Abstract
Automated tracking methods facilitate screening for and characterization of abnormal locomotion or more complex behaviors in Drosophila. We developed the Iowa Fly Locomotion and Interaction Tracker (IowaFLI Tracker), a MATLAB based video analysis system, to identify and track multiple flies in a small arena. We report altered motor activity in the K+ and Na+ channel mutants, Hk1 and parats1, which had previously been shown to display abnormal larval locomotion. Environmental factors influencing individual behavior, such as available “social space,” were studied by using IowaFLI Tracker to simultaneously track multiple flies in the same arena. We found that crowding levels affect individual fly activity, with the total movement of individual flies attenuated around a particular density. This observation may have important implications in the design of activity chambers for studying particular kinds of social interactions. IowaFLI Tracker also directly quantifies social interactions by tracking the amount of time individuals are in proximity to one another—visualized as an ‘interactogram.’ This feature enables the development of a ‘target-preference’ assay to study male courtship behavior where males are presented with a choice between two immobilized, decapitated females, and their locomotion and interactions quantified. We used this assay to study the chemosensory mutants olfD (paraolfD, sbl2) and Gr32a and their preferences towards virgin or mated females. Male olfD flies showed reduced courtship levels, with no clear preference towards either, while Gr32a males preferentially courted with virgin females over mated females in this assay. These initial results demonstrate that IowaFLI Tracker can be employed to explore motor coordination and social interaction phenomena in behavioral mutants of Drosophila.
Keywords: IowaFLI Tracker, DIAS, Crowding, ‘Interactogram’, ‘Target-Preference’ Assay, Courtship, Ion Channels, para, Hk, olfD, Gr32a, Pheromone receptor
Introduction
Coordinated movements in Drosophila melanogaster, such as larval crawling (Troncoso et al., 1987; Wang et al., 1997; Wang et al., 2002; Fox et al., 2006), adult walking (Gotz & Wenking, 1973; Strauss & Heisenberg, 1990; Strauss, 2002) and flight (Frye & Dickinson, 2004) are required for many types of complex behaviors including foraging (Pereira & Sokolowski, 1993), aggression (Nilsen et al., 2004; Ueda & Wu, 2009), courtship (Greenspan & Ferveur, 2000), and escape (Trimarchi & Schneiderman, 1995). Automated analyses of fly locomotion using computer-based video methods have been utilized for several decades and can be employed while studying these behaviors. Computer-assisted analysis of larval locomotion using the Dynamic Image Analysis System (DIAS) revealed involvement of several genes in modulating larval locomotion (Wang et al., 1997). DIAS has subsequently been used to track adult walking (Rodan et al., 2002) and other groups have developed similar methods to track fly walking (Martin et al., 1999; Martin, 2004; Grover et al., 2008; Gilestro & Cirelli, 2009; Branson et al., 2009; Seeling et al., 2010).
This paper presents several applications of a set of MATLAB scripts, the Iowa Fly Locomotion and Interaction Tracker (IowaFLI Tracker), which tracks both locomotion and interactions between multiple adult Drosophila individuals in order to analyze how particular genes and environmental conditions influence behavior. Our approach was to develop a versatile system that requires relatively inexpensive components and can provide information on both individual fly locomotion patterns and social interactions among multiple flies.
To initiate quantification of adult locomotion in the large collection of Drosophila mutants, we chose to extend our previous study on two ion channel mutants Hyperkinetic (Hk) and paralytic (para) that show altered larval locomotion to their adult locomotion which had not been quantified in a similar manner (Wang et al., 1997). The gene Hyperkinetic encodes a K+ channel β subunit, and the mutant allele Hk1 displays vigorous leg shaking under ether anesthesia (Kaplan & Trout, 1969; Chouinard et al., 1995). The second mutant used, parats1, is a reversible temperature-sensitive paralytic (>29 °C) allele of the gene that encodes a Na+ channel (Suzuki et al., 1971; Siddiqi & Benzer, 1976; Loughney et al., 1989). Because these mutants directly affect nerve and muscle excitability, it is important to quantitatively describe how motor coordination and locomotion are affected in adults, as well as larvae.
In developing IowaFLI Tracker’s ability to track multiple flies simultaneously, we found this system was suitable for exploring the role “social” factors play on an individual’s activity. A significant amount of interest has been generated in understanding how these factors that influence locomotion and activity in Drosophila (Ruan & Wu, 2008; Ueda & Wu, 2009; Sokolowski, 2010). By altering the number of individuals in an arena, we examined the role of available “social space” (i.e. crowding, Simon, et al., 2012) on an individual’s locomotion.
Social interactions could be directly identified by finding sequential frames in which flies were in close proximity to each other, and visualized as an ‘interactogram.’ Based on this approach, we developed a ‘target-preference’ paradigm amenable to video analysis via IowaFLI Tracker that allows for direct analysis of courtship preference and used it to study how chemosensory mutants olfD (paraolfD, sbl2; Siddiqi, 1987; Ayyub et al., 1990), and Gr32a (Miyamoto & Amerein, 2008) interacted towards mated and virgin females.
Our research has benefited by the pioneering work of Prof. Obaid Siddiqi and his continued contributions to Drosophila neurogenetics. This work has been inspired in part by his early work on adult and larval physiology as well as his more recent work in the neurogenetics of chemosensory behavior. Some of the mutants used in this study find roots in his early work (Siddiqi & Benzer, 1976; Ayyub et al., 1990). His life-long contributions to Drosophila neurogenetics will impact researchers in the field for generations to come.
Materials and Methods
Drosophila Stocks
Genotypes used include previously described lines: wild-type Canton-S (CS), Hk1, parats1 (Wang, et. al, 1997), olfD (Siddiqi, 1987; Ruan & Wu, 2008), and Gr32a (a gift from Hubert Amerlin, see Miyamoto & Amerein, 2008). All fly stocks were raised on standard cornmeal media at room temperature (23°C). Flies used for the locomotion and male-male interaction assays (Figures 2 - 4) were between 3 and 6 days old; and males used in the target preference assay (Figures 5 - 6) were between 4 and 8 days old. All flies were collected over CO2 anesthesia 24 hours prior to use, unless otherwise indicated.
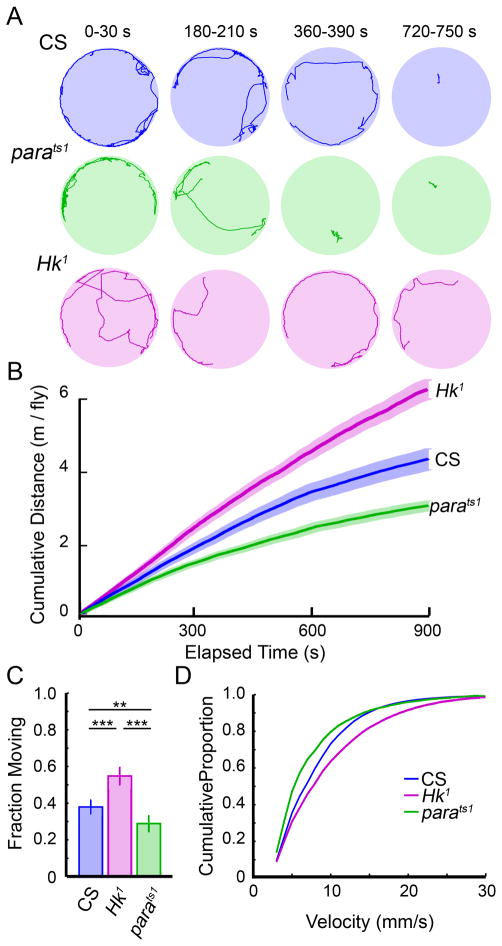
Figure 2. Locomotion in two Drosophila ion channel mutants: parats1, and Hk1.
(A) Four male flies were placed in an arena and tracked for 15 minutes. Shown are sample tracks from individual flies at four 30 s intervals. (B) The mean cumulative distance traveled for individuals from each of the three genotypes. Shaded regions represent S.E.M. Hk1 and parats1 showed significantly altered patterns of locomotion with increased and decreased total distances traveled respectively when compared to CS (p < 0.01, Mann-Whitney U Test, n=40, 32, 24 flies for CS, Hk1 and parats1 respectively) (C) The proportion of time individuals were moving, defined to be when their velocity was <3 mm/s. Error bars represent standard error of proportion (** p < 0.01, *** p < 0.001, Mann-Whitney U Test). (D) The cumulative distribution of fly velocities indicate the proportion of time a fly is moving (velocity > 3 mm/s), but is traveling slower than the corresponding velocity. The velocity profiles for CS, Hk1, and parats1 were not significantly different, (Kolmogorov-Smirnov test, p > 0.05).
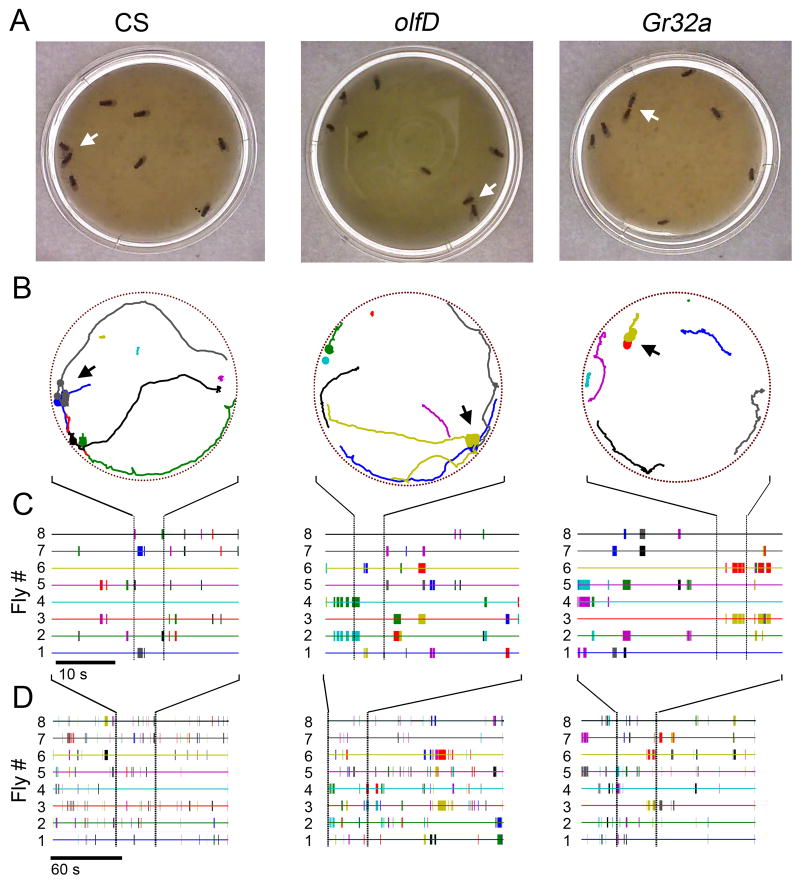
Figure 4. Interactions among male chemosensory mutants.
(A) Sample frames of interactions among eight individuals of CS, olfD, and Gr32a genotype. White arrows indicate ‘tapping,’ ‘tapping’ and ‘head-butting’, respectively in the frames. (B) Tracks of locomotor activity in the 5 seconds around the time of the events in (A). The width of a fly’s track is expanded during a period of interaction, and the arrows indicate the interactions shown in (A). Tracks of individual flies are identified with different colors. (C) Representative pair-wise ‘interactograms’ over a 30 s period for the three genotypes. History of interactions for each individual are identified with the same color of the track in (B) and interacting events are marked with hatches of different colors to identify the interacting partner. (D) Expanded period of interactogram (at a compressed resolution to cover 150 s) of the corresponding panels in (C).
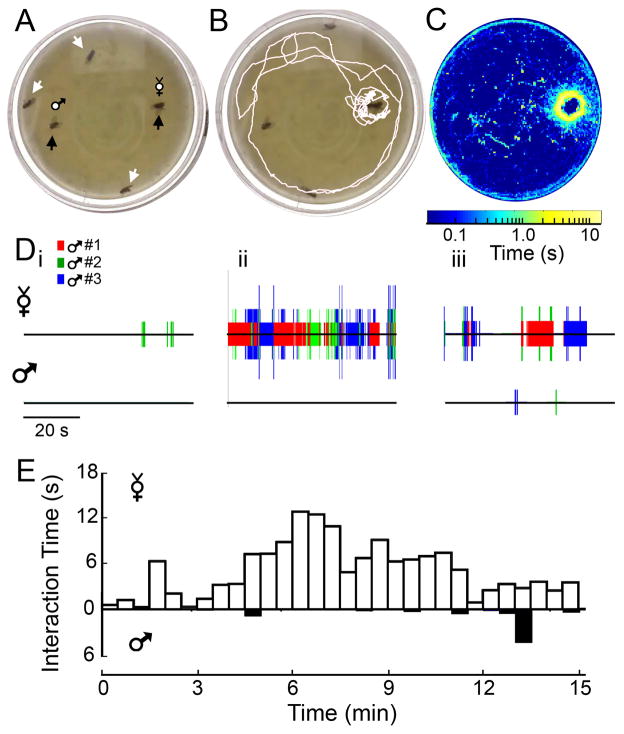
Figure 5. The ‘target-preference’ assay.
(A) Sample frame from the preference assay. Three male flies (white arrows) were introduced to a chamber with two pinned and decapitated ‘targets’ (virgin female on right, male on left, black arrows). (B) Sample 30 s track of a male fly. (C) A 15-min log-place map of fly locations the sample video. (D) 1-min interactograms for each target at (i) 3-min, (ii) 6-min, and (iii) 12-min in the video. Colors indicate the identity of interacting male, and ticks are stacked during periods when multiple males interact with the target. (E) Plot of the average time an individual fly spends interacting with a target during 30-s intervals through the 15-min videos (n = 15 flies).
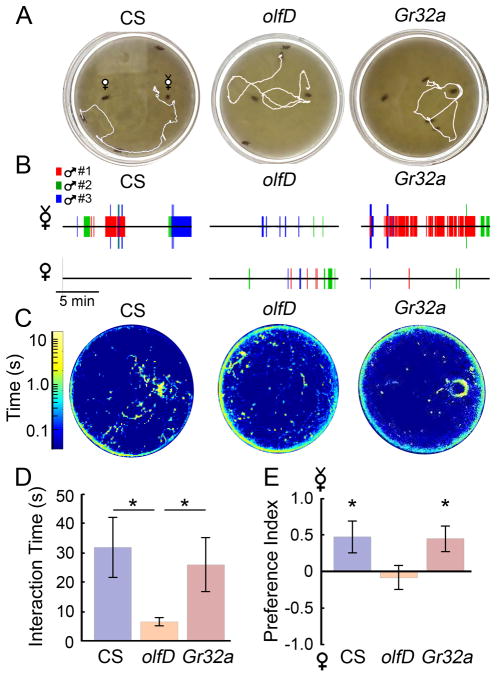
Figure 6. Preference towards virgin target flies in the chemosensory mutants olfD and Gr32a.
(A) Sample 30-s tracks of a male CS, olfD, and Gr32a fly approaching the virgin target flies (on right). (B) 15-min interactograms from the sample videos shown in (A). (C) Sample log-place map from videos in (A). (D) Bar graphs of the average amount of time male flies interact with targets during a 15-min video. Male olfD mutants spent far less time interacting than either CS or Gr32a males (Student’s T test, *p < 0.05, n= 15, 30, and 15 flies respectively for CS, olfD, and Gr32a). (E) The preference index of the mutants studied. Values greater than 0 indicate preference towards virgin targets, while those less than 0 indicate a preference towards mated targets. CS and Gr32a males showed a significant preference towards virgin targets (Student’s T-Test, *p < 0.05).
Fly Imaging
For most experiments, male flies were placed in a disposable 35 mm petri-dish (Becton Dickinson, Falcon 351008, Franklin Lakes, NJ, USA) filled with approximately 7 ml of standard cornmeal media, leaving approximately 2 mm of headspace. In this configuration, flies are free to walk, but flight is suppressed. We also examined behavior in a smaller chamber (a 28 mm diameter beaker) filled with standard cornmeal media with a small amount of live yeast smeared on the center (Ueda & Wu, 2009). A sliding transparent ceiling, consisting of a clear polyethylene film placed over one end of a hollow cylinder of the same diameter, was adjusted to be 2 mm above the food surface to enclose the arena. All chambers used for video recording experiments were pre-conditioned by placing at least 6 CS males in the arena for at least 12 hours.
As illustrated in Figure 1A, recording chambers were placed on a light-box (DW Viewbox, DW Group, LTD, Milton Keynes, UK), and video recordings were taken by a Logitech webcam (c905, Logitech, Newark, CA, USA) approximately 8 cm above the chamber. Videos were recorded at 640 x 480 resolution and at 30 frames-per-second. A custom-written MATLAB (version r2010b, MathWorks, Natick, MA, USA) routine was used to control video acquisition, although it is not required for subsequent image analysis. A 15 minute video recording session was started almost immediately after flies were introduced to the chamber (< 10 s), and the entire video was used for subsequent analysis.
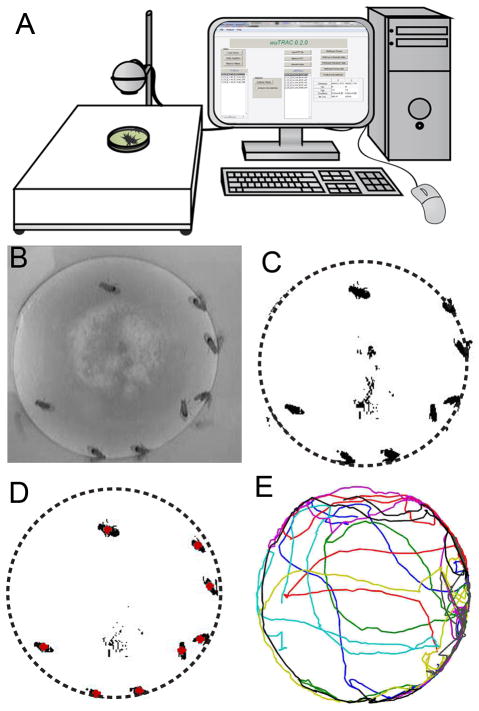
Figure 1. Overview of the IowaFLI Tracker system.
(A) Flies are placed in a 35 mm petri dish containing standard Drosophila medium. The chamber is illuminated by a light box, and a webcam images the arena. (B) A sample image taken by a webcam used to capture fly locomotion. (C) The same image after intensity thresholding. Dotted lines indicate chamber perimeter. (D) Image after static background subtraction. Red points indicate the area-center for the eight largest objects—which correspond to the eight flies in the arena. See Materials and Methods and Supplemental Information for details on how these are established. (E) Sample 30 s plot of fly tracks. Colors indicate individual’s identity.
Target Preference Assay
Males and mated females that were used as targets were co-housed together ensure mating; virgin targets were collected within eight hours of eclosion over CO2 anesthesia and housed separately. All target flies used in the assay were 2–3 days old. Immediately prior to video recordings, target flies were briefly anesthetized on ice, decapitated (to reduce female movements, Ferveur & Sureau, 1996), and a minuten pin was placed through the 1st or 2nd abdominal segment. Only flies which were able to regain a standing posture unaided and showed scratching reflexes after decapitation were pinned and used in subsequent experiments. The pinned target flies were placed on opposite sides of the 35 mm petri-dish, approximately 10 mm away from the wall with their abdomens facing inward. Three male “seekers” were aspirated into the arena and video acquisition began immediately after introduction of the three male flies. Target preference assays were done over several days.
Image processing
All image processing was done using IowaFLI Tracker, a suite of custom-written scripts in MATLAB (with Image Processing Toolbox, r2010b) running on a PC. Prior to image analysis, image scaling parameters were defined in order to convert from pixel units to dimensional units and to correct for aspect ratio error introduced by camera alignment. Additionally, the coordinates of each fly in the initial video frame were selected—this information is later used to assign an identity to each fly. All frames were first converted into 8 bit grayscale images (Figure 1B). Subsequently, each video frame was converted to a black-white (binary) image based on user-selected threshold for each video to achieve clear differentiation between the flies and background features (Figure 1C, Figure S1). A ‘background’ frame was calculated for each video by selecting every 900th black-white frame throughout out the recording period (15 min) and identifying all pixels which were always black. To simplify analysis, we keep track only of the centroids of each fly to compute their locomotion parameters and to extract interaction information among individuals. To determine fly locations, the video’s background frame was subtracted from each processed frame and the objects (sets of adjacent pixels) were found and sorted by size. In most cases, the large objects in an arena corresponded to individual flies—other objects observed were much smaller and considered noise. For each object, the center-of-area (centroid) was calculated and represented a fly’s location (Figure 1D). The identity of each fly was tracked by comparing the set of fly locations in the previous frame to those in the current frame (or in the case of the first frame, compared to the initial, user-defined, coordinates).
In the vast majority of frames we analyzed, this procedure successfully identified tracks of individual flies and their interaction history. However, in relatively rare instances when the two objects merge, usually when two flies are touching, IowaFLI Tracker relied on an additional procedures to track individuals (See Supplementary Information and Figure S2). We found that based on a selection of fifteen videos previously used in analyzing locomotion in CS, Hk1, and parats1 (Figure 2), on average, there were less than twelve instances per video where human intervention was required to resolved the identity of flies after an interaction (Supplemental Table 1), and that in a 35 mm chamber containing multiple flies (up to eight), the rate at which IowaFLI Tracker mistook the identities of flies following a short-range interaction was low enough that human supervision and manual correction for such errors was not a technical hurdle.
The output of IowaFLI Tracker is an array of position vectors that may be used to calculate several kinetic parameters, and may be exported as a text or Excel file. Additionally, within IowaFLI Tacker, several other parameters were calculated and plotted. The cumulative distance traveled was determined by summing the translocations in a fly’s centroid over time. Similarly, a fly’s velocity was computed by multiplying the changes in its centroid between frames by the frame rate (30 Hz). In addition, we measured the fraction of time spent moving by counting frames in which the fly’s velocity was greater than 3 mm/s (slightly larger than one-body length per second). For each genotype, IowaFLI Tracker can display the velocity spread about the median and skewness of the distribution in a cumulative velocity plot. This is done by collecting the velocity data from the entire sample population of each genotype. A parameter of particular significance is the inter-fly distance, which was calculated for each frame and used as the basis for determining fly-fly interactions. In this report, flies were defined to be interacting in a sequence of frames if their centroids were within 2.5 mm of each other (approximately one body length). However, the user is able to modify the criteria for interaction. We demonstrate the use of IowaFLI Tracker to document the sequences and quantify the durations of fly-fly interactions.
In addition to the above technical description, several general operational guidelines are provided in the Supplemental Information, in which the input and output parameters are defined and explained within the conceptual framework.
Results
Quantitative Measurement of Altered Locomotion in Ion Channel Mutants
A straightforward use for our system is to detect and describe modified locomotion in mutant flies. We chose to examine the ion channel mutants Hk1 and parats1 that have previously been well-characterized in the context of larval locomotion (Wang, et. al, 1997). We placed four males in a chamber and recorded their movements for 15 minutes. Sample tracks of 30 seconds from a representative fly of each genotype are plotted in Figure 2A at 0, 3, 6, and 12 minutes. To assess overall activity we plotted the cumulative distance traveled versus time (Figure 2B), and found that Hk1 showed significantly increased total locomotion as compared to CS flies (mean travel distances over 15 min of 6.19 m vs 4.33 m, Mann-Whitney U test, p < 0.01). Interestingly, parats1 showed significantly decreased activity (3.05 m, Mann-Whitney U test, p < 0.01) compared to the other genotypes—even though the experiments were done at permissive temperatures for parats1. These results correlate with the observations of larval locomotion where the pause duration between bouts of locomotion is increased in parats1 mutants (at permissive temperatures) and decreased in Hk1 mutants (Wang et al., 1997).
We found that many flies did not move over portions of the recording period. We measured the fraction of time moving with a criterion for fly movement when its velocity exceeded 3 mm/s, slightly more than their body-length per second. CS flies spent approximately 38% of the time moving, while Hk1, true to their namesake, were significantly more mobile (55 %), and parats1 flies were least mobile (29 %, Figure 2C). While the proportion of time spent moving varied across the three genotypes, the velocity distribution profiles of Hk1 and parats1 (Figure 2D, see Materials and Methods for computational details) were not significantly different from that of CS (Kolmogrov-Smirnov test, p>0.05), with median velocities of 6.86, 7.87, and 5.37 mm/s for CS, Hk1 and parats1 respectively. These velocities are similar to the value reported in a study by Martin et al. (2004), where CS flies showed a mean velocity of around 10 mm/s in a single-fly open-field walking assay. However, both of these values were substantial lower than the velocity reported in the “Buridan Paradigm” (Gotz, 1980), where a single, wing-clipped, CS fly continuously walks back-and-forth towards unreachable visual targets placed on opposite sides of an arena, producing a mean walking speed of approximately 16 mm/s over a 15 minute period (Strauss et al., 1992). Taken together, these observations highlight the distinction between two different walking behaviors, one with clear visual targets while the other without distinct visual targets during exploration.
Social Space Effects on Drosophila Activity
Because IowaFLI Tracker was designed to track multiple flies in an arena, the influence of fly-fly interactions, as well as the effect of altering crowding conditions, on locomotion was of particular interest to us. In addition to the 35 mm petri dish arenas used in previous experiments, we also looked at a 28 mm arena that contained Drosophila media as well as a small amount of yeast paste in the center, and was designed to facilitate studying ‘aggressive’ behaviors (Ueda & Wu, 2009). Both arenas had similar circular planar conformations and headroom amounts (2 mm). To explore the effects of social factors on the movement of individuals, we recorded videos with 1, 2, 4, 8 and 12 flies in the 35 mm arena, and 1, 2, 4, 6, and 8 flies in the 28 mm arena.
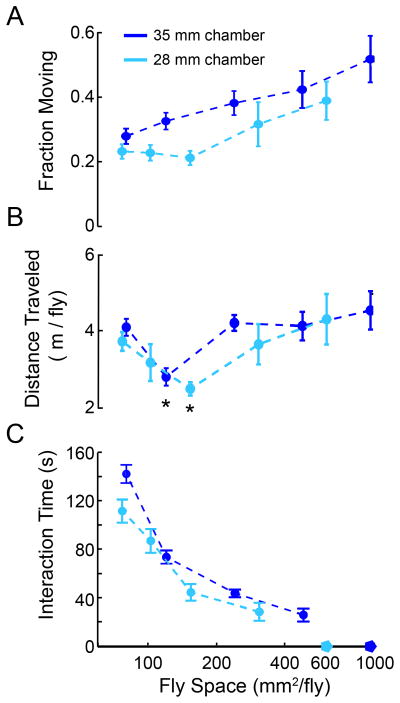
Interestingly, the trends of crowding effects on locomotion seemed to be consistent on fly locomotion in the two arenas when the ‘fly space,’ defined to be the surface area of the arena divided by the number of flies in the arena, was corrected for. We found that, despite the differences in arena construction, the proportion of time individuals were moving (velocity > 3 mm/s) decreased as the available space decreased (Figure 3A). Perhaps surprisingly, in both types of chamber, the total distance traveled did not follow this monotonic trend. We found that as the fly space decreased, individual distances traveled also decreased down to a local minimum before it increased again when the fly space is below about 120 mm2/fly (Figure 3B). When available fly space was between 120 and 150 mm2/fly, corresponding to 8 flies in the 35 mm arena, and 4 flies in the 28 mm arena, the total travel distance was most attenuated (Tukey Post-hoc Test, p<0.05). Given our results on the role of crowding on fly locomotion, we were interested in directly tracking social interactions between flies. We used IowaFLI Tracker to determine when individuals came into close proximity to one another (when a fly was within 2.5 mm, about one body length, of another individual). While this does not provide direct information about the “type” of interaction, it does enable an assessment of how fly space affects the frequency of social interactions. As shown in Figure 3C, in both arenas, the time spent interacting with other flies increases as the fly space decreases, as expected. However, our results based on two different arena designs provide rather similar activity parameters.
Figure 3. Fly activity levels depend on available “social space”.
(A) A varying number of CS flies were introduced to each chamber (1, 2, 4, 8, 12 flies for 35 mm chamber, dark blue line; 1, 2, 4, 6, 8 flies for 28 mm chamber, light blue line), and their activity was recorded for 15 minutes. Show are the fraction of time that the flies were moving (velocity > 3 mm/s) during the video (error bars represent standard error of proportion, n= 12, 18, 40, 72, 72 flies respectively for 35 mm chamber 1, 2, 4, 8 and 12 fly videos; n=16, 10, 56, 54, 64 flies respectively for 28 mm chamber 1, 2, 4, 6 and 8 fly videos ). (B). Plotted are the average distance traveled versus the per-fly surface area was plotted. Error bars represent S.E.M. (*p<0.05, ANOVA, Tukey Post Hoc HSD). (C) The amount of time an individual fly spends interacting with other flies (within 2 mm of another fly) during the video. (mean ± SEM). It should be noted that when one fly is in an arena (denoted by pentagrams), no interactions occur.
An ‘Interactogram’ of Male-Male Short-Range Interaction in Chemosensory Mutants
Based on identification of individual time periods in which flies are in close proximity, IowaFLI Tracker enabled us to track the location, duration and frequency of interactions among the flies in an arena for the entirety of the observation period. Figure 4A-B shows examples of male-male interactions among CS flies as well as the chemosensory mutants olfD and Gr32a. The entire history of fly interactions can be mapped out in this manner over space and time, documented as an ‘interactogram’ (Figure 4C-D). In each interacting group of flies, individuals are identified by different colors and the interacting events are marked by cross-hatches of colors corresponding to the interacting partners. These interactions may be correlated with fly positions to elucidate the spatial information regarding social interaction patterns (Figure 4B). While we only looked at male flies of a single genotype in an arena, this approach may be extended to a number of experiments, including studying interactions between genotypes or among individuals raised under various environmental conditions.
An Open-Field ‘Target-Preference’ Assay of Courtship Behaviors in Chemosensory Mutants
Courtship is perhaps the best studied ‘social interaction’ in Drosophila (Spieth, 1974; Greenspan & Ferveur, 2000; Hall, 2002). In spite of a large body of literature on courtship behavior based on observations using ‘mating wheels’ or similar designs (Hotta & Benzer, 1976; Siegel & Hall, 1979), there have been fewer studies on a male fly’s courtship preference when presented with multiple mates. Using IowaFLI Tracker in this manner, in principle, could enable an incisive quantification of the temporal and spatial characteristics of fly courtship choice behaviors. We first investigated the suitability of using IowaFLI tracker to track the behavior of three males (‘seekers’) in an arena presented with two decapitated ‘target’ flies, a virgin female and a young male (Figure 5A). Target flies for the preference assay were decapitated and pinned to reduce their courtship rejection movements and locomotion during short-range interactions. Previous reports have indicated that males will still vigorously court with the decapitated targets (Ferveur & Sureau, 1996; Miyamoto & Amerein, 2008). It should be noted that in our simplified design, male flies did not copulate with a target even following vigorous courting.
We used IowaFLI Tracker to determine the positions of the males over time as well as identify the periods that they were in proximity to each target and to one another. Figure 5B shows a male fly’s track in the arena within a 30-s period and Figure 5C depicts the position map for all three male flies over the entire 15 minute video. Figure 5D displays the ‘interactograms’ at three 30-s time periods (at 3, 6 and 12 min) for the two targets with the identity of approaching males coded by color. During interactions with a target virgin, courtship behaviors (e.g. ‘licking’, ‘orienting,’ and ‘singing’, Spieth, 1974) were observed very frequently, which could in principle be annotated on an ‘interactogram.’ Furthermore, a ‘preference index’ may be calculated as (T1-T2)/(T1+T2) for each fly, where T1 is the amount of time interacting with the first target, and similarly T2 is the amount of time interacting with the second target (values < 0 indicate a preference towards T2, whereas values > 0 indicate preference towards T1). When presented with a choice between male and virgin female targets, IowaFLI Tracker clearly demonstrated that male flies had a preference towards virgin female targets (Figures. 5B-D), with a statistically significant preference index of 0.43 ± 0.2 (mean ± S.E.M, Student’s T test p <0.05, n = 15 flies). IowaFLI Tracker records the behavior over time and can reveal time-dependent characteristics such as priming or latency, onset kinetics, sustainment or decay, and recurrence of fly interactive behaviors. In Figure 5E, we examined the ensemble of 5 trials of different groups, and found that there is a clear latency to courtship (~4 min), and a characteristic peaking time (median ~6:30 min), followed by a gradual decay in the frequency of courtship activity in our ‘target-preference’ assay.
One particularly interesting aspect of courtship is the role of chemosensory cues in male courtship behavior. Normally, CS flies court more vigorously with virgin females than with previously mated ones (Scott et al., 1988), and several cuticular long-chain hydrocarbon pheromones have been identified to play a role (Ferveur, 1997; Everaets et al., 2010). Altering production of these hydrocarbons in mated females leads to increased attractiveness to males (Billeter et al., 2009). The pheromone 7-tricosene is an example of an inhibitory pheromone that is sufficient to inhibit courtship behaviors (Scott, 1986). Loss-of-function mutations of the chemoreceptor Gr32a in males also increase the frequency of courtship towards mated females and even other males (Miyamoto & Amerein, 2008). We used the courtship ‘target-preference’ assay to study male CS, olfD, and Gr32a responses to two different targets, a mated and a virgin female, on opposite sides of the arena. As Figure 6 shows, CS males had a strong preference to interact with virgin targets versus mated ones (preference index = 0.47 ± 0.21, p<0.05). In contrast, olfD mutant males did not have a strong preference towards either option (0.08 ± 0.16, p > 0.05), and we noted that these males showed a decreased amount of courtship in general, consistent with prior observations (Tompkins, 1984). Interestingly, Gr32a males showed a pattern of interaction similar to that of CS males, with a clear preference towards virgin targets (0.45 ± 0.18, p < 0.05), despite previous implications towards increased courtship towards mated females (Miyamoto & Amerein, 2008).
Discussion
Historically, a computer-assisted motion analysis software system, DIAS (Dynamic Image Analysis System) was first developed at the University of Iowa to analyze cell motility in cell culture systems (Soll, 1995). DIAS was initially utilized in Drosophila neurobiology to study growth cone motility in neuronal cultures (Kim & Wu, 1991; Wu, 1998), and was subsequently found to be suitable for tracking larval locomotion to reveal subtle dynamic differences in motor control in a variety of behavioral mutants (Wang, et. al, 1997; Wang, et al., 2002; Caldwell, et. al., 2003; Fox, et al., 2006; Scantlebury, et al., 2007; Pulver, et al., 2009). Such applications of DIAS involve analysis of dynamic changes in object shapes. DIAS, initially developed for analog video input, requires preprocessing prior to analysis and it would be labor-intensive to quantify social interactions among individuals while still tracking their shapes. In our analysis we tracked only the centroids of each fly which provides essential information for locomotion parameters and interaction events among individuals. We aim at an effective yet simple approach to the need of quantitative description of Drosophila behaviors, particularly social interactions. By using an ordinary PC to run MATLAB and to control an off-the-shelf webcam, we have constructed a versatile tool suitable for basic quantitative analysis of fly behavior.
In developing IowaFLI Tracker, we first extended the study of the Na+ channel mutant parats1 and the K+ channel mutant Hk1 that have previously been characterized for larval locomotion defects (Wang, et. al, 1997). We found that like their behavior as larvae, adult flies of these two mutants display contrasting motor activity levels (Figure 2). Based on the preliminary results, our system has the promise to characterize mutations for changes in several locomotion parameters under different environmental conditions, such as crowding factors that affect social interactions.
For example, IowaFLI Tracker can be applied to study the concept of “fly space”—the amount of space afforded to individual flies in a population (the total arena area divided by the number of flies), and how it influences individual fly locomotion and group interactions. Simon et al. (2012) have previously developed a computer-assisted paradigm to study social space in Drosophila and show that flies tend to congregate in groups. As Figure 3B shows, the total travel distance for individual flies in two arenas of different sizes is similar when allowing for sufficient social space. As the number of flies in the arena increases, a minimal total travel distance was found at approximately 120 mm2/fly. Upon further crowding, fly travel distance significantly increased again and individuals displayed higher amounts of interactions, spending more time within proximity among each other (Figure 3C). Taken together, these findings are consistent with the idea that there is a particular density at which flies tend to space themselves (Sexton & Stalker 1961; Connolly, 1968; Simon et al., 2012), a characteristic social space (Figure 3B), and that a compressed social space with further crowding induces interactive behaviors (Figure 3C). It should be noted that a variety of arena shapes and sizes have been used in studies of Drosophila behavior to obtain quantitative indices in different reports. It will be important to consider “social space” effects in interpreting such parameters and in designing arena-based assays to study particular types of behavior.
The IowaFLI Tracker also facilitates the construction of an ‘interactogram’ to document the complete time-series of interactions among flies. This can be extended to serve the function of an ethogram when interaction events of particular interest on the’ interactogram’ are qualified. This can be done during video acquisition or upon replay with the time sequence identified on the ‘interactogram’. Other, more sophisticated computer-assisted systems have been developed to serve this function at a greater detail (Branson et al., 2009; Dankert et al., 2009).
We present here an exploration into a new design of choice paradigm, the ‘target-preference’ assay for mate-choice behavior by using IowaFLI Tracker (Figure 5). We used three male flies in the experiments to develop and validate tracking of multiple individuals in an arena. An immediate extension in future work would be to investigate the potential social effects by varying the number of males in the arena and detecting any non-additive effects on their individual mate-preference behavior. Our initial work focused on two established chemosensory mutants, olfD and Gr32a. Our results show a decreased amount of courtship in olfD males, which is consistent with previous observations of their behavior in other paradigms (Tompkins, 1984). Interestingly, Gr32a males do show a preference towards virgin females over mated ones (Figure 6). This contrasts with a previous report where loss of the Gr32a chemoreceptor results in an increased courtship index of Gr32a males towards mated females (Miyamoto & Amerein, 2008). The ‘target-preference’ assay reported here provides a direct test of a male fly’s preference towards two distinct immobile targets, while the courtship index quantifies the fraction of time spent on courting when a male is presented with a freely moving single female. These two paradigms can provide distinct, yet complimentary, information on Drosophila courtship behavior.
Supplementary Material
Acknowledgments
We would like to thank Dr. Toshi Kitamoto and Mr. Ryan Jewell for their input during early stages of this work, members of the Wu Lab for their critical reading of the manuscript, and Dr. Hubert Amerin for his generous gift of Gr32a stocks. This research was supported by NIH grants GM88804 and GM 80255 to C.F.W. J.I. was a recipient of an Iowa Center for Research by Undergraduates award and a Carver College of Medicine Research Fellowship.
IowaFLI Tracker is intended for research and educational use, and is openly available under a Creative Commons, Attribution, Non-commercial, ShareAlike 3.0 Unported license (CC BY-NC-SA, http://creativecommons.org/licenses/by-nc-sa/3.0/) from the website: www.journalofneurogenetics.org. Subsequent reports utilizing IowaFLI Tracker must refer to it as “IowaFLI Tracker,” and should cite this article as: Iyengar, A., Imoehl, J., Ueda, A., Nirschl, J., Wu, C.-F. (2012) Automated quantification of locomotion, social interaction and mate preference in Drosophila mutants. J Neurogenet, published volume, inclusive pages.
References
- Ayyub C, Parajape J, Rodrigues V, Siddiqi O. Genetic of Olfactory Behavior in Drosophila. J Neurogenet. 1990;6:243–262. doi: 10.3109/01677069009107114. [DOI] [PubMed] [Google Scholar]
- Billeter JC, Atallah J, Krupp J, Milar J, Levine J. Specialized cells tag sexual and species identity in Drosophila melanogaster. Nature. 2009;461:987–991. doi: 10.1038/nature08495. [DOI] [PubMed] [Google Scholar]
- Branson K, Robie A, Bender J, Perona P, Dickinson M. High-throughput ethomics in large groups of Drosophila. Nat Methods. 2009;6:451–457. doi: 10.1038/nmeth.1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell J, Miller M, Wing S, Soll D, Eberl D. Dynamic analysis of larval locomotion in Drosophila chordotonal organ mutants. Proc Natl Acad Sci USA. 2003;100:16053–16058. doi: 10.1073/pnas.2535546100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly K. The social facilitation of preening behavior in Drosophila melanogaster. Anim Behav. 1968;16:385–391. doi: 10.1016/0003-3472(68)90023-7. [DOI] [PubMed] [Google Scholar]
- Chouinard S, Wilson G, Schlimgen A, Ganetzky B. A potassium channel beta subunit related to the aldo-keto reductase superfamily is encoded by the Drosophila Hyperkinetic locus. Proc Natl Acad Sci USA. 1995;92:6763–6767. doi: 10.1073/pnas.92.15.6763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dankert H, Wang L, Hoopfer E, Anderson D, Perona P. Automated monitoring and analysis of social beahvior in Drosophila. Nat Meth. 2009;6:297–303. doi: 10.1038/nmeth.1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everaets C, Farine JP, Cobb M, Ferveur JF. Drosophila cuticular hydrocarbons revisited: mating status alters cuticular profiles. PLoS ONE. 2010;5:e9607. doi: 10.1371/journal.pone.0009607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferveur JF. The pheromonal role of cuticular hydrocarbons in Drosophila melanogaster. BioEssays. 1997;19:353–356. doi: 10.1002/bies.950190413. [DOI] [PubMed] [Google Scholar]
- Ferveur JF, Sureau G. Simultaneous influence on male courtship of stimulatory and inhibitory pheromones produced by live sex-mosaic Drosophila melanogaster. Proc R Soc B. 1996;263:967–973. doi: 10.1098/rspb.1996.0143. [DOI] [PubMed] [Google Scholar]
- Fox L, Soll D, Wu CF. Coordination and modulation of locomotion pattern generators in Drosophila larvae: Effects of altered biogenic amine levels by the Tyramine beta Hydroxlyase mutation. J Neurosci. 2006;26:1486–1498. doi: 10.1523/JNEUROSCI.4749-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye M, Dickinson M. Closing the loop between neurobiology and flight behavior in Drosophila. Curr Opinion Neurobio. 2004;14:729–736. doi: 10.1016/j.conb.2004.10.004. [DOI] [PubMed] [Google Scholar]
- Gilestro G, Cirelli C. pySolo: a complete suite for sleep analysis in Drosophila. Bioinformatics. 2009;25:1466–1467. doi: 10.1093/bioinformatics/btp237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotz K. Visual guidance in Drosophila. In: Siddiqi O, Babu P, Hall L, Hall J, editors. Development and neurobiology of Drosophila. New York: Plenum; 1980. pp. 391–407. [Google Scholar]
- Gotz K, Wenking H. Visual control of locomotion in the walking fruitfly Drosophila. J Comp Physiol. 1973;85:235–266. [Google Scholar]
- Greenspan R, Ferveur JF. Courtshp in Drosophila. Annu Rev Genet. 2000;34:205–232. doi: 10.1146/annurev.genet.34.1.205. [DOI] [PubMed] [Google Scholar]
- Grover D, Tower J, Tavare S. O fly, where art thou? J R Soc Interface. 2008;5:1181–1191. doi: 10.1098/rsif.2007.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall J. Courtship lite: A personal history of reproductive behavioral neurogenetics in Drosophila. J Neurogenet. 2002;16:135–163. doi: 10.1080/01677060215307. [DOI] [PubMed] [Google Scholar]
- Hotta Y, Benzer S. Courtship in Drosophila mosaics: Sex-specific foci for sequential action patterns. Proc Natl Acad Sci USA. 1976;73:4154–4158. doi: 10.1073/pnas.73.11.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan W, Trout W. The behaivor of four neurological mutants of Drosophila. Genetics. 1969;61:399–409. doi: 10.1093/genetics/61.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YT, Wu CF. Distinctions in growth cone morphology and motility between monopolar and multipolar neurons in Drosophila CNS cultures. J Neurobiol. 1991;22:263–275. doi: 10.1002/neu.480220306. [DOI] [PubMed] [Google Scholar]
- Loughney K, Kreber R, Ganetzky B. Molecular analysis of the para locus, a sodium channel gene in Drosophila. Cell. 1989;58:1143–1154. doi: 10.1016/0092-8674(89)90512-6. [DOI] [PubMed] [Google Scholar]
- Martin JR, Raabe T, Heisenberg M. Central complex substructures are required for the maintenance of locomotor activity in Drosophila melanogaster. J Comp Physiol A. 1999;185:277–288. doi: 10.1007/s003590050387. [DOI] [PubMed] [Google Scholar]
- Martin JR. A portrait of locomotor behaviour in Drosophila determined by a video-tracking paradigm. Behav Proc. 2004;67:207–219. doi: 10.1016/j.beproc.2004.04.003. [DOI] [PubMed] [Google Scholar]
- Miyamoto T, Amerein H. Suppression of male courtship by a Drosophila pheromone receptor. Nat Neurosci. 2008;11:874–876. doi: 10.1038/nn.2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen SP, Chan YB, Huber R, Kravitz E. Gender-selective patterns of aggressive behavior in Drosophila melanogaster. Proc Natl Acad Sci USA. 2004;101:12342–12347. doi: 10.1073/pnas.0404693101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira H, Sokolowski M. Mutations in the larval foraging gene affect adult locomotor behavior after feeding in Drosophila melanogaster. Proc Natl Acad Sci USA. 1993;90:5044–5046. doi: 10.1073/pnas.90.11.5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulver S, Pashkovski S, Hornstein NG, Griffith L. Temporal dynamics of neuronal activation by Channelrhodopsin-2 and TRPA1 determine behavioral output in Drosophila larvae. J Neurophysiol. 2009;101:3075–3088. doi: 10.1152/jn.00071.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodan A, Kiger J, Heberlein U. Functional dissection of neuroanatomical loci regulating ethanol sensitivity in Drosophila. J Neurosci. 2002;22:9490–9501. doi: 10.1523/JNEUROSCI.22-21-09490.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan H, Wu CF. Social interaction-mediated lifespan extension of Drosophila Cu/Zn superoxide dismutase mutants. Proc Natl Acad Sci USA. 2008;105:7506–7510. doi: 10.1073/pnas.0711127105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scantlebury N, Sajic R, Campos A. Kinematic analysis of Drosophila larval locomotion in response to intermittent light pulses. Behav Genet. 2007;37:513–524. doi: 10.1007/s10519-007-9146-3. [DOI] [PubMed] [Google Scholar]
- Scott D. Sexual mimicry regulates the attractiveness of mated Drosophila melanogaster females. Proc Natl Acad Sci USA. 1986;83:8429–8433. doi: 10.1073/pnas.83.21.8429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott D, Richmond R, Carlson D. Pheromones exchanged during mating: a mechanism for mate assessment in Drosophila. Anim Behav. 1988;36:1164–1173. [Google Scholar]
- Seeling J, Chiappe M, Lott G, Dutta A, Osbourne J, Reiser M, Jayaraman V. Two-photon calicum imaging from head-fixed Drosophila during optomotor walking behavior. Nat Methods. 2010;7:535–540. doi: 10.1038/nmeth.1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton O, Stalker H. Spacing patterns of female Drosophila paramelanica. Anim Behav. 1961;9:77–78. [Google Scholar]
- Siddiqi O. Neurogenetics of olfaction in Drosophila melanogaster. Trend Genet. 1987;3:137–142. [Google Scholar]
- Siddiqi O, Benzer S. Neurophysiological defects in temperature-sensitive paralytic mutants of Drosophila melanogaster. Proc Natl Acad Sci USA. 1976;73:3253–3257. doi: 10.1073/pnas.73.9.3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel R, Hall J. Conditioned responses in courtship behavior of normal and mutant Drosophila. Proc Natl Acad Sci USA. 1979;76:3430–3434. doi: 10.1073/pnas.76.7.3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon A, Chou M, Salazar E, Nicholson T, Saini N, Metchev S, Krantz D. A simple assay to study social behavior in Drosophila: measurement of social space within a group. Genes Brain Behav. 2012;11:243–252. doi: 10.1111/j.1601-183X.2011.00740.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolowski M. Social interactions in "simple" model systems. Neuron. 2010;65:780–794. doi: 10.1016/j.neuron.2010.03.007. [DOI] [PubMed] [Google Scholar]
- Soll D. The use of computers in understanding how animal cells crawl. Internatl Rev Cytology. 1995;163:43–103. [PubMed] [Google Scholar]
- Spieth H. Courtship Behavior in Drosophila. Annu Rev Entomology. 1974;19:385–405. doi: 10.1146/annurev.en.19.010174.002125. [DOI] [PubMed] [Google Scholar]
- Strauss R. The central complex and genetic dissection of locomotor behavior. Curr Opinion Neurobiol. 2002;12:633–638. doi: 10.1016/s0959-4388(02)00385-9. [DOI] [PubMed] [Google Scholar]
- Strauss R, Hanesch U, Kinkelin M, Wolf R, Heisenberg M. No-Bridge of Drosophila melanogaster: Portrait of a structural brain mutant of the central complex. J Neurogenet. 1992;8:125–155. doi: 10.3109/01677069209083444. [DOI] [PubMed] [Google Scholar]
- Strauss R, Heisenberg M. Coordination of legs during straight walking and turning in Drosophila melanogaster. J Comp Physiol A. 1990;167:403–412. doi: 10.1007/BF00192575. [DOI] [PubMed] [Google Scholar]
- Suzuki D, Grigliatti T, Williamson R. Temperature-sensitive mutations in Drosophila melanogaster, VII. A mutation (parats) causing reversible adult paralysis. Proc Natl Acad Sci USA. 1971;68:890–893. doi: 10.1073/pnas.68.5.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompkins L. Genetic analysis of sex appeal in Drosophila. Behav Genet. 1984;14:411–440. doi: 10.1007/BF01065443. [DOI] [PubMed] [Google Scholar]
- Trimarchi J, Schneiderman A. Different neural pathways coordinate Drosophila flight initiations evoked by visual and olfactory stimuli. J Exp Biol. 1995;198:1099–1104. doi: 10.1242/jeb.198.5.1099. [DOI] [PubMed] [Google Scholar]
- Troncoso B, Godoy-Herrera R, Mora W. The development of larval movement patterns in Drosophila. Heredity. 1987;58:321–329. [Google Scholar]
- Ueda A, Wu CF. Effects of social isolation on neuromuscular excitability and aggressive behaviors in Drosophila: Altered responses by Hk and gsts1, two mutation implicated in redox regulation. J Neurogenet. 2009;23:378–394. doi: 10.3109/01677060903063026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Sylwester A, Reed D, Wu DAJ, Soll D, Wu CF. Morphometric description of the wandering behavior in Drosophila larvae: aberant locomotion in Na+ and K+ channel mutants revealed by computer-assisted motion analysis. J Neurogenet. 1997;11:231–254. doi: 10.3109/01677069709115098. [DOI] [PubMed] [Google Scholar]
- Wang J, Soll D, Wu CF. Morphometric description of the wandering behavior in Drosophila larvae: a phenotypic analysis of K+ channel mutants. J Neurogenet. 2002;16:45–63. doi: 10.1080/01677060213106. [DOI] [PubMed] [Google Scholar]
- Wu C-F. Neuronal Growth Cone Motility: Contributions from Neurogenetic Analyses of Cultured Drosophila Neurons. In: Soll D, Wessels D, editors. Motion Analysis of Living Cells. New York: Wiley-Liss; 1998. pp. 235–261. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.