Abstract
The epicardium encapsulates the heart and functions as a source of multipotent progenitor cells and paracrine factors essential for cardiac development and repair. Injury of the adult heart results in reactivation of a developmental gene program in the epicardium, but the transcriptional basis of epicardial gene expression has not been delineated. We established a mouse embryonic heart organ culture and gene expression system that facilitated the identification of epicardial enhancers activated during heart development and injury. Epicardial activation of these enhancers depends on a combinatorial transcriptional code centered on CCAAT/enhancer binding protein (C/EBP) transcription factors. Disruption of C/EBP signaling in the adult epicardium reduced injury-induced neutrophil infiltration and improved cardiac function. These findings reveal a transcriptional basis for epicardial activation and heart injury, providing a platform for enhancing cardiac regeneration.
During embryogenesis, the epicardium secretes mitogenic factors to promote cardiomyocyte proliferation and provides multipotent progenitor cells to form the coronary vasculature and the fibrous architecture of the heart (1). Cells of the adult epicardium are typically quiescent but are rapidly activated in response to cardiac injury, promoting cell cycle reentry and embryonic gene expression (2–10). Although several recent lineage-tracing experi ments demonstrated the presence of multipotent cardiovascular progenitor cells within the activated adult epicardium (3, 5, 8, 9), there has been a lack of functional studies that directly manipulate gene expression specifically in the adult epicardium to evaluate its contribution to cardiac regeneration and repair. Here, we report the transcriptional mechanisms underlying epicardial activation during cardiac development and repair, and a functional link between the adult epicardium and cardiac remodeling following ischemic injury.
Identification of enhancer elements that exhibit activity in the mouse embryonic epicardium
To decipher the transcriptional basis of epicardial activation, we sought to identify cis-regulatory DNA sequences sufficient to confer epicardial expression during development and injury. We therefore designed a mouse embryonic heart organ culture and transfection assay to facilitate delivery of reporter plasmids to the epicardium (Fig. 1A) and later rapid screening of epicardial enhancer elements in luciferase reporter assays.
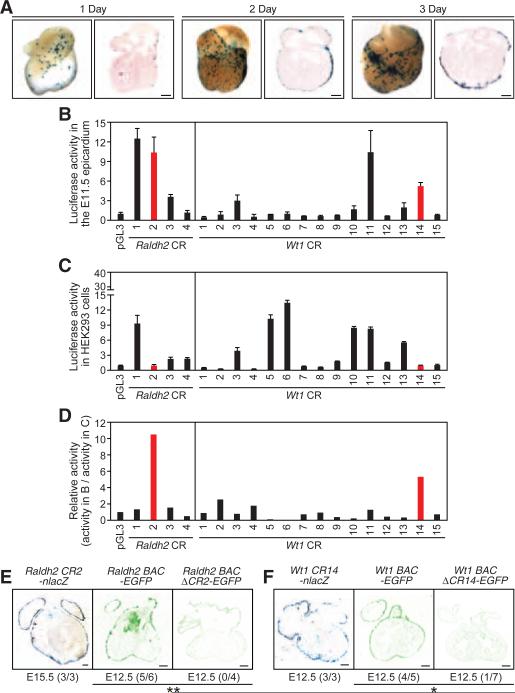
Fig. 1.
Functional screen and identification of epicardial enhancers. (A) Epicardial lacZ expression in E11.5 mouse hearts 1 to 3 days after transfection of a CMV-lacZ plasmid. Whole-mount and transverse-section views are presented. (B and C) Enhancer activity of each conserved region (CR) in the epicardium [(B), n = 2 to 4 hearts] and in HEK293 cells [(C), n = 3] (mean ± SEM). (D) Relative activity. The data for Raldh2 CR2 and Wt1 CR14 are highlighted in red. (E and F) Raldh2 CR2 and Wt1 CR14 are sufficient and necessary to direct epicardial gene expression. Displayed are transgenic hearts that express a nuclear lacZ (nlacZ) driven by an enhancer (left), an EGFP reporter in a control BAC (middle), or an EGFP reporter in an enhancer-deleted BAC (right). The number of embryos that show epicardial reporter activity out of the total number of transgenic embryos is shown. Scale bars, 200 μm. *P < 0.05; **P < 0.01.
Epicardium development is evolutionarily conserved from fish to mammals (11, 12). In both zebrafish and mice, retinaldehyde dehydrogenase 2 (RALDH2), Wilms tumor 1 (WT1), transcription factor 21 (TCF21), and T-box 18 (TBX18) transcription factors are highly enriched in the embryonic epicardium (6, 11–14), and their expression is reactivated in the adult epicardium after injury (2, 6) (fig. S1). We reasoned that the epicardial enhancers might reside in evolutionarily conserved regions (CRs) associated with epicardial marker genes. Among the 39 CRs that encompass 740 kb of genomic regions (fig. S2 and table S1), we identified sequences Raldh2 CR2 and Wt1 CR14 as promising epicardial enhancer candidates, showing robust activity in the embryonic day 11.5 (E11.5) epicardium but not in human embryonic kidney 293 (HEK293) cells that lack expression of epicardial marker genes (Fig. 1, B to D, and fig. S3).
Raldh2 CR2 and Wt1 CR14 are localized in introns 35 and 15 kb downstream of the transcriptional start site, respectively (fig. S4). We next examined whether these enhancers drive epicardial gene expression in vivo. We engineered enhancer-lacZ transgenic mice and observed robust β-galactosidase expression throughout the embryonic epicardium (Fig. 1, E and F). We also generated bacterial artificial chromosome (BAC) transgenic mice that express an enhanced green fluorescent protein (egfp) reporter gene in a 225-kb mouse Raldh2 BAC or a 186-kb mouse Wt1 BAC (fig. S5). Deletion of putative enhancer sequences resulted in a drastic loss of GFP expression in the epicardium (Fig. 1, E and F) and other tissues (fig. S6). Together, these results identify Raldh2 CR2 and Wt1 CR14 as enhancers that are sufficient and necessary to direct gene expression in the epicardium.
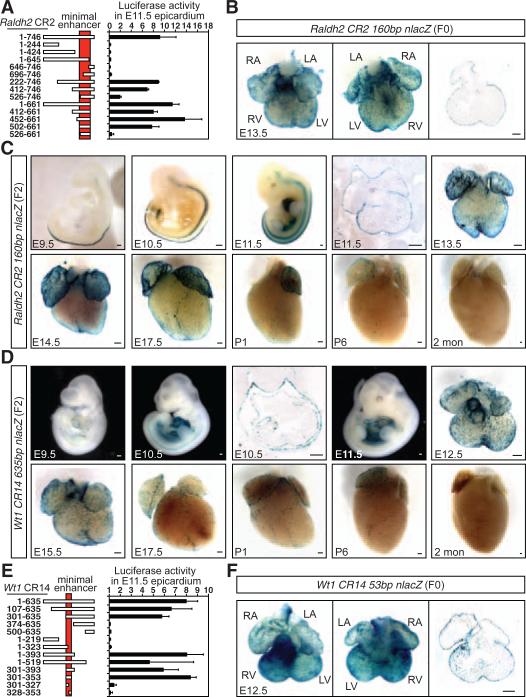
Both RALDH2 and WT1 are important regulators of epicardial functions and heart development (1, 15–17). Deletion analyses of Raldh2 CR2 revealed a 160–base pair (bp) region that directed robust epicardial enhancer activity (Fig. 2, A and B). In two independent transgenic mouse lines that we examined, the epicardial activity of the 160-bp Raldh2 CR2 enhancer initiated at E11.5 (18), persisted throughout gestation, and decreased postnatally (Fig. 2C and fig. S7). We observed a similar temporal pattern of epicardial activity for the 635-bp Wt1 CR14 enhancer (Fig. 2D) (5, 6). Reporter expression was also detected in mesothelial cells and other tissues (figs. S8 and S9). Further deletion mapping of Wt1 CR14 revealed a region of 53 bp that showed complete activity in the epicardium (Fig. 2, E and F, and fig. S9E).
Fig. 2.
Dynamic activity of Raldh2 and Wt1 epicardial enhancers. (A) Epicardial enhancer activity of Raldh2 CR2 maps to a 160-bp region (mean ± SEM, n = 2 to 4 hearts). (B) The Raldh2 CR2 minimal enhancer directs epicardial expression in vivo. The staining pattern represents six out of seven F0 transgenic embryos analyzed. (C) Temporal and spatial expression of the 160-bp Raldh2 CR2-nlacZ transgene in a stable mouse line with an emphasis on the activity in the heart. (D) Dynamic activity of the 635-bp Wt1 CR14 enhancer in a stable mouse line. (E) Mapping of the epicardial enhancer activity of Wt1 CR14 to a 53-bp sequence (n = 2 to 4 hearts). (F) Epicardial expression of a 53-bp Wt1 CR14-nlacZ transgenic heart, which represents three out of six transgenic embryos analyzed. LA, left atrium; RA, right atrium; LV, left ventricle; RV, right ventricle. Scale bars, 200 μm.
Activation of epicardial enhancers by a C/EBP-dependent transcriptional combinatorial code
To search for the transcription factors that regulate the epicardial enhancers of Raldh2 and Wt1, we first isolated lacZ+ epicardial cells from E11.5 Tcf21lacZ hearts (19), performed a microarray analysis of gene expression, and assembled a list of epicardium-enriched transcription factors (fig. S10). Second, analyses of the 160-bp Raldh2 CR2 and the 53-bp Wt1 CR14 minimal enhancer sequences by transcription element search system (TESS) revealed that both enhancers contain putative C/EBP binding sites (fig. S11), and C/EBPβ was enriched in epicardial cells in our microarray analysis (fig. S10).
The C/EBP family of basic leucine zipper (bZIP) transcription factors consists of six members in mammals (C/EBPα, β, δ, ε, γ, and ζ) (20). Quantitative analyses of C/EBP family gene expression in E11.5 Tcf21lacZ epicardial cells revealed the expression of most members and enrichment of Cebpb and Cebpd transcripts in the embryonic epicardium (Fig. 3, A and B). Epicardial expression of C/EBPβ mRNA and proteins was further confirmed (fig. S12).
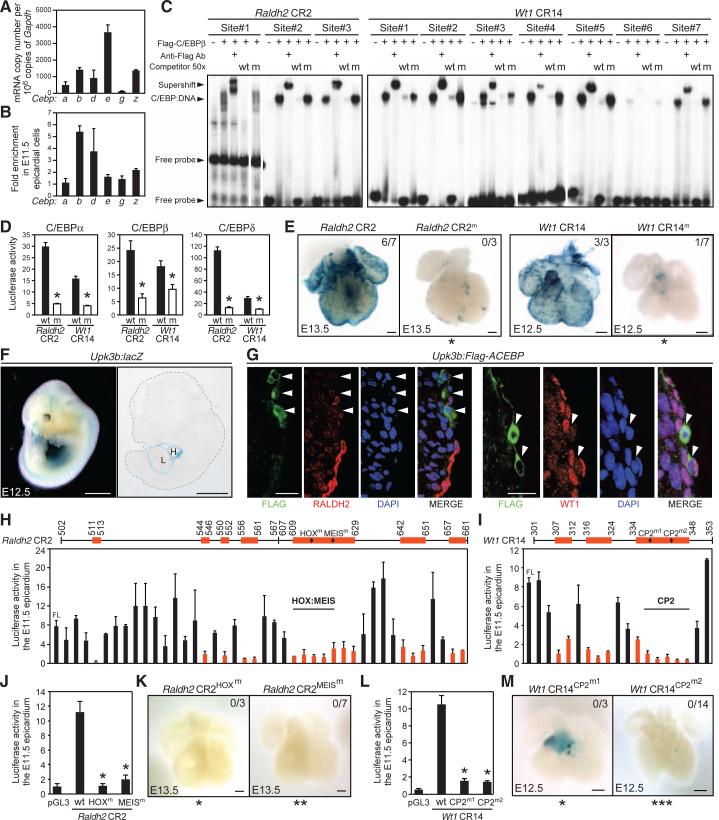
Fig. 3.
A C/EBP-dependent combinatorial code for Raldh2 and Wt1 activation in the embryonic epicardium. (A and B) Quantitative analyses of mRNA abundance and relative enrichment of C/EBP family members in E11.5 epicardial cells (n = 3). (C) Binding of C/EBPβ proteins to predicted sites. (D) Activation of the wild-type (wt) but not mutant (m) enhancers by C/EBP proteins in HEK293 cells (n = 3). (E) Mutations of C/EBP sites reduce epicardial enhancer activity in vivo. (F) X-gal stain of a transgenic embryo. H, heart; L, Liver. (G) Immunostaining of E12.5 transgenic hearts. DAPI, 4′,6-diamidino-2-phenylindole (H and I) Three–base-pair deletion scanning analyses of minimal enhancers (n = 2 to 6 hearts). The numbers on the top denote the positions of the critical regions (red) revealed by functional mapping. (J to M) Point mutations in either HOX, MEIS, or CP2 binding sites abolish epicardial enhancer activity in heart organ cultures [(J and L), n = 3 hearts] and transgenic embryos (K and M). For (E), (K), and (M), the number of epicardial lacZ+ embryos out of the number of transgenic embryos analyzed is presented in the upper right corner. Scale bars: 2 mm (F), 200 μm [(E), (K), and (M)], 100 μm (G). All error bars are SEM. In statistical analyses, all mutants were compared with wild-type controls. *P < 0.05; **P < 0.01; ***P < 0.005.
We investigated whether C/EBP transcription factors bind and activate the Raldh2 and Wt1 enhancers. C/EBP proteins recognize a consensus sequence of (A/C)TTNCNN(A/C)A (21). The Raldh2 CR2 and Wt1 CR14 enhancers contain multiple putative C/EBP binding sites (fig. S11). Electrophoresis mobility shift assays (EMSAs) showed robust binding of C/EBPβ to 9 (out of 10) predicted sites (Fig. 3C and fig. S13). Furthermore, overexpression of C/EBPα, C/EBPβ, and C/EBPδ in HEK293 cells activated the 160-bp Raldh2 CR2 enhancer and the 635-bp Wt1 CR14 enhancer by two- to sixfold compared with mutant enhancers that could not bind C/EBP (Fig. 3D and fig. S14). In addition, we observed reduced expression of Raldh2 and Wt1 in primary epicardial cells in which Cebpa, Cebpb, or Cebpd were knocked down individually (fig. S15). Collectively, these data suggest that C/EBPα, C/EBPβ, and C/EBPδ may have redundant functions in regulating Raldh2 and Wt1 gene expression.
We next determined whether C/EBP binding is required for enhancer activity in the embryonic epicardium. Mutations of C/EBP sites resulted in a substantial reduction of enhancer activity in transgenic embryos and heart organ cultures (Fig. 3E and figs. S16 and S17). These data support the conclusion that the C/EBP binding sites are essential for the activity of the Raldh2 CR2 and Wt1 CR14 enhancers in the epicardium during development.
To express a dominant negative C/EBP (ACEBP) in the embryonic epicardium in vivo, we searched for another independent promoter or enhancer that exhibits epicardial activity during development. We identified Uroplakin 3b (Upk3b) as an E11.5 epicardium-enriched gene in our microarray analysis and found its expression to be highly restricted to the mesothelial cells that cover the internal organs, including the heart (fig. S18). We characterized the promoter of Upk3b and demonstrated that it indeed directed specific expression of a β-galactosidase reporter in mesothelial cells at E12.5 (Fig. 3F). Next, we verified that ACEBP inhibited transcriptional activity of C/EBP family members through cytoplasmic sequestration (fig. S19). We then generated transgenic mouse embryos that expressed a Flag-tagged ACEBP under the control of the Upk3b promoter and examined gene expression in the epicardium. Analysis of E12.5 transgenic hearts revealed a substantial loss of RALDH2 and WT1 expression in ACEBP-expressing epicardial cells (Fig. 3G). These results suggest that C/EBP transcription factors are required for Raldh2 and Wt1 epicardial enhancer activity and gene expression in the embryonic epicardium.
To further delineate the transcriptional code governing the Raldh2 and Wt1 epicardidal enhancers, we generated deletion mutants with either 20- or 3-bp deletions across the minimal enhancer regions and examined their activity in ex vivo epicardial luciferase assays. Such deletion scanning analyses revealed a critical region in the Raldh2 CR2 that contains a conserved HOX:MEIS composite binding site (Fig. 3H and fig. S20, A to C) and a region in the Wt1 CR14 that contains a predicted binding site for the CP2 subfamily of Grainyhead (GRH) transcription factors (Fig. 3I and fig. S23, A and B). We next examined the abundance and expression pattern of MEIS, HOX, and GRH families of transcription factors and found that several members are expressed and enriched in E11.5 epicardial cells (figs. S20 to S24). In addition, we performed chromatin immunoprecipitation (ChIP) analyses in primary epicardial cells, which detected the binding of MEIS1/2, HOX2B, C/EBPα, and C/EBPδ to the endogenous Raldh2 CR2 enhancer and the binding of several CP2 family and C/EBP family proteins to the Wt1 CR14 locus (fig. S25). We further observed simultaneous binding and strong synergistic activation of the Wt1 CR14 enhancer by C/EBP and CP2 family proteins (fig. S25). Moreover, mutant enhancers with point mutations that abolish either HOX or MEIS binding in the Raldh2 CR2 or mutations that abolish CP2 binding in the Wt1 CR14 region showed diminished epicardial activity in both organ cultures and transgenic embryos (Fig. 3, J to M). Altogether, our data suggest a combinatorial code of C/EBP, HOX, and MEIS in regulating the epicardial activity of the Raldh2 CR2, and a role of C/EBP and the CP2 subfamily of Grainyhead transcription factors in governing the Wt1 CR14 enhancer activity.
C/EBP-dependent reactivation of epicardial enhancer activity and gene expression in adult injured hearts
If reexpression of the developmental gene program in the adult epicardium after myocardial injury involves the reactivation of embryonic epicardial enhancers, the Raldh2 CR2 and Wt1 CR14 enhancers should be reactivated after injury. We examined our stable transgenic mouse lines that harbor a Raldh2 or Wt1 epicardial enhancer-driving nlacZ and subjected these adult animals to either permanent or transient coronary artery ligation surgery to induce myocardial infarction (MI) or ischemia reperfusion (IR), respectively. One day after MI, intense β-galactosidase activity was detected in the epicardium overlying the infarct zone and the border zone, and the expression was maintained for at least 7 days after injury (Fig. 4, A and B). Expression was most pronounced at the border zone, but much weaker and scattered expression in regions of the epicardium outside the infarct was also detected. We observed a similar pattern of activity for both enhancers after IR injury (fig. S27).
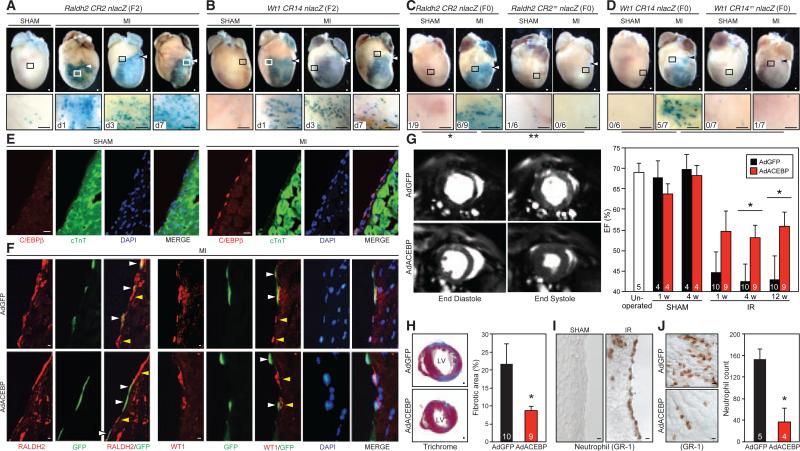
Fig. 4.
Epicardial C/EBP signaling regulates injury-induced epicardial gene activation and cardiac remodeling in adult hearts. (A and B) Reactivation of Raldh2 and Wt1 enhancers after myocardial infarction (MI). Magnified views of boxed areas are shown below. Arrowheads mark coronary ligatures. (C and D) Diminished injury responses in transgenic mice carrying C/EBP-binding mutant enhancers. The number of epicardial lacZ+ hearts out of the number of transgenic hearts analyzed is shown. (E) Epicardial induction of C/EBPβ proteins 3 days after MI. (F) Reduced RALDH2 and WT1 expression in AdACEBP-infected epicardial cells 3 days after MI. White arrowheads point at infected cells (GFP+) and yellow arrowheads mark uninfected epicardial cells (GFP–). (G) Magnetic resonance images (left) and ejection fraction (EF) measurements (right) of AdGFP- versus AdACEBP-injected hearts at different weeks (w) after IR surgery. (H) Analysis of fibrotic area (stained in blue in the trichrome stain) of heart sections from AdGFP- versus AdACEBP-injected mice 12 weeks after IR surgery. (I) Immunohistochemical stains of GR-1 on heart sections reveal enrichment of neutrophils around the epicardium 1 day after IR injury. (J) Neutrophil counts in the infarct area of AdGFP- versus AdACEBP-injected hearts 1 day after IR. LV, left ventricle. For (G), (H), and (J), the number in the column denotes the total number of animals analyzed. Scale bars, 200 μm for (A) to (D) and (H); 20 μm for (E), (F), (I), and (J). *P < 0.05; ** P < 0.01.
We further examined whether C/EBP transcription factors are involved in reactivation of the Raldh2 and Wt1 epicardial enhancers and gene expression following myocardial injury. First, we generated transgenic mice that express lacZ under the control of wild-type Raldh2 and Wt1 epicardial enhancers or mutant enhancers that lack C/EBP binding sites. After cardiac ischemic injury, lacZ expression was largely absent in mice with the C/EBP binding mutant enhancers (Fig. 4, C and D). Next, we examined the expression of C/EBP family members after cardiac injury. Robust up-regulation of Cebpb mRNA and proteins specifically in the epicardium was observed after both MI and IR injury (Fig. 4E and fig. S28). We also found strong induction of Cebpa and weak expression of Cebpd in cells of the epicardium and in the infarct area (fig. S28). Furthermore, to examine the function of C/EBP in vivo, we transduced the adult epicardium with GFP- or ACEBP-expressing adenovirus (AdGFP or AdACEBP). Viruses were carefully injected into the pericardial space, and examination of heart cross-sections revealed restricted viral expression in the epicardium (fig. S29). In the epicardial cells infected with AdACEBP, the injury-induced expression of RALDH2 and WT1 was attenuated (Fig. 4F). These results support the conclusion that C/EBP proteins mediate injury-induced activation of Raldh2 and Wt1 epicardial enhancers and gene expression in the adult epicardium.
Inhibition of C/EBP signaling in the adult epicardium confers cardioprotection
We investigated whether expression of ACEBP in the adult epicardium could result in measurable changes in the function of ischemic hearts. Cardiac function following ischemia reperfusion was assessed by ejection fraction (EF) using magnetic resonance imaging. At both 4 and 12 weeks, cardiac function was significantly improved in the AdACEBP-injected mice compared with that of AdGFP-injected mice (Fig. 4G). The EF for AdACEBP versus AdGFP was 53.1 ± 3.1% versus 42.6 ± 4.2% at 4 weeks (P < 0.05) and 55.9 ± 3.5% versus 43.0 ± 5.7% at 12 weeks (P < 0.05). Moreover, AdACEBP-infected hearts showed a pronounced 2.5-fold reduction in the fibrotic area, compared with AdGFP-infected hearts after ischemia reperfusion (Fig. 4H). These data indicate that disruption of C/EBP activation in the epicardium of injured hearts can lead to improved contractile function and decreased myocardial fibrosis. Although at the earliest time point of analysis (1 week after surgery), the EF of AdACEBP-injected hearts (54.6 ± 5.0%) was not statistically significantly different from that of AdGFP-infected hearts (44.7 ± 5.1%, P = 0.091) (Fig. 4G), the ~10% improvement of EF in the AdACEBP-treated group was already present, suggesting that the key process affected by AdACEBP infection likely occurred within the first week after IR injury.
Inflammation is one of the earliest responses after tissue injury. Neutrophil influx is associated with expansion of IR-induced cardiac injury, and interventions targeting neutrophil infiltration confer cardioprotection (22, 23). Therefore, we examined neutrophil recruitment in injured hearts 1 day after IR. Intriguingly, we observed that many GR-1–positive neutrophils localized on the epicardial surface and in the subepicardial space in addition to the infarct area (Fig. 4I and fig. S30). Moreover, the neutrophil count in the AdACEBP-injected hearts was only 25% of that in the AdGFP-injected hearts (Fig. 4J). These results suggest an important function of C/EBP-mediated epicardial activation in promoting leukocyte recruitment and inflammatory responses. Notably, inactivation of C/EBPβ in lung epithelial cells was also recently found to result in blunted neutrophil influx and pulmonary inflammation in response to cigarette smoke (24), suggesting a general role of C/EBP in the epithelium in regulating the innate immune response during tissue injury repair.
Discussion
Our work reveals a transcriptional basis of epicardial activation in both heart development and the injury response. Our data support a model (fig. S31) in which members of the C/EBP family of transcription factors are activated in the epicardium in response to both developmental cues and injury signals, and function together with HOX, MEIS, and Grainyhead transcription factors to establish a transcriptional code for embryonic gene expression in the epicardium. Grainyhead transcription factors have been implicated in epithelial barrier formation and wound healing in fruit flies and mice (25, 26). Our study also suggests a potentially important role of the Grainyhead family in the injury response and function of the epithelial cells that encapsulate the heart. In addition, the present work uncovers a previously unappreciated role of the epicardium in regulation of the inflammatory response and neutrophil infiltration after injury. In this regard, manipulations that inhibit neutrophil recruitment in animals undergoing reperfusion following MI diminish infarct size (22, 23). Other inflammatory cells also contribute appreciably to cardiac damage and repair during pathological remodeling (22, 23). Future investigation of the potential role of retinoic acid (27, 28) and other epicardium-secreted cytokines and chemokines (5) as C/EBP downstream targets in regulating inflammation may uncover novel molecular targets and facilitate strategies to reduce reperfusion damage and enhance cardiac repair.
Supplementary Material
Acknowledgments
We thank J. Cabrera for graphical assistance; J. Shelton and J. A. Richardson for histology; A. Mobley for assistance with fluorescence-activated cell sorting; the UT Southwestern microarray core facility; and K. Kangasniemi, K. Sagiyama, and M. Takahashi for cardiac magnetic resonance imaging. We are grateful to G. Begemann, S. Jane, P. Johnson, C. Lagman, R. MacDonald, R. Roeder, J. Rosen, G. Stark, C. Vinson, M. Wolfgang, and C. Zahnow for plasmids and reagents. We thank J. Chen, N. Munshi, P. Yi, and members of the Olson laboratory for scientific discussion and critical reading of the manuscript. G.N.H. is an O'Donnell Foundation Fellow of the Life Sciences Research Foundation and a recipient of the NIH Pathway to Independence Award (1K99HL114738). Supported by NIH grant HL100401-01, the Donald W. Reynolds Center for Clinical Cardiovascular Research, the American Heart Association, the Jon Holden DeHaan Foundation, the Leducq Foundation, and the Robert A. Welch Foundation (grant 1-0025) (E.N.O.). Microarray data have been deposited in Gene Expession Omnibus (accession no. GSE41959).
Footnotes
Supplementary Materials
References and Notes
- 1.Sucov HM, Gu Y, Thomas S, Li P, Pashmforoush M. Pediatr. Cardiol. 2009;30:617. doi: 10.1007/s00246-009-9391-8. [DOI] [PubMed] [Google Scholar]
- 2.Lepilina A, et al. Cell. 2006;127:607. doi: 10.1016/j.cell.2006.08.052. [DOI] [PubMed] [Google Scholar]
- 3.Kikuchi K, et al. Development. 2011;138:2895. doi: 10.1242/dev.067041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kikuchi K, et al. Dev. Cell. 2011;20:397. doi: 10.1016/j.devcel.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou B, et al. J. Clin. Invest. 2011;121:1894. doi: 10.1172/JCI45529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Limana F, et al. J. Mol. Cell. Cardiol. 2010;48:609. doi: 10.1016/j.yjmcc.2009.11.008. [DOI] [PubMed] [Google Scholar]
- 7.Porrello ER, et al. Science. 2011;331:1078. doi: 10.1126/science.1200708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smart N, et al. Nature. 2007;445:177. doi: 10.1038/nature05383. [DOI] [PubMed] [Google Scholar]
- 9.Smart N, et al. Nature. 2011;474:640. doi: 10.1038/nature10188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Winter EM, et al. Circulation. 2007;116:917. doi: 10.1161/CIRCULATIONAHA.106.668178. [DOI] [PubMed] [Google Scholar]
- 11.Serluca FC. Dev. Biol. 2008;315:18. doi: 10.1016/j.ydbio.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 12.Winter EM, Gittenberger-de Groot AC. Cell. Mol. Life Sci. 2007;64:692. doi: 10.1007/s00018-007-6522-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou B, et al. Nature. 2008;454:109. doi: 10.1038/nature07060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cai CL, et al. Nature. 2008;454:104. doi: 10.1038/nature06969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen TH, et al. Dev. Biol. 2002;250:198. doi: 10.1006/dbio.2002.0796. [DOI] [PubMed] [Google Scholar]
- 16.von Gise A, et al. Dev. Biol. 2011;356:421. doi: 10.1016/j.ydbio.2011.05.668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martínez-Estrada OM, et al. Nat. Genet. 2010;42:89. doi: 10.1038/ng.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moss JB, et al. Dev. Biol. 1998;199:55. doi: 10.1006/dbio.1998.8911. [DOI] [PubMed] [Google Scholar]
- 19.Lu J, et al. Proc. Natl. Acad. Sci. U.S.A. 2000;97:9525. [Google Scholar]
- 20.Schrem H, Klempnauer J, Borlak J. Pharmacol. Rev. 2004;56:291. doi: 10.1124/pr.56.2.5. [DOI] [PubMed] [Google Scholar]
- 21.Osada S, Yamamoto H, Nishihara T, Imagawa M. J. Biol. Chem. 1996;271:3891. doi: 10.1074/jbc.271.7.3891. [DOI] [PubMed] [Google Scholar]
- 22.Timmers L, et al. Cardiovasc. Res. 2012;94:276. doi: 10.1093/cvr/cvs018. [DOI] [PubMed] [Google Scholar]
- 23.Frangogiannis NG. Pharmacol. Res. 2008;58:88. doi: 10.1016/j.phrs.2008.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Didon L, et al. Am. J. Respir. Crit. Care Med. 2011;184:233. doi: 10.1164/rccm.201007-1113OC. [DOI] [PubMed] [Google Scholar]
- 25.Ting SB, et al. Science. 2005;308:411. [Google Scholar]
- 26.Mace KA, Pearson JC, McGinnis W. Science. 2005;308:381. doi: 10.1126/science.1107573. [DOI] [PubMed] [Google Scholar]
- 27.Kim CH. Vitam. Horm. 2011;86:83. doi: 10.1016/B978-0-12-386960-9.00004-6. [DOI] [PubMed] [Google Scholar]
- 28.Hall JA, Grainger JR, Spencer SP, Belkaid Y. Immunity. 2011;35:13. doi: 10.1016/j.immuni.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.