Abstract
Objective
To implement the SNOMED CT electronic specifications for reporting quality measures and to identify critical issues that affect implementation.
Background
The Centers for Medicare and Medicaid (CMS) have issued the electronic specifications for reporting quality measures requiring vendors and hospital systems to use standardized data elements to provide financial incentives for eligible providers.
Methods
The electronic specifications from CMS were downloaded and extracted. All SNOMED CT codes were examined individually as part of the creation of a mapping table for distribution by a vendor for incorporation into electronic health record systems. A qualitative and quantitative evaluation of the SNOMED CT codes was done as a follow up to the mapping project.
Results
A total of 10643 SNOMED codes were examined for the 44 measures. The approved SNOMED CT code sets contain aberrancies in content such as incomplete IDs, the use of description IDs instead of concept IDs, inactive codes, morphology and observable codes for clinical findings and the inclusion of non-human content.
Conclusion
Implementers of these approved specifications must do additional rigorous review and make edits in order to avoid incorporating errors into their EHR products and systems.
Keywords: SNOMED CT, electronic health records, national health policy
1. Introduction
In July 2010, the Centers for Medicare & Medicaid Services (CMS) issued a final rule [1] to implement provisions of the American Recovery and Reinvestment Act of 2009 (Recovery Act) [2] in order to provide incentive payments to eligible healthcare professionals who are meaningful users of certified electronic health record (EHR) technology. “As part of the criteria for satisfying meaningful use, clinical quality measures results (numerators, denominators, and exclusions) must be reported to CMS” [3]. The format for reporting these quality (or performance) measures includes the use of standardized data elements that were assigned to meet the expectations for interoperability, data sharing, and aggregate measurements.
Effective with this rule, was the publication of the electronic specifications for clinical quality measures (CQM) to be used for reports for 2011 and 2012. These specifications were produced to enable implementers to begin integrating these specifications within their products and have been made available for download via the CMS website [3]. These electronic specifications include standardized code lists or value sets that have been deemed as acceptable values for the decision-making components of a given quality measure. The expectation is that vendors, hospitals and others responsible for submitting reports to CMS for reimbursement will be able to use these new standardized electronic data elements within their EHR systems to enable eligible providers to receive incentive payments.
The electronic specifications published by CMS are based on quality data sets (QDS) that were established by the National Quality Forum (NQF) through the Health Information Technology Expert Panel (HITEP) [4] that defined the data requirements in the context of EHR usage. These quality data sets were further expanded into an electronic standard known as the “eMeasure” [5] to enable EHR vendors to more directly manage the quality measurements within their products and included standardized code lists from an adopted vocabulary standard such as SNOMED CT, RXNORM, LOINC, ICD9CM, ICD10CM, etc.
A total of 44 NQF-endorsed quality (performance) measures were “retooled” into an electronic “eMeasure” format based on the data elements described within the original paper-based measures. The developers of the individual measures or measure stewards (e.g. American Medical Association, National Committee for Quality Assurance) worked with the NQF to transform all of the existing data elements into quality data sets (QDS) so that every data element within a given measure had a corresponding QDS. For example, the QDS for the data element “systolic blood pressure” included a standard concept (i.e. “systolic blood pressure”), a QDS data type (i.e. “physical exam finding”), a standard vocabulary (i.e. SNOMED CT) with version (i.e. July 2009), and a standard code list or value set with numeric codes (i.e. 12929001, 163030003, 251070002, etc.). The standard code list or value set was deemed to contain the acceptable values for the measurement.
2. Objectives
In order to comply with the final rule by CMS, vendors have been actively working to enable implementation of the published electronic specifications into their products so that eligible professionals could participate in the incentive program. In this paper, all of the SNOMED CT codes that were published for quality reporting of the 44 measures were analyzed as part of the creation of a mapping table for the implementation of these specifications into a vendor’s product [6]. In the process, notations were made as to the appropriateness of the codes for inclusion within a measure and the suitability for implementation. Additional steps were required in order to avoid errors and to enable end-user implementation.
3. Methods
A qualitative and quantitative approach was used to evaluate the electronic specifications for the CMS quality measures in terms of the appropriateness of the selected SNOMED CT codes as well as the number of aberrancies noted.
First, the electronic specifications including supplemental specifications for the 44 quality measures were downloaded from the CMS website [3]. Each measure had its own folder that included two files – a human-readable PDF document and an NQF retooled measure spreadsheet. The PDF document described the measure and its rationale as well as the criteria used to identify the population and data criteria. Numerator, denominator and exclusion criteria were defined as well as the calculations used for the measure. The spreadsheet included the standard concepts for the measure with corresponding standard category and data type. These were aligned with the selected standard taxonomy, version used and standard code list. For example, measure #13, “Hypertension: Blood Pressure Measurement“, had a standard concept of “Diastolic Blood Pressure”, a standard category of “physical exam” and a data type of “physical exam finding”. These were aligned with the standard taxonomy of “SNOMED CT”, version “July 2009”, and a standard code list that included 24 SNOMED CT codes.
Next, all of the SNOMED CT codes were extracted and aligned with each quality measure in a local Access database. For example, measure #34 “Colorectal Cancer Screening” had 43 associated SNOMED codes that represented “allowable” values for the standard concept of “colorectal cancer”. This translated into 43 rows in a database, each aligned with the measure number 34.
Next, the SNOMED CT fully specified name (from the January 2011 release) was added in order to view the description of the code as well as the corresponding SNOMED CT category for each concept.
All of the codes were examined individually as part of the creation of a mapping table for distribution by a vendor [6] to enable end-users to link from their product to the CMS designated standard code lists for reporting purposes. As each code was reviewed and mapped, a notes column was added to the database to record any aberrancy that may have had an impact on implementation by end-users (►Fig. 1). These included notations to make end-users aware of codes that were not mapped because they were deemed to be inappropriate or codes that needed additional data such as the actual numeric value for an observable entity (e.g. blood pressure value). The notations included references to incomplete concept ID numbers, the use of description ID’s instead of concept IDs, the inclusion of non-human concepts, morphology codes and observables as well as SNOMED CT codes that were no longer current with the designated concept status of “limited”, “duplicate”, “movedto” and “ambiguous”. Upon completion, a table was created to summarize the aberrancies noted for each measure.
Figure 1.
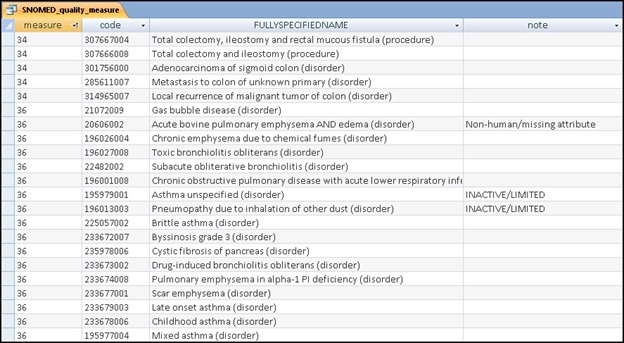
Local database record of aberrancies in the SNOMED CT quality measure code sets
Tools that were used in the process of this analysis included the Clue browser [7] that was used for individual look up of codes as well as cross checking of the final aberrancies using a separate SNOMED CT database that included the 3 core tables (concept, relationship and description) from the January 2011 release as well as a review of the SNOMED CT non-human subset text file available from the official US download site [8]. Some of the human-readable pdf files for the individual measures were also reviewed for the analysis.
4. Results
A total of 10643 SNOMED codes were obtained from the downloaded specifications [3] for the 44 measures. Some SNOMED CT codes were used more than once within a given measure for the various QDS entries. For example, in measure #12, “111880001” was listed as a value for the QDS_data_type “diagnosis inactive” and “diagnosis active” for the concept 111880001| acute HIV infection (disorder). Other measures used some of the same SNOMED CT codes. The majority of the SNOMED CT concepts were clinical findings (85%) [n = 9092] and procedures (15%)[n = 1348]; other categories included observable entities [n = 125], situations [n = 38], physical objects [n = 10], morphologic abnormalities [n = 7], substances [n = 7] and product [n = 1]; there were 15 incomplete ID numbers that did not correspond to SNOMED CT concepts.
For example, “4627003” was included in the standard code list for 7 different measures and did not correspond to any SNOMED ID. All of these measures related to diabetes so it is possible that a single digit was omitted for an intended concept such as “74627003”| diabetic complication (disorder).
►Table 1 summarizes the findings of the aberrancies noted upon completion of the mapping exercise and analysis of the notations. In addition to those with incomplete ID numbers [n = 15], there were some with only description ID’s [n = 51] that needed to be transposed into SNOMED concept IDs’. For example, for measures #18 and 24, the code “58080013” corresponded to the description ID for the concept 34801009| ectopic pregnancy (disorder).There were 125 codes from the SNOMED CT observable entities hierarchy. The majority [n = 114] of these were designated in the CMS spreadsheets as allowable values for standard concepts (systolic blood pressure [n = 66], diastolic blood pressure [n = 43], heart rate [n = 2], and BMI [n = 3] with the QDS data type of “physical exam finding”); others included ejection fraction [n = 6] as part of the QDS data type of “diagnostic study result” or Gleason score [n =2] as a “laboratory test result”. Two additional concepts – pregnancy [n = 1] and estimated date of conception [n = 2] were assigned to QDS data types of “diagnosis active” and “patient characteristic” respectively.
Table 1.
Summary of aberrancies in SNOMED CT quality measure code sets
| Measure number | Number of SNOMED CT codes | Incomplete ID | Description ID | Concept Status | Non-human | Observable entity | Morphologic abnormality | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Limited | Moved to | Ambiguous | Duplicate | w/attribute | w/o attribute | ||||||
| 1 | 65 | 2 | 1 | ||||||||
| 2 | 97 | ||||||||||
| 4 | 72 | ||||||||||
| 12 | 395 | 10 | 1 | 1 | |||||||
| 13 | 164 | 4 | 42 | ||||||||
| 14 | 317 | 10 | 1 | ||||||||
| 18 | 133 | 23 | 3 | 23 | |||||||
| 24 | 37 | 23 | 2 | ||||||||
| 27 | 26 | ||||||||||
| 28a | 49 | ||||||||||
| 28b | 33 | ||||||||||
| 31 | 50 | 1 | |||||||||
| 32 | 21 | ||||||||||
| 33 | 53 | 1 | 4 | ||||||||
| 34 | 73 | 4 | |||||||||
| 36 | 109 | 2 | 1 | 1 | |||||||
| 38 | 899 | 12 | 1 | 3 | |||||||
| 41 | 10 | 1 | |||||||||
| 43 | 0 | ||||||||||
| 47 | 4 | ||||||||||
| 52 | 2662 | 10 | 1 | 8 | 11 | ||||||
| 55 | 204 | 2 | 13 | 4 | 1 | ||||||
| 56 | 148 | 2 | 13 | 4 | 1 | ||||||
| 59 | 153 | 2 | 13 | 4 | 1 | ||||||
| 61 | 169 | 2 | 13 | 4 | 22 | 1 | |||||
| 62 | 788 | 2 | 13 | 2 | 5 | 5 | 1 | ||||
| 64 | 154 | 2 | 13 | 4 | 1 | ||||||
| 67 | 353 | 7 | 2 | ||||||||
| 68 | 152 | 8 | |||||||||
| 70 | 341 | 5 | 1 | 3 | 1 | ||||||
| 73 | 177 | 8 | 22 | ||||||||
| 74 | 102 | 3 | |||||||||
| 75 | 192 | 8 | |||||||||
| 81 | 189 | 1 | 3 | ||||||||
| 83 | 290 | 2 | 1 | 3 | 4 | ||||||
| 84 | 963 | 2 | 3 | 2 | |||||||
| 86 | 4 | ||||||||||
| 88 | 66 | ||||||||||
| 89 | 70 | ||||||||||
| 105 | 117 | ||||||||||
| 385 | 70 | 1 | 1 | ||||||||
| 387 | 273 | 2 | |||||||||
| 389 | 166 | 1 | 2 | ||||||||
| 421 | 80 | 2 | |||||||||
| 575 | 153 | 2 | 13 | 4 | 1 | ||||||
| S | 10643 | 15 | 51 | 194 | 2 | 7 | 41 | 16 | 17 | 125 | 7 |
A review of the accompanying documentation (i.e. the pdf file) for measure #13 “Hypertension: Blood Pressure Measurement” indicated that these values were intended to be measures for numerator data such as the recorded physical exam finding of “systolic blood pressure”. Similarly, for measure #389, “Prostate Cancer: Avoidance of Overuse of Bone Scan for Staging Low Risk Prostate Cancer Patients” a Gleason score numerical value (e.g. Gleason score ≤6) was intended to be used from a laboratory test result for denominator data.
There were 7 value sets that included the morphology code “38542009 |nodular glomerulosclerosis (morphologic abnormality)” as part of the QDS datatype “diagnosis active”. For example, measure #55 “Diabetes: Eye exam” included this morphology code for the standard concept of “diabetes” with the QDS data type of “diagnosis active”. For mapping purposes, this code was transposed to a SNOMED CT clinical finding “63510008 | nodular type diabetic glomerulosclerosis (disorder).”
As the latest version of SNOMED CT (January 2011) was used for the mapping project and analysis, several codes from the official download were no longer current and included those with a changed concept status of a) Limited [n = 194]; b) Movedto [n = 2], c) Ambiguous [n = 7], and d) Duplicate [n = 41]. Of note, during the analysis, some of the quality measures were found to use SNOMED CT codes sets from different versions. For example, measure #14 “Prenatal care: Anti-D Immune Globulin” had value sets using SNOMED CT versions from January 2008, July 2009 and July 2010 for different data elements.
Non-human codes were also found among the code lists [n = 33]. Some of those non-human codes had a designated IS_A relationship to a non-human concept [n = 16] visible through the browser[7] and were defined as such within the SNOMED CT relationship table. For example, “79816004 | canine hemorrhagic gastroenteritis (disorder)” contained the IS_A relationship of “non-human disorder by body site” that was clearly visible when viewing the concept individually through a browser. There were 17 non-human codes that did not have a relationship designated as non-human. For example, “79624007| canine infectious cyclic thrombocytopenia (disorder)” had an IS_A relationship to “thrombocytopenic disorder” but no designated non-human relationship when viewed through the browser. However both groups of codes (i.e. those with and without the non-human designation) were contained in the non-human subset of SNOMED codes that is provided with the official release of SNOMED CT files biannually.
5. Discussion
Since the initiation of the final rule by CMS, implementers have been working to incorporate the published electronic specifications into their products so that end-users and hospitals could qualify for the 2011 and 2012 incentive program. In this paper, it was demonstrated that the implementation of the electronic specifications required the evaluation of thousands of individual codes in order to assess the appropriateness and context for inclusion for linkage into an existing EHR product. In the process of evaluating the SNOMED CT codes that were part of the electronic specifications, aberrancies were noted that if implemented would include errors with potential adverse results for reporting quality. Careful review of the designated quality data sets was necessary in order to exclude inappropriate codes. They included incomplete code ID’s, the use of description IDs instead of concept IDs, morphology and observable entity codes for clinical findings, inactive codes (e.g. limited, ambiguous, movedto, duplicate) and the inclusion of non-human content.
SNOMED CT has been chosen [9] as one of the major standard terminologies for the backbone of the Electronic Health Record in the United States. It is a comprehensive clinical terminology produced by the IHTSDO [10] that includes 19 top-level categories with interconnecting attributes [11–12] that define its structure and meaning and can enable description-logic [13–14] reasoning. Efforts at implementing SNOMED CT into existing EHR systems have been described previously [15–18]. While it is fairly granular and robust for capturing clinical data, it is also quite large and somewhat complex for users who may be more familiar with the long-standing classification systems. Recent work has also highlighted some of the anomalies within the structure of the SNOMED CT hierarchies making it problematic for use within some applications [19].
Some of the SNOMED CT codes sets used in these electronic specifications appeared to be inappropriate for the data type and category. Those included the use of concepts from SNOMED CT categories “body structure” (includes morphology codes) and “observable entity”. The morphology codes may have been mistaken for a clinical finding since they often have a lexical match in both categories. However, their meaning and implementation would be very different. While “38542009 | nodular glomerulosclerosis (morphologic abnormality)” would be appropriate for defining a disease (as an attribute), it would not be valid for the QDS data type “diagnosis active”. Instead, a clinical finding “63510008 | nodular type diabetic glomerulosclerosis (disorder)” would be used.
Similarly, the large number of observable entities that were used for physical findings and laboratory test results suggests some confusion in assigning these as the actual allowable values. “Concepts in this hierarchy (observable entity) can be thought of as representing a question or procedure which can produce an answer or a result” [20]. In short, observable entities in SNOMED CT are placeholders for values such as a numeric value. Using the SNOMED CT concept “271649006| systolic blood pressure (observable entity)” as an “allowable” value for a physical finding would be incomplete in that this is only the name of an observation and not the observation result. The accompanying value (e.g. 140 mmHg) that is the result of observation would be a physical finding. Similarly for a Gleason score, the measure lists the data type as a “laboratory test result” and requires a numerical entry for the score, so a code from the value set (i.e. “372278000| Gleason score [observable entity]”) that is just the name of the observation would not be correct for reporting. SNOMED CT does provide, however, results for Gleason scores within its clinical finding category (e.g. “84556003 | Gleason grade score 6 out of 10 [finding]”).
Over 240 codes designated in the electronic specifications were found to be inactive with a change in concept status (i.e. limited, movedto, ambiguous, duplicate) since they were published. Monitoring concept status changes have become an integral part of terminology management with the need to archive and update various code sets over time [21–22] along with any structural changes to the terminology standard that are made during ongoing revisions [17]. Besides version control by EHR implementers, the issuing of code sets that contain different versions within a single quality measure adds an additional level of necessary scrutiny.
Another concern for errors was the inclusion of non-human content. Concepts (n = 16) such as “79816004 | canine hemorrhagic gastroenteritis (disorder)” had a defined IS_A relationship of “Non-human disorder by body site” if viewed through the browser or as defined in the SNOMED CT relationship table. Similarly, “41615007| Marek's disease (disorder)” or “4109002| Sweeney (disorder)” contained a non-human relationship. However, their fully specified name was not explicit in including a non-human species as part of the description so it may have been more likely that they were mistaken for human diseases. Another group of non-human codes [n = 17] were included in the electronic specifications that did not have a visible relationship as “non-human”. While concepts such as “79624007| canine infectious cyclic thrombocytopenia (disorder)” included the more obvious “canine” in the description, “81549005| pyramidal disease (disorder)”, a “disorder of hoof” was less obvious. Of note, however, all of these non-human concepts were included along with the biannual published release of SNOMED CT in the text file known as the “non-human subset”. The inclusion of non-human content within SNOMED CT may not be general knowledge. At this time, there is no planned release of a “human-only” subset that could be used to avoid similar errors in the future. However, documentation has been improved to help end-users better understand the current structure of SNOMED CT. In any case, the inclusion of non-human content is problematic for electronic health records. If these codes were implemented directly and not excluded, an end-user could have selected “Sweeney disease” from a drop-down menu or linked to a table for reporting that would knowingly be inaccurate.
As SNOMED CT is evolving [23], implementation issues must be considered. It has been noted previously that editorial policies can greatly impact implementation [17]. In this instance, editorial edits [24] may be helpful so that all non-human content has an affiliated non-human relationship as well as a more explicit fully specified name. That would enable each of these concepts to be clearly visible as non-human through a browser. The creation of a human-only subset for the SNOMED CT release is another option that could help minimize errors in the future.
One explanation for the inclusion of non-human concepts may have been the process used in creating value sets [25–27] for the various data elements. One approach to generating and updating value sets has been to use “intensional” definitions where components of an entire node or subnode can be extracted programmatically – without review of individual concepts. It is assumed that all “child concepts” of the “parent concept” have similar inherited properties [28] that make them valid. As it relates to this paper, this could explain why non-human concepts may have been included as part of a subnode extraction. Creating value sets by using “extensional” definitions is more time consuming and requires individual review and exclusions of certain concepts as deemed by domain experts who customize each list.
In conclusion, better documentation, better education and better tooling is needed in order to avoid pitfalls in implementing SNOMED CT for quality reporting from EHRs. Some emphasis should be placed on ways to make implementation easier to work with existing EHR products that are already entrenched in existing systems.
Clinical Relevance Statement
The electronic specifications issued by CMS are required for the reporting of quality measures from electronic health records. The specifications for the SNOMED CT data elements that have been approved contain aberrancies such as incomplete IDs, the use of description IDs instead of concept IDs, inactive codes, morphology and observable codes for clinical findings and the inclusion of non-human content. Implementers of these approved specifications must do additional rigorous review and make edits in order to avoid incorporating errors into their EHR products and systems.
Conflicts of Interest
None
Protection of Human and Animal Subjects
No human and/or animal subjects were included in this project
Acknowledgements
The author would like to thank Medicomp Systems for their contributions related to SNOMED CT implementation.
References
- 1.Centers for Medicare and Medicaid Services, Department of Health and Human Services. 42 CFR Parts 412, 413, 422 et al. Medicare and Medicaid programs; electronic health record incentive program; final rule; published July 28, 2010. http://edocket.access.gpo.gov/2010/pdf/2010–17207.pdf Accessed October 4, 2011
- 2.Public Law 111 – 5 –American Recovery and Reinvestment Act of 2009, February 17, 2009. http://www.gpo.gov/fdsys/pkg/PLAW-111publ5/pdf/PLAW-111publ5.pdf Accessed October 4, 2011
- 3.CMS: Electronic specifications quality measures. http://www.cms.gov/QualityMeasures/03_Electronic Specifications.asp Accessed October 4, 2011
- 4.NQF: Health IT Expert Panel II (HITEP II). http://www.qualityforum.org/projects/hitep2.aspx Accessed October 4, 2011
- 5.NQF: Electronic quality measures (eMeasures). http://www.qualityforum.org/Projects/e-g/eMeasures/ Electronic_Quality_Measures.aspx Accessed October 4, 2011
- 6.Medicomp Systems Available athttp://www.medicomp.com Accessed October 4, 2011
- 7.The Clinical Information Consultancy, CliniClue Xplore http://www.cliniclue.com/cliniclue_xplore Accessed October 4, 2011
- 8.United States National Library of Medicine, Unified Medical Language System® (UMLS®), Downloads, 2011http://www.nlm.nih.gov/research/umls/licensedcontent/downloads.html Accessed October 4, 2011
- 9.U.S. Department of Health and Human Services (April 26, 2007) “HHS joins international partners to promote electronic health records standards”, News Releasehttp://www.hhs.gov/news/press/2007pres/04/pr20070426a.html Accessed October 4, 2011
- 10.International Health Terminology Standards Development Organisation (IHTSDO) http://www.ihtsdo.org/ Accessed October 4, 2011
- 11.International Health Terminology Standards Development Organisation. SNOMED CT. SNOMED Clinical terms®. 2011 Chapter 4. Attributes used in. User Guide January. International Release. [Google Scholar]
- 12.Cornet R.Definitions and qualifiers in SNOMED CT; Methods Inf Med 2009; 48 (2): 178–183 [PubMed] [Google Scholar]
- 13.Nadkarni PM, Marenco LA. Implementing description-logic rules for SNOMED-CT attributes through a table-driven approach. J Am Med Inform Assoc 2010; 17 (2): 182–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cornet R, Abu-Hanna A.Auditing description-logic-based medical terminological systems by detecting equivalent concept definitions. Int J Med Inform 2008; 77(5): 336–345 [DOI] [PubMed] [Google Scholar]
- 15.Wade G, Rosenbloom ST. Experiences mapping a legacy interface terminology to SNOMED CT. BMC Med Inform Decision Making 2008; 8 (Suppl. 1): S3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rosenbloom ST, Brown SH, Froehling D, Bauer BA, et al. Using SNOMED CT to represent two interface terminologies. J Am Med Inform Assoc 2009; 16(1): 81–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wade G, Rosenbloom ST. The impact of SNOMED CT revisions on a mapped interface terminology: terminology development and implementation issues. J Biomed Inform 2009; 42(3): 490–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu J, Lane K, Lo E, Lam M, Truong T, Veillette C.Addressing SNOMED CT implementation challenges through multi-disciplinary collaboration. Stud Health Technol Inform 2010; 160 (Pt 2): 981–985 [PubMed] [Google Scholar]
- 19.Rector AL, Brandt S, Schneider T.Getting the foot out of the pelvis: modeling problems affecting use of SNOMED CT hierarchies in practical applications. J Am Med Inform Assoc 2011; 18(4): 432–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.International Health Terminology Standards Development Organisation. SNOMED Clinical terms®. 2011 5.5. Observable entity. User Guide January. International Release. [Google Scholar]
- 21.Ingenerf J, Beisiegel T.A version management system for SNOMED CT. Stud Health Technol Inform 2008; 136: 827–832 [PubMed] [Google Scholar]
- 22.Lee D, Cornet R, Lau F.Implications of SNOMED CT versioning, Int J Med Inform 2011; 80(6): 442–453 [DOI] [PubMed] [Google Scholar]
- 23.Schulz S, Suntisrivaraporn B, Baader F, Boeker M.SNOMED reaching its adolescence: Ontologists’ and logicians’ health check. Int J Med Inform 2009; 78 (Suppl. 1): S86-S94 [DOI] [PubMed] [Google Scholar]
- 24.International Health Terminology Standards Development Organisation: SNOMED CT® Editorial Guide, January2011International Release [Google Scholar]
- 25.Pathak J, Jiang G, Dwarkanath SO, Buntrock JD, Chute CG: Adopting graph traversal techniques for context-driven value sets extraction from biomedical knowledge sources. Proc IEEE Int Conf Semant Comput 2008: 460–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rector A, Qamar R, Marley T.Binding ontologies and coding systems to electronic health records and messages. 2nd International Workshop on Formal Biomedical Knowledge Representation; 2006: 11–19 CEUR WS Proceedings [Google Scholar]
- 27.Lee DH, Lau FY, Quan H.A method for encoding clinical datasets with SNOMED CT. BMC Med Inform Decis Mak 2010; 10: 53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bodenreider O, Smith B, Kumar A, Burgun A.Investigating subsumption in SNOMED CT: An exploration into large description logic-based biomedical terminologies. Artif Intell Med 2007; 39(3): 183–195 [DOI] [PMC free article] [PubMed] [Google Scholar]


