression of its receptors occur in many cells, activation is typically paracrine and restricted to sites where TGF-β1 is released from latency (Figure 1). TGF-β1 activation has been shown to be influenced by plasmin, matrix metalloproteinases, thrombospondin-1, pH, and reactive oxygen species (Annes et al., 2003). However, TGF-β1 activation can also be controlled by certain integrins that can bind to the Arg-Gly-Asp (RGD) sequence within LAPs, resulting in a force-dependent conformational change that frees the active form from its prodomain (Munger et al., 1999; Shi et al., 2011). For the entire superfamily, a large repertoire of soluble extracellular agonists and antagonists within tissues provide further complexity in regulating ligand access to their receptors (Derynck and Miyazono, 2008).
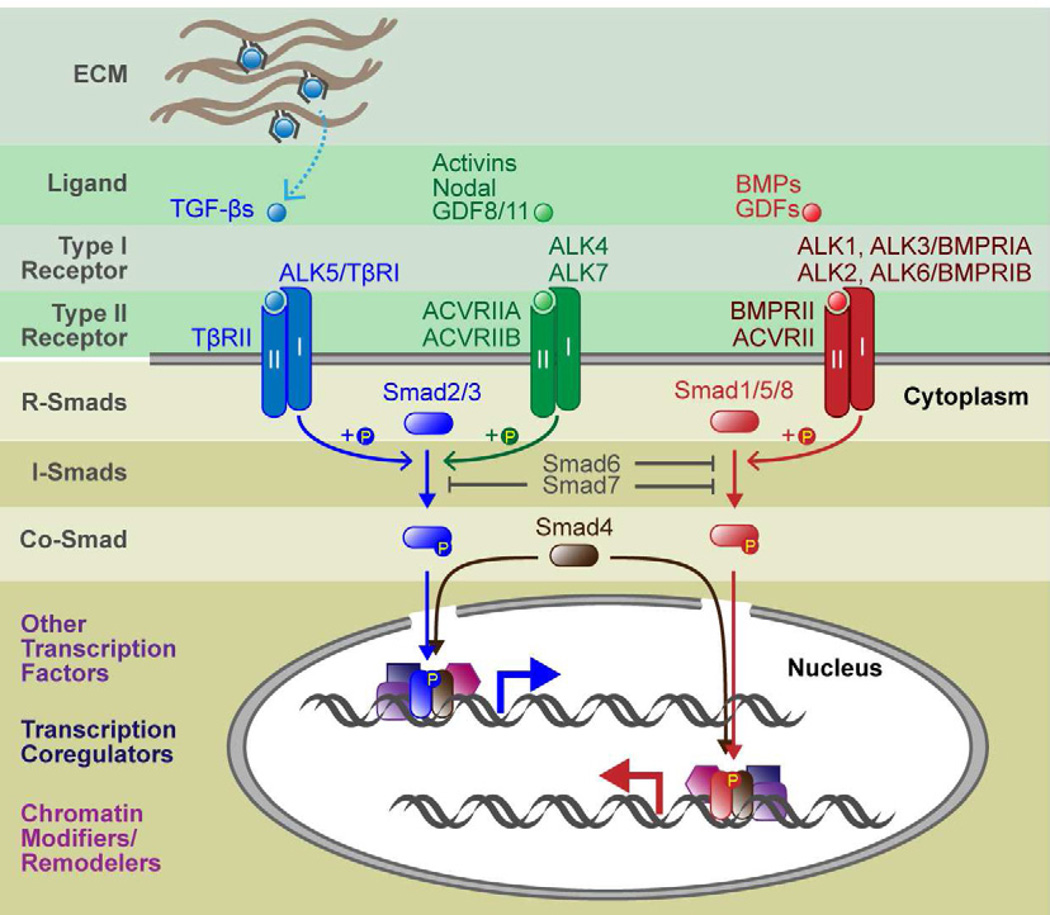
Figure 1. Canonical Signaling Pathways of TGF-β Superfamily.
When TGF-β propeptides are processed, the ligand associates noncovalently with the processed peptide. For some ligands, including the classical TGF-βs, activity is blocked by the peptide, and hence has been given the term latency associated peptide (LAP). LAPs can bind to latent TGF-β binding proteins (LTBPs) and other proteins within the extracellular matrix (ECM), and various activation mechanisms (dashed arrow) are then needed to extricate them from the ligand. Once liberated, ligands bind to specific combinations of type I and type II receptor serine/threonine kinases. In the ligand-receptor complexes, type II receptors phosphorylates and activates type I receptors, which in turn phosphorylate R-Smads and initiate intracellular signaling. R-Smad activation bifurcates the superfamily into two major branches: TGF-βs, activins and Nodal primarily use Smad2 and 3; BMPs and GDFs use Smad1, 5 and 8. Inhibitory Smads (Smad6 and 7) function by blocking type I receptor-mediated phosphorylation of Smad1/5/8 (Smad6) or both Smad2/3 and Smad1/5/8 (Smad7). Once phosphorylated, R-Smads couple with Co-Smad (Smad4) and translocate to the nucleus to act as a bipartite transcription factor. Generally, R-Smad-Smad4 transcription complexes have weak affinities to DNA and hence bind to other transcription factors for stable and specific binding to gene enhancers/promoters. R-Smad-Smad4 also associates with transcription coregulators, chromatin modifiers, and other chromatin remodelers.
Active TGF-β ligands signal by binding and bringing together two transmembrane serine-threonine kinases, known as receptor types I and II (Derynck, 1994) (Figure 1). In vertebrates, there are seven type I receptors [Activin-receptor like kinases (ALKs) 1–7] and five type II receptors (Schmierer and Hill, 2007). Superfamily ligands bind to and signal through specific type I and type II receptor complexes. Upon ligand activation, a type II receptor phosphorylates its type I receptor partner, which then propagates the signal by phosphorylating intracellular downstream substrates. The key canonical effectors are regulatory R-Smads 1,2,3,5 and 8. Smad2/3 become phosphorylated by ALK4/5/7 upon TGF-β, Nodal or Activin signaling. By contrast, Smad1/5/8 are typically phosphorylated by ALK1/2/3/6, downstream of BMP or GDF signaling, although some GDFs, such as GDF-8/myostatin and GDF11, function through Smad2/3, not Smad1/5/8. Inhibitory Smads (I-Smads: Smad6 and 7) can interfere with both of these branches by directly binding to R-Smads and blocking their modification (Figure 1).
Once phosphorylated, R-Smads can complex with a common mediator Smad (co-Smad: Smad4), translocate to the nucleus and form an active bipartite transcription factor. Smads require chromatin to assemble the basal transcription machinery, and thus are thought to affect transcription through modulation of chromatin structure (Ross et al., 2006). They can either activate or repress transcription through their interactions with chromatin-modifying co-activators such as p300/CBP histone acetyl-transferases (HATs) or co-repressors such as histone deacetylases (HDACs) or ATPases of the SWI/SNF chromatin-remodeling complex (Ross and Hill, 2008). Adding diversity to their transcriptional responses are their distinctive DNA sequence specificities. Complexes of Smad4 and activated (phosphorylated) Smad2/3 bind to AGAC or its complement GTCT, known as a Smad-binding element (SBE) (Dennler et al., 1998; Zawel et al., 1998), whereas Smad4-pSmad1/5/8 complexes preferentially bind to GGCGCC or GGAGCC, which is also known as the BMP-response element (BRE) (Katagiri et al., 2002; Korchynskyi and Dijke, 2002; Morikawa et al., 2011).
While the majority of TGF-β signaling routes go through phosphorylated R-Smads, not all responses involve Smad4. An interesting case in point is the catalytic subunit of a well-known transactivator of nuclear factor κB (NF-κB), IκB kinase (IKKα). In cultured epidermal keratinocytes, IKKα recruits pSmad2/3 to a specific promoter region and drives differentiation (Descargues et al., 2008). A Smad4-independent role for pR-Smads has also been implicated in microRNA maturation in the nucleus (Davis et al., 2008).
While less understood, pR-Smad-independent TGF-β signaling pathways have also been identified. One is E3 ligase TRAF6 (TNF receptor-associated factor 6), which can be activated directly by TGF-β receptor signaling. TRAF6 then modifies and activates TAK1 (TGF-β activated kinase-1), which can activate various kinases to elicit a diverse array of context-dependent responses (Sorrentino et al., 2008; Yamashita et al., 2008). Activated TGF-β receptors can also phosphorylate Shc, thereby recruiting Grb and Sos to the membrane, activating Ras GTPase and ERK MAPK signaling (Lee et al., 2007). Ligand-activated type II receptor TβRII can even signal independently of TβRI. One of its targets is Par6, which when phosphorylated, can spark events resembling an epithelial-mesenchymal transition (EMT) (Ozdamar et al., 2005). TGF-β1 signaling can also affect actin dynamics by activating the small GTPase RhoA (Bhowmick et al., 2001), and also Pak2 kinase, normally downstream of Cdc42 and Rac1 (Wilkes et al., 2003).
Endowed with the capacity to trigger such a remarkable cornucopia of downstream signaling pathways and cellular responses, it is no surprise that TGF-β superfamily members are potent morphogens in development. Their roles begin early, when positional information first specifies endoderm, mesoderm, and ectoderm (Zorn and Wells, 2007), and then patterns the body axes (De Robertis and Kuroda, 2004; Schier and Talbot, 2005). Their involvement continues during embryogenesis, as tissues and organs form. The TGF-β superfamily also functions in maintaining tissue homeostasis and regeneration, which are the life-sustaining features that are fueled by adult stem cells. The challenge for researchers is to understand the extrinsic and intrinsic factors that dictate how such signaling will influence a stem cell in its niche, and how these processes go awry in human disorders such as cancers and metastases, where TGF-β signaling pathways are known to be deregulated. By examining and comparing TGF-β superfamily signaling in different stem cell populations, glimpses as to how this might happen start to emerge. In this review, we witness elaborate molecular machineries that are build upon orchestrated crosstalk with intersecting signaling pathways to produce cell type-specific, context-dependent roles of TGF-β superfamily members.
TGF-β Superfamily Signaling in Embryonic Stem Cells
Roles of TGF-β Superfamily in Self-renewal and Pluripotency
Embryonic stem cells (ESCs) are pluripotent cells, initially derived from the inner cell mass (ICM) of mouse blastocyst embryos and grown as a cell line in tissue culture. While mouse ESCs (mESCs) provide a crucial tool for manipulating embryos to study mouse genetics, development, and physiology (Evans, 2011), human ESCs (hESCs) hold promise for regenerative medicine (Thomson et al., 1998). However, hESCs appear to have been derived from a later developmental stage than mESCs (Tesar et al., 2007; Brons et al., 2007). Thus, caution must be exercised in comparing data from mouse and human for either ESCs or induced pluripotent stem cells (iPSCs), which are ESC-like cells derived from genetic manipulation of adult tissue cells (Takahashi and Yamanaka, 2006).
Whether mouse or human, ESCs are remarkable for their capacity to be maintained and propagated in vitro, where they can be differentiated to form a myriad of cell types (Smith, 2001; Murry and Keller, 2008). ESC fate decisions and maintenance are controlled by both intrinsic and extrinsic factors. Internally, epigenetic processes such as DNA methylation and histone modifications contribute to ESC fate determination (Christophersen and Helin, 2010), while core transcription factors, including Tcf3, Nanog, Oct4, and Sox2, form auto-regulatory circuits that maintain self-renewal and pluripotency (Boyer et al., 2005; Chen et al., 2008; Kim et al., 2008). Externally however, mESCs and hESCs rely on different signaling pathways to self-renew and maintain pluripotency. mESCs rely on leukemia inhibitory factor (LIF) and BMP signaling (Smith, 2001; Qi et al., 2004), whereas hESCs rely on fibroblast growth factor (FGF) and Nodal/Activin signaling (Besser, 2004; Vallier et al., 2005). At least for mESCs, BMP and LIF combined alleviate the need for serum and feeder cells (Ying et al., 2003). Specific small molecule inhibitors of ERK and the GSK3β kinase (2i conditions) yield a similar outcome, indicating that under conditions where ERK levels are low, silencing GSK3β activity may be able to substitute for activating STAT3 (Ying et al., 2008). Analogously, serum-free defined medium supplemented with TGF-β1 or Nodal and FGF2 on a vitronectin substrate, can sustain in vitro growth of both hESCs and hiPSCs (Chen et al., 2011).
Although mESC fate decisions are influenced by external BMP/LIF and internal ERK activity, the details of these interconnections are still unfolding. Recently, Chen and colleagues demonstrated that BMP4 is required to significantly inhibit ERK activity (Li et al., 2012), which is consistent with a previous report (Qi et al., 2004). Digging deeper into mechanism, the authors provided evidence that BMP4 may exert these effects directly through canonical pSmad1/5-Smad4 activation of the gene encoding ERK-specific dual-specificity phosphatase 9 (Dusp9). Underscoring the physiological relevance of these findings, Dusp9 is expressed in E3.5 embryos of wild-type but not Smad4−/− mice. Moreover in vitro, Dusp9 knockdown uncouples BMP’s braking power on ERK, resulting in enhanced ERK phosphorylation/activation and decreased ESC self-renewal. Conversely, Dusp9 overexpression mimics BMP signaling to dampen ERK activity and sustain mESC self-renewal (Li et al., 2012). Together, these findings make a compelling case for Dusp9 as an essential BMP4 mediator of ERK inhibition in the natural process of mESC self-renewal (Figure 2A).
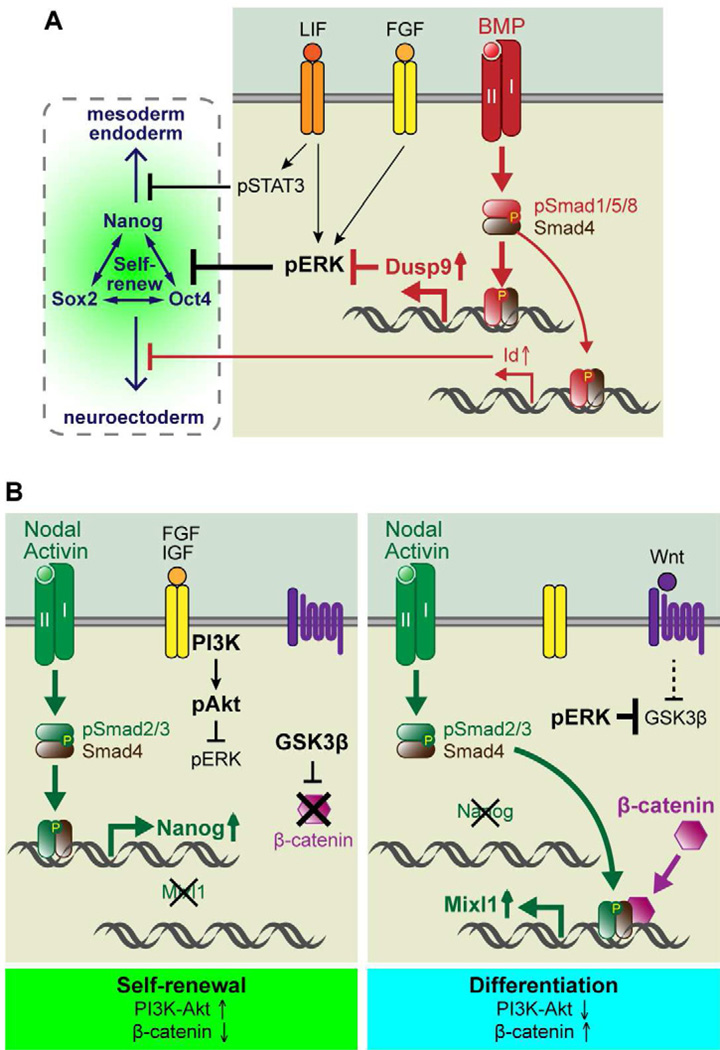
Figure 2. Signaling Crosstalk between TGF-β and Other Pathways in Mouse and Human ESCs.
(A) Self-renewal mechanisms in mouse ESCs. LIF and BMPs cooperate to maintain self-renewal. Tyrosine kinase receptor signaling, e.g. FGFs, typically activate downstream effectors ERK-MAPK, and induce differentiation. BMPs induce canonical Smad1/5/8 signaling pathways and one of their key targets for ESCs was recently shown to be the gene encoding ERK-specific phosphatase Dusp9. Dusp9 negatively affects ERK activity and hence support ESC self-renewal. Through the simultaneous induction of another BMP target gene, Id, and through activation of the STAT3 pathway by LIF, these two factors further promote ESC self-renewal by inhibiting mesendoderm and neuroectoderm differentiation, respectively.
(B) Self-renewal mechanisms in human ESCs. Nodal/Activin and hyperactive PI3K/Akt signaling cooperate to maintain self-renewal. Nodal/Activin induce canonical Smad2/3 signaling pathways and a key target for ESCs is Nanog. Hyperactive PI3K/Akt signaling can be achieved by some growth factors, such as FGF and IGFs, and this leads to suppression of ERK activity. When PI3K/Akt signaling diminishes, ERK inhibition is relieved, which in turn, suppresses the GSK3β kinase. This results in stabilization of β-catenin, which apparently associates with pSmad2/3-Smad4 to influence the target genes that become transcribed, thereby switching the ESC from a self-renewing to differentiating status.
An enigma is that BMP has opposite effects on hESC fate control, where it represses self-renewal and promotes differentiation (Zhang et al., 2008; Bernardo et al., 2011). In this regard, two points are noteworthy. First, as classical morphogens, BMPs and Nodals are known to elicit different developmental outcomes depending upon concentration. Secondly, the developmental differences between mESCs and hESCs could affect their differential responses.
Roles of TGF-β Superfamily in Balancing ESC Self-Renewal and Differentiation
During embryogenesis, progenitors respond to temporally well-defined regiments of morphogens which activate transcriptional networks and progressively seal cell fates. In many cases, including those involving TGF-β superfamily members, ERKs and canonical Wnts, the morphogens and signaling pathways that function in promoting lineage specification and differentiation are the same ones that are essential for the maintenance or self-renewal of cultured ESCs (McLean et al., 2007; Sumi et al., 2008). In the past few years, researchers have begun to uncover how relative activities of common signaling pathways can be recapitulated in vitro to determine whether ESCs should self-renew or differentiate.
Nodal/Activin signaling is critical for self-renewal and pluripotency of both hESCs and mESCs, where it leads to nuclear appearance of pSmad2/3 and transactivation of the key pluripotency gene Nanog (Xu et al., 2008; Vallier et al., 2009). Moreover, mouse epiblasts lacking either Nodal or Smad2/Smad3 are unusually small and express very little Oct4 (Conlon et al., 1994; Dunn et al., 2004). Analogously, treatment of hESCs with the Nodal receptor kinase inhibitor SB431542 results in decreased expression of pluripotency genes (James et al., 2005; Vallier et al., 2005). In hESCs, Nodal/pSmad2/3 signaling and GSK3β inhibition are sufficient to maintain the undifferentiated state (Besser, 2004). Thus, although the underlying mechanisms might differ in details, TGF-β superfamily signaling in ESCs converges squarely in the stream of events that reinforce self-renewal and pluripotency.
Despite its crucial role in ESC pluripotency, Nodal/Activin signaling also functions in vitro and in vivo to drive differentiation of pluripotent stem cells toward mesendoderm, the common progenitor of definitive mesoderm and endoderm lineages (D’Amour et al., 2005; Arnold and Robertson, 2009). Studying hESCs, Vallier and colleagues recently reported how Nodal/Activin signaling might maintain pluripotency without inducing mesendoderm differentiation (Chng et al., 2010). They identified Smad-interacting protein 1 (SIP1) as a strongly upregulated gene upon SB431542-mediated inhibition of Nodal/Activin. When Nodal/Activin signaling is active, Smad2/3 together with Nanog and Oct4 repress SIP1 expression. When Nodal/Activin signaling is dampened, elevated SIP1 further suppresses residual Nodal/Activin, thereby diminishing the mesendoderm-inducing effects of Nodal/Activin and BMP signaling. These changes favor the neuroectodermal “default” pathway (Camus et al., 2006), which is consistent with the finding that the synergistic action of BMP inhibitor (Noggin) and Nodal/Activin/TGF-β inhibitor (SB431542) is sufficient to induce neural induction in hESCs and iPSCs (Chambers et al., 2009). This model reveals how extracellular signals can cooperate with the core pluripotency transcriptional network to balance neuroectoderm and mesendoderm differentiation.
A recent study on hESCs by Dalton and colleagues provides a curious example of how this decision can be affected by complex crosstalk between Nodal/Activin/Smad2/3, MAPK/ERK and canonical Wnt/GSK3β/β-catenin pathways (Singh et al., 2012). They show that certain growth factors such as Fgf2 or Igf1/heregulin superactivate PI3K/Akt, and this leads to paradoxical suppression rather than enhancement of ERK activity. Such growth factors can thus act in conjunction with Nodal/Activin/Smad2/3/Nanog signaling to maintain self-renewal (Figure 2B, left). Conversely, reducing PI3K/Akt in ESCs exposed to Nodal favors mesendoderm differentiation (Figure 2B, right). At first glance, the effects of Nodal and PI3K on pluripotency seem similar to those on BMP4 and ERK suppression. However, the results differ in that robust PI3K/Akt elevates rather than suppresses GSK3β kinase levels, and in the face of active canonical Wnt/β-catenin signaling, Nodal/Activin induces genes that drive mesendoderm differentiation.
Typically, self-renewal is favored when GSK3β activity is suppressed, because it leads to enhanced stabilization of β-catenin, a key transcriptional co-factor of the canonical Wnt signaling pathway (Besser, 2004; Ying et al., 2008; Berge et al., 2011; Wray et al., 2011). Given the crosstalk between TGF-β superfamily and Wnt signaling pathways and the powerful effects of both pathways on self-renewal and differentiation, it is increasingly apparent that the combinatorial activities of other signaling pathways can dictate different outcomes.
Transcriptional networks downstream of TGF-β superfamily signaling
Recent genome-wide analyses of Smad proteins bound to chromatin have greatly advanced our understanding of TGF-β superfamily signaling. On their own, Smads bind with low affinity to DNA, and require interactions with other transcription factors to stabilize their binding to chromatin (Ross and Hill, 2008). In ESCs, Oct4, Sox2 and Nanog bind to a number of key pluripotency genes (Takahashi and Yamanaka, 2006; Graf and Enver, 2009). Interestingly, in response to BMP signaling, Smad1 (presumably with Smad4) binds to and regulates many of the pluripotency genes targeted by the Oct4/Nanog/Sox2 transcriptional complex (Chen et al., 2008). Similarly, in response to Nodal/Activin signaling, Smad3 (presumably with Smad4) forms a physical complex with Oct4 and directly regulates many of the same genes bound by Oct4 (Mullen et al., 2011). Additionally, Oct4 deficiency causes a dramatic reduction in Smad3 occupancy across the genome, and genes normally co-occupied by Oct4 and Smad3 lose their responsiveness to Nodal/Activin signaling. Since in ESCs, key pluripotency genes have a more open chromatin state, the ability of ESC master regulators to recruit pSmad3-Smad4 may also facilitate the weaker binding pSmad3-Smad4 complexes to recognize and bind to their cognate binding elements.
Another intriguing facet is that depending upon the cell type and its associated master regulators, Smad3 changes its repertoire of interacting partners and target genes. Thus, in myotubes, Smad3 co-occupies sites with Myod1 on muscle genes, while in pro-B cells, Smad3 couples with PU.1 on lymphocyte-specific genes (Figure 3A). These findings suggest that master transcription factors expressed by stem cells and progenitors function by priming key lineage-specific genes, which can then become activated in response to canonical TGF-β superfamily signaling. Since the abundance of cell type-specific master transcription factors increases upon differentiation, the master regulators ultimately dominate the competition for interactions with Smad2/3 (Young, 2011).
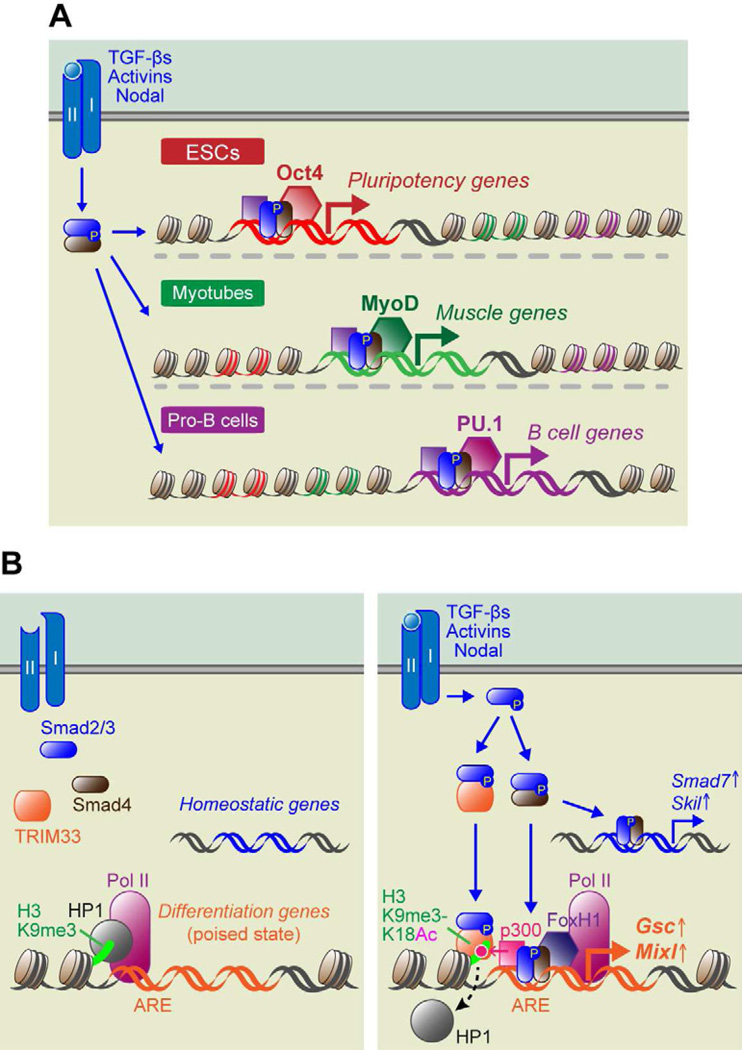
Figure 3. Molecular Mechanisms of ESC Differentiation and Lineage-specific Roles of TGF-β Signaling.
(A) In ESCs, activated pSmad2/3-Smad4 complexes are recruited by the master regulator Oct4 to genes that maintain pluripotency. In myotubes and Pro-B lymphocyte progenitors, activated Smad complexes are recruited by different lineage-specific master regulators, MyoD and PU.1, respectively, which are bound to lineage-specific genes governing muscle and B-cell differentiation.
(B) During mesendodermal fate specification of ESCs, signaling through TGF-βs, Activins and Nodal results in the generation of two distinct transcriptional complexes: pSmad2/3-Smad4 and pSmad2/3-TRIM33. Expression of homeostatic genes, such as Smad7 and Skil, are mediated only by pSmad2/3-Smad4. In contrast, master regulator genes of differentiation, such as Gsc and Mixl, exist in a transcriptionally poised state, as represented by the presence not only of RNA polymerase II at the transcription start site, but also the repressive histone modification, H3K9me3 and its binding associate HP1. In order to activate these poised genes, cooperative actions must occur between pSmad2/3-TRIM33 and pSmad2/3-Smad4 and an additional transcription cofactor FoxH1. In some way, this elicits an active chromatin conformation as revealed by histone acetyltransferase p300 and the added acetylation mark at lysine 18 on H3.
Genes that control master regulators of differentiation are often maintained in a quiescent but “poised” state, which can be rapidly transcribed in response to differentiation signals. The promoters of two master regulator genes for early embryonic development, goosecoid (Gsc) and Mixl1, contain poised features, which include RNA polymerase II (Pol II) paused at the transcription start site, and kept from active transcription by a chromatin compacting complex of H3K9me3 and HP1 (Figure 3B, left). Recently, the Massagué group reported that in response to Nodal/Activin signaling, pSmad2/3 forms a complex with tripartite motif 33 (TRIM33, also known as TIF1γ/ectodermin), previously identified as a TGF-β1-induced pSmad2/3 binding partner in hematopoietic stem cells (HSCs) (He et al., 2006). In this case, pSmad2/3-TRIM33 complexes appeared to translocate to the nucleus where they recognized histone marks, displaced HP1 and allowed binding of Smad4-pSmad2/3 to Activin Response Elements (AREs) within the Gsc and Mixl1 promoters (Xi et al., 2011). In turn, the complex recruited additional transcriptional regulators, further generating the requisite active chromatin conformation and relieving Gsc and Mixl1 from their poised states (Figure 3B, right). By contrast, Activin/Nodal regulated genes involved in essential cellular functions and homeostasis do not exist in this poised state and consequently responded to TGF-β signaling by either activating or repressing their transcription. An additional point worth mentioning is that TRIM33 depletion in mESCs markedly inhibited the expression of mesendodermal but not ectodermal markers, further consistent with the established role of Nodal/Activin in mesendodermal fate specification.
While this model invoking TRIM33 is intriguing, studies by Piccolo and colleagues suggest that TRIM33 binds to Smad4 and not Smad2/3, and that it functions as a ubiquitin ligase for Smad4 (Dupont et al., 2005; Agricola et al., 2011). Moreover, at least two other hypothetical models could explain Nodal/Activin-induced triggering of ESC differentiation: the first is through the crosstalk with Wnt signaling described above (Singh et al, 2012), and the other is through physical interactions between pSmad2/3-Smad4 and histone demethylase JMJD3/KDM6B, resulting in a loss of H3K27me3 repressive marks in Gsc and other Nodal/Activin target gene loci (Dahle et al, 2010; Kim et al., 2011). Given the intensity of research in this arena, the precise transcriptional governance of ESC differentiation by signaling through TGF-β superfamily members will surely continue to evolve.
TGF-β Superfamily Signaling in Tissue-Specific Stem Cells
Most if not all adult tissues harbor resident stem cells that are dedicated to life-long maintenance and wound repair. Some are perpetually active, such as intestinal stem cells, while others are more sparingly used, such as stem cells of hair follicles, hematopoietic system and mammary gland. Irrespective of cycling behaviors, adult stem cells typically give rise to intermediate progenitors, sometimes referred to as “transit amplifying” (TA) cells, which then progress to differentiate to make functional tissue. TGF-β superfamily members figure prominently in most if not all of these steps, and broadly in most if not all tissues. Studies in the past decade have begun to shed light on some of the reasons why.
At high levels, TGF-βs often inhibit cell proliferation in a reversible manner (Massagué, 2008). This raises particular intrigue for the stem cell field, where many adult stem cells not only display TGF-β receptors on their cell surface but also survive for months in a quiescent state (Cotsarelis et al., 1990; Cheshier et al., 1999; Tumbar et al., 2004). In contrast to senescence and terminal differentiation, quiescent stem cells can re-enter the cell cycle in response to specific environmental cues. An ability of TGF-β superfamily members to balance active proliferation and reversible cell cycle exit may be important to maintain reservoirs of stem cells that are able to respond quickly to changes in tissue physiology.
Hematopoietic Stem Cells
Each day, billions of new blood cells are made to replace the billions lost. This stream of fresh new cells in our blood originates from multipotent hematopoietic stem cells (HSCs), which give rise to all myeloid and lymphoid lineages (Wang and Wagers, 2011). Adult HSCs reside as rare cells in the bone marrow, where they sit atop a hierarchy of progenitors that become progressively lineage restricted (Zhang et al., 2003; Orkin and Zon, 2008). However, HSCs show heterogeneous behavior at the clonal level (Lemischka et al., 1986; Smith et al., 1991) and recent evidence suggests that the hematopoietic system is actually maintained by a consortium of functionally distinct HSC subtypes with different self-renewal and differentiation potentials (Sieburg et al., 2006; Dykstra et al., 2007).
Goodell and colleagues recently characterized two distinct HSC subpopulations and discovered that they respond differently to TGF-β signaling (Challen et al., 2010). One subtype was relatively quiescent, longer-lived and biased towards differentiating into myeloid lineages, while the other subtype was more proliferative, shorter-lived, and biased towards lymphoid differentiation. Each subtype was successful in long-term, multi-lineage reconstitution of the entire blood system, and thus merited HSC classification. Moreover, each population maintained inherent bias towards generating their designated hematopoietic branch in serial transplantations.
Interestingly, TGF-β exacerbated these functional differences in vitro and in vivo. At high levels, TGF-βs inhibited proliferation of both myeloid-biased (My-) and lymphoid-biased (Ly-) HSCs. However at lower concentrations, TGF-βs stimulated My-HSC but still impaired Ly-HSC proliferation in vitro, and when recombinant TGF-β1 was administered to mice during hematopoietic reconstitution in vivo, these differential effects were recapitulated (Figure 4A). Although the exact mechanisms underlying these distinct cellular responses to TGF-β signaling are not yet known, the TGF-β-induced differences in behavior were accompanied by subtype-specific differences in expression of the master regulators of myeloid and lymphoid differentiation, namely PU.1 (granulocyte/macrophage lineage) and Ikaros (B and T cell lineage) (Challen et al., 2010). In light of these new findings, it is intriguing that aging HSCs have a myeloid-differentiation bias compared to their young counterparts (Sudo et al., 2000; Kim et al., 2003). In the future, it will be interesting to see if these age-related differences are rooted in local or systemic TGF-β production, and if so, whether subtype balance might be restored by lowering TGF-β levels.
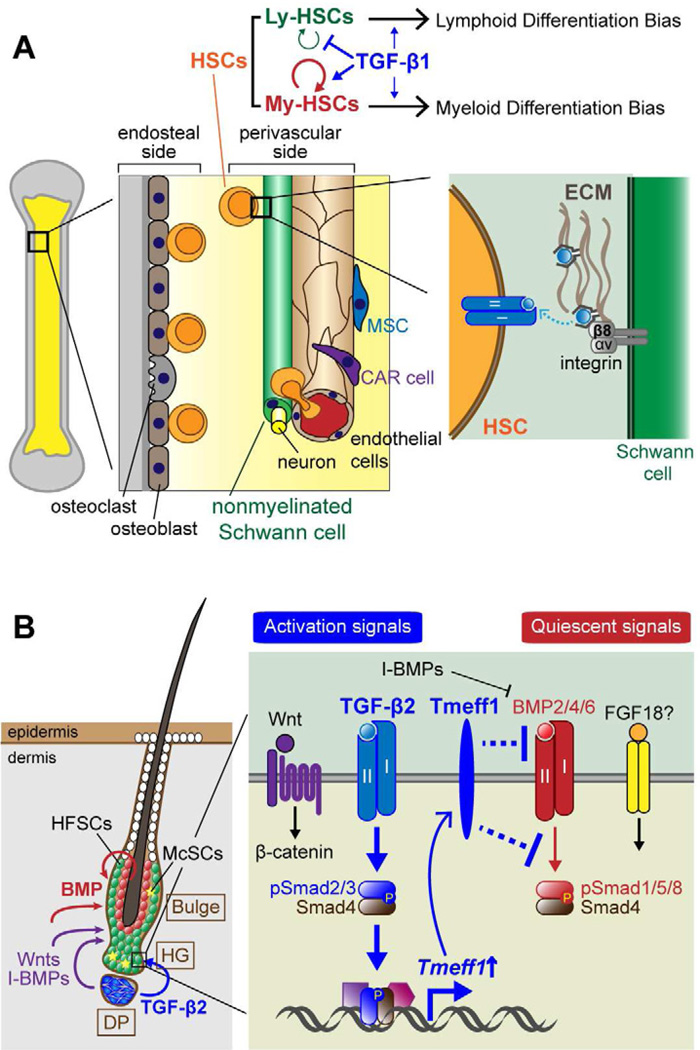
Figure 4. Diverse functions of TGF-β signaling in the control of adult stem cell behaviors.
(A) The bone marrow niche consists of a variety of cell types, which include hematopoietic stem cells (HSCs), mesenchymal stem cells, osteoblasts, endothelial cells, CXCL12-abundant reticular (CAR) cells, sympathetic neurons and nonmyelinating Schwann cells. HSCs located on the endosteal side of the niche tend to be quiescent, a feature necessary for their self-renewal and maintenance of stemness, while HSCs residing on the perivascular side are more active, priming them for differentiation. HSC quiescence is dependent upon active TGF-β, which appears to be released from its associated LAP through a mechanism involving β8-integrin, expressed on the surface of the nonmyelinated Schwann cells. TGF-β signaling also exacerbates the functional differences between the myeloid and lymphoid HSC subtypes. TGF-β enhances My-HSC self-renewal at low levels and drives myeloid differentiation at higher levels. By contrast, even at low levels, TGF-β induces Ly-HSC differentiation and inhibits their self-renewal.
(B) In the hair follicle, epithelial hair follicle stem cells (HFSCs) and melanocyte stem cells (McSCs) are located in a niche referred to as the bulge and hair germ (HG). Demarcating the dermis and epithelium is a basement membrane, and although the dermis contributes to niche signaling, HFSCs are restricted to the follicle side of the basement membrane. Sandwiched between the HFSC layer and the club hair is a layer of inner bulge cells (red circles) that emit high levels of BMP6 and FGF18 which maintain HFSC quiescence. Another key niche component is a transient one, the dermal papilla (DP), which only sits at the base of the bulge during the resting phase of the hair cycle, when HFSCs are in their most quiescent state. During this phase, crosstalk between HFSCs and DP change their transcriptional states and ultimately lead to TGF-β2 production by the DP, and pSmad2/3-Smad4 signaling by the adjacent HFSCs. A key downstream TGF-β2 signaling target is the gene encoding Tmeff1, which inhibits BMP signaling and lowers the threshold necessary to initiate HFSC self-renewal and fate specification necessary to launch a new round of hair growth.
TGF-β signaling has long been implicated in regulating HSC quiescence (Fortunel et al., 2000; Yamazaki et al., 2009). In part, TGF-βs seem to prevent HSC re-entry into the cell cycle by upregulating transcription of the cyclin-dependent kinase (CDK) inhibitor p57Kip2 and suppressing PI3K/Akt signaling (Yamazaki et al., 2006). Niche cells in the bone marrow produce latent TGF-β. Since HSCs cannot activate TGF-β by themselves (Yamazaki et al., 2009), it has remained a mystery as to how active TGF-β exists in the HSC niche.
Recently, Nakauchi and colleagues used antibodies specific to the latent and active TGF-β forms to reveal that the nonmyelinating Schwann cells ensheathing sympathetic nerves in bone marrow are responsible for activation (Yamazaki et al., 2011). Although many details of the pathway remain to be determined, integrins αvβ8 expressed by Schwann cells most likely facilitate proteolytic degradation of the latent peptide by activating matrix metalloproteinases (Mu et al., 2002; Annes et al., 2003). To ascertain how TGF-β controls HSC quiescence, the authors generated lymphocyte-deficient, TβRII conditional knockout mice and infused bone marrow cells from these mice into lethally-irradiated recipients (Yamazaki et al., 2011). HSCs lacking TβRII showed reduced pSmad2/3, increased cycling, and reduced long-term repopulating activity in HSCs. Indeed, the contribution of TβRII-deficient HSCs to peripheral blood myeloid cells gradually declined over time. Furthermore, when nonmyelinating Schwann cells in bone marrow were depleted by denervation of sympathetic nerves and then used as recipients in repopulation analyses, pSmad2/3 was reduced in HSCs and their overall numbers declined. The ability to convert the latent form of TGF-β to its active state adds to the multiplicity of ways in which niche cells might control quiescence of their stem cells (Figure 4A).
By studying the zebrafish hematopoietic system, the Zon group recently showed that analogous to ESCs, HSCs utilize both BMP and Wnt signaling to regulate lineage selection and differentiation (Trompouki et al., 2011). Moreover in responding to these signaling cues, HSCs apply many of the transcriptional paradigms used by ESCs. Thus, Smad1 and Tcf3 selectively co-occupied at enhancer elements of HSC lineage-distinctive genes in concert with cell type-specific master regulators: GATA1 for the erythroid lineage and C/EBPα, which often co-functions with PU.1, for the myeloid lineage. In addition, signaling greatly enhanced activation of these blood cell type-specific enhancer elements, most likely through recruitment of p300 histone acetyltransferase. Moreover at the start of hematopoietic regeneration, BMP and Wnt signaling resulted in colocalization of Smad1 and Tcf3 to master regulator-bound genes that define progenitor cell fate. As regeneration continued and progenitor cells began to differentiate, different master regulators redirected Smad1 and Tcf3 to genes expressed at more mature hematopoietic stages (Trompouki et al., 2011).
Hair Follicle Stem Cells
In adult mice, hair follicles undergo dynamic, synchronized bouts of growth (anagen), degeneration (catagen) and rest (telogen). During the resting phase, which can last for months, epithelial hair follicle stem cells (HFSCs) are quiescent and reside in a niche, referred to as the bulge (Fuchs, 2007). The single layer of polarized bulge HFSCs is sandwiched between an outer basement membrane and an inner layer of keratin 6-expressing, differentiated cells that anchor the old hair (Hsu et al., 2011). A small cluster of “primed” HFSCs (the hair germ, HG), reside at the bulge base and will be the first progenitors activated at the start of each new hair cycle. The HG directly overlies the dermal papilla (DP), which serves as the essential mesenchymal signaling center for HFSCs (Cotsarelis et al., 1990). Adding to the complexity of the bulge stem cell niche are smooth muscle fibers (Fujiwara et al., 2011), sensory neurons (Brownell et al., 2011), adipocytes (Festa et al., 2011), blood vessels, and a sheath of dermal fibroblasts. A small number of melanocyte stem cells (McSCs) also reside in the bulge, particularly in the HG, and coordinate their quiescence, activation and lineage commitments with the HFSCs (Nishimura et al., 2002).
Throughout the resting phase, HFSC quiescence is maintained in part by BMPs provided from the inner layer of non-stem niche cells (Hsu et al., 2011) and from surrounding dermal fibroblasts and adipocytes (Plikus et al., 2008). When the Bmpr1a gene is conditionally ablated during the resting phase, HFSCs become active and overproduce, and tumor-like structures form from the bulge (Zhang et al., 2006; Kobielak et al., 2007). Thus in the normal niche, BMP signaling must be transiently lowered in order to activate proliferation and lineage commitment in a small number of HFSCs, but then it must be elevated again to enable the niche to restore quiescence within its residents. BMP signaling also functions in the TA cells of the follicle, but in this case, elevated signaling is associated with terminal differentiation (Kobielak et al., 2003).
The hair cycle is fueled by epithelial-mesenchymal crosstalk that takes place at the base of the bulge stem cell niche during the resting phase. It culminates in the accumulation of the necessary threshold of activating factors to transition the stem cells to begin a new round of hair growth (Greco et al., 2009). The process involves Wnt-activating cues (Merrill et al., 2001; Enshell-Seijffers et al., 2010; Rabbani et al., 2011) as well as BMP inhibitory factors (Kulessa et al., 2000; Botchkarev et al., 2001; Zhang et al., 2006). Many of these factors, including Rspo1/2 (Wnt-activating) and Sostdc1 (BMP-inhibitory), are made by the DP (Greco et al., 2009).
Recently, we uncovered an additional player, TGF-β2, which is primarily produced by DP but triggers signaling and nuclear pSmad2 in the overlying HG cells just prior to their activation (Oshimori and Fuchs, 2012). Unexpectedly, TGF-β signaling appeared to stimulate HFSC proliferation by counteracting quiescence in the niche. Moreover, TβRII-deficient HFSCs displayed elevated pSmad1 and BMP signaling and delayed hair cycle entry. Interestingly, the antagonistic effects of TGF-β signaling on BMP signaling appeared to be rooted in at least one underlying target gene, Tmeff1, which was previously shown to block BMP2-mediated mesoderm induction in Xenopus embryos (Chang et al., 2003). Tmeff1 is normally upregulated concomitant with TGF-β2 signaling and inhibition of BMP signaling in activated HG progenitors (Oshimori and Fuchs, 2012). In TβRII mutant progenitors, Tmeff1 was diminished, and when Tmeff1 was knocked down in wild-type HFSCs, TGF-β2’s suppression of BMP signaling was abrogated, and hair follicle regeneration was significantly delayed (Figure 4B). Finally, Tmeff1 diminished the response of wild-type HFSCs to BMP signaling in vitro, in a fashion similar to that of TGF-β2.
Similar to the effects of TGF-βs observed for My-HSCs (Challen et al., 2010), HFSCs were stimulated by low concentrations of TGF-β2 both in vivo and in vitro, but higher concentrations resulted in cell cycle arrest (Oshimori and Fuchs, 2012). In the future, it will be interesting to see the extent to which the opposing stem cell behaviors elicited by different TGF-β levels arise from the antagonizing effects that TGF-β signaling can have on BMP signaling, and whether additional antagonistic effects will emerge between other members of the TGF-β superfamily. That said, it is already evident that not all stem cells respond similarly to TGF-β signaling, since for Ly-HSCs, TGF-βs even at low levels showed growth inhibition (Challen et al., 2010). Similarly in the hair follicle, niche-derived TGF-β signaling appears to function in McSC quiescence, and loss of TβRII in McSCs results in their ectopic activation and differentiation (Nishimura et al., 2010), which does not happen when TβRII is targeted in HFSCs (Oshimori and Fuchs, 2012).
An additional note in comparing HFSCs and HSCs is that analogous to HSCs, a handful of key transcription factors and epigenetic regulators have been shown to function as master regulators of HFSC renewal and maintenance (Blanpain and Fuchs, 2009; Lien et al., 2011; Chen et al., 2012b). Although a few of the factors, e.g. Lhx2, are expressed by both HSCs and HFSCs, most are not, suggesting that the main role of master regulators in stem cells is to govern lineage-specific programs rather than universal features of stemness. Moreover, although the diverse roles of BMPs, Wnts and TGF-βs in activating tissue-specific lineage programs in stem cells is in agreement with the ability of their transcriptional effectors to interact with these tissue-specific master regulators, their significantly broader expression among stem cells renders them candidates for regulating general stem cell features as well.
Lessons Learned from Studying TGF-β Superfamily Signaling in Other Adult Stem Cell Populations
Comprehensive coverage of TGF-β superfamily signaling in all adult stem cells is beyond the scope of the current review. However, in addition to reinforcing many of the paradigms discussed thus far, some intriguing new twists on these signaling networks have emerged from other systems. Here, we highlight just a few of the many lessons learned from studying TGF-β signaling in other adult stem cell populations.
Neural Stem Cells
The central nervous system develops from self-renewing, multipotent neuroepithelial cells that give rise to neural progenitor cells and eventually to neurons, astrocytes, and oligodendrocytes. The choice between self-renewal versus differentiation of neuroepithelial cells determines growth and size of the developing brain (Götz and Huttner, 2005). As in many stem cells, Wnt/β-catenin signaling promotes cell-cycle progression of neuroepithelial cells (Chenn and Walsh, 2002). Recently, Sommer and colleagues showed that TGF-β signaling controls brain size by antagonizing canonical Wnt signaling and negatively regulating neuroepithelial cell self-renewal (Falk et al., 2008).
Neurogenesis continues throughout adult life, and is fueled by at least two specialized neural stem cell (NSC) niches: the subventricular zone (SVZ) of the lateral ventricles and the subgranular zone (SGZ) of the hippocampal dentate gyrus (Zhao et al., 2008). In the SGZ, quiescent NSCs (Sox2+ radial cells) co-exist with their progenitors (Sox2+ nonradial cells). Recently, Gage and colleagues found that quiescent NSCs express ALK3/BMPRIA, while their progeny express ALK6/BMPRIB (Mira et al., 2010). Although pSmad1 was detected in both NSCs and progenitors, BMP signaling was transiently inhibited in NSCs undergoing proliferation. Moreover, when BMP signaling was blocked in the brain niche, NSCs initially proliferated, but this eventually waned. Interestingly, even though the number of progenitors diminished, the number of NSCs remained constant. These observations are compatible with a model in which the production of progenitors by asymmetrically dividing NSCs becomes compromised after a certain number of mitotic rounds, resembling NSC behavior in aged hippocampus (Hattiangady and Shetty, 2008). Taken together, these findings argue that canonical BMP signaling downstream of ALK3/BMPRIA regulates NSC quiescence, which is required to support continuous neurogenesis throughout the adult life. It will be interesting in the future to identify the source of BMP ligands and antagonists in the brain, and to elucidate the significance of using different BMP receptors (BMPR1A and BMPR1B) during normal homeostasis.
Skeletal muscle stem cells
Skeletal muscle development, growth, and regeneration rely on muscle stem cells (MuSCs). Fetal myogenesis is important for the growth and maturation of muscles. Duprez and colleagues recently revealed that BMP4 signals from emerging tendons impact the behavior of a subpopulation of dividing MuSCs at the tips of fetal skeletal muscle (Wang et al., 2010). In contrast to NSCs, enhanced BMP signaling via ALK3/BMPRIA seemed to promote MuSC proliferation, as judged by increased numbers of fetal MuSCs and muscle fibers. Conversely, blocking BMP signaling resulted in fewer MuSCs. When taken together with the markedly elevated BMP expression by tendons, these results are consistent with prior observations noting increased satellite (MuSC) cell proliferation at sites close to tendons (Tsujimura et al., 2006), and longitudinal growth at the ends of skeletal muscle fibers (Williams and Goldspink, 1971).
In the adult, quiescent MuSCs reside between muscle fibers and surrounding basement membrane. Upon muscle damage, quiescent MuSCs begin proliferating, differentiate into myoblasts, and fuse to form de novo multinucleated myofibers (Collins et al., 2005). However, as the muscle ages, its ability to regenerate diminishes and eventually fails. It has been suggested that the age-related decline in muscle tissue regeneration may result from diminished activation of Notch pathway, which is known to antagonize Wnt signaling and enable MuSCs to proliferate (Conboy et al., 2003; Brack et al., 2007).
Recently, Conboy and colleagues found that factors secreted by aged myofibers result in higher levels of TGF-β and nuclear pSmad3 signaling in MuSCs, whether old or young (Carlson et al., 2008). These tantalizing findings prompt the speculation that with age, signaling activity in the MuSC niche shifts from active Notch to active TGF-β/pSmad3. Consistent with this notion, endogenous Notch and pSmad3 have opposing effects on MuSC proliferation. Through mechanisms not fully understood, active Notch reduces pSmad3 occupancy on key TGF-β-dependent target genes, such as CDK inhibitors (Carlson et al., 2008). To this end, attenuation of canonical TGF-β signaling in old, injured muscle restores MuSC activity in vivo. These findings provide compelling evidence that this age-specific interplay between active Notch and TGF-β/pSmad3 signaling controls CDK inhibitor levels in MuSCs and in turn governs tissue regenerative capacity upon muscle injury.
Recently, synergistic crosstalk between Notch and TGF-β signaling was also discovered in prostate epithelial homeostasis (Valdez et al., 2012). In prostate basal stem/progenitor cells, Notch appears to serve not only as a downstream effector but also an amplifier for the TGF-β-induced cytostatic program.
Bone stem cells
Alterations in bone remodeling occur in many human bone diseases. The normal adult bone undergoes continual remodeling by precisely coordinating the activities of osteoblasts, which deposit the calcified bone matrix, and osteoclasts, which resorb bone (Zaidi, 2007). Factors released from bone matrix during osteoclastic bone resorption are thought to orchestrate migration of mesenchymal stem cells (MSCs) to the resorptive surfaces of the bone. Cao and colleagues demonstrated that osteoclastic bone resorption releases stored TGF-β1, which recruits bone MSCs to active sites of remodeling through canonical pSmad signaling (Tang et al., 2009). Interestingly, either over-activation or inhibition of TGF-β signaling caused significant reductions in MSCs at bone remodeling surfaces. Moreover, hyperactivation of TGF-β signaling uncoupled bone resorption and formation processes. Probing into mechanism, local TGF-β1 levels appear to determine how many bone MSCs will be recruited to resorptive sites, which secrete osteotropic factors such as BMPs, IGFs and PDGFs that promote MSC differentiation into osteoblasts.
Cancer Stem Cells
Cancers develop from normal cells that eventually gain the ability to proliferate aberrantly and become malignant. Many of the mutations leading to cancer affect signaling pathways involving TGF-β superfamily members (Bierie and Moses, 2006; Massagué, 2008). In addition, for cancers arising in individuals who are not genetically at high risk, tissue-specific stem cells may be preferential targets for initial oncogenic mutations, because they are long-lived and hence have time to accumulate the myriad of mutations that will ultimately lead to cancer (Lobo et al., 2007). Cancer stem cells (CSCs) are cells within a cancer that have the capacity to be serially transplanted at the single cell level and initiate new cancers with characteristics of the parental tumor. The concept of CSCs reveals a new framework of cancer therapeutic strategies (Clevers, 2011). CSCs have been identified in a number of different cancers of both hematopoietic and solid tissue lineages (Driessens et al., 2012; Schepers et al., 2012; Chen et al., 2012a).
Schober and Fuchs recently identified and characterized CSCs purified from the tumor-stroma interface in a number of malignant squamous cell carcinomas (SCCs) (Schober and Fuchs, 2011). The SCCs investigated included those whose skin epithelium is compromised for TGF-β signaling and which showed enhanced integrin/focal adhesion kinase (FAK) signaling and tumor susceptibility (Guasch et al., 2007), and those which lack integrin/FAK signaling and exhibit increased levels of tumor regression (McLean et al., 2004). TβRII-null CSCs formed highly aggressive, less differentiated SCCs, and exhibited a dramatic increase in the ability to initiate secondary tumor formation. Surprisingly, these phenotypes were almost completely compromised in CSCs from SCCs lacking both TβRII and FAK (Schober and Fuchs, 2011). Thus, CSC proliferation and expansion at the tumor-stroma interface where TGF-β and integrin/FAK signaling intersect, is dependent upon the extent to which CSCs can respond to stromal TGF-β1 and counterbalance the elevated integrin/FAK signaling that otherwise occurs during malignant transformation.
Chronic myeloid leukemia (CML) is a cancer of white blood cells caused by a constitutively active tyrosine kinase, BCR-ABL. It is typified by clonal expansion of HSCs. Although treatments with tyrosine kinase inhibitors dramatically improve survival rate, leukemia-initiating cells (LICs) can escape treatments and drive CML recurrence. In CML cell lines, BCR-ABL activates PI3K/Akt signaling leading to nuclear export and diminished activity of Foxo transcription factors. If Akt phosphorylation is suppressed in LICs, Foxo3a becomes nuclear, a feature that correlates with long-term maintenance, survival and malignant potential of LICs (Naka et al., 2010). As in normal HSCs (Yamazaki et al., 2006), TGF-β selectively inhibits Akt activation and facilitates nuclear Foxo3a in CML LICs and not non-LICs, underscoring the possible therapeutic importance of small molecule inhibitors of TGF-β signaling. Indeed, in mouse CML models, the TβRI inhibitor Ly364947 combined with the tyrosine kinase inhibitor imatinib significantly reduced recipent lethality and decreased CML infiltration in lung, suggesting that CML LICs are even more sensitive than normal HSCs to TGF-β inhibitors (Naka et al., 2010).
Agonistic effects of TGF-β signaling are also found in malignant gliomas, where TGF-β works as an oncogenic factor and is also considered a therapeutic target (Akhurst, 2006; Seoane, 2008). Indeed, when patient-derived gliomas are dissociated into single cells and cultured, TGF-β enhanced the number and size of neurospheres, suggesting that TGF-β signaling increased self-renewal in glioma initiating cells (GICs) (Peñuelas et al., 2009). Interestingly in this case, the effects of TGF-βs on GIC self-renewal were traced to the LIF/STAT signaling pathway, and the gene encoding LIF appeared to be a direct pSmad2/3-Smad4 target. Moreover, TGF-β did not induce LIF nor increase the self-renewal capacity of normal NSCs, again underscoring the striking differences in how TGF-βs effect self-renewal in normal versus CSCs. Miyazono and colleagues also demonstrated that glioma cells produce autocrine TGF-β signaling and TGF-β-Sox4-Sox2 pathway is important for maintenance of stemness of GICs (Ikushima et al., 2009). BMP has also been reported to be involved in both suppression and promotion of GIC tumorigenicity (Piccirillo et al., 2006; Lee et al., 2008).
The agonistic effects of TGF-βs on CSCs of leukemias and gliomas are in stark contrast to the antagonistic effects of TGF-βs on CSCs from squamous cell carcinomas. The ability of different CSCs to respond differently to TGF-βs underscores the potential dangers in using TGF-β agonists or antagonists in the clinic. In addition, even for a single stem cell type, TGF-βs can have dual role in oncogenesis. Thus in many normal and premalignant cells, TGF-β enforces homeostasis and suppresses tumor progression either directly, through cell-autonomous tumor-suppressive effects or indirectly, through effects on the stroma. However, when cancer cells lose TGF-β tumor-suppressive responses, they can use TGF-βs to their advantage to initiate immune evasion, produce growth factors, transform into an invasive phenotype, and metastasize to establish and expand elsewhere in the body (Massagué, 2008).
In addition to cell proliferation control of tumor cells and CSCs, TGF-βs also play distinct roles in epithelial-mesenchymal transitions (EMTs). EMTs affect critical steps of morphogenesis by interconverting epithelial cell types into cells with mesenchymal attributes, allowing polarized cells of an epithelial sheet to delaminate, assume a spindle-like mesenchymal shape, migrate from their site of origin, and invade surrounding tissue (Thiery et al., 2009). In addition, some epithelial cells that pass through an EMT acquire self-renewing traits typical of stem cells (Mani et al., 2008), and when activated in carcinoma cells, EMTs can lead to metastasis and high-grade malignancy (Yang and Weinberg, 2008). Interestingly, immortalized human mammary epithelial cells with mesenchymal features display autocrine TGF-β1 as well as canonical and non-canonical Wnt signaling (Scheel et al., 2011). In vitro, these signaling pathways cooperatively enhance mammosphere formation and other stemness characteristics, and upon Ras-oncogenic transformation and transplantation, signs of increased tumor-initiation and metastasis emerge. In some cell types, BMP signaling has been found to antagonize TGF-β1-driven EMTs (Zeisberg et al., 2003), providing another example of mutual antagonism between the TGF-β and BMP branches of superfamily signaling.
Conclusions and perspectives
A decade ago, it was becoming increasingly clear that most if not all adult tissues have stem cells. A stream of new insights have followed, shedding light on how stem cells decide when to make tissue, when to stop making it and when to self-renew. At the helm of these decisions is the intricate balance between proliferation and cell fate signals that stem cells receive from their niche microenvironments. Not surprisingly, the TGF-β superfamily features prominently in making the choice, often in consultation with other intersecting pathways, e.g. Wnts and receptor tyrosine kinases. But each stem cell niche is different. Thus, even though every stem cell may be endowed with receptors that enable them to respond to TGF-βs/Nodal, BMPs, Wnts and growth factors, their responses often differ, in part because of intrinsic differences established during the course of development, and in part because the particular constellation of extracellular cues within each niche is distinct. When coupled with the ability of TGF-β and BMP branches of the superfamily to antagonize each other as well as intersect with and influence receptor tyrosine kinase and Wnt pathways, even subtle differences, perhaps only in signal doses, can have profound effects.
While the complexities underlying TGF-β superfamily regulation of stem cell behavior may seem daunting, sifting through the biochemical mechanisms has been aided by recent advances in genome-wide analyses of Smad binding to chromatin. Major new insights have come from the finding that the choice of genes to be regulated by pSmad2/3-Smad4 is guided by the master regulators expressed by a particular stem cell lineage. Similar findings have come for Tcf3, a key component in the Wnt pathway, suggesting that this paradigm may hold for other signaling pathways that control stem cell behavior and fate commitment. Indeed, when coupled with sequence and context-specific differences in pR-Smad-Smad4 binding to its targets, this model could explain not only how stem cells receive and translate different dosages and combinations of extracellular cues, but also how a particular signaling pathway perceives the temporal lineage of a tissue-specific stem cell, and why a cell’s life history matters. The concept may be especially pertinent in explaining why stem/progenitor cells at distinct developmental stages may respond differently to a collection of extracellular cues. Moreover, given that new master regulators may themselves be targets of a signaling pathway, it begins to show why the order in which a cell receives a series of signals often matters critically. In this regard, spatial and temporal profiling of signaling at the single-cell level, by for example the reporter system we used (Oshimori and Fuchs, 2012), will help to understand the complex mechanisms of TGF-β,
As we emphasized in this review, new levels of regulation within the TGF-β superfamily continue to emerge, and with them come new appreciations for the questions still unanswered and areas remaining to be explored. While we have learned that the strength of TGF-β superfamily signaling often dictates cell fate, it is not clear how different signaling levels are sensed by different stem cells and how this is impacted by their niche. How does signaling dosage impact R-Smad-Smad4 target gene selection and/or expression? Similarly, the sources and cells responsible for activation of TGF-β ligands have helped to provide new insights into their roles in governing the stem cell niche, but our knowledge on TGF-β ligand bioavailability is still limited. How broadly are mechanisms of latency and ligand activation utilized in biology? Are they operative in stem cell niches, and if so which niches and under what circumstances? Another intriguing facet of TGF-β signaling is the existence of non-canonical signaling among the TGF-β superfamily. How broadly are such mechanisms used by different superfamily members, and under what occasions? Do switches in canonical and non-canonical TGF-β superfamily signaling pathways function in regulating the balance between self-renewal and fate commitment? Thus in addition to the tremendous advances made in our knowledge of stem cells and TGF-β superfamily signaling, there are many exciting and fascinating discoveries awaiting the field in the decade to come. Given that stem cells and TGF-β signaling stand precariously at the intersection between normal tissue biology and cancer, the new insights awaiting to be gained will no doubt find their way to clinical applications.
Acknowledgments
We thank S. Beronja and Y.C. Hsu for valuable discussions. N.O. was supported by the Human Frontier Science Program (HFSP) and is currently supported by the Japan Society for the Promotion of Science. E.F. is an investigator of the Howard Hughes Medical Institute. The authors’ research that led up to this review was supported by grants to E.F. from National Institutes of Health (R01-AR31737) and the Emerald Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agricola E, Randall RA, Gaarenstroom T, Dupont S, Hill CS. Recruitment of TIF1γ to chromatin via its PHD finger-bromodomain activates its ubiquitin ligase and transcriptional repressor activities. Mol Cell. 2011;43:85–96. doi: 10.1016/j.molcel.2011.05.020. [DOI] [PubMed] [Google Scholar]
- Akhurst RJ. Large- and small-molecule inhibitors of transforming growth factor-beta signaling. Curr Opin Investig Drugs. 2006;7:513–521. [PubMed] [Google Scholar]
- Annes JP, Munger JS, Rifkin DB. Making sense of latent TGFbeta activation. J Cell Sci. 2003;116:217–224. doi: 10.1242/jcs.00229. [DOI] [PubMed] [Google Scholar]
- Arnold SJ, Robertson EJ. Making a commitment: cell lineage allocation and axis patterning in the early mouse embryo. Nat Rev Mol Cell Biol. 2009;10:91–103. doi: 10.1038/nrm2618. [DOI] [PubMed] [Google Scholar]
- Berge ten D, Kurek D, Blauwkamp T, Koole W, Maas A, Eroglu E, Siu RK, Nusse R. Embryonic stem cells require Wnt proteins to prevent differentiation to epiblast stem cells. Nat Cell Biol. 2011;13:1070–1075. doi: 10.1038/ncb2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardo AS, Faial T, Gardner L, Niakan KK, Ortmann D, Senner CE, Callery EM, Trotter MW, Hemberger M, Smith JC, et al. BRACHYURY and CDX2 mediate BMP-induced differentiation of human and mouse pluripotent stem cells into embryonic and extraembryonic lineages. Cell Stem Cell. 2011;9:144–155. doi: 10.1016/j.stem.2011.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besser D. Expression of nodal, lefty-a, and lefty-B in undifferentiated human embryonic stem cells requires activation of Smad2/3. J. Biol. Chem. 2004;279:45076–45084. doi: 10.1074/jbc.M404979200. [DOI] [PubMed] [Google Scholar]
- Bhowmick NA, Ghiassi M, Bakin A, Aakre M, Lundquist CA, Engel ME, Arteaga CL, Moses HL. Transforming growth factor-beta1 mediates epithelial to mesenchymal transdifferentiation through a RhoA-dependent mechanism. Mol. Biol. Cell. 2001;12:27–36. doi: 10.1091/mbc.12.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierie B, Moses HL. Tumour microenvironment: TGFbeta: the molecular Jekyll and Hyde of cancer. Nat Rev Cancer. 2006;6:506–520. doi: 10.1038/nrc1926. [DOI] [PubMed] [Google Scholar]
- Blanpain C, Fuchs E. Epidermal homeostasis: a balancing act of stem cells in the skin. Nat Rev Mol Cell Biol. 2009;10:207–217. doi: 10.1038/nrm2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botchkarev VA, Botchkareva NV, Nakamura M, Huber O, Funa K, Lauster R, Paus R, Gilchrest BA. Noggin is required for induction of the hair follicle growth phase in postnatal skin. Faseb J. 2001;15:2205–2214. doi: 10.1096/fj.01-0207com. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brack AS, Conboy MJ, Roy S, Lee M, Kuo CJ, Keller C, Rando TA. Increased Wnt signaling during aging alters muscle stem cell fate and increases fibrosis. Science. 2007;317:807–810. doi: 10.1126/science.1144090. [DOI] [PubMed] [Google Scholar]
- Brons IG, Smithers LE, Trotter MW, Rugg-Gunn P, Sun B, Chuva de Sousa Lopes SM, Howlett SK, Clarkson A, Ahrlund-Richter L, Pedersen RA, Vallier L. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448:191–195. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- Brownell I, Guevara E, Bai CB, Loomis CA, Joyner AL. Nerve-derived sonic hedgehog defines a niche for hair follicle stem cells capable of becoming epidermal stem cells. Cell Stem Cell. 2011;8:552–565. doi: 10.1016/j.stem.2011.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camus A, Perea-Gomez A, Moreau A, Collignon J. Absence of Nodal signaling promotes precocious neural differentiation in the mouse embryo. Dev Biol. 2006;295:743–755. doi: 10.1016/j.ydbio.2006.03.047. [DOI] [PubMed] [Google Scholar]
- Carlson ME, Hsu M, Conboy IM. Imbalance between pSmad3 and Notch induces CDK inhibitors in old muscle stem cells. Nature. 2008;454:528–532. doi: 10.1038/nature07034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challen GA, Boles NC, Chambers SM, Goodell MA. Distinct hematopoietic stem cell subtypes are differentially regulated by TGF-beta1. Cell Stem Cell. 2010;6:265–278. doi: 10.1016/j.stem.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Eggen BJL, Weinstein DC, Brivanlou AH. Regulation of nodal and BMP signaling by tomoregulin-1 (X7365) through novel mechanisms. Dev Biol. 2003;255:1–11. doi: 10.1016/s0012-1606(02)00075-1. [DOI] [PubMed] [Google Scholar]
- Chen G, Gulbranson DR, Hou Z, Bolin JM, Ruotti V, Probasco MD, Smuga-Otto K, Howden SE, Diol NR, Propson NE, et al. Chemically defined conditions for human iPSC derivation and culture. Nat Methods. 2011;8:424–429. doi: 10.1038/nmeth.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Li Y, Yu T-S, McKay RM, Burns DK, Kernie SG, Parada LF. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012a;488:522–526. doi: 10.1038/nature11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Heller E, Beronja S, Oshimori N, Stokes N, Fuchs E. An RNA interference screen uncovers a new molecule in stem cell self-renewal and long-term regeneration. Nature. 2012b;485:104–108. doi: 10.1038/nature10940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- Chenn A, Walsh CA. Regulation of cerebral cortical size by control of cell cycle exit in neural precursors. Science. 2002;297:365–369. doi: 10.1126/science.1074192. [DOI] [PubMed] [Google Scholar]
- Cheshier SH, Morrison SJ, Liao X, Weissman IL. In vivo proliferation and cell cycle kinetics of long-term self-renewing hematopoietic stem cells. Proc Natl Acad Sci USA. 1999;96:3120–3125. doi: 10.1073/pnas.96.6.3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chng Z, Teo A, Pedersen RA, Vallier L. SIP1 mediates cell-fate decisions between neuroectoderm and mesendoderm in human pluripotent stem cells. Cell Stem Cell. 2010;6:59–70. doi: 10.1016/j.stem.2009.11.015. [DOI] [PubMed] [Google Scholar]
- Christophersen NS, Helin K. Epigenetic control of embryonic stem cell fate. J Exp Med. 2010;207:2287–2295. doi: 10.1084/jem.20101438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H. The cancer stem cell: premises, promises and challenges. Nat. Med. 2011;17:313–319. doi: 10.1038/nm.2304. [DOI] [PubMed] [Google Scholar]
- Collins CA, Olsen I, Zammit PS, Heslop L, Petrie A, Partridge TA, Morgan JE. Stem cell function, self-renewal, and behavioral heterogeneity of cells from the adult muscle satellite cell niche. Cell. 2005;122:289–301. doi: 10.1016/j.cell.2005.05.010. [DOI] [PubMed] [Google Scholar]
- Conboy IM, Conboy MJ, Smythe GM, Rando TA. Notch-mediated restoration of regenerative potential to aged muscle. Science. 2003;302:1575–1577. doi: 10.1126/science.1087573. [DOI] [PubMed] [Google Scholar]
- Conlon FL, Lyons KM, Takaesu N, Barth KS, Kispert A, Herrmann B, Robertson EJ. A primary requirement for nodal in the formation and maintenance of the primitive streak in the mouse. Development. 1994;120:1919–1928. doi: 10.1242/dev.120.7.1919. [DOI] [PubMed] [Google Scholar]
- Cotsarelis G, Sun TT, Lavker RM. Label-retaining cells reside in the bulge area of pilosebaceous unit: implications for follicular stem cells, hair cycle, and skin carcinogenesis. Cell. 1990;61:1329–1337. doi: 10.1016/0092-8674(90)90696-c. [DOI] [PubMed] [Google Scholar]
- Dahle Ø, Kumar A, Kuehn MR. Nodal signaling recruits the histone demethylase Jmjd3 to counteract polycomb-mediated repression at target genes. Sci Signal. 2010;3:ra48. doi: 10.1126/scisignal.2000841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Amour KA, Agulnick AD, Eliazer S, Kelly OG, Kroon E, Baetge EE. Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat Biotechnol. 2005;23:1534–1541. doi: 10.1038/nbt1163. [DOI] [PubMed] [Google Scholar]
- Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Robertis EM, Kuroda H. Dorsal-ventral patterning and neural induction in Xenopus embryos. Annu. Rev. Cell Dev. Biol. 2004;20:285–308. doi: 10.1146/annurev.cellbio.20.011403.154124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennler S, Itoh S, Vivien D, Dijke ten P, Huet S, Gauthier JM. Direct binding of Smad3 and Smad4 to critical TGF beta-inducible elements in the promoter of human plasminogen activator inhibitor-type 1 gene. Embo J. 1998;17:3091–3100. doi: 10.1093/emboj/17.11.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derynck R. TGF-beta-receptor-mediated signaling. Trends Biochem. Sci. 1994;19:548–553. doi: 10.1016/0968-0004(94)90059-0. [DOI] [PubMed] [Google Scholar]
- Derynck R, Miyazono K. The TGF-[beta] family. Cold Spring Harbor Laboratory Pr; 2008. [Google Scholar]
- Descargues P, Sil AK, Sano Y, Korchynskyi O, Han G, Owens P, Wang X-J, Karin M. IKKalpha is a critical coregulator of a Smad4-independent TGFbeta-Smad2/3 signaling pathway that controls keratinocyte differentiation. Proc Natl Acad Sci USA. 2008;105:2487–2492. doi: 10.1073/pnas.0712044105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driessens G, Beck B, Caauwe A, Simons BD, Blanpain C. Defining the mode of tumour growth by clonal analysis. Nature. 2012 doi: 10.1038/nature11344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn NR, Vincent SD, Oxburgh L, Robertson EJ, Bikoff EK. Combinatorial activities of Smad2 and Smad3 regulate mesoderm formation and patterning in the mouse embryo. Development. 2004;131:1717–1728. doi: 10.1242/dev.01072. [DOI] [PubMed] [Google Scholar]
- Dupont S, Zacchigna L, Cordenonsi M, Soligo S, Adorno M, Rugge M, Piccolo S. Germ-layer specification and control of cell growth by Ectodermin, a Smad4 ubiquitin ligase. Cell. 2005;121:87–99. doi: 10.1016/j.cell.2005.01.033. [DOI] [PubMed] [Google Scholar]
- Dykstra B, Kent D, Bowie M, McCaffrey L, Hamilton M, Lyons K, Lee S-J, Brinkman R, Eaves C. Long-term propagation of distinct hematopoietic differentiation programs in vivo. Cell Stem Cell. 2007;1:218–229. doi: 10.1016/j.stem.2007.05.015. [DOI] [PubMed] [Google Scholar]
- Enshell-Seijffers D, Lindon C, Kashiwagi M, Morgan BA. beta-catenin activity in the dermal papilla regulates morphogenesis and regeneration of hair. Dev Cell. 2010;18:633–642. doi: 10.1016/j.devcel.2010.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans M. Discovering pluripotency, 30 years of mouse embryonic stem cells. Nat Rev Mol Cell Biol. 2011;12:680–686. doi: 10.1038/nrm3190. [DOI] [PubMed] [Google Scholar]
- Falk S, Wurdak H, Ittner LM, Ille F, Sumara G, Schmid M-T, Draganova K, Lang KS, Paratore C, Levéen P, et al. Brain area-specific effect of TGF-beta signaling on Wnt-dependent neural stem cell expansion. Cell Stem Cell. 2008;2:472–483. doi: 10.1016/j.stem.2008.03.006. [DOI] [PubMed] [Google Scholar]
- Festa E, Fretz J, Berry R, Schmidt B, Rodeheffer M, Horowitz M, Horsley V. Adipocyte lineage cells contribute to the skin stem cell niche to drive hair cycling. Cell. 2011;146:761–771. doi: 10.1016/j.cell.2011.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortunel N, Hatzfeld J, Kisselev S, Monier MN, Ducos K, Cardoso A, Batard P, Hatzfeld A. Release from quiescence of primitive human hematopoietic stem/progenitor cells by blocking their cell-surface TGF-beta type II receptor in a short-term in vitro assay. Stem Cells. 2000;18:102–111. doi: 10.1634/stemcells.18-2-102. [DOI] [PubMed] [Google Scholar]
- Fuchs E. Scratching the surface of skin development. Nature. 2007;445:834–842. doi: 10.1038/nature05659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara H, Ferreira M, Donati G, Marciano DK, Linton JM, Sato Y, Hartner A, Sekiguchi K, Reichardt LF, Watt FM. The basement membrane of hair follicle stem cells is a muscle cell niche. Cell. 2011;144:577–589. doi: 10.1016/j.cell.2011.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Götz M, Huttner WB. The cell biology of neurogenesis. Nat Rev Mol Cell Biol. 2005;6:777–788. doi: 10.1038/nrm1739. [DOI] [PubMed] [Google Scholar]
- Graf T, Enver T. Forcing cells to change lineages. Nature. 2009;462:587–594. doi: 10.1038/nature08533. [DOI] [PubMed] [Google Scholar]
- Gray AM, Mason AJ. Requirement for activin A and transforming growth factor--beta 1 pro-regions in homodimer assembly. Science. 1990;247:1328–1330. doi: 10.1126/science.2315700. [DOI] [PubMed] [Google Scholar]
- Greco V, Chen T, Rendl M, Schober M, Pasolli HA, Stokes N, Cruz-Racelis Dela J, Fuchs E. A two-step mechanism for stem cell activation during hair regeneration. Cell Stem Cell. 2009;4:155–169. doi: 10.1016/j.stem.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guasch G, Schober M, Pasolli HA, Conn EB, Polak L, Fuchs E. Loss of TGFbeta signaling destabilizes homeostasis and promotes squamous cell carcinomas in stratified epithelia. Cancer Cell. 2007a;12:313–327. doi: 10.1016/j.ccr.2007.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattiangady B, Shetty AK. Aging does not alter the number or phenotype of putative stem/progenitor cells in the neurogenic region of the hippocampus. Neurobiol. Aging. 2008;29:129–147. doi: 10.1016/j.neurobiolaging.2006.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W, Dorn DC, Erdjument-Bromage H, Tempst P, Moore MAS, Massagué J. Hematopoiesis controlled by distinct TIF1gamma and Smad4 branches of the TGFbeta pathway. Cell. 2006;125:929–941. doi: 10.1016/j.cell.2006.03.045. [DOI] [PubMed] [Google Scholar]
- Hsu Y-C, Fuchs E. A family business: stem cell progeny join the niche to regulate homeostasis. Nat Rev Mol Cell Biol. 2012;13:103–114. doi: 10.1038/nrm3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu Y-C, Pasolli HA, Fuchs E. Dynamics between stem cells, niche, and progeny in the hair follicle. Cell. 2011;144:92–105. doi: 10.1016/j.cell.2010.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikushima H, Todo T, Ino Y, Takahashi M, Miyazawa K, Miyazono K. Autocrine TGF-beta signaling maintains tumorigenicity of glioma-initiating cells through Sry-related HMG-box factors. Cell Stem Cell. 2009;5:504–514. doi: 10.1016/j.stem.2009.08.018. [DOI] [PubMed] [Google Scholar]
- James D, Levine AJ, Besser D, Hemmati-Brivanlou A. TGFbeta/activin/nodal signaling is necessary for the maintenance of pluripotency in human embryonic stem cells. Development. 2005;132:1273–1282. doi: 10.1242/dev.01706. [DOI] [PubMed] [Google Scholar]
- Katagiri T, Imada M, Yanai T, Suda T, Takahashi N, Kamijo R. Identification of a BMP-responsive element in Id1, the gene for inhibition of myogenesis. Genes Cells. 2002;7:949–960. doi: 10.1046/j.1365-2443.2002.00573.x. [DOI] [PubMed] [Google Scholar]
- Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Moon H-B, Spangrude GJ. Major age-related changes of mouse hematopoietic stem/progenitor cells. Ann. N. Y. Acad. Sci. 2003;996:195–208. doi: 10.1111/j.1749-6632.2003.tb03247.x. [DOI] [PubMed] [Google Scholar]
- Kim SW, Yoon SJ, Chuong E, Oyolu C, Wills AE, Gupta R, Baker J. Chromatin and transcriptional signatures for Nodal signaling during endoderm formation in hESCs. Dev Biol. 2011;357:492–504. doi: 10.1016/j.ydbio.2011.06.009. [DOI] [PubMed] [Google Scholar]
- Kobielak K, Pasolli HA, Alonso L, Polak L, Fuchs E. Defining BMP functions in the hair follicle by conditional ablation of BMP receptor IA. J Cell Biol. 2003;163:609–623. doi: 10.1083/jcb.200309042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobielak K, Stokes N, la Cruz de J, Polak L, Fuchs E. Loss of a quiescent niche but not follicle stem cells in the absence of bone morphogenetic protein signaling. Proc Natl Acad Sci USA. 2007;104:10063–10068. doi: 10.1073/pnas.0703004104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korchynskyi O, Dijke ten P. Identification and functional characterization of distinct critically important bone morphogenetic protein-specific response elements in the Id1 promoter. J. Biol. Chem. 2002;277:4883–4891. doi: 10.1074/jbc.M111023200. [DOI] [PubMed] [Google Scholar]
- Kulessa H, Turk G, Hogan BL. Inhibition of Bmp signaling affects growth and differentiation in the anagen hair follicle. Embo J. 2000;19:6664–6674. doi: 10.1093/emboj/19.24.6664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapraz F, Röttinger E, Duboc V, Range R, Duloquin L, Walton K, Wu S-Y, Bradham C, Loza MA, Hibino T, et al. RTK and TGF-beta signaling pathways genes in the sea urchin genome. Dev Biol. 2006;300:132–152. doi: 10.1016/j.ydbio.2006.08.048. [DOI] [PubMed] [Google Scholar]
- Lee J, Son MJ, Woolard K, Donin NM, Li A, Cheng CH, Kotliarova S, Kotliarov Y, Walling J, Ahn S, et al. Epigenetic-mediated dysfunction of the bone morphogenetic protein pathway inhibits differentiation of glioblastoma-initiating cells. Cancer Cell. 2008;13:69–80. doi: 10.1016/j.ccr.2007.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MK, Pardoux C, Hall MC, Lee PS, Warburton D, Qing J, Smith SM, Derynck R. TGF-beta activates Erk MAP kinase signalling through direct phosphorylation of ShcA. Embo J. 2007;26:3957–3967. doi: 10.1038/sj.emboj.7601818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemischka IR, Raulet DH, Mulligan RC. Developmental potential and dynamic behavior of hematopoietic stem cells. Cell. 1986;45:917–927. doi: 10.1016/0092-8674(86)90566-0. [DOI] [PubMed] [Google Scholar]
- Li MO, Flavell RA. TGF-beta: a master of all T cell trades. Cell. 2008;134:392–404. doi: 10.1016/j.cell.2008.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Fei T, Zhang J, Zhu G, Wang L, Lu D, Chi X, Teng Y, Hou N, Yang X, et al. BMP4 Signaling Acts via dual-specificity phosphatase 9 to control ERK activity in mouse embryonic stem cells. Cell Stem Cell. 2012;10:171–182. doi: 10.1016/j.stem.2011.12.016. [DOI] [PubMed] [Google Scholar]
- Lien W-H, Guo X, Polak L, Lawton LN, Young RA, Zheng D, Fuchs E. Genome-wide maps of histone modifications unwind in vivo chromatin states of the hair follicle lineage. Cell Stem Cell. 2011;9:219–232. doi: 10.1016/j.stem.2011.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo NA, Shimono Y, Qian D, Clarke MF. The biology of cancer stem cells. Annu. Rev. Cell Dev. Biol. 2007;23:675–699. doi: 10.1146/annurev.cellbio.22.010305.104154. [DOI] [PubMed] [Google Scholar]
- Mani SA, Guo W, Liao M-J, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massagué J. TGFbeta in Cancer. Cell. 2008;134:215–230. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massagué J, Xi Q. TGF-β control of stem cell differentiation genes. FEBS Lett. 2012;586:1953–1958. doi: 10.1016/j.febslet.2012.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean AB, D'Amour KA, Jones KL, Krishnamoorthy M, Kulik MJ, Reynolds DM, Sheppard AM, Liu H, Xu Y, Baetge EE, et al. Activin a efficiently specifies definitive endoderm from human embryonic stem cells only when phosphatidylinositol 3-kinase signaling is suppressed. Stem Cells. 2007;25:29–38. doi: 10.1634/stemcells.2006-0219. [DOI] [PubMed] [Google Scholar]
- McLean GW, Komiyama NH, Serrels B, Asano H, Reynolds L, Conti F, Hodivala-Dilke K, Metzger D, Chambon P, Grant SGN, et al. Specific deletion of focal adhesion kinase suppresses tumor formation and blocks malignant progression. Genes Dev. 2004;18:2998–3003. doi: 10.1101/gad.316304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrill BJ, Gat U, DasGupta R, Fuchs E. Tcf3 and Lef1 regulate lineage differentiation of multipotent stem cells in skin. Genes Dev. 2001;15:1688–1705. doi: 10.1101/gad.891401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mira H, Andreu Z, Suh H, Lie DC, Jessberger S, Consiglio A, San Emeterio J, Hortigüela R, Marqués-Torrejón MA, Nakashima K, et al. Signaling through BMPR-IA regulates quiescence and long-term activity of neural stem cells in the adult hippocampus. Cell Stem Cell. 2010;7:78–89. doi: 10.1016/j.stem.2010.04.016. [DOI] [PubMed] [Google Scholar]
- Morikawa M, Koinuma D, Tsutsumi S, Vasilaki E, Kanki Y, Heldin CH, Aburatani H, Miyazono K. ChIP-seq reveals cell type-specific binding patterns of BMP-specific Smads and a novel binding motif. Nucleic Acids Res. 2011;39:8712–8727. doi: 10.1093/nar/gkr572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu D, Cambier S, Fjellbirkeland L, Baron JL, Munger JS, Kawakatsu H, Sheppard D, Broaddus VC, Nishimura SL. The integrin alpha(v)beta8 mediates epithelial homeostasis through MT1-MMP-dependent activation of TGF-beta1. J Cell Biol. 2002;157:493–507. doi: 10.1083/jcb.200109100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen AC, Orlando DA, Newman JJ, Lovén J, Kumar RM, Bilodeau S, Reddy J, Guenther MG, Dekoter RP, Young RA. Master transcription factors determine cell-type-specific responses to TGF-β signaling. Cell. 2011;147:565–576. doi: 10.1016/j.cell.2011.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munger JS, Sheppard D. Cross talk among TGF-β signaling pathways, integrins, and the extracellular matrix. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a005017. a005017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munger JS, Huang X, Kawakatsu H, Griffiths MJ, Dalton SL, Wu J, Pittet JF, Kaminski N, Garat C, Matthay MA, et al. The integrin alpha v beta 6 binds and activates latent TGF beta 1: a mechanism for regulating pulmonary inflammation and fibrosis. Cell. 1999;96:319–328. doi: 10.1016/s0092-8674(00)80545-0. [DOI] [PubMed] [Google Scholar]
- Murry CE, Keller G. Differentiation of embryonic stem cells to clinically relevant populations: lessons from embryonic development. Cell. 2008;132:661–680. doi: 10.1016/j.cell.2008.02.008. [DOI] [PubMed] [Google Scholar]
- Naka K, Hoshii T, Muraguchi T, Tadokoro Y, Ooshio T, Kondo Y, Nakao S, Motoyama N, Hirao A. TGF-beta-FOXO signalling maintains leukaemia-initiating cells in chronic myeloid leukaemia. Nature. 2010;463:676–680. doi: 10.1038/nature08734. [DOI] [PubMed] [Google Scholar]
- Nishimura EK, Jordan SA, Oshima H, Yoshida H, Osawa M, Moriyama M, Jackson IJ, Barrandon Y, Miyachi Y, Nishikawa S-I. Dominant role of the niche in melanocyte stem-cell fate determination. Nature. 2002;416:854–860. doi: 10.1038/416854a. [DOI] [PubMed] [Google Scholar]
- Nishimura EK, Suzuki M, Igras V, Du J, Lonning S, Miyachi Y, Roes J, Beermann F, Fisher DE. Key roles for transforming growth factor beta in melanocyte stem cell maintenance. Cell Stem Cell. 2010;6:130–140. doi: 10.1016/j.stem.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkin SH, Zon LI. Hematopoiesis: an evolving paradigm for stem cell biology. Cell. 2008;132:631–644. doi: 10.1016/j.cell.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshimori N, Fuchs E. Paracrine TGF-β Signaling Counterbalances BMP-Mediated Repression in Hair Follicle Stem Cell Activation. Cell Stem Cell. 2012;10:63–75. doi: 10.1016/j.stem.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozdamar B, Bose R, Barrios-Rodiles M, Wang H-R, Zhang Y, Wrana JL. Regulation of the polarity protein Par6 by TGFbeta receptors controls epithelial cell plasticity. Science. 2005;307:1603–1609. doi: 10.1126/science.1105718. [DOI] [PubMed] [Google Scholar]
- Peñuelas S, Anido J, Prieto-Sánchez RM, Folch G, Barba I, Cuartas I, García-Dorado D, Poca MA, Sahuquillo J, Baselga J, et al. TGF-beta increases glioma-initiating cell self-renewal through the induction of LIF in human glioblastoma. Cancer Cell. 2009;15:315–327. doi: 10.1016/j.ccr.2009.02.011. [DOI] [PubMed] [Google Scholar]
- Piccirillo SG, Reynolds BA, Zanetti N, Lamorte G, Binda E, Broggi G, Brem H, Olivi A, Dimeco F, et al. Bone morphogenetic proteins inhibit the tumorigenic potential of human brain tumour-initiating cells. Nature. 2006;444:761–765. doi: 10.1038/nature05349. [DOI] [PubMed] [Google Scholar]
- Plikus MV, Mayer JA, la Cruz de D, Baker RE, Maini PK, Maxson R, Chuong C-M. Cyclic dermal BMP signalling regulates stem cell activation during hair regeneration. Nature. 2008;451:340–344. doi: 10.1038/nature06457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X, Li TG, Hao J, Hu J, Wang J, Simmons H, Miura S, Mishina Y, Zhao GQ. BMP4 supports self-renewal of embryonic stem cells by inhibiting mitogen-activated protein kinase pathways. Proc Natl Acad Sci USA. 2004;101:6027–6032. doi: 10.1073/pnas.0401367101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabbani P, Takeo M, Chou W, Myung P, Bosenberg M, Chin L, Taketo MM, Ito M. Coordinated Activation of Wnt in Epithelial and Melanocyte Stem Cells Initiates Pigmented Hair Regeneration. Cell. 2011;145:941–955. doi: 10.1016/j.cell.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross S, Hill CS. How the Smads regulate transcription. Int. J. Biochem. Cell Biol. 2008;40:383–408. doi: 10.1016/j.biocel.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Ross S, Cheung E, Petrakis TG, Howell M, Kraus WL, Hill CS. Smads orchestrate specific histone modifications and chromatin remodeling to activate transcription. Embo J. 2006;25:4490–4502. doi: 10.1038/sj.emboj.7601332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheel C, Eaton EN, Li SH-J, Chaffer CL, Reinhardt F, Kah K-J, Bell G, Guo W, Rubin J, Richardson AL, et al. Paracrine and Autocrine Signals Induce and Maintain Mesenchymal and Stem Cell States in the Breast. Cell. 2011;145:926–940. doi: 10.1016/j.cell.2011.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepers AG, Snippert HJ, Stange DE, van den Born M, van Es JH, van de Wetering M, Clevers H. Lineage Tracing Reveals Lgr5+ Stem Cell Activity in Mouse Intestinal Adenomas. Science. 2012 doi: 10.1126/science.1224676. [DOI] [PubMed] [Google Scholar]
- Schier AF, Talbot WS. Molecular genetics of axis formation in zebrafish. Annu. Rev. Genet. 2005;39:561–613. doi: 10.1146/annurev.genet.37.110801.143752. [DOI] [PubMed] [Google Scholar]
- Schmierer B, Hill CS. TGFbeta-SMAD signal transduction: molecular specificity and functional flexibility. Nat Rev Mol Cell Biol. 2007;8:970–982. doi: 10.1038/nrm2297. [DOI] [PubMed] [Google Scholar]
- Schober M, Fuchs E. Tumor-initiating stem cells of squamous cell carcinomas and their control by TGF-β and integrin/focal adhesion kinase (FAK) signaling. Proc Natl Acad Sci USA. 2011;108:10544–10549. doi: 10.1073/pnas.1107807108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengle G, Ono RN, Sasaki T, Sakai LY. Prodomains of transforming growth factor beta (TGFbeta) superfamily members specify different functions: extracellular matrix interactions and growth factor bioavailability. J. Biol. Chem. 2011;286:5087–5099. doi: 10.1074/jbc.M110.188615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seoane J. The TGFBeta pathway as a therapeutic target in cancer. Clin Transl Oncol. 2008;10:14–19. doi: 10.1007/s12094-008-0148-2. [DOI] [PubMed] [Google Scholar]
- Shi M, Zhu J, Wang R, Chen X, Mi L, Walz T, Springer TA. Latent TGF-β structure and activation. Nature. 2011;474:343–349. doi: 10.1038/nature10152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieburg HB, Cho RH, Dykstra B, Uchida N, Eaves CJ, Muller-Sieburg CE. The hematopoietic stem compartment consists of a limited number of discrete stem cell subsets. Blood. 2006;107:2311–2316. doi: 10.1182/blood-2005-07-2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh AM, Reynolds D, Cliff T, Ohtsuka S, Mattheyses AL, Sun Y, Menendez L, Kulik M, Dalton S. Signaling network crosstalk in human pluripotent cells: a Smad2/3-regulated switch that controls the balance between self-renewal and differentiation. Cell Stem Cell. 2012;10:312–326. doi: 10.1016/j.stem.2012.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AG. Embryo-derived stem cells: of mice and men. Annu. Rev. Cell Dev. Biol. 2001;17:435–462. doi: 10.1146/annurev.cellbio.17.1.435. [DOI] [PubMed] [Google Scholar]
- Smith LG, Weissman IL, Heimfeld S. Clonal analysis of hematopoietic stem-cell differentiation in vivo. Proc Natl Acad Sci USA. 1991;88:2788–2792. doi: 10.1073/pnas.88.7.2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorrentino A, Thakur N, Grimsby S, Marcusson A, Bulow vonV, Schuster N, Zhang S, Heldin C-H, Landström M. The type I TGF-beta receptor engages TRAF6 to activate TAK1 in a receptor kinase-independent manner. Nat Cell Biol. 2008;10:1199–1207. doi: 10.1038/ncb1780. [DOI] [PubMed] [Google Scholar]
- Sudo K, Ema H, Morita Y, Nakauchi H. Age-associated characteristics of murine hematopoietic stem cells. J Exp Med. 2000;192:1273–1280. doi: 10.1084/jem.192.9.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumi T, Tsuneyoshi N, Nakatsuji N, Suemori H. Defining early lineage specification of human embryonic stem cells by the orchestrated balance of canonical Wnt/beta-catenin, Activin/Nodal and BMP signaling. Development. 2008;135:2969–2979. doi: 10.1242/dev.021121. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Tang Y, Wu X, Lei W, Pang L, Wan C, Shi Z, Zhao L, Nagy TR, Peng X, Hu J, et al. TGF-beta1-induced migration of bone mesenchymal stem cells couples bone resorption with formation. Nat. Med. 2009;15:757–765. doi: 10.1038/nm.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesar PJ, Chenoweth JG, Brook FA, Davies TJ, Evans EP, Mack DL, Gardner RL, McKay RD. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- Thiery JP, Acloque H, Huang RYJ, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Trompouki E, Bowman TV, Lawton LN, Fan ZP, Wu D-C, Dibiase A, Martin CS, Cech JN, Sessa AK, Leblanc JL, et al. Lineage Regulators Direct BMP and Wnt Pathways to Cell-Specific Programs during Differentiation and Regeneration. Cell. 2011;147:577–589. doi: 10.1016/j.cell.2011.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsujimura T, Kinoshita M, Abe M. Response of rabbit skeletal muscle to tibial lengthening. J Orthop Sci. 2006;11:185–190. doi: 10.1007/s00776-005-0991-8. [DOI] [PubMed] [Google Scholar]
- Tumbar T, Guasch G, Greco V, Blanpain C, Lowry WE, Rendl M, Fuchs E. Defining the epithelial stem cell niche in skin. Science. 2004;303:359–363. doi: 10.1126/science.1092436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallier L, Alexander M, Pedersen RA. Activin/Nodal and FGF pathways cooperate to maintain pluripotency of human embryonic stem cells. J Cell Sci. 2005;118:4495–4509. doi: 10.1242/jcs.02553. [DOI] [PubMed] [Google Scholar]
- Vallier L, Mendjan S, Brown S, Chng Z, Teo A, Smithers LE, Trotter MWB, Cho CH-H, Martinez A, Rugg-Gunn P, et al. Activin/Nodal signalling maintains pluripotency by controlling Nanog expression. Development. 2009;136:1339–1349. doi: 10.1242/dev.033951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Noulet F, Edom-Vovard F, Tozer S, Le Grand F, Duprez D. Bmp signaling at the tips of skeletal muscles regulates the number of fetal muscle progenitors and satellite cells during development. Dev Cell. 2010;18:643–654. doi: 10.1016/j.devcel.2010.02.008. [DOI] [PubMed] [Google Scholar]
- Wang LD, Wagers AJ. Dynamic niches in the origination and differentiation of haematopoietic stem cells. Nat Rev Mol Cell Biol. 2011;12:643–655. doi: 10.1038/nrm3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkes MC, Murphy SJ, Garamszegi N, Leof EB. Cell-type-specific activation of PAK2 by transforming growth factor beta independent of Smad2 and Smad3. Mol. Cell. Biol. 2003;23:8878–8889. doi: 10.1128/MCB.23.23.8878-8889.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams PE, Goldspink G. Longitudinal growth of striated muscle fibres. J Cell Sci. 1971;9:751–767. doi: 10.1242/jcs.9.3.751. [DOI] [PubMed] [Google Scholar]
- Wray J, Kalkan T, Gomez-Lopez S, Eckardt D, Cook A, Kemler R, Smith A. Inhibition of glycogen synthase kinase-3 alleviates Tcf3 repression of the pluripotency network and increases embryonic stem cell resistance to differentiation. Nat Cell Biol. 2011;13:838–845. doi: 10.1038/ncb2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MY, Hill CS. Tgf-beta superfamily signaling in embryonic development and homeostasis. Dev Cell. 2009;16:329–343. doi: 10.1016/j.devcel.2009.02.012. [DOI] [PubMed] [Google Scholar]
- Xi Q, Wang Z, Zaromytidou A-I, Zhang XH-F, Chow-Tsang L-F, Liu JX, Kim H, Barlas A, Manova-Todorova K, Kaartinen V, et al. A poised chromatin platform for TGF-β access to master regulators. Cell. 2011;147:1511–1524. doi: 10.1016/j.cell.2011.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R-H, Sampsell-Barron TL, Gu F, Root S, Peck RM, Pan G, Yu J, Antosiewicz-Bourget J, Tian S, Stewart R, et al. NANOG is a direct target of TGFbeta/activin-mediated SMAD signaling in human ESCs. Cell Stem Cell. 2008;3:196–206. doi: 10.1016/j.stem.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita M, Fatyol K, Jin C, Wang X, Liu Z, Zhang YE. TRAF6 mediates Smad-independent activation of JNK and p38 by TGF-beta. Mol Cell. 2008;31:918–924. doi: 10.1016/j.molcel.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki S, Ema H, Karlsson G, Yamaguchi T, Miyoshi H, Shioda S, Taketo MM, Karlsson S, Iwama A, Nakauchi H. Nonmyelinating Schwann cells maintain hematopoietic stem cell hibernation in the bone marrow niche. Cell. 2011;147:1146–1158. doi: 10.1016/j.cell.2011.09.053. [DOI] [PubMed] [Google Scholar]
- Yamazaki S, Iwama A, Takayanagi S-I, Eto K, Ema H, Nakauchi H. TGF-beta as a candidate bone marrow niche signal to induce hematopoietic stem cell hibernation. Blood. 2009;113:1250–1256. doi: 10.1182/blood-2008-04-146480. [DOI] [PubMed] [Google Scholar]
- Yamazaki S, Iwama A, Takayanagi S-I, Morita Y, Eto K, Ema H, Nakauchi H. Cytokine signals modulated via lipid rafts mimic niche signals and induce hibernation in hematopoietic stem cells. Embo J. 2006;25:3515–3523. doi: 10.1038/sj.emboj.7601236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Weinberg RA. Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev Cell. 2008;14:818–829. doi: 10.1016/j.devcel.2008.05.009. [DOI] [PubMed] [Google Scholar]
- Ying QL, Nichols J, Chambers I, Smith A. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell. 2003;115:281–292. doi: 10.1016/s0092-8674(03)00847-x. [DOI] [PubMed] [Google Scholar]
- Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, Cohen P, Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young RA. Control of the embryonic stem cell state. Cell. 2011;144:940–954. doi: 10.1016/j.cell.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi M. Skeletal remodeling in health and disease. Nat. Med. 2007;13:791–801. doi: 10.1038/nm1593. [DOI] [PubMed] [Google Scholar]
- Zawel L, Dai JL, Buckhaults P, Zhou S, Kinzler KW, Vogelstein B, Kern SE. Human Smad3 and Smad4 are sequence-specific transcription activators. Mol Cell. 1998;1:611–617. doi: 10.1016/s1097-2765(00)80061-1. [DOI] [PubMed] [Google Scholar]
- Zeisberg M, Hanai J-I, Sugimoto H, Mammoto T, Charytan D, Strutz F, Kalluri R. BMP-7 counteracts TGF-beta1-induced epithelial-to-mesenchymal transition and reverses chronic renal injury. Nat. Med. 2003;9:964–968. doi: 10.1038/nm888. [DOI] [PubMed] [Google Scholar]
- Zhang J, He XC, Tong W-G, Johnson T, Wiedemann LM, Mishina Y, Feng JQ, Li L. Bone morphogenetic protein signaling inhibits hair follicle anagen induction by restricting epithelial stem/progenitor cell activation and expansion. Stem Cells. 2006;24:2826–2839. doi: 10.1634/stemcells.2005-0544. [DOI] [PubMed] [Google Scholar]
- Zhang J, Niu C, Ye L, Huang H, He X, Tong W-G, Ross J, Haug J, Johnson T, Feng JQ, et al. Identification of the haematopoietic stem cell niche and control of the niche size. Nature. 2003;425:836–841. doi: 10.1038/nature02041. [DOI] [PubMed] [Google Scholar]
- Zhang P, Li J, Tan Z, Wang C, Liu T, Chen L, Yong J, Jiang W, Sun X, Du L, et al. Short-term BMP-4 treatment initiates mesoderm induction in human embryonic stem cells. Blood. 2008;111:1933–1941. doi: 10.1182/blood-2007-02-074120. [DOI] [PubMed] [Google Scholar]
- Zhao C, Deng W, Gage FH. Mechanisms and functional implications of adult neurogenesis. Cell. 2008;132:645–660. doi: 10.1016/j.cell.2008.01.033. [DOI] [PubMed] [Google Scholar]
- Zorn AM, Wells JM. Molecular basis of vertebrate endoderm development. Int. Rev. Cytol. 2007;259:49–111. doi: 10.1016/S0074-7696(06)59002-3. [DOI] [PubMed] [Google Scholar]