Abstract
Aeromonas hydrophila is both a human and animal pathogen, and the cytotoxic enterotoxin (Act) is a crucial virulence factor of this bacterium because of its associated hemolytic, cytotoxic, and enterotoxic activities. Previously, to define the role of some regulatory genes in modulating Act production, we showed that deletion of a glucose-inhibited division gene (gidA) encoding tRNA methylase reduced Act levels, while overproduction of DNA adenine methyltransferase (Dam) led to a concomitant increase in Act-associated biological activities of a diarrheal isolate SSU of A. hydrophila. Importantly, there are multiple GATC binding sites for Dam within an upstream sequence of the gidA gene and one such target site in the act gene upstream region. We showed the dam gene to be essential for the viability of A. hydrophila SSU, and, therefore, to better understand the interaction of the encoding genes, Dam and GidA, in act gene regulation, we constructed a gidA in-frame deletion mutant of Escherichia coli GM28 (dam+) and GM33 (Δdam) strains. We then tested the expressional activity of the act and gidA genes by using a promoterless pGlow-TOPO vector containing a reporter green fluorescent protein (GFP). Our data indicated that in GidA+ strains of E. coli, constitutive methylation of the GATC site(s) by Dam negatively regulated act and gidA gene expression as measured by GFP production. However, in the ΔgidA strains, irrespective of the presence or absence of constitutively active Dam, we did not observe any alteration in the expression of the act gene signifying the role of GidA in positively regulating Act production. To determine the exact mechanism of how Dam and GidA influence Act, a real-time quantitative PCR (RT-qPCR) assay was performed. The analysis indicated an increase in gidA and act gene expression in the A. hydrophila Dam-overproducing strain, and these data matched with Act production in the E. coli GM28 strain. Thus, the extent of DNA methylation caused by constitutive versus overproduction of Dam, as well as possible conformation of DNA influence the expression of act and gidA genes in A. hydrophila SSU. Our results indicate that the act gene is under the control of both Dam and GidA modification methylases, and Dam regulates Act production via GidA.
Keywords: GATC Dam target sites, Promoter activity, tRNA uridine 5 carboxymethylaminomethyl, modification enzyme
1. Introduction
Among various Aeromonas species, A. hydrophila is an aquatic environmental and food-borne microorganism which poses a health risk (Edberg et al., 2007). A diarrheal isolate SSU of A. hydrophila studied in our laboratory is involved in human infections, and its major virulence factor, the cytotoxic enterotoxin (Act), leads to septicemia, gastroenteritis and mortality in a mouse model (Chopra and Houston, 1999; Sha et al., 2001, 2002). We reported that a glucose-inhibited division protein, GidA, modulated the expression of the act gene (Sha et al., 2004).
GidA protein was first described in the Escherichia coli K12 strain, and disruption of the gidA gene affected cell division when grown in a medium containing glucose by interrupting chromosomal replication, resulting in a cell elongation phenotype (von Meyenburg et al., 1982). Further studies showed that GidA is a flavin adenine dinucleotide (FAD) binding enzyme (White et al., 2001) and also a tRNA modification methylase that catalyzes the addition of the carboxymethylaminomethyl group onto the C5 carbon atom of uridine at position 34 (U34) of RNAs (Urbonavicius et al., 2005; Yim et al., 2006). It was found that the post-transcriptional modification of tRNA represents a significant element controlling gene expression (Gustilo et al., 2008).
The deletion of a gidA gene attenuated the pathogenesis of some bacteria, such as Streptococcus pyogenes, Pseudomonas aeruginosa, and Salmonella enterica serovar Typhimurium (Cho and Caparon, 2008; Gupta et al., 2009; Shippy et al., 2011). In particular, it was noted that gidA gene mutant had a nearly normal global transcription profile, when compared to that of parental S. pyogenes strain, but the mutant produced significantly reduced levels of multiple virulence factors due to impaired translational efficiencies (Cho and Caparon, 2008). The gidA mutation in a plant pathogen P. syringae led to pleiotropic effects, resulting in diverse phenotypic traits, swarming, presence of fluorescent pigment and virulence, which indicated a possible regulatory role for GidA in moderating translational fidelity (Kinscherf and Willis, 2002).
On the other hand, DNA adenine methyltransferase (Dam) methylates the adenine base of 5′-GATC-3′ specific sites and is a global gene regulator modifying DNA. Thus, Dam plays an important role in controlling various processes in prokaryotic cells, such as transcriptional regulation, mismatch repair, host–pathogen interactions, and binding of the replication initiation complex to the methylated origin of replication (oriC) site (Casadesus and Low, 2006; Chatti and Landoulsi, 2008; Low and Casadesus, 2008; Marinus and Casadesus, 2009). It is known that initiation of replication of the chromosome occurs at a unique site, namely the oriC. Among the promoters possibly involved in the transcriptional activation of replication initiation included those for the gidAB genes, as transcription of gid genes was needed for activation of oriC (Bates et al., 1997). Further, regulation of DNA methylation activity through promoter methylation and compatibility between methylation of a promoter with its active transcription have been reported (Barnard et al., 2004; Marinus and Casadesus, 2009). Dam methylation can control promoter transcription, and transcriptional repression by Dam appears to be more common than transcriptional activation.
Our previous results showed that overproduction of Dam in A. hydrophila SSU increased Act-associated biological activities; however, a decrease in such activities was noted in the gidA gene mutant of the corresponding parental strain which produced a constitutive level of Dam (Erova et al., 2006a, 2006b). These data indicated that the extent of DNA methylation which was governed by the amount of Dam present dictated the levels of GidA and Act in A. hydrophila SSU. Dam and GidA proteins are highly conserved in many prokaryotes, and because the dam gene is essential for the viability of the A. hydrophila SSU strain, but not of E. coli, this fact allowed us to use E. coli K12 as a model to address the hypothesis regarding act gene regulation by both enzymes. We show data which further our understanding on the cross-talk between the decisive regulators of gene expression, namely dam and gidA, on Act production in A. hydrophila SSU. Our previous data indicated that Act is the most potent virulence factor among the three enterotoxins of A. hydrophila SSU, and GidA and Dam are involved in its control (Erova et al., 2006a; Sha et al., 2004). In the present study, we explored how these regulatory proteins, Dam and GidA, influence Act production. These studies are important, as modulation of bacterial virulence genes in general could be under the control of Dam and GidA and needs further investigation.
2. Materials and methods
2.1. Strains, plasmids, and growth conditions
Bacterial strains and plasmids used in this study are listed in Table 1. A. hydrophila and E. coli cultures were grown at 37 °C in Luria–Bertani (LB) broth and LB agar plates (Sambrook et al., 1989). The antibiotics ampicillin (Ap), chloramphenicol (Cm), and rifampin (Rif) were used at concentrations of 100, 20 and 200 μg/ml, respectively.
Table 1.
Strains and plasmids used in this study.
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| A. hydrophila SSU-R strains | Rifampin resistance strain, Rifr | Laboratory |
| Parental | pBAD plasmid, Rifr Apr | Erova et al. (2006b) |
| Dam-overproducing | pBAD-dam plasmid, Rifr Apr | Erova et al. (2006b) |
| E. coli K12 strains | ||
| TOP10 | F− mcrA Δ mrr-hsdRMS-mcrBC Φ80lacZΔM15 ΔlacX74 recA1 deoR araD139 Δ(ara-leu)7697 galU galK rpsL (Strr) endA1 nupG | Invitrogen |
| GM28 | F− sup-85 (Am) | Marinus and Morris(1973) |
| GM33 | F− dam-3 sup-85 (Am) | Marinus and Morris(1973) |
| GM28 (pGlow-UpgidA) | GM28 with pGlow-UpgidA plasmid | This study |
| GM33 (pGlow-UpgidA) | GM33 with pGlow-UpgidA plasmid | This study |
| GM28 (pGlow-Upact) | GM28 with pGlow-Upact plasmid | This study |
| GM33 (pGlow-Upact) | GM33 with pGlow-Upact plasmid | This study |
| GM28 (pGlow-TOPO) | GM28 with pGlow-TOPO vector | This study |
| GM33 (pGlow-TOPO) | GM33 with pGlow-TOPO vector | This study |
| GM28 gidA mutant | In-frame gidA mutant of GM28 | This study |
| GM33 gidA mutant | In-frame gidA mutant of GM33 | This study |
| GM28 gidA mutant (pGlow-Upact) | GM28 gidA mutant carrying pGlow-Upact plasmid | This study |
| GM33 gidA mutant (pGlow-Upact) | GM33 gidA mutant carrying pGlow-Upact plasmid | This study |
| GM28 gidA mutant (pGlow-UpgidA) | GM28 gidA mutant carrying pGlow-UpgidA plasmid | This study |
| GM33 gidA mutant (pGlow-UpgidA) | GM33 gidA mutant carrying pGlow-UpgidA plasmid | This study |
| Plasmids | ||
| pGlow-TOPO | Promoterless vector, Apr Nmr | Invitrogen |
| pGlow-UpgidA | pGlow carrying upstream region of A. hydrophila gidA gene, Apr Nmr | This study |
| pGlow-Upact | pGlow carrying upstream region of A. hydrophila act gene, Apr Nmr | This study |
| pKD46 | λ Red recombinase plasmid, Apr | Datsenko and Wanner(2000) |
| pKD3 | cat cassette template with FRT sites, Apr Cmr | Datsenko and Wanner(2000) |
| pCP20 | FLP recombinase plasmid, Apr Cmr | Datsenko and Wanner(2000) |
| pBAD/Thio-E | Prokaryotic expression vector, Apr | Invitrogen |
| pBAD-act | act gene of A. hydrophila cloned into pBAD/Thio-E, Apr | This study |
2.2. DNA isolation and polymerase chain reaction (PCR)
Plasmid DNA was isolated by using a QIAprep Spin Miniprep Kit (Qiagen, Valencia, CA). Genomic DNA (gDNA) was isolated by using a DNeasy Tissue Kit (Qiagen). The primers (Table 2) were synthesized by Integrated DNA Technologies, Inc., Coralville, IA. Amplified PCR products were purified by using a QIAquick PCR Purification Kit (Qiagen). PCR assays from single bacterial colonies were performed as described previously (Erova et al., 2006b).
Table 2.
Primers used in this study.
| Primer | Sequence (5′–3′) | Purpose |
|---|---|---|
| UpgidA-N | TCGCCTGAACCGGGAGCGGGAGATC | PCR amplification of A. hydrophila region upstream of gidA gene start codon |
| UpgidA-C | ATTGGGGATCACCTTGATCTGCACG | |
| Upact-N | CGGGGTACCGAGCCATGTTATCCCG | PCR amplification of A. hydrophila region upstream of act gene start codon |
| Upact-C | CCGGAATTCCATAGCAACCCCAATA | |
| T7-F | TAATACGACTCACTATAGGG | DNA sequencing of the upstream region in pGlow-PCRa |
| GFP-R | GGGTAAGCTTTCCGTATGTA | |
| F-pKD3 | GCCCGGGCTTCAATCCATTTTCATACCGCGTTATG | PCR amplification of a Cmr cassette for λ Red recombination system |
| CGAGGCAATCACCATGTGTAGGCTGGAGCTGCTTC | ||
| R-pKD3 | GCGGGTGCTTACCAGGCATTTTTAATGCGTTATGC | |
| GCTACGACGCAGCATTATCCTCCTTAGTTCCTATT | ||
| UpF-Cm | TACCCAGGATCCCAGGTCTTTCTCA | PCR verification of gidA knockout mutant in GM28 and GM33 E. coli strains |
| UpR-Cm | AGAGGTTCCAACTTTCACCATAATG | |
| DownF-Cm | ACGCCACATCTTGCGAATATATGTG | |
| DownR-Cm | AGCGAAATACCTGCGTCTTTCAGCA | |
| gidA-N | GACATGAACTACCGCGACGTC | RT-qPCR for gidA gene |
| gidA-C | ATGAACCAGCAGGATGGAGATG | |
| act-N | GCAATATCGAAATCGGTGCGC | RT-qPCR for act gene |
| act-C | GCTGAAGCCAAGCCCGGAGAGC | |
| gap-1N | AAGCTGACCGGCAACGC | RT-qPCR for gap-1 control gene |
| gap-1C | CAGATAGGCATTGAGGCTCTCC | |
| actN-NcoI | CATGCCATGGATGCAAAAACTAAAAATAACTGGCT | Cloning of the act gene into pBAD/Thio-E vector |
| actC-PmeI | AGCTTTGTTTAAACTTATTGATTGGCTGCTGGCGT |
Underlining indicates FRT and restriction endonuclease sites.
pGlow-UpgidA and pGlow-Upact plasmids.
2.3. Cloning of A. hydrophila SSU upstream regions of gidA and act genes into pGlow-TOPO promoterless vector
For assaying expressional activity and to study how methylation in upstream regions of gidA and act genes affects expression of these genes, we cloned the upstream sequences into a pGlow-TOPO promoterless vector (Invitrogen, Carlsbad, CA). Upstream sequences of the gidA and act genes of A. hydrophila SSU contained promoter regions, which we identified by using the SoftBerry program (www.softberry.com). To obtain PCR products corresponding to the upstream regions of these genes, UpgidA-N/UpgidA-C and Upact-N/Upact-C primers were employed (Table 2) along with gDNA of A. hydrophila as a template. The amplified products (517-bp and 295-bp, respectively) were cloned into a pGlow-TOPO plasmid, and the recombinant plasmids were transformed into TOP10-competent cells. Recombinant plasmids were sequenced from both strands with T7-F and green fluorescent protein (GFP)-R primers (Table 2) to examine the sequence of an inserted PCR product and to confirm the correct fusion with the gene encoding GFP. The correct recombinant plasmids, pGlow-UpgidA and pGlow-Upact, were transformed into E. coli GM28 and GM33 strains which either harbored dam and gidA genes or were deleted for the dam gene and harbored gidA gene (Table 1). We measured transcriptional activities of promoters as a measure of GFP production in relative fluorescence units (RFU) normalized to the total protein concentration after subtraction of the background. The promoterless pGlow-TOPO vector alone in E. coli GM28 and GM33 strains was used as a control.
2.4. Construction of gidA deleted in-frame mutants of E. coli GM28 and GM33 strains
To obtain gidA in-frame knockout mutants, we used the λ Red recombinase system (Baba et al., 2006; Datsenko and Wanner, 2000) with some modifications. The competent E. coli GM28 (dam+) and GM33 (Δdam) cells were transformed with pKD46 plasmid carrying λ Red recombinase, and cultures were grown at 30 °C because this helper plasmid has a temperature-sensitive replication. In order to generate a PCR product for electroporation of the above transformants, we constructed primers (Table 2) that had homologous regions adjacent to a gidA gene and that of a pKD3 plasmid carrying the Cmr gene cassette, which was flanked by FRT (flippase [FLP] recognition target) sites. To eliminate unamplified methylated template DNA, we used DpnI restriction enzyme to digest purified PCR products and then electroporated them into GM28 and GM33 strains carrying pKD46 plasmid which were cultivated in the presence of 10 mM l-arabinose to induce the pKD46 λ Red expression system. To cure the pKD46 plasmid from E. coli cells, we incubated mixtures after electroporation for 1 h at 37 °C and selected for colonies that were Cmr and Aps. To remove the Cmr gene from mutants, E. coli GM28 and GM33 strains were transformed with pCP20 plasmid that shows temperature-sensitive (30 °C) replication and thermal (43 °C) induction of FLP synthesis. After screening at 43 °C, the identified Cms and Aps single mutant colonies without pCP20 were verified by PCR.
To verify insertion of the Cmr gene cassette into chromosomal DNA of E. coli, Cmr colonies were PCR confirmed by using two pairs of primers, UpF-Cm/UpR-Cm and DownF-Cm/DownR-Cm (Table 2). The oligos UpF-Cm and DownR-Cm were designed based on flanking sequences of the E. coli gidA gene on chromosomal DNA. The second pair of primers, UpR-Cm and DownF-Cm, was designed based on the Cmr gene cassette sequence. As shown in Fig. IA (Supplementary data), PCR products of Up- and Down-regions corresponded to the calculated sizes of 931-bp and 777-bp, respectively. A mutant thus generated still contained the Cmr cassette. To confirm the gidA knockout mutant without antibiotic resistance (Fig. IB, Supplementary data) after curing of pCP20 plasmid, single colonies were tested by PCR with three pairs of primers (UpF-Cm/UpR-Cm, DownF-Cm/DownR-Cm and UpF-Cm/DownR-Cm) (Table 2). Primers UpF-Cm/UpR-Cm and DownF-Cm/DownR-Cm did not amplify the DNA product, a finding which indicated that the Cmr gene cassette was eliminated. As expected, when we used UpF-Cm and DownR-Cm primers, a PCR product (216-bp) was amplified which represented stretches of up- and down-stream sequences from the gidA gene.
Fig. 1.
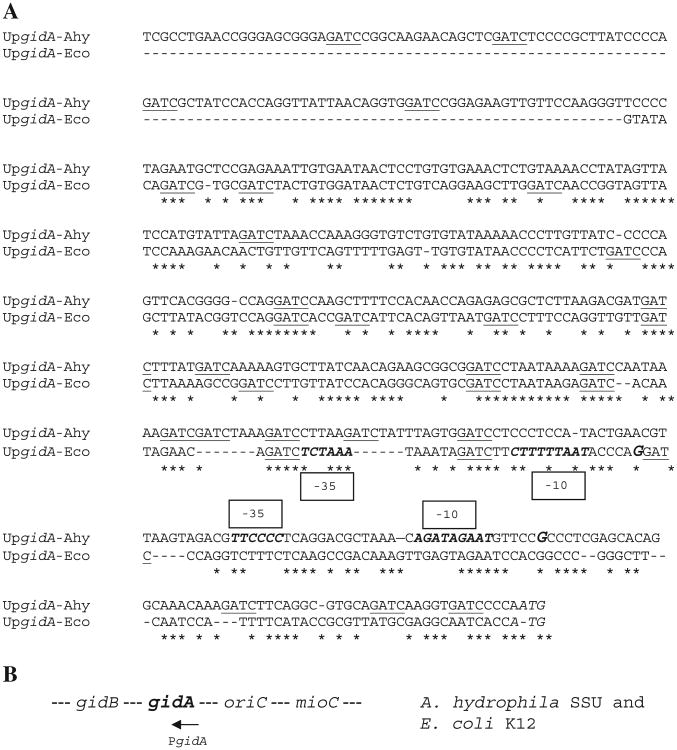
A. DNA sequence alignment of gidA gene upstream regions from A. hydrophila SSU and E. coli K12 strains. Asterisks denote conserved nucleotides. The underlined sequences represent GATC sites for Dam methylation. The potential transcriptional start sites (G) and putative -10, -35 boxes are indicated in italics and bold letters. B. Schematic diagram showing gidA regions of A. hydrophila and E. coli. The figure is not drawn to scale.
2.5. Transformation and electroporation of E. coli strains
Chemical-competent E. coli GM28 and GM33 cells for transformation were prepared by using the Z-Competent E. coli transformation kit from Zymo Research, Orange, CA. Competent E. coli GM28 (pKD46) and GM33 (pKD46) cells for electroporation of a PCR product were prepared by using 10% glycerol, as we previously described (Erova et al., 2006b). Table 1 contains the final strains with different recombinant plasmids in either dam- and/or gidA-positive and negative backgrounds that were generated for this study.
2.6. Dam methylation status of DNA from A. hydrophila SSU and E. coli strains
To verify an identical Dam methylation status of DNA from A. hydrophila SSU and E. coli GM28 and GM33 strains, DNA was isolated from these strains and digested with DpnI and DpnII restriction endonucleases which recognize either methylated or unmethylated DNA (Roberts et al., 2003).
2.7. Preparation of E. coli cell extracts and a gene expression assay
To prepare cell extracts from E. coli GM28 and GM33 strains containing constructed recombinant plasmids, we grew the cells at 37 °C in LB medium with Ap until a middle log phase. We then harvested cells and washed them with 1× Dulbecco's phosphate-buffered saline (DPBS). The cells were disrupted by sonication on ice by using two 10-s pulses, cell extracts were recovered after centrifugation, and the total amount of protein in extracts was determined by using the Bradford protein assay (Bradford, 1976). Extracts (200 μl) were assayed in duplicate by using 96-well microtiter plates (Costar, black, transparent bottom) and read employing an FLX800 Microplate Fluorescence Reader (Bio-Tek Instruments, Inc., Winooski, VT). GFP levels under the control of act and gidA promoters and upstream DNA sequences were measured with excitation at 400 nm and emission at 508 nm. Background fluorescence of the extract was determined by using E. coli cells with the pGlow-TOPO vector alone.
2.8. Real-time (RT)-qPCR assay
Total RNA from parental A. hydrophila SSU and Dam-overproducing strains containing pBAD and pBAD-damAhSSU plasmids, respectively (Erova et al., 2006b), was isolated by using the RiboPure-Bacteria Kit (Ambion, Inc., Austin, TX), and 20 μg of total RNA was processed for RT-qPCR assay. RT-qPCR was performed in the LightCycler thermal cycler system (Roche Diagnostics, Indianapolis, IN) using SYBR Green I dye (Qiagen), as previously described (Fadl et al., 2006). Sequences of primers used for amplification of the gidA gene, act gene, and endogenous control gap-1 gene (glyceraldehyde-3-phosphate dehydrogenase, GAPDH) are listed in Table 2. RT-qPCR assays were run in parallel and a fold-change value was determined by using the comparative threshold method (Livak and Schmittgen, 2001) after normalization of gidA and act genes to gap-1.
2.9. Cloning of the A. hydrophila act gene under the araBAD promoter in E. coli GM28 and GM33 strains
To delineate Act production of A. hydrophila in E. coli, pBAD-act recombinant plasmid (Table 1) was generated by using actN-NcoI and actC-PmeI primers (Table 2) and replacing the NcoI-PmeI fragment of pBAD/Thio-E vector under its arabinose-inducible PBAD promoter. The plasmid was then subjected to transformation into E. coli GM28 and GM33 strains and their ΔgidA mutants. To induce expression of the act gene from recombinant plasmid, the medium was supplemented with l-arabinose (0.2%) overnight.
2.10. Measurement of hemolytic activity associated with Act of A. hydrophila SSU
The hemolytic activity associated with Act was measured by examining the release of hemoglobin from rabbit erythrocytes (Colorado Serum Co., Denver, CO) in E. coli strains, as described previously for A. hydrophila (Erova et al., 2006b). E. coli strains with different genotypes containing the pBAD-act recombinant plasmid were grown overnight in LB medium with Ap and arabinose, and cell extracts were prepared as above (section 2.7). For hemolytic activity assay, 100 μl of 1× DPBS was added to each of the wells of a 96-well microtiter plate. The cell extracts were added to the first well in each row of a microtiter plate followed by serial 2-fold dilution and the addition of 100 μl of 3% rabbit erythrocytes. The plate was incubated at 37 °C for 1 h and observed for hemolytic activity associated with Act. The supernatants were taken from those wells that showed partial lysis of red blood cells, and the hemoglobin release was recorded at 540 nm by using a VERSAmax microplate reader (Sunnyvale, CA).
2.11. Statistics
Wherever applicable, at least three independent experiments were performed. The data were analyzed by using the Student's t test, and P values of ≤0.05 were considered significant.
3. Results
3.1. DNA methylation by E. coli and A. hydrophila SSU Dam
GidA is a highly conserved protein among different Gram-negative bacteria. The amino acid sequence for GidA from A. hydrophila SSU has 98% identity with GidA from A. hydrophila ATCC7966T; 96% identity with GidA of A. salmonicida A449; 76% with Vibrio fischeri; 75% with S. enterica; and 74% identity with the GidA protein of E. coli K12 (Blattner et al., 1997; McClelland et al., 2001; Reith et al., 2008; Ruby et al., 2005; Seshadri et al., 2006). The gidA gene upstream region of A. hydrophila SSU (517-bp) and E. coli K12 (381-bp) strains contained 18 and 14 GATC Dam target sites, respectively (Fig. 1). The examination of upstream regions of the A. hydrophila act gene indicated the presence of one GATC sequence.
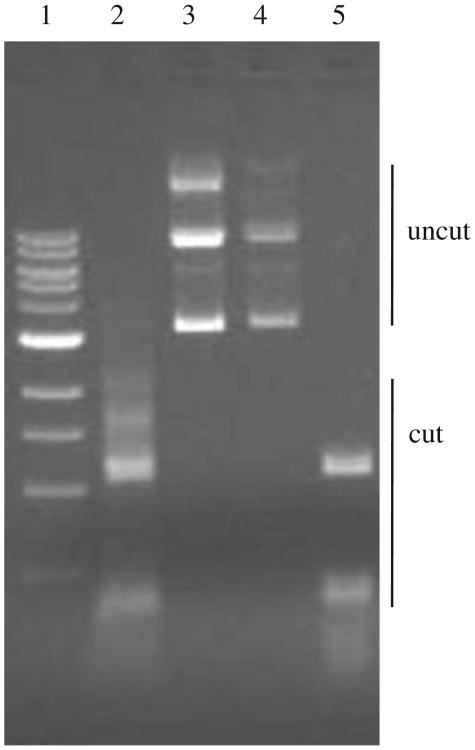
It is known that DpnI enzyme digests Dam-methylated DNA, and DpnII does not digest Dam-methylated DNA. To show that E. coli GM28 (Dam+) and A. hydrophila SSU (Dam+) strains have identical Dam activity with GATC specificity and that GM33 strain is deficient in the dam gene, plasmid DNA (pGlow-UpgidA) isolated from E. coli strains was treated with DpnI and DpnII restriction endonucleases. The pGlow-UpgidA plasmid from the GM28 strain which harbors dam was digested with DpnI but not by DpnII (Fig. 2, lanes 2 and 3). The opposite trend was noted when this plasmid DNA isolated from E. coli GM33 strain (Δdam) was digested with DpnI and DpnII enzymes (Fig. 2, lanes 4 and 5). Identical digestions were observed for pGlow-Upact plasmid DNA (data not shown). A. hydrophila gDNA showed a similar digestion pattern with DpnI and DpnII restriction endonucleases (data not shown) as the pGlow-UpgidA and pGlow-Upact plasmids from E. coli GM28 strain, with DpnI and not DpnII digesting gDNA.
Fig. 2.
DpnI and DpnII cleavage patterns of plasmid DNA isolated from E. coli GM28 (dam+) and GM33 (Δdam) strains containing pGlow-TOPO vector with the upstream region of the gidA gene of A. hydrophila SSU. Lane 1, 1 kb DNA Ladder (New England BioLabs); lanes 2 and 3, isolated pGlow-UpgidA plasmid DNA from E. coli GM28 strain treated with DpnI enzyme (lane 2) or DpnII (lane 3); lanes 4 and 5, from E. coli GM33 strain containing pGlow-UpgidA plasmid digested with DpnI and DpnII, respectively. A. hydrophila gDNA showed similar patterns with DpnI and DpnII restriction endonucleases as DNA from E. coli GM28 strain (lanes 2 and 3).
3.2. Effect of methylation by Dam on act (Upact) and gidA (UpgidA) expression as a measure of GFP in E. coli dam+ and Δdam genotypes
Table 3 gives the results of the quantitative analysis of relative act and gidA expressional activities by using gene-encoding GFP as a tran-scriptional reporter in dam+ and Δdam E. coli cells. Our data indicated that GFP levels under the control of act and gidA gene promoters and their upstream sequences which can be methylated from the pGlow-TOPO vector in E. coli were dependent on the DNA methylation status. For example, transcriptional activities of Upact (91.3±1.7 RFU/mg protein) and UpgidA (62.2±1.4 RFU/mg protein) -regions in terms of GFP production were approximately two times lower in the GM28 strain (dam+) when compared to that in GM33 strain (Δdam) (183.5 ± 3.1 and 131.7± 1.1 RFU/mg protein, respectively). These results indicated that constitutive methylation by Dam suppressed the activities of both act and gidA genes as a measure of GFP production in GM28 strain.
Table 3.
Level of gene expression as measured by using GFP assaya of upstream region of A. hydrophila act and gidA genes in different Dam background strains of E. coli.
| Genotype of E. coli | Mean activity (RFU/mg protein) ± SD |
|---|---|
| dam+ gidA+ (pGlow-Upact) | 91.3 ±1.7 |
| Δdam gidA+ (pGlow-Upact) | 183.5 ± 3.1b |
| dam+ gidA+ (pGlow-UpgidA) | 62.2 ±1.4 |
| Δdam gidA+ (pGlow-UpgidA) | 131.7±1.1b |
| dam+ ΔgidA (pGlow-Upact) | 51.7 ±0.93 |
| Δdam ΔgidA (pGlow-Upact) | 52.1 ± 0.21 |
| dam+ ΔgidA (pGlow-UpgidA) | 60.1 ±1.0 |
| Δdam ΔgidA (pGlow-UpgidA) | 125.8 ±1.0b |
Activity as GFP emission in relative fluorescence unit (RFU) was normalized to the same protein concentration (1 mg) for each indicated E. coli strain cell extract.
Statistically significant value (P≤0.05) using the Student's t test.
3.3. Effect of methylation by Dam on the expression of act (Upact) and gidA (UpgidA) in E. coli dam+ ΔgidA and Δdam ΔgidA mutants
To determine the expression of act and gidA gene upstream regions as a function of GFP production in ΔgidA mutants, we deleted the gidA gene from the E. coli GM28 and GM33 strains. The pGlow-Upact and pGlow-UpgidA plasmids were transformed in generated E. coli mutant strains, and their cell extracts were used for measuring expressional activities, as described above. Table 3 shows no significant differences in the levels of GFP produced through the Upact region in dam+ ΔgidA and Δdam ΔgidA mutants (51.7±0.93 versus 52.1±0.21 RFU/mg protein), indicating the role of GidA in modulating Act production regardless of the methylation status of the Upact. However, we observed a two-fold difference in the UpgidA activity in terms of GFP production between E. coli GM28 dam+ ΔgidA and GM33 Δdam ΔgidA strains (60.1±1.0 versus 125.8 ± 1.0 RFU/mg protein), as we observed also for dam+ gidA+ and Δdam gidA+ backgrounds, indicating the suppressive activity of methylation by Dam on GidA production. This would cause decreased production of Act, as we indeed noted (91.3±1.7 [dam+ gidA+] versus 183.5 ± 3.1 [Δdam gidA+] RFU/mg protein) (Table 3).
3.4. RT-qPCR analysis
We used RT-qPCR analysis to study whether altered Dam production affected the differential gidA and act gene expression in parental versus Dam-overproducing A. hydrophila SSU strains. The transcriptional expression of gidA and act genes was 1.9-fold and 2.1-fold higher in Dam-overproducing strain than in the parental strain with constitutive expression of the dam gene, respectively. These data verified that gidA and act gene expression was up-regulated by overproduction of Dam. Additionally, increases in the levels of GFP were observed in E. coli Δdam strains when reporter protein GFP was produced from a plasmid under the control of gidA (131.7±1.1 RFU/mg protein) or act promoter and upstream regions (183.5±3.1 RFU/mg protein) (Table 3). Our results tend to suggest that the hypermethyla-tion of GATC sites upstream of gidA and act genes in A. hydrophila increased Act production, while DNA methylation of these sites by the constitutive production of Dam would have an opposite effect, i.e., reduced Act production.
3.5. Effect of dam and gidA genes on Act-associated hemolytic activity in E. coli
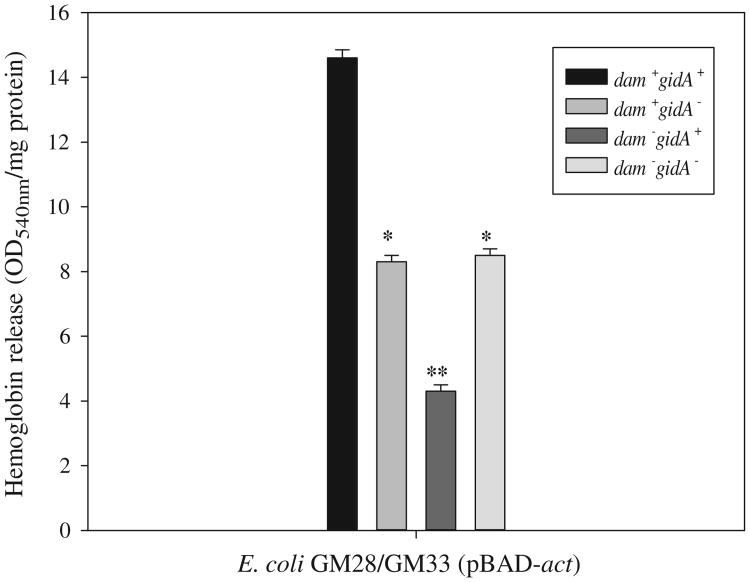
To demonstrate the utility of using E. coli K12 as a model to study act gene expression of A. hydrophila SSU, we tested the hemolytic activity of Act from pBAD-act recombinant plasmid in E. coli GM28 (dam+), GM33 (Δdam) and ΔgidA mutants of these strains. The highest hemolytic activity (14.6±0.25) associated with Act (Fig. 3) was noted in the E. coli GM28 parental strain (which harbored dam and gidA genes), which dropped significantly (8.3±0.2) in the ΔgidA mutant strain of GM28 in the presence of Dam. As expected, the E. coli GM33 mutant strain (Δdam and ΔgidA) exhibited hemolytic activity (8.5±0.2) associated with Act similar to that noted for the E. coli GM28 ΔgidA strain (positive for Dam), further providing evidence of the role of GidA in regulating Act production. We observed a further drop in hemolytic activity (4.3±0.2) of Act in the E. coli GM33 strain, which harbored gidA, but not the dam gene (Fig. 3). E. coli strains with pBAD vector alone did not exhibit any hemolytic activity. These data are suggestive that methylation by Dam could vary due to conformation of DNA in the upstream region of the target gene due to the specific genome architecture of specific pathogens.
Fig. 3.
Act-associated hemolytic activity in cell extracts of E. coli GM28 (pBAD-act) and GM33 (pBAD-act) in the gidA+ and ΔgidA background was evaluated by measuring the release of hemoglobin from red blood cells. E. coli strains with pBAD vector alone did not exhibit any hemolytic activity (data not shown). * denotes statistically significant difference between dam+ gidA+ versus dam+ gidA− mutants; ** denotes statistically significance between dam+ gidA− versus dam− gidA+ mutant.
4. Discussion
The gidA gene is conserved among prokaryotes, which indicate that it has a key function within the cell. Indeed, when the E. coli gidA mutant was grown on glucose-containing media, it produced long filamentous cells indicating that gidA transcription was involved in the initiation of chromosomal replication (Asai et al., 1990; Ogawa and Okazaki, 1994). These results implied that GidA might function to connect glucose metabolism, ribosome function, chromosome replication, and cell division. Recent studies implicated GidA in a number of biological and pathogenic processes, possibly pointing to its global regulatory role. P. aeruginosa GidA selectively controlled quorum-sensing gene expression post-transcriptionally via RhlR-dependent and -independent pathways (Gupta et al., 2009). Likewise, gidA mutant of S. pyogenes produced significantly reduced levels of multiple virulence factors due to impaired translational efficiencies compared to those of the parental strain (Cho and Caparon, 2008).
The mechanisms that lead to bacterial cell phase variation are classified either as genetic or epigenetic, and Dam belongs to the latter category in that Dam mediates phase variation to control expression and production of some outer surface structures in E. coli, e.g., an outer-membrane protein Ag43 (van der Woude, 2008). The methylation state of GATC sites in the dnaA promoter appeared to be of little consequence to the timing of initiation, as elimination of GATC sites in dnaA promoter did not result in an initiation timing defect (Wilkinson et al., 2006).
The connection between DNA methylation and promoter activity was found for the herpes simplex virus thymidine kinase gene, when the promoters were methylated in vitro (Levine et al., 1991). These authors showed a possible involvement of protein mediators in the inhibition of promoter activity by methylation associated with a transcriptional inactivity. Well-known pap promoter controls the expression of pyelonephritis-associated pili in uropathogenic E. coli (UPEC) in response to methylation by Dam (Barnard et al., 2004). Another study demonstrated that the differential methylation by Dam of GATC sequences in the pap promoter regulated the expression of pili genes necessary for UPEC cellular adhesion (Peterson and Reich, 2006). These investigators concluded that GATC flanking sequences might be critical for expression of the pap promoter and other Dam-regulated genes by affecting the activity of Dam at such sites.
Our earlier published data showed that GidA positively regulated Act production at the translational level and indicated no significant differences in the act gene transcription level between the gidA mutant and WT strain of A. hydrophila (Sha et al., 2004). This observation was confirmed later by Nordman et al. (2007), as E. coli gidA transcription levels, which were measured by RNA slot-blot analysis in the gidA mutant, were also found to be similar to those seen in WT bacterium.
The GATC Dam methylation sites are unevenly distributed in the genome of E. coli, and the oriC has a GATC-rich region (Barras and Marinus, 1988). This observation is in agreement with the role of Dam in DNA replication initiation. We noted multiple GATC Dam sites in the upstream region of a gidA gene and provided evidence that the expression of the gidA gene could be controlled by Dam. It is plausible that Dam might affect the act gene directly and/or through the GidA regulator. To test that, we cloned the upstream regions of the gidA and act genes of A. hydrophila into a pGlow-TOPO vector. This promoterless vector gave us the possibility to fuse the promoter region of a gene of interest with the GFP reporter to assay the activity of an upstream region. We observed higher levels of A. hydrophila GidA and Act as a function of GFP production in the dam mutant of E. coli GM33 than in GM28 strain (Table 3). These results are consistent with previous findings about the inverse correlation between methylation and promoter activity showing that the gidA transcript level was about 2.5-fold higher in the dam mutant compared to that in the E. coli PC2 parental strain (Bogan and Helmstetter, 1997). E. coli lacking Dam showed an increase in transcription of the genes belonging to the SOS regulon, and a comparison of global gene expression in Δdam, ΔseqA, and Dam-overproducing strains showed that the profiles for SeqA-deficient and Dam-overproducing cells were almost identical and distinct from that of Δdam cells (Lobner-Olesen et al., 2003). Similarly, our data provided evidence that levels of GFP produced under the control of act and gidA gene promoters and upstream sequences were higher in Δdam E. coli compared to that in dam+ E. coli (Table 3). On the other hand, the dam mutant of S. Typhimurium exhibited decreased virulence and also pointed to GATC-binding sites as possibly having a role in regulating the virulence of Salmonella and other related bacteria (Chatti and Landoulsi, 2008).
A decreased level of GidA can result in the absence of tRNA modification that may alter the expression of the act gene. Therefore, we generated gidA deletion mutants of E. coli GM28 (dam+) and E. coli GM33 (Δdam) and obtained a double Δdam ΔgidA mutant viable strain for the first time for E. coli K12. Most interestingly, in gidA mutant strains, expression of the act gene in the dam mutant strain remained at the same low level (52.1±0.21 RFU/mg protein), as in the dam+ strain (51.7±0.93 RFU/mg protein). Thus, in the absence of the gidA gene, Dam does not affect act gene expression. However, gidA promoter activity, as measured by GFP production in E. coli from the plasmid in gidA mutant strains, was still dependent on the extent of DNA methylation, showing suppression of the activity in the dam+ strain (60.1±1.0 RFU/mg protein) compared to the Δdam strain (125.8±1.0 RFU/mg protein) (Table 3). This investigation showed a connection between the amount of Dam produced (constitutive versus overproduction from the plasmid) which modulated the gidA gene expression level and then GidA leading to regulation of Act production. Our data suggested that the constitutive expression of the dam gene resulted in the decreased expression of both gidA and act genes in E. coli (Table 3), while overproduction of Dam led to an increase in GidA and Act production in A. hydrophila (Erova et al., 2006b).
We noted a significant decrease (57%) in hemolytic activity associated with Act in the ΔgidA mutant of E. coli GM28 (pBAD-act) (Fig. 3), which matched with our earlier published data in A. hydrophila SSU showing that deletion of the gidA gene led to reduced alkaline phosphatase activity associated with the act∷phoA reporter construct in comparison to the WT strain (Sha et al., 2004). From these results, the contribution of GidA in modulating Act activity was readily evident, and, thus, our data are consistent regarding the modulation of Act by Dam and GidA in E. coli versus A. hydrophila. Importantly, we observed a further decrease in Act-associated hemolytic activity in the E. coli Adam gidA+ GM33 strain, compared to dam+ ΔgidA− strain of E. coli GM28 (Fig. 3). These data led us to suggest that methylation by Dam of the gidA upstream sequence contributes significantly to Act regulation (Fig. 4). These intriguing observations also indicated that methylation caused by E. coli Dam of A. hydrophila gidA upstream sequence in E. coli could be somewhat different when compared to the methylation caused by A hydrophila Dam in the homologous strain. Finally, methylation by Dam (hypo- or hyper-) of the gidA gene upstream region can affect the expression of the gidA gene, which in turn, would regulate the act gene in A. hydrophila versus E. coli.
Fig. 4.

A schematic showing the pathway of Act regulation in A. hydrophila SSU by Dam and GidA. Dam methylates adenine residue of the GATC sites of the gidA upstream sequence (UpgidA) to modulate gidA expression. GidA with tRNA methylase activity, in turn, possibly through as yet an unknown gene (x), leads to Act production.
To investigate the interaction of Dam and GidA on the regulation of act, we used RT-qPCR analysis to understand how, in fact, the methylation level of DNA by Dam can affect gidA and act gene expression. The results indicated that in the A. hydrophila SSU Dam-overproducing strain, the transcriptional level of the gidA gene was increased two times, but the transcriptional level of the act gene also increased two times, which was unexpected, based on our earlier data that GidA modulates Act at the translational level (Sha et al., 2004). Based on the presented data, we can speculate how GidA regulates act gene expression. GidA is a global regulatory protein and, thus, modulates the expression of many known and unknown genes, and one of them may affect Act production (Fig. 4). This regulation, by gidA for example, can exist at the translational level by tRNA modifications (Gustilo et al., 2008; Saier, 1995).
This study provided evidence that the intermediate protein GidA is involved in the mechanism related to the influence of Dam on bacterial pathogenesis by modulating the activity of the promoter region associated with transcription and possibly indicates that different virulence genes may share a common regulatory mechanism whose activity is dependent on Dam methylation. Specifically for A. hydrophila, it appears that once the organism reaches the target site in the human/animal host, Dam is overproduced, which will, in turn, increase Act production, leading to significant tissue damage or diarrhea depending upon the route of bacterial entry. However, in the natural environmental niche, constitutive levels of Dam will suppress Act production to conserve energy. In the future, we will examine modifying adenine residues of GATC site(s) in the upstream region of the gidA gene to show whether this affects Act production in dam+ and Δdam background strains of E. coli.
Supplementary Material
Acknowledgments
We thank Martin G. Marinus (University of Massachusetts Medical School, Worcester, MA) for providing E. coli K12 GM28 and GM33 strains and UTMB Molecular Genomics Core for the RT-qPCR assay. This study was supported by grant AI41611 from NIH NIAID. We thank Mardelle J. Susman for editorial assistance.
Abbreviations
- Act
Aeromonas cytotoxic enterotoxin
- Dam
DNA adenine methyltransferase
- tRNA
transfer RNA
- GidA
glucose-inhibited division A protein
- GATC site
guanine-adenine-thymine-cytosine site
- RT-qPCR
real-time quantitative polymerase chain reaction
- GFP
green fluorescent protein
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- DPBS
Dulbecco's phosphate-buffered saline
- FAD
Flavin adenine dinucleotide
- OriC
origin of replication
- Ap
Ampicillin
- Cm
Chloramphenicol
- Rif
Rifampin
- gDNA
genomic DNA
- RFU
relative fluorescence units
- FRT
flippase [FLP] recognition target
- Cms
chloramphenicol sensitive
- Aps
ampicillin sensitive
- pap
pyelonephritis-associated pili
- seqA gene
sequestration A gene
Footnotes
Supplementary materials related to this article can be found online at doi:10.1016/j.gene.2012.02.024.
References
- Asai T, Takanami M, Imai M. The AT richness and gid transcription determine the left border of the replication origin of the E. coli chromosome. EMBO J. 1990;9:4065–4072. doi: 10.1002/j.1460-2075.1990.tb07628.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2:0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnard A, Wolfe A, Busby S. Regulation at complex bacterial promoters: how bacteria use different promoter organizations to produce different regulatory outcomes. Curr Opin Microbiol. 2004;7:102–108. doi: 10.1016/j.mib.2004.02.011. [DOI] [PubMed] [Google Scholar]
- Barras F, Marinus MG. Arrangement of Dam methylation sites (GATC) in the Escherichia coli chromosome. Nucleic Acids Res. 1988;16:9821–9838. doi: 10.1093/nar/16.20.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates DB, Boye E, Asai T, Kogoma T. The absence of effect of gid or mioC transcription on the initiation of chromosomal replication in Escherichia coli. Proc Natl Acad Sci U S A. 1997;94:12497–12502. doi: 10.1073/pnas.94.23.12497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattner FR, Plunkett G, III, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, Gregor J, Davis NW, Kirkpatrick HA, Goeden MA, Rose DJ, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- Bogan JA, Helmstetter CE. DNA sequestration and transcription in the oriC region of Escherichia coli. Mol Microbiol. 1997;26:889–896. doi: 10.1046/j.1365-2958.1997.6221989.x. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Casadesus J, Low D. Epigenetic gene regulation in the bacterial world. Microbiol Mol Biol Rev. 2006;70:830–856. doi: 10.1128/MMBR.00016-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatti A, Landoulsi A. GATC sites might regulate virulence of Salmonella typhimurium. Foodborne Pathog Dis. 2008;5:555–557. doi: 10.1089/fpd.2008.0099. [DOI] [PubMed] [Google Scholar]
- Cho KH, Caparon MG. tRNA modification by GidA/MnmE is necessary for Streptococcus pyogenes virulence: a new strategy to make live attenuated strains. Infect Immun. 2008;76:3176–3186. doi: 10.1128/IAI.01721-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra AK, Houston CW. Enterotoxins in Aeromonas-associated gastroenteritis. Microbes Infect. 1999;1:1129–1137. doi: 10.1016/s1286-4579(99)00202-6. [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edberg SC, Browne FA, Allen MJ. Issues for microbial regulation: Aeromonas as a model. Crit Rev Microbiol. 2007;33:89–100. doi: 10.1080/10408410601172180. [DOI] [PubMed] [Google Scholar]
- Erova TE, Fadl AA, Sha J, Khajanchi BK, Pillai LL, Kozlova EV, Chopra AK. Mutations within the catalytic motif of DNA adenine methyltransferase (Dam) of Aeromonas hydrophila cause the virulence of the Dam-overproducing strain to revert to that of the wild-type phenotype. Infect Immun. 2006a;74:5763–5772. doi: 10.1128/IAI.00994-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erova TE, Pillai L, Fadl AA, Sha J, Wang S, Galindo CL, Chopra AK. DNA adenine methyltransferase influences the virulence of Aeromonas hydrophila. Infect Immun. 2006b;74:410–424. doi: 10.1128/IAI.74.1.410-424.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadl AA, Galindo CL, Sha J, Erova TE, Houston CW, Olano JP, Chopra AK. Deletion of the genes encoding the type III secretion system and cytotoxic enterotoxin alters host responses to Aeromonas hydrophila infection. Microb Pathog. 2006;40:198–210. doi: 10.1016/j.micpath.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Gupta R, Gobble TR, Schuster M. GidA posttranscriptionally regulates rhl quorum sensing in Pseudomonas aeruginosa. J Bacteriol. 2009;191:5785–5792. doi: 10.1128/JB.00335-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustilo EM, Vendeix FA, Agris PF. tRNA's modifications bring order to gene expression. Curr Opin Microbiol. 2008;11:134–140. doi: 10.1016/j.mib.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinscherf TG, Willis DK. Global regulation by gidA in Pseudomonas syringae. J Bacteriol. 2002;184:2281–2286. doi: 10.1128/JB.184.8.2281-2286.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine A, Cantoni GL, Razin A. Inhibition of promoter activity by methylation: possible involvement of protein mediators. Proc Natl Acad Sci U S A. 1991;88:6515–6518. doi: 10.1073/pnas.88.15.6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lobner-Olesen A, Marinus MG, Hansen FG. Role of SeqA and Dam in Escherichia coli gene expression: a global/microarray analysis. Proc Natl Acad Sci U S A. 2003;100:4672–4677. doi: 10.1073/pnas.0538053100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low DA, Casadesus J. Clocks and switches: bacterial gene regulation by DNA adenine methylation. Curr Opin Microbiol. 2008;11:106–112. doi: 10.1016/j.mib.2008.02.012. [DOI] [PubMed] [Google Scholar]
- Marinus MG, Casadesus J. Roles of DNA adenine methylation in host–pathogen interactions: mismatch repair, transcriptional regulation, and more. FEMS Microbiol Rev. 2009;33:488–503. doi: 10.1111/j.1574-6976.2008.00159.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinus MG, Morris NR. Isolation of deoxyribonucleic acid methylase mutants of Escherichia coli K-12. J Bacteriol. 1973;114:1143–1150. doi: 10.1128/jb.114.3.1143-1150.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland M, Sanderson KE, Spieth J, Clifton SW, Latreille P, Courtney L, Porwollik S, Ali J, Dante M, Du F, Hou S, Layman D, Leonard S, Nguyen C, Scott K, Holmes A, Grewal N, Mulvaney E, Ryan E, Sun H, Florea L, Miller W, Stoneking T, Nhan M, Waterston R, Wilson RK. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature. 2001;413:852–856. doi: 10.1038/35101614. [DOI] [PubMed] [Google Scholar]
- Nordman J, Skovgaard O, Wright A. A novel class of mutations that affect DNA replication in E. coli. Mol Microbiol. 2007;64:125–138. doi: 10.1111/j.1365-2958.2007.05651.x. [DOI] [PubMed] [Google Scholar]
- Ogawa T, Okazaki T. Cell cycle-dependent transcription from the gid and mioC promoters of Escherichia coli. J Bacteriol. 1994;176:1609–1615. doi: 10.1128/jb.176.6.1609-1615.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson SN, Reich NO. GATC flanking sequences regulate Dam activity: evidence for how Dam specificity may influence pap expression. J Mol Biol. 2006;355:459–472. doi: 10.1016/j.jmb.2005.11.003. [DOI] [PubMed] [Google Scholar]
- Reith ME, Singh RK, Curtis B, Boyd JM, Bouevitch A, Kimball J, Munholland J, Murphy C, Sarty D, Williams J, Nash JH, Johnson SC, Brown LL. The genome of Aeromonas salmonicida subsp. salmonicida A449: insights into the evolution of a fish pathogen. BMC Genomics. 2008;9:427. doi: 10.1186/1471-2164-9-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts RJ, Vincze T, Posfai J, Macelis D. REBASE: restriction enzymes and methyltransferases. Nucleic Acids Res. 2003;31:418–420. doi: 10.1093/nar/gkg069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby EG, Urbanowski M, Campbell J, Dunn A, Faini M, Gunsalus R, Lostroh P, Lupp C, McCann J, Millikan D, Schaefer A, Stabb E, Stevens A, Visick K, Whistler C, Greenberg EP. Complete genome sequence of Vibrio fischeri: a symbiotic bacterium with pathogenic congeners. Proc Natl Acad Sci U S A. 2005;102:3004–3009. doi: 10.1073/pnas.0409900102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saier MH., Jr Differential codon usage: a safeguard against inappropriate expression of specialized genes? FEBS Lett. 1995;362:1–4. doi: 10.1016/0014-5793(95)00185-c. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. A Laboratory Manual. Wiley & Co.; Cold Spring Harbor, New York: 1989. Molecular Cloning. [Google Scholar]
- Seshadri R, Joseph SW, Chopra AK, Sha J, Shaw J, Graf J, Haft D, Wu M, Ren Q, Rosovitz MJ, Madupu R, Tallon L, Kim M, Jin S, Vuong H, Stine OC, Ali A, Horneman AJ, Heidelberg JF. Genome sequence of Aeromonas hydrophila ATCC 7966 T: jack of all trades. J Bacteriol. 2006;188:8272–8282. doi: 10.1128/JB.00621-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha J, Lu M, Chopra AK. Regulation of the cytotoxic enterotoxin gene in Aeromonas hydrophila: characterization of an iron uptake regulator. Infect Immun. 2001;69:6370–6381. doi: 10.1128/IAI.69.10.6370-6381.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha J, Kozlova EV, Chopra AK. Role of various enterotoxins in Aeromonas hydrophila-induced gastroenteritis: generation of enterotoxin gene-deficient mutants and evaluation of their enterotoxic activity. Infect Immun. 2002;70:1924–1935. doi: 10.1128/IAI.70.4.1924-1935.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha J, Kozlova EV, Fadl AA, Olano JP, Houston CW, Peterson JW, Chopra AK. Molecular characterization of a glucose-inhibited division gene, gidA, that regulates cytotoxic enterotoxin of Aeromonas hydrophila. Infect Immun. 2004;72:1084–1095. doi: 10.1128/IAI.72.2.1084-1095.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shippy DC, Eakley NM, Bochsler PN, Fadl AA. Biological and virulence characteristics of Salmonella enterica serovar Typhimurium following deletion of glucose-inhibited division (gidA) gene. Microb Pathog. 2011;50:303–313. doi: 10.1016/j.micpath.2011.02.004. [DOI] [PubMed] [Google Scholar]
- Urbonavicius J, Skouloubris S, Myllykallio H, Grosjean H. Identification of a novel gene encoding a flavin-dependent tRNA:m5U methyltransferase in bacteria— evolutionary implications. Nucleic Acids Res. 2005;33:3955–3964. doi: 10.1093/nar/gki703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Woude MW. Some types of bacterial phase variation are epigenetic. Microbe. 2008;3:21–26. [Google Scholar]
- von Meyenburg K, Jorgensen BB, Nielsen J, Hansen FG. Promoters of the atp operon coding for the membrane-bound ATP synthase of Escherichia coli mapped by Tn10 insertion mutations. Mol Gen Genet. 1982;188:240–248. doi: 10.1007/BF00332682. [DOI] [PubMed] [Google Scholar]
- White DJ, Merod R, Thomasson B, Hartzell PL. GidA is an FAD-binding protein involved in development of Myxococcus xanthus. Mol Microbiol. 2001;42:503–517. doi: 10.1046/j.1365-2958.2001.02659.x. [DOI] [PubMed] [Google Scholar]
- Wilkinson TG, II, Kedar GC, Lee C, Guzman EC, Smith DW, Zyskind JW. The synchrony phenotype persists after elimination of multiple GATC sites from the dnaA promoter of Escherichia coli. J Bacteriol. 2006;188:4573–4576. doi: 10.1128/JB.00089-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yim L, Moukadiri I, Bjork GR, Armengod ME. Further insights into the tRNA modification process controlled by proteins MnmE and GidA of Escherichia coli. Nucleic Acids Res. 2006;34:5892–5905. doi: 10.1093/nar/gkl752. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.