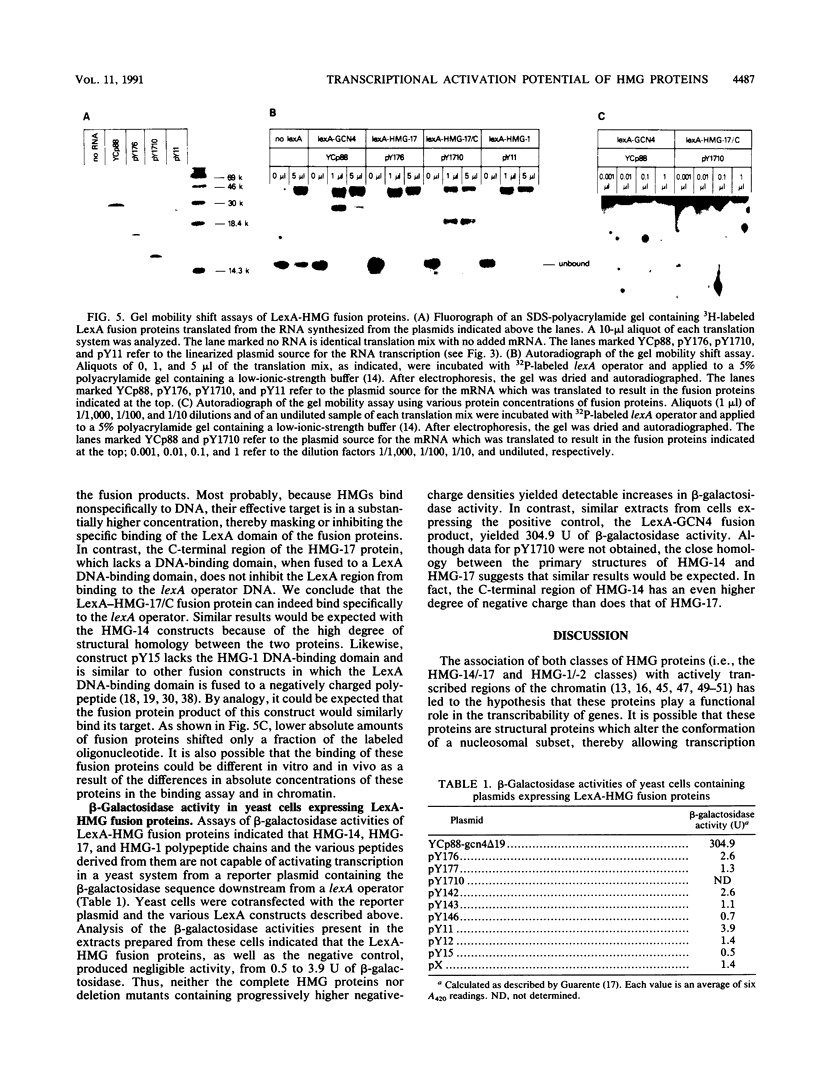
Abstract
Chromosomal proteins HMG-14, HMG-17, and HMG-1 are among the most abundant, ubiquitous, and evolutionarily conserved nonhistone proteins. Analysis of their structure reveals features which are similar to those of certain transcription factors. The distribution of charged amino acid residues along the polypeptide chains is asymmetric: positive charges are clustered toward the N-terminal region, while negative charges are clustered toward the C-terminal region. The residues in the C-terminal region have the potential to form alpha helices with negatively charged surfaces. The abilities of HMG-14, -17, and -1 to function as transcriptional activators were studied in Saccharomyces cerevisiae cells expressing LexA-HMG fusion proteins (human HMG-14 and -17 and rat HMG-1) which bind to reporter molecules containing the beta-galactosidase gene downstream from a lexA operator. Fusion constructs expressing deletion mutants of HMG-14, -17, and -1 were also tested. Analysis of binding to the lexA operator with in vitro-synthesized fusion proteins shows that there are more sites for HMG-14, -17, and -1 binding than for LexA binding and that only the fusion constructs which contain the C-terminal, acidic domains of HMG-17 bind the lexA operator specifically. None of the LexA-HMG fusion protein constructs elevate the level of beta-galactosidase activity in transfected yeast cells. Thus, although HMG-14, -17, and -1 are structurally similar to acidic transcriptional activators, these chromosomal proteins do not function as activators in this test system.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abercrombie B. D., Kneale G. G., Crane-Robinson C., Bradbury E. M., Goodwin G. H., Walker J. M., Johns E. W. Studies on the conformational properties of the high-mobility-group chromosomal protein HMG 17 and its interaction with DNA. Eur J Biochem. 1978 Mar;84(1):173–177. doi: 10.1111/j.1432-1033.1978.tb12154.x. [DOI] [PubMed] [Google Scholar]
- Bustin M., Lehn D. A., Landsman D. Structural features of the HMG chromosomal proteins and their genes. Biochim Biophys Acta. 1990 Jul 30;1049(3):231–243. doi: 10.1016/0167-4781(90)90092-g. [DOI] [PubMed] [Google Scholar]
- Carballo M., Puigdomènech P., Palau J. DNA and histone H1 interact with different domains of HMG 1 and 2 proteins. EMBO J. 1983;2(10):1759–1764. doi: 10.1002/j.1460-2075.1983.tb01654.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cary P. D., King D. S., Crane-Robinson C., Bradbury E. M., Rabbani A., Goodwin G. H., Johns E. W. Structural studies on two high-mobility-group proteins from calf thymus, HMG-14 and HMG-20 (ubiquitin), and their interaction with DNA. Eur J Biochem. 1980 Dec;112(3):577–580. doi: 10.1111/j.1432-1033.1980.tb06123.x. [DOI] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Prediction of the secondary structure of proteins from their amino acid sequence. Adv Enzymol Relat Areas Mol Biol. 1978;47:45–148. doi: 10.1002/9780470122921.ch2. [DOI] [PubMed] [Google Scholar]
- Dodgson J. B., Browne D. L., Black A. J. Chicken chromosomal protein HMG-14 and HMG-17 cDNA clones: isolation, characterization and sequence comparison. Gene. 1988 Mar 31;63(2):287–295. doi: 10.1016/0378-1119(88)90532-x. [DOI] [PubMed] [Google Scholar]
- Dorbic T., Wittig B. Chromatin from transcribed genes contains HMG17 only downstream from the starting point of transcription. EMBO J. 1987 Aug;6(8):2393–2399. doi: 10.1002/j.1460-2075.1987.tb02517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorbic T., Wittig B. Isolation of oligonucleosomes from active chromatin using HMG17-specific monoclonal antibodies. Nucleic Acids Res. 1986 Apr 25;14(8):3363–3376. doi: 10.1093/nar/14.8.3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druckmann S., Mendelson E., Landsman D., Bustin M. Immunofractionation of DNA sequences associated with HMG-17 in chromatin. Exp Cell Res. 1986 Oct;166(2):486–496. doi: 10.1016/0014-4827(86)90493-3. [DOI] [PubMed] [Google Scholar]
- Earnshaw W. C. Anionic regions in nuclear proteins. J Cell Biol. 1987 Oct;105(4):1479–1482. doi: 10.1083/jcb.105.4.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebina Y., Takahara Y., Kishi F., Nakazawa A., Brent R. LexA protein is a repressor of the colicin E1 gene. J Biol Chem. 1983 Nov 10;258(21):13258–13261. [PubMed] [Google Scholar]
- Einck L., Bustin M. Inhibition of transcription in somatic cells by microinjection of antibodies to chromosomal proteins. Proc Natl Acad Sci U S A. 1983 Nov;80(22):6735–6739. doi: 10.1073/pnas.80.22.6735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Einck L., Bustin M. The intracellular distribution and function of the high mobility group chromosomal proteins. Exp Cell Res. 1985 Feb;156(2):295–310. doi: 10.1016/0014-4827(85)90539-7. [DOI] [PubMed] [Google Scholar]
- Fried M., Crothers D. M. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981 Dec 11;9(23):6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnier J., Osguthorpe D. J., Robson B. Analysis of the accuracy and implications of simple methods for predicting the secondary structure of globular proteins. J Mol Biol. 1978 Mar 25;120(1):97–120. doi: 10.1016/0022-2836(78)90297-8. [DOI] [PubMed] [Google Scholar]
- Guarente L. Yeast promoters and lacZ fusions designed to study expression of cloned genes in yeast. Methods Enzymol. 1983;101:181–191. doi: 10.1016/0076-6879(83)01013-7. [DOI] [PubMed] [Google Scholar]
- Hope I. A., Mahadevan S., Struhl K. Structural and functional characterization of the short acidic transcriptional activation region of yeast GCN4 protein. Nature. 1988 Jun 16;333(6174):635–640. doi: 10.1038/333635a0. [DOI] [PubMed] [Google Scholar]
- Hope I. A., Struhl K. Functional dissection of a eukaryotic transcriptional activator protein, GCN4 of yeast. Cell. 1986 Sep 12;46(6):885–894. doi: 10.1016/0092-8674(86)90070-x. [DOI] [PubMed] [Google Scholar]
- Hurstel S., Granger-Schnarr M., Schnarr M. Contacts between the LexA repressor--or its DNA-binding domain--and the backbone of the recA operator DNA. EMBO J. 1988 Jan;7(1):269–275. doi: 10.1002/j.1460-2075.1988.tb02809.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igo-Kemenes T., Hörz W., Zachau H. G. Chromatin. Annu Rev Biochem. 1982;51:89–121. doi: 10.1146/annurev.bi.51.070182.000513. [DOI] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan D. J., Duncan C. H. Full length cDNA sequence for bovine high mobility group 1 (HMG1) protein. Nucleic Acids Res. 1988 Nov 11;16(21):10375–10375. doi: 10.1093/nar/16.21.10375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinschmidt J. A., Scheer U., Dabauvalle M. C., Bustin M., Franke W. W. High mobility group proteins of amphibian oocytes: a large storage pool of a soluble high mobility group-1-like protein and involvement in transcriptional events. J Cell Biol. 1983 Sep;97(3):838–848. doi: 10.1083/jcb.97.3.838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Landsman D., Bustin M. Chromosomal proteins HMG-14 and HMG-17. Distinct multigene families coding for similar types of transcripts. J Biol Chem. 1986 Dec 5;261(34):16087–16091. [PubMed] [Google Scholar]
- Landsman D., Soares N., Gonzalez F. J., Bustin M. Chromosomal protein HMG-17. Complete human cDNA sequence and evidence for a multigene family. J Biol Chem. 1986 Jun 5;261(16):7479–7484. [PubMed] [Google Scholar]
- Landsman D., Srikantha T., Westermann R., Bustin M. Chromosomal protein HMG-14. Complete human cDNA sequence and evidence for a multigene family. J Biol Chem. 1986 Dec 5;261(34):16082–16086. [PubMed] [Google Scholar]
- Landsman D., Zavou S., Soares N., Goodwin G. H., Bustin M. Mouse non-histone chromosomal protein HMG-17 cDNA sequence. Nucleic Acids Res. 1988 Nov 11;16(21):10386–10386. doi: 10.1093/nar/16.21.10386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Ptashne M. Deletion analysis of GAL4 defines two transcriptional activating segments. Cell. 1987 Mar 13;48(5):847–853. doi: 10.1016/0092-8674(87)90081-x. [DOI] [PubMed] [Google Scholar]
- Malik N., Smulson M., Bustin M. Enrichment of acetylated histones in polynucleosomes containing high mobility group protein 17 revealed by immunoaffinity chromatography. J Biol Chem. 1984 Jan 25;259(2):699–702. [PubMed] [Google Scholar]
- Mardian J. K., Paton A. E., Bunick G. J., Olins D. E. Nucleosome cores have two specific binding sites for nonhistone chromosomal proteins HMG 14 and HMG 17. Science. 1980 Sep 26;209(4464):1534–1536. doi: 10.1126/science.7433974. [DOI] [PubMed] [Google Scholar]
- Mitchell P. J., Tjian R. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science. 1989 Jul 28;245(4916):371–378. doi: 10.1126/science.2667136. [DOI] [PubMed] [Google Scholar]
- Paonessa G., Frank R., Cortese R. Nucleotide sequence of rat liver HMG1 cDNA. Nucleic Acids Res. 1987 Nov 11;15(21):9077–9077. doi: 10.1093/nar/15.21.9077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptashne M. How eukaryotic transcriptional activators work. Nature. 1988 Oct 20;335(6192):683–689. doi: 10.1038/335683a0. [DOI] [PubMed] [Google Scholar]
- Reeves R. Transcriptionally active chromatin. Biochim Biophys Acta. 1984 Sep 10;782(4):343–393. doi: 10.1016/0167-4781(84)90044-7. [DOI] [PubMed] [Google Scholar]
- Sadowski I., Ma J., Triezenberg S., Ptashne M. GAL4-VP16 is an unusually potent transcriptional activator. Nature. 1988 Oct 6;335(6190):563–564. doi: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]
- Sandeen G., Wood W. I., Felsenfeld G. The interaction of high mobility proteins HMG14 and 17 with nucleosomes. Nucleic Acids Res. 1980 Sep 11;8(17):3757–3778. doi: 10.1093/nar/8.17.3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shick V. V., Belyavsky A. V., Mirzabekov A. D. Primary organization of nucleosomes. Interaction of non-histone high mobility group proteins 14 and 17 with nucleosomes, as revealed by DNA-protein crosslinking and immunoaffinity isolation. J Mol Biol. 1985 Sep 20;185(2):329–339. doi: 10.1016/0022-2836(85)90407-3. [DOI] [PubMed] [Google Scholar]
- Sigler P. B. Transcriptional activation. Acid blobs and negative noodles. Nature. 1988 May 19;333(6170):210–212. doi: 10.1038/333210a0. [DOI] [PubMed] [Google Scholar]
- Singh J., Dixon G. H. High mobility group proteins 1 and 2 function as general class II transcription factors. Biochemistry. 1990 Jul 3;29(26):6295–6302. doi: 10.1021/bi00478a026. [DOI] [PubMed] [Google Scholar]
- Srikantha T., Landsman D., Bustin M. Cloning of the chicken chromosomal protein HMG-14 cDNA reveals a unique protein with a conserved DNA binding domain. J Biol Chem. 1988 Sep 25;263(27):13500–13503. [PubMed] [Google Scholar]
- Struhl K. Promoters, activator proteins, and the mechanism of transcriptional initiation in yeast. Cell. 1987 May 8;49(3):295–297. doi: 10.1016/0092-8674(87)90277-7. [DOI] [PubMed] [Google Scholar]
- Tremethick D. J., Molloy P. L. Effects of high mobility group proteins 1 and 2 on initiation and elongation of specific transcription by RNA polymerase II in vitro. Nucleic Acids Res. 1988 Dec 9;16(23):11107–11123. doi: 10.1093/nar/16.23.11107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt F., Molloy P. L. High mobility group proteins 1 and 2 stimulate binding of a specific transcription factor to the adenovirus major late promoter. Nucleic Acids Res. 1988 Feb 25;16(4):1471–1486. doi: 10.1093/nar/16.4.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H., Groudine M. Chromosomal subunits in active genes have an altered conformation. Science. 1976 Sep 3;193(4256):848–856. doi: 10.1126/science.948749. [DOI] [PubMed] [Google Scholar]
- Weisbrod S. T. Properties of active nucleosomes as revealed by HMG 14 and 17 chromatography. Nucleic Acids Res. 1982 Mar 25;10(6):2017–2042. doi: 10.1093/nar/10.6.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisbrod S. Active chromatin. Nature. 1982 May 27;297(5864):289–295. doi: 10.1038/297289a0. [DOI] [PubMed] [Google Scholar]
- Weisbrod S., Groudine M., Weintraub H. Interaction of HMG 14 and 17 with actively transcribed genes. Cell. 1980 Jan;19(1):289–301. doi: 10.1016/0092-8674(80)90410-9. [DOI] [PubMed] [Google Scholar]
- Wen L., Huang J. K., Johnson B. H., Reeck G. R. A human placental cDNA clone that encodes nonhistone chromosomal protein HMG-1. Nucleic Acids Res. 1989 Feb 11;17(3):1197–1214. doi: 10.1093/nar/17.3.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann R., Grossbach U. Localization of nuclear proteins related to high mobility group protein 14 (HMG 14) in polytene chromosomes. Chromosoma. 1984;90(5):355–365. doi: 10.1007/BF00294162. [DOI] [PubMed] [Google Scholar]




