Abstract
We found that cells of Saccharomyces cerevisiae have an elevated level of the NAD-dependent glutamate dehydrogenase (NAD-GDH; encoded by the GDH2 gene) when grown with a nonfermentable carbon source or with limiting amounts of glucose, even in the presence of the repressing nitrogen source glutamine. This regulation was found to be transcriptional, and an upstream activation site (GDH2 UASc) sufficient for activation of transcription during respiratory growth conditions was identified. This UAS was found to be separable from a neighboring element which is necessary for the nitrogen source regulation of the gene, and strains deficient for the GLN3 gene product, required for expression of NAD-GDH during growth with the activating nitrogen source glutamate, were unaffected for the expression of NAD-GDH during growth with activating carbon sources. Two classes of mutations which prevented the normal activation of NAD-GDH in response to growth with nonfermentable carbon sources, but which did not affect the nitrogen-regulated expression of NAD-GDH, were found and characterized. Carbon regulation of GDH2 was found to be normal in hxk2, hap3, and hap4 strains and to be only slightly altered in a ssn6 strain; thus, in comparison with the regulation of previously identified glucose-repressed genes, a new pathway appears to be involved in the regulation of GDH2.
Full text
PDF

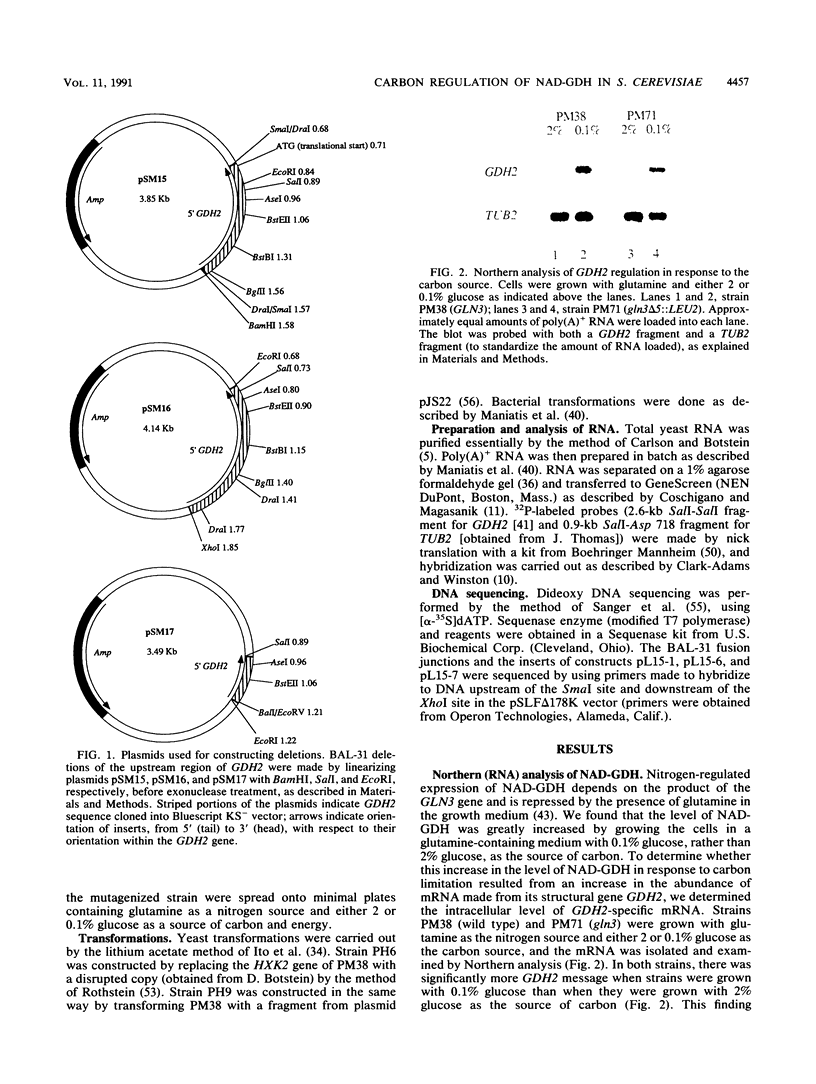

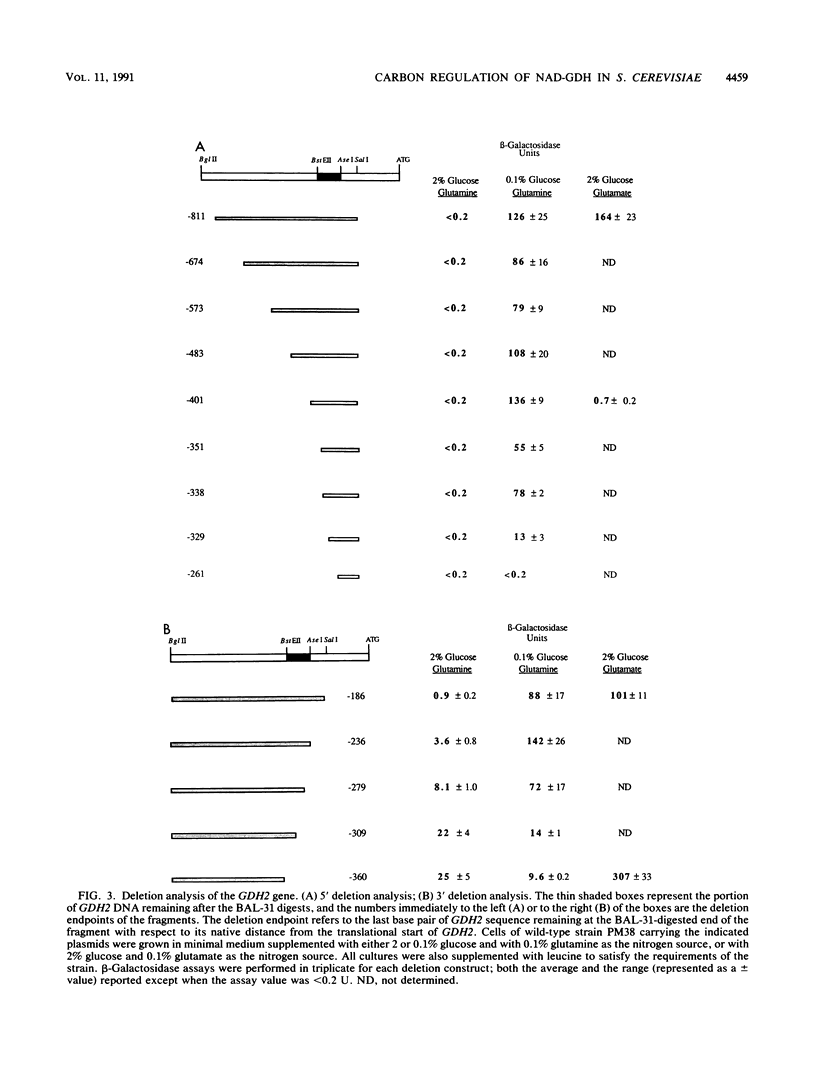
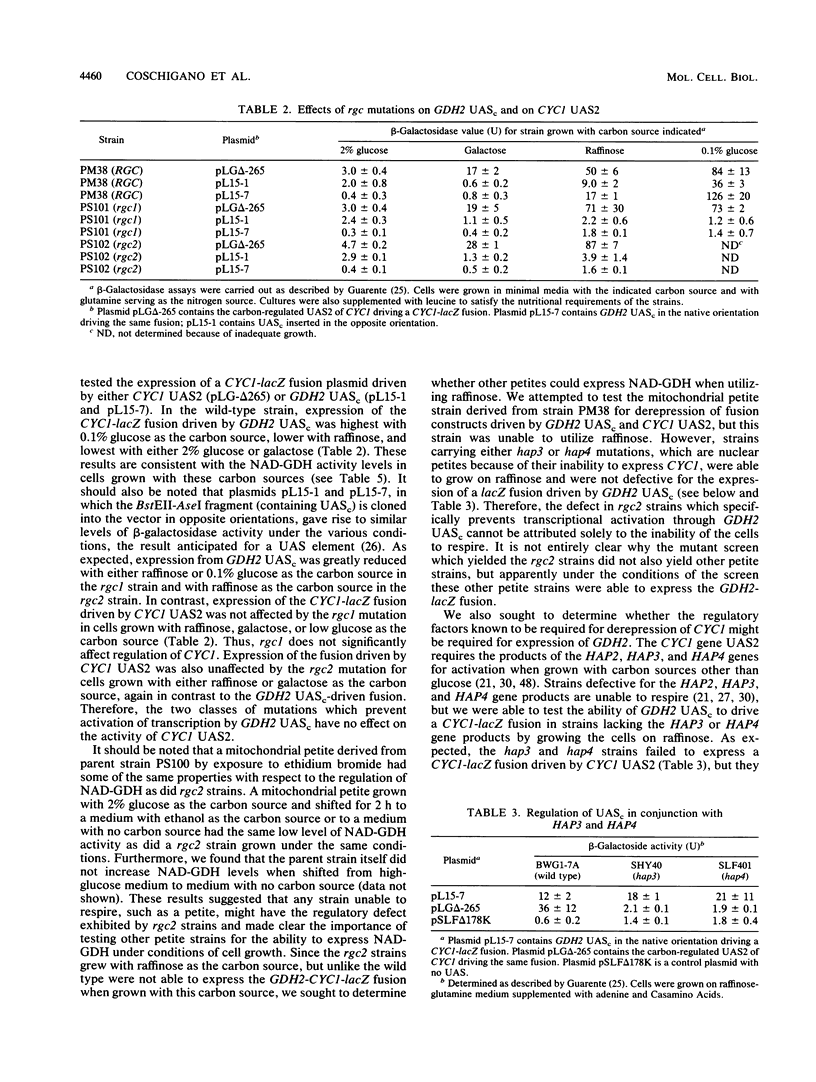




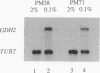
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Benjamin P. M., Wu J. I., Mitchell A. P., Magasanik B. Three regulatory systems control expression of glutamine synthetase in Saccharomyces cerevisiae at the level of transcription. Mol Gen Genet. 1989 Jun;217(2-3):370–377. doi: 10.1007/BF02464906. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Calderón J., Mora J. Glutamine assimilation pathways in Neurospora crassa growing on glutamine as sole nitrogen and carbon source. J Gen Microbiol. 1989 Oct;135(10):2699–2707. doi: 10.1099/00221287-135-10-2699. [DOI] [PubMed] [Google Scholar]
- Carlson M., Botstein D. Two differentially regulated mRNAs with different 5' ends encode secreted with intracellular forms of yeast invertase. Cell. 1982 Jan;28(1):145–154. doi: 10.1016/0092-8674(82)90384-1. [DOI] [PubMed] [Google Scholar]
- Carlson M., Osmond B. C., Botstein D. Mutants of yeast defective in sucrose utilization. Genetics. 1981 May;98(1):25–40. doi: 10.1093/genetics/98.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson M., Osmond B. C., Neigeborn L., Botstein D. A suppressor of SNF1 mutations causes constitutive high-level invertase synthesis in yeast. Genetics. 1984 May;107(1):19–32. doi: 10.1093/genetics/107.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson M. Regulation of sugar utilization in Saccharomyces species. J Bacteriol. 1987 Nov;169(11):4873–4877. doi: 10.1128/jb.169.11.4873-4877.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celenza J. L., Carlson M. A yeast gene that is essential for release from glucose repression encodes a protein kinase. Science. 1986 Sep 12;233(4769):1175–1180. doi: 10.1126/science.3526554. [DOI] [PubMed] [Google Scholar]
- Ciriacy M. Isolation and characterization of yeast mutants defective in intermediary carbon metabolism and in carbon catabolite derepression. Mol Gen Genet. 1977 Jul 20;154(2):213–220. doi: 10.1007/BF00330840. [DOI] [PubMed] [Google Scholar]
- Clark-Adams C. D., Winston F. The SPT6 gene is essential for growth and is required for delta-mediated transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1987 Feb;7(2):679–686. doi: 10.1128/mcb.7.2.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coschigano P. W., Magasanik B. The URE2 gene product of Saccharomyces cerevisiae plays an important role in the cellular response to the nitrogen source and has homology to glutathione s-transferases. Mol Cell Biol. 1991 Feb;11(2):822–832. doi: 10.1128/mcb.11.2.822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courchesne W. E., Magasanik B. Regulation of nitrogen assimilation in Saccharomyces cerevisiae: roles of the URE2 and GLN3 genes. J Bacteriol. 1988 Feb;170(2):708–713. doi: 10.1128/jb.170.2.708-713.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crouse G. F., Frischauf A., Lehrach H. An integrated and simplified approach to cloning into plasmids and single-stranded phages. Methods Enzymol. 1983;101:78–89. doi: 10.1016/0076-6879(83)01006-x. [DOI] [PubMed] [Google Scholar]
- Dantzig A. H., Wiegmann F. L., Jr, Nason A. Regulation of glutamate dehydrogenases in nit-2 and am mutants of Neurospora crassa. J Bacteriol. 1979 Mar;137(3):1333–1339. doi: 10.1128/jb.137.3.1333-1339.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsman J. C., Grivell L. A. Expression of the gene encoding subunit II of yeast QH2: cytochrome c oxidoreductase is regulated by multiple factors. Curr Genet. 1990 Jun;17(6):459–464. doi: 10.1007/BF00313072. [DOI] [PubMed] [Google Scholar]
- Drillien R., Aigle M., Lacroute F. Yeast mutants pleiotropically impaired in the regulation of the two glutamate dehydrogenases. Biochem Biophys Res Commun. 1973 Jul 17;53(2):367–372. doi: 10.1016/0006-291x(73)90671-2. [DOI] [PubMed] [Google Scholar]
- Dubois E. L., Grenson M. Absence of involvement of glutamine synthetase and of NAD-linked glutamate dehydrogenase in the nitrogen catabolite repression of arginase and other enzymes in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1974 Sep 9;60(1):150–157. doi: 10.1016/0006-291x(74)90185-5. [DOI] [PubMed] [Google Scholar]
- Dubois E., Vissers S., Grenson M., Wiame J. M. Glutamine and ammonia in nitrogen catabolite repression of Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1977 Mar 21;75(2):233–239. doi: 10.1016/0006-291x(77)91033-6. [DOI] [PubMed] [Google Scholar]
- Entian K. D. Genetic and biochemical evidence for hexokinase PII as a key enzyme involved in carbon catabolite repression in yeast. Mol Gen Genet. 1980;178(3):633–637. doi: 10.1007/BF00337871. [DOI] [PubMed] [Google Scholar]
- Forsburg S. L., Guarente L. Identification and characterization of HAP4: a third component of the CCAAT-bound HAP2/HAP3 heteromer. Genes Dev. 1989 Aug;3(8):1166–1178. doi: 10.1101/gad.3.8.1166. [DOI] [PubMed] [Google Scholar]
- Forsburg S. L., Guarente L. Mutational analysis of upstream activation sequence 2 of the CYC1 gene of Saccharomyces cerevisiae: a HAP2-HAP3-responsive site. Mol Cell Biol. 1988 Feb;8(2):647–654. doi: 10.1128/mcb.8.2.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangloff S. P., Marguet D., Lauquin G. J. Molecular cloning of the yeast mitochondrial aconitase gene (ACO1) and evidence of a synergistic regulation of expression by glucose plus glutamate. Mol Cell Biol. 1990 Jul;10(7):3551–3561. doi: 10.1128/mcb.10.7.3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein A., Lampen J. O. Beta-D-fructofuranoside fructohydrolase from yeast. Methods Enzymol. 1975;42:504–511. doi: 10.1016/0076-6879(75)42159-0. [DOI] [PubMed] [Google Scholar]
- Grenson M., Dubois E., Piotrowska M., Drillien R., Aigle M. Ammonia assimilation in Saccharomyces cerevisiae as mediated by the two glutamate dehydrogenases. Evidence for the gdhA locus being a structural gene for the NADP-dependent glutamate dehydrogenase. Mol Gen Genet. 1974;128(1):73–85. doi: 10.1007/BF00267295. [DOI] [PubMed] [Google Scholar]
- Guarente L., Hoar E. Upstream activation sites of the CYC1 gene of Saccharomyces cerevisiae are active when inverted but not when placed downstream of the "TATA box". Proc Natl Acad Sci U S A. 1984 Dec;81(24):7860–7864. doi: 10.1073/pnas.81.24.7860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L., Lalonde B., Gifford P., Alani E. Distinctly regulated tandem upstream activation sites mediate catabolite repression of the CYC1 gene of S. cerevisiae. Cell. 1984 Feb;36(2):503–511. doi: 10.1016/0092-8674(84)90243-5. [DOI] [PubMed] [Google Scholar]
- Guarente L., Mason T. Heme regulates transcription of the CYC1 gene of S. cerevisiae via an upstream activation site. Cell. 1983 Apr;32(4):1279–1286. doi: 10.1016/0092-8674(83)90309-4. [DOI] [PubMed] [Google Scholar]
- Guarente L. Yeast promoters and lacZ fusions designed to study expression of cloned genes in yeast. Methods Enzymol. 1983;101:181–191. doi: 10.1016/0076-6879(83)01013-7. [DOI] [PubMed] [Google Scholar]
- HALPERN Y. S., UMBARGER H. E. Utilization of L-glutamic and 2-oxoglutaric acid as sole sources of carbon by Escherichia coli. J Gen Microbiol. 1961 Oct;26:175–183. doi: 10.1099/00221287-26-2-175. [DOI] [PubMed] [Google Scholar]
- HIERHOLZER G., HOLZER H. REPRESSION DER SYNTHESE VON DPN-ABHAENGIGER GLUTAMINSAEUREDEHYDROGENASE IN SACCHAROMYCES CEREVISIAE DURCH AMMONIUMIONEN. Biochem Z. 1963 Dec 3;339:175–185. [PubMed] [Google Scholar]
- Hahn S., Guarente L. Yeast HAP2 and HAP3: transcriptional activators in a heteromeric complex. Science. 1988 Apr 15;240(4850):317–321. doi: 10.1126/science.2832951. [DOI] [PubMed] [Google Scholar]
- Hahn S., Pinkham J., Wei R., Miller R., Guarente L. The HAP3 regulatory locus of Saccharomyces cerevisiae encodes divergent overlapping transcripts. Mol Cell Biol. 1988 Feb;8(2):655–663. doi: 10.1128/mcb.8.2.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes D. S., Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981 Jun;114(1):193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinghorn J. R., Pateman J. A. NAD and NADP l-glutamate dehydrogenase activity and ammonium regulation in Aspergillus nidulans. J Gen Microbiol. 1973 Sep;78(1):39–46. doi: 10.1099/00221287-78-1-39. [DOI] [PubMed] [Google Scholar]
- Lehrach H., Diamond D., Wozney J. M., Boedtker H. RNA molecular weight determinations by gel electrophoresis under denaturing conditions, a critical reexamination. Biochemistry. 1977 Oct 18;16(21):4743–4751. doi: 10.1021/bi00640a033. [DOI] [PubMed] [Google Scholar]
- Lobo Z., Maitra P. K. Genetics of yeast hexokinase. Genetics. 1977 Aug;86(4):727–744. doi: 10.1093/genetics/86.4.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardo A., Carine K., Scheffler I. E. Cloning and characterization of the iron-sulfur subunit gene of succinate dehydrogenase from Saccharomyces cerevisiae. J Biol Chem. 1990 Jun 25;265(18):10419–10423. [PubMed] [Google Scholar]
- Ma H., Botstein D. Effects of null mutations in the hexokinase genes of Saccharomyces cerevisiae on catabolite repression. Mol Cell Biol. 1986 Nov;6(11):4046–4052. doi: 10.1128/mcb.6.11.4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller S. M., Magasanik B. Role of NAD-linked glutamate dehydrogenase in nitrogen metabolism in Saccharomyces cerevisiae. J Bacteriol. 1990 Sep;172(9):4927–4935. doi: 10.1128/jb.172.9.4927-4935.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell A. P., Ludmerer S. W. Identification of a glutaminyl-tRNA synthetase mutation Saccharomyces cerevisiae. J Bacteriol. 1984 May;158(2):530–534. doi: 10.1128/jb.158.2.530-534.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell A. P., Magasanik B. Biochemical and physiological aspects of glutamine synthetase inactivation in Saccharomyces cerevisiae. J Biol Chem. 1984 Oct 10;259(19):12054–12062. [PubMed] [Google Scholar]
- Mitchell A. P., Magasanik B. Purification and properties of glutamine synthetase from Saccharomyces cerevisiae. J Biol Chem. 1983 Jan 10;258(1):119–124. [PubMed] [Google Scholar]
- Mitchell A. P., Magasanik B. Regulation of glutamine-repressible gene products by the GLN3 function in Saccharomyces cerevisiae. Mol Cell Biol. 1984 Dec;4(12):2758–2766. doi: 10.1128/mcb.4.12.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olesen J., Hahn S., Guarente L. Yeast HAP2 and HAP3 activators both bind to the CYC1 upstream activation site, UAS2, in an interdependent manner. Cell. 1987 Dec 24;51(6):953–961. doi: 10.1016/0092-8674(87)90582-4. [DOI] [PubMed] [Google Scholar]
- Pinkham J. L., Guarente L. Cloning and molecular analysis of the HAP2 locus: a global regulator of respiratory genes in Saccharomyces cerevisiae. Mol Cell Biol. 1985 Dec;5(12):3410–3416. doi: 10.1128/mcb.5.12.3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repetto B., Tzagoloff A. Structure and regulation of KGD1, the structural gene for yeast alpha-ketoglutarate dehydrogenase. Mol Cell Biol. 1989 Jun;9(6):2695–2705. doi: 10.1128/mcb.9.6.2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Rodriguez L., Lampen J. O., MacKay V. L. SUC1 gene of Saccharomyces: a structural gene for the large (glycoprotein) and small (carbohydrate-free) forms of invertase. Mol Cell Biol. 1981 May;1(5):469–474. doi: 10.1128/mcb.1.5.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross J., Reid G. A., Dawes I. W. The nucleotide sequence of the LPD1 gene encoding lipoamide dehydrogenase in Saccharomyces cerevisiae: comparison between eukaryotic and prokaryotic sequences for related enzymes and identification of potential upstream control sites. J Gen Microbiol. 1988 May;134(5):1131–1139. doi: 10.1099/00221287-134-5-1131. [DOI] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz J., Carlson M. Molecular analysis of SSN6, a gene functionally related to the SNF1 protein kinase of Saccharomyces cerevisiae. Mol Cell Biol. 1987 Oct;7(10):3637–3645. doi: 10.1128/mcb.7.10.3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F., Stewart J. W., Margoliash E., Parker J., Campbell W. The structural gene for yeast cytochrome C. Proc Natl Acad Sci U S A. 1966 Jun;55(6):1498–1504. doi: 10.1073/pnas.55.6.1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suissa M., Suda K., Schatz G. Isolation of the nuclear yeast genes for citrate synthase and fifteen other mitochondrial proteins by a new screening method. EMBO J. 1984 Aug;3(8):1773–1781. doi: 10.1002/j.1460-2075.1984.tb02045.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright R. M., Poyton R. O. Release of two Saccharomyces cerevisiae cytochrome genes, COX6 and CYC1, from glucose repression requires the SNF1 and SSN6 gene products. Mol Cell Biol. 1990 Mar;10(3):1297–1300. doi: 10.1128/mcb.10.3.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]