Abstract
Cysteine oxidation mediates oxidative stress toxicity and signaling. It has been long proposed that the thioredoxin (Trx) system, which consists of Trx and thioredoxin reductase (Trr), is not only involved in recycling classical Trx substrates, such as ribonucleotide reductase, but it also regulates general cytoplasmic thiol homeostasis. To investigate such a role, we have performed a proteome-wide analysis of cells expressing or not the two components of the Trx system. We have compared the reversibly oxidized thiol proteomes of wild-type Schizosaccharomyces pombe cells with mutants lacking Trx or Trr. Specific Trx substrates are reversibly-oxidized in both strain backgrounds; however, in the absence of Trr, Trx can weakly recycle its substrates at the expense of an alternative electron donor. A massive thiol oxidation occurs only in cells lacking Trr, with 30% of all cysteine-containing peptides being reversibly oxidized; this oxidized cysteine proteome depends on the presence of Trxs. Our observations lead to the hypothesis that, in the absence of its reductase, the natural electron donor Trx becomes a powerful oxidant and triggers general thiol oxidation. Antioxid. Redox Signal. 18, 1549–1556.
Introduction
Oxidative stress can introduce wide changes in the cell's proteome, many of which are the cause of cytotoxicity. Cysteine residues in proteins can suffer oxidation by hydrogen peroxide (H2O2) to form sulfenic acid, which is then stabilized by disulfide linkage with another thiol group, or hyper-oxidized to sulfinic and sulfonic acid forms. In general, only sulfenic acid and disulfides are reversible modifications. Two eukaryotic cellular compartments, the endoplasmic-reticulum and the mitochondrial inter-membrane space, and the bacterial periplasm possess specific systems to catalyze disulfide formation to mediate polypeptide folding and/or protein activity (for reviews, see 1, 5).
The cysteine residues of many enzymes cycle from a thiol to a disulfide form as part of their catalytic cycles, and thioredoxin (Trx) acts as an electron donor to recycle these oxidized enzymes. Additionally, Trxs have been proposed to maintain solvent-exposed cysteines in the reduced form. We have challenged this hypothesis by comparing the oxidized thiol proteome of cells lacking Trx with that of wild-type cells. Unexpectedly, the absence of Trx per se does not lead to general thiol oxidation. On the contrary, lack of thioredoxin reductase transforms the natural electron donor Trx into a potent and general thiol oxidant.
In general, most solvent-exposed cysteines in the cytoplasm are in their thiol/reduced form. Exceptions to this rule are proteins accumulating disulfides as part of the protein's enzymatic activity [e.g., ribonucleotide reductases, peroxiredoxins, 3′-phosphoadenosine-5′-phosphosulfate (PAPS) reductase, methionine sulfoxide reductases etc.] (for a review, see 8). Both the strong reducing cytosolic environment and the recycling of catalytic disulfides in proteins such as ribonucleotide reductase have been attributed to potent redox buffers, the thioredoxin (Trx) and glutathione (GSH)-glutaredoxin (Grx) systems. Trxs and Grxs share a CXXC active site motif in a structure called the Trx-fold. Briefly, these robust disulfide reductases exert their role mainly, but not exclusively, through thiol-disulfide exchange reactions, with the formation of unstable mixed disulfide intermediates with the disulfide-containing protein; recycling of oxidized Trx and Grx require the participation of Trx reductase (Trr) and GSH/GSH reductase, respectively, with NADPH as the final electron donor in both cases (8). The participation of these Trx family members in general thiol homeostasis is, however, a matter of debate. Cells devoid of Trr were soon reported to trigger oxidation of specific protein targets in the cytoplasm of Escherichia coli; this basal disulfide accumulation was largely eliminated in cells lacking both Trr and Trxs (9). In this particular system, the reporter of disulfide formation was a cytoplasmic-targeted alkaline phosphatase, a naturally periplasmic protein whose activity requires the formation of a disulfide bond. It would obviously be of great importance to generate an inventory of oxidized proteins in cells lacking only Trx or Trr, to confirm or dismiss the experiments described using only alkaline phosphatase, a protein naturally interacting with the periplasmic Trx family member DsbA, as a reporter.
Over the last few years, many proteomic approaches were developed to define the oxidized thiol proteome of different cell types or conditions (for a review, see 4). We have recently optimized a gel-free approach to characterize and compare two oxi-proteomes (6), and used it to investigate the role of Trx and Trr in both the recycling of Trx substrates and in the redox control of general thiol reduction in the cytoplasm.
Proteomic analysis of reversible cysteine oxidation in wild-type and Trx mutants using isotope-coded affinity tag reagents
The proteomic approach we have recently optimized (6) is based on the following premises. We first blocked reduced thiols in extracts with iodoacetamide, and we sequentially reduced and labeled reversibly oxidized thiols with heavy or light isotope-coded affinity tag (ICAT) reagents, which are biotin-based iodoacetamide derivatives, so that two different samples can be compared at once after combination of the labeled extracts, trypsin digestion, biotin-based purification of peptides containing oxidized cysteines, and liquid chromatography and analyzed by mass spectrometry (LC-MS/MS) analysis (Fig. 1A). For the same protein extracts, we quantified individual relative protein concentrations with stable-isotope dimethyl labeling. Only a fraction, 72%–78%, of the proteins with reversibly oxidized cysteine-containing peptides were also quantified with dimethyl labeling from the total protein extracts, since only 19%–30% of the most abundant proteins in the Schizosaccharomyces pombe proteome were detected from total extracts using mass spectrometry.
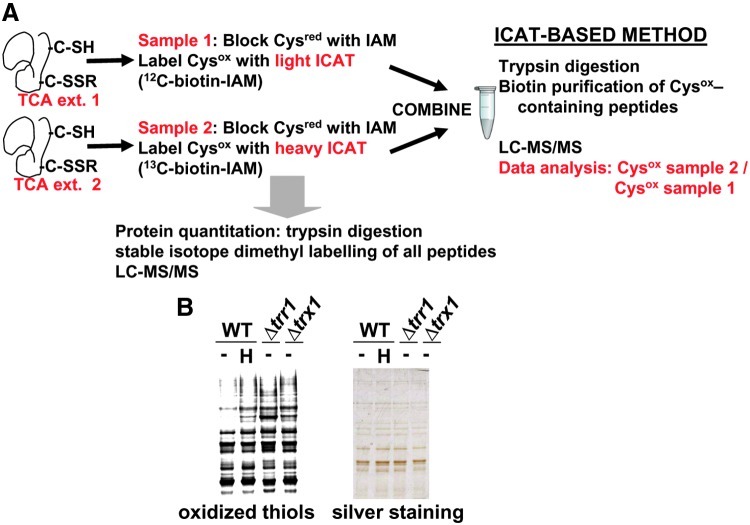
FIG. 1.
Isotope-coded affinity tag (ICAT) strategy for studying the in vivo status of reversibly oxidized cysteines in different strain backgrounds and conditions. (A) Schematic representation of the ICAT methodology. Trichloroacetic acid (TCA) protein extracts were obtained for each pair of samples to be analyzed at a time. Thiols (Cysred) in the extracts were alkylated with iodoacetamide (IAM). Upon reduction of oxidized thiols (Cysox), resulting thiols were alkylated with either light (12C-biotin-IAM) or heavy (13C-biotin-IAM) ICAT reagent. Labeled protein extracts were then mixed and digested with trypsin. ICAT-labeled peptides were affinity purified through streptavidin columns, fractionated by liquid chromatography, and analyzed by mass spectrometry (LC-MS/MS). To quantify individual proteins by dimethyl labeling, small fractions of protein extracts were digested with trypsin, and resulting peptides were labeled at their amino groups with light or heavy formaldehyde (dimethyl labeling). Resulting peptides were mixed and fractionated by LC-MS/MS. (B) Labeling of reversibly oxidized cysteines for 1D electrophoresis. Free thiols in TCA protein extracts of untreated (−) or treated (0.2 mM H2O2 for 30 s; H) cultures of strains 972 (WT), SG167 (Δtrr1), and MJ15 (Δtrx1) were alkylated with iodoacetamide. Upon reduction of oxidized thiols, resulting thiols were alkylated with a fluorescently labeled iodoacetamide derivative. Samples were analyzed by fluorescent 1D gel electrophoresis (oxidized thiols) and with silver staining, as a control of protein loading. (To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars.)
We used this methodology to measure the enrichment ratios across multiple peptides between extracts of untreated wild-type fission yeast cells and three different environmental or genetic conditions: extracts from H2O2-treated wild-type cultures, or from cells lacking Trx1 (the main cytoplasmic Trx in S. pombe) or Trr1 (the only Trr of fission yeast). As shown in Figure 1B, all three conditions triggered general reversible thiol oxidation as determined by 1D electrophoresis of fluorescently labeled oxidized thiols, with Δtrr1 cells displaying massive oxidation.
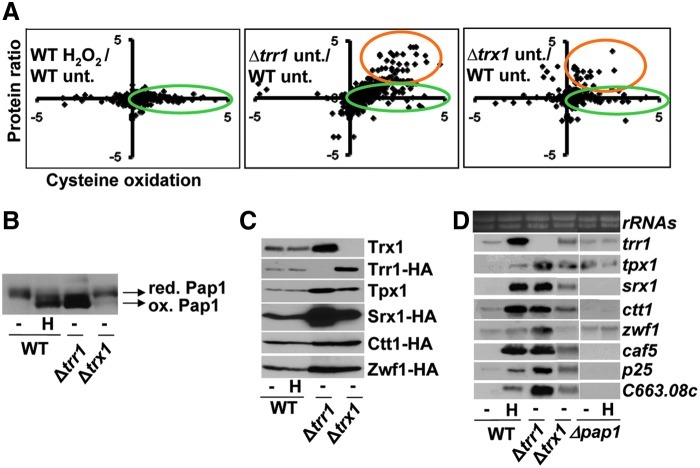
For those peptides displaying ratios of both cysteine oxidation and protein levels, we plotted thiol oxidation ratios against protein ratios. As expected, peptides from H2O2-treated wild-type cultures did not present variations in protein ratios, since only 30 s treatments were applied, without significant changes in the proteome (Fig. 2A, left panel). On the contrary, cells lacking Trr1 displayed a large number of peptides that where overexpressed in extracts, whereas Δtrx1 cells did only display minor protein enrichment over wild-type (Fig. 2A, middle and right panels, highlighted with orange ovals). We noticed that in both strain backgrounds, many of the proteins overexpressed are dependent on the transcription factor Pap1, a H2O2-responding protein that is constitutively active/oxidized in cells lacking Trr1 and partially oxidized in cells lacking Trx1 (Fig. 2B). mRNAs and proteins dependent on activated Pap1, such as srx1, ctt1, or tpx1 (coding for sulfiredoxin, catalase, or peroxiredoxin, respectively), are constitutively expressed in these strains (Fig. 2C, D).
FIG. 2.
Over-expression of proteins in Δtrr1 and in Δtrx1 cells is partially dependent on the transcription factor Pap1. (A) ICAT data representation: cysteine oxidation versus protein expression in the three ICAT studied pairs. Cysteine oxidation from four experimental conditions was analyzed pairwise using the ICAT method depicted in Figure 1A. In each panel, the Log2 ratio of cysteine oxidation is plotted in a scatter diagram versus its Log2 protein ratio (72%–78% of the cysteine-containing peptides displayed protein values by dimethyl labeling). Left panel: 972 treated with 0.2 mM H2O2 for 30 s versus 972 untreated (WT H2O2/WT unt.). Center panel: NG25 untreated versus 972 untreated (Δtrr1 unt./WT unt.). Right panel: MJ15 untreated versus 972 untreated (Δtrx1 unt./WT unt.). Green circles represent peptides with increased cysteine oxidation and orange circles represent peptides from over expressed proteins. (B–D) The activity of the transcription factor Pap1 explains increased protein levels in some strain backgrounds. (B) In vivo oxidation of Pap1. Cultures of strains IC2 (WT), IC71 (Δtrr1), and MJ2 (Δtrx1) were treated (H) or not (−) with 0.2 mM H2O2 for 5 min. TCA extracts were obtained and analyzed by nonreducing electrophoresis. Reduced/inactive (red.) and oxidized/active (ox.) Pap1 forms are indicated with arrows. (C) Protein levels of some Pap1-dependent targets. Cultures of strains expressing or not tagged proteins were treated (H) or not (−) with 0.2 mM H2O2 for 5 min. TCA extracts were obtained, and specific Pap1-dependent proteins were analyzed from extracts of strains: 972, SG167 (Δtrr1) and MJ15 (Δtrx1) for Trx1 and Tpx1; MJ8 (trr1-HA WT), SG167 (Δtrr1), and SG202 (trr1-HA Δtrx1) for Trr1-HA; SB50 (srx1-HA WT), SB69 (srx1-HA Δtrr1), and SB68 (srx1-HA Δtrx1) for Srx1-HA; JF17 (ctt1-HA WT), SG200 (ctt1-HA Δtrr1), and SG198 (ctt1-HA Δtrx1) for Ctt1-HA; and SG18 (zwf1-HA WT), in SB32 (zwf1-HA Δtrr1), and in SG37 (zwf1-HA Δtrx1) for Zwf1-HA. Western blots were performed using polyclonal anti-Tpx1 or anti-Trx1 antibodies, or monoclonal anti-HA antibodies. (D) Transcriptional analysis of Pap1-dependent genes. RNA from strains 972 (WT), IC1 (Δpap1), IC71 (Δtrr1), and MJ2 (Δtrx1), untreated (−) or treated with 0.2 mM H2O2 for 15 min (H), was obtained and analyzed by Northern blot with probes for trr1, tpx1, srx1, ctt1, zwf1, caf5, p25, and SPCC663.08c. Ribosomal RNA (rRNAs) was used as a loading control.
General oxidation of thiols in Δtrr1 cells is dependent on the presence of oxidized cytoplasmic Trx1 and Trx3 and/or Tpx1
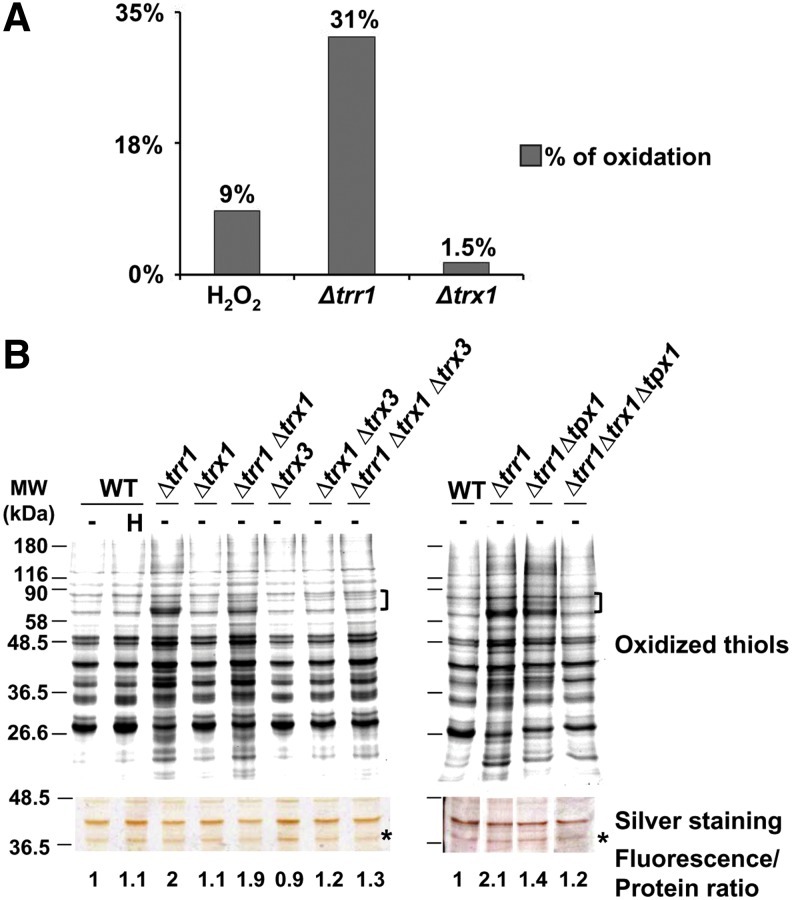
We show in Figure 3A the percentage of peptides whose ratio of cysteine oxidation versus protein levels are over 1.5-fold; we also included peptides not displaying values on protein concentration, excluding those regulated by Pap1 (3). Reversible thiol oxidation by peroxides or in the absence of Trx1 is close to 1.5%, whereas it goes up to 31% in cells lacking Trr1 (Fig. 3A). A double mutant Δtrr1 Δtrx1 displayed a slightly lower percentage of oxidized thiols as observed by 1D electrophoresis of fluorescently labeled thiol-oxidized proteins (Fig. 3B). Since S. pombe expressed another reported cytoplasmic Trx, Trx3, we generated a triple mutant Δtrr1 Δtrx1 Δtrx3, which indeed displayed a significant reduction of general thiol oxidation (Fig. 3B). An almost identical reduction of thiol oxidation in a Δtrr1 background was accomplished by double deletion of trx1 and tpx1, coding for the major peroxiredoxin in S. pombe (Fig. 3B), probably the most abundant substrate of Trxs in fission yeast (our own unpublished data). We conclude that the absence of the Trx system per se does not trigger general thiol oxidation, but elimination of the only Trr, Trr1, does so by means of the accumulation of oxidized Trxs (Trx1 or Trx3) or their main and very abundant substrate, Tpx1.
FIG. 3.
General oxidation of thiols in Δtrr1 cells is dependent on the presence of oxidized cytoplasmic Trx1 and Trx3 and/or Tpx1. (A) Percentage of oxidized cysteines in the different strains according to ICAT data. For each peptide in each biological condition (wild-type treated with H2O2; untreated Δtrr1; or untreated Δtrx1) a ratio of oxidation was always obtained comparing to untreated wild-type cells (WT unt.). An oxidation average ratio was calculated for those peptides having values>1.5-fold in 2 out of 3 biological replicates (for wild-type treated with H2O2 and untreated Δtrr1 samples), or>1.5-fold in 2 out of 2 biological replicates (for Δtrx1 samples). For those peptides having values of protein quantification by dimethyl labeling, a ratio was calculated as oxidation average ratio/protein levels, and only those having this ratio>1.5-fold are included in this graph. For peptides not displaying values on protein concentration, we eliminated those regulated by Pap1, and those having an oxidation average ratio>1.5-fold are included in this graph. Bars represent the percentage of cysteine-containing peptides, which fulfill the previous criteria for each experimental condition. (B) Role of the Trx/Trr system in the homeostasis of cysteine protein oxidation. Free thiols from TCA protein extracts from untreated (−) or treated (0.2 mM H2O2, 30 s; H) strains 972 (WT), SG167 (Δtrr1), MJ15 (Δtrx1), PG22 (Δtrr1 Δtrx1), SG189 (Δtrx3), IC76 (Δtrx1 Δtrx3), SG185 (Δtrr1 Δtrx1 Δtrx3), SG164 (Δtrr1 Δtpx1), and SG170 (Δtrr1 Δtrx1 Δtpx1) were processed as described in Figure 1B. The intensity of selected fluorescent labeled proteins, indicated in the figure with left braces, was quantified with ImageQuant. The intensity of selected proteins of the silver staining gel, indicated with an asterisk, was quantified with ImageJ. Fluorescence to protein ratios were calculated and are indicated in the figure. (To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars.)
Trx substrates are more reversibly oxidized in cells lacking either Trx1 or Trr1 than in wild-type cells
We next analyzed the inventories of ICAT-labeled peptides reversibly oxidized with respect to untreated wild-type extracts following the criteria described above for Figure 3A. As shown in Table 1, cells lacking Trx1 clearly accumulated oxidized peptides of bona fide substrates of Trx (8), such as PAPS reductase (Met16), peroxiredoxin (Tpx1), and methionine sulfoxide reductases (Msr1 and Msr2), among others. Several Trx substrates reversibly oxidized in Δtrx1 extracts also accumulated in cells lacking Trr1 (Met16 in Table 1 and data not shown). As expected, cells lacking Trr1 showed accumulation of oxidized Trx1. Many other proteins, which may have accessible and reactive cysteine residues and are not classical substrates of Trx, are also reversibly oxidized in this strain.
Table 1.
Reversibly Oxidized Peptides Identified from Δtrr1 and Δtrx1 Cells Using the Isotope-Coded Affinity Tag Methodology
| Protein name | Descriptiona | Cysb | Average cys oxidation ratioc | Protein ratiod | Ratio cys oxidation/protein levels |
|---|---|---|---|---|---|
| 1.1- ICAT labeled peptides identified from Δtrx1 cells | |||||
| Thioredoxin substrates | |||||
| Met16 | PAPS reductase | 241 | 11.2 | - | - |
| Mxr1 | peptide methionine sulfoxide reductase | 159 | 5.5 | - | - |
| Msr2 | methionine sulfoxide reductase | 67 | 3.1 | - | - |
| Tpx1 | thioredoxin peroxidase | 169 | 10.9 | 5.4 | 2.0 |
| Gpx1 | glutathione peroxidase | 82 | 1.8 | 1.1 | 1.6 |
| 1.2- ICAT labeled peptides identified from Δtrr1 cells | |||||
| Antioxidant enzymes/pathways | |||||
| Trx1 | thioredoxin | 16 | 26.9 | 7.7 | 3.5 |
| Thioredoxin substrates | |||||
| Met16 | PAPS reductase | 241 | 11.2 | 2.4 | 4.8 |
| Protein folding & catabolism | |||||
| SPAC24C9.08 | vacuolar carboxypeptidase | 87 | 3.5 | - | - |
| Erv1 | sulfhydryl oxidase | 26 | 2.9 | - | - |
| SPBC16G5.09 | serine carboxypeptidase | 330 | 2.6 | - | - |
| Ero12 | ER oxidoreductin | 269 | 2.3 | - | - |
| Ppi1 | cyclophilin peptidyl-prolyl cis-trans isomerase | 38 | 4.8 | 0.9 | 5.1 |
| Sti1 | chaperone activator | 486 | 2.6 | 1.3 | 2.0 |
| Metabolic pathways components | |||||
| Cellular amino acid metabolic process | |||||
| SPBC8D2.18c | adenosylhomocysteinase | 196 | 9.2 | 1.8 | 5.2 |
| Leu2 | 3-isopropylmalate dehydratase | 437 | 5.3 | 1.4 | 3.7 |
| SPAC9E9.06c | threonine synthase | 490 | 3.4 | 1.0 | 3.3 |
| SPBC3B8.03 | saccharopine dehydrogenase | 95 | 2.5 | 1.0 | 2.5 |
| Trp2 | tryptophan synthase | 416 | 3.2 | 1.6 | 2.0 |
| Carbohydrate metabolic process | |||||
| Bgl2 | glucan 1,3-beta-glucosidase precursor | 55 | 2.8 | - | - |
| SPBC12C2.11 | glutamine-fructose-6-phosphate transaminase | 2 | 2.6 | - | - |
| Translation | |||||
| Sum3 | Moc2 RNA helicase | 287 | 4.1 | 0.7 | 5.7 |
| Rps27 | ribosomal protein | 77 | 1.6 | 0.7 | 2.3 |
| OTHER | |||||
| SPAC10F6.17c | mitochondrial pyruvate dehydrogenase | 157 | 5.0 | - | - |
| SPAPB8E5.04c | Niemann-Pick disease hE1 homolog | 118 | 3.4 | - | - |
| SPAC6C3.02c | mitochondrial CHCH domain protein | 145,154 | 2.7 | - | - |
| SPCC550.01c | CHCH domain protein | 27 | 2.6 | - | - |
| SPBC337.07c | carboxypeptidase | 426 | 2.4 | - | - |
| Mae2 | malic enzyme | 475 | 6.1 | 1.4 | 4.2 |
| SPBC4.06 | mitochondrial acid phosphatase | 103 | 5.4 | 1.5 | 3.6 |
| Byr3 | zinc finger protein | 63 | 1.6 | 0.6 | 2.9 |
| Tim40 | TIM22 inner membrane protein import complex | 220 | 1.8 | 0.9 | 2.0 |
In this list, only peptides having oxidation ratios>1.5 in 2 out of 2 biological replicates for Δtrx1 cells or>1.5 in 2 out of 3 biological replicates for Δtrr1 cells were considered. For those peptides fulfilling the previous criteria, only peptides having a ratio of cysteine oxidation versus protein concentration>1.5 for the condition Δtrx1 versus wild-type or>2 for the condition Δtrr1 versus wild-type are shown. In the case of peptides not displaying protein concentration, only those not being Pap1 dependent and with oxidation ratios>1.5 (Δtrx1 versus wild-type) or>2 (Δtrr1 versus wild-type) are shown. Cysteine 798 of ribonucleotide reductase/Cdc22 appeared reversibly oxidized in one biological replicate of each one of the sample pairs, Δtrx1 versus wild-type (average Cys oxidation ratio of 7.0) and Δtrr1 vs. wild-type (average Cys oxidation ratio of 4.7).
Proteins with more than one peptide are shown in italics (only the peptide with the highest oxidation ratio is shown)
Position of the cysteine residue in each correspondent protein sequence modified with ICAT reagent
Average of the oxidation ratios obtained for each peptide in 2 out of 2 biological replicates for the couple Δtrx1 versus wild-type untreated and in 2 out of 3 biological replicates for the couple Δtrr1 versus wild-type untreated
Average ratio of protein levels calculated with the ratios of protein levels of peptides belonging to the same protein in the couples Δtrx1 versus wild-type untreated and Δtrr1 versus wild-type untreated
ICAT, isotope-coded affinity tag; PAPS, 3′-phosphoadenosine-5′-phosphosulfate
In the absence of Trr1, Trx1 can weakly recycle its substrates at the expense of an alternative electron donor
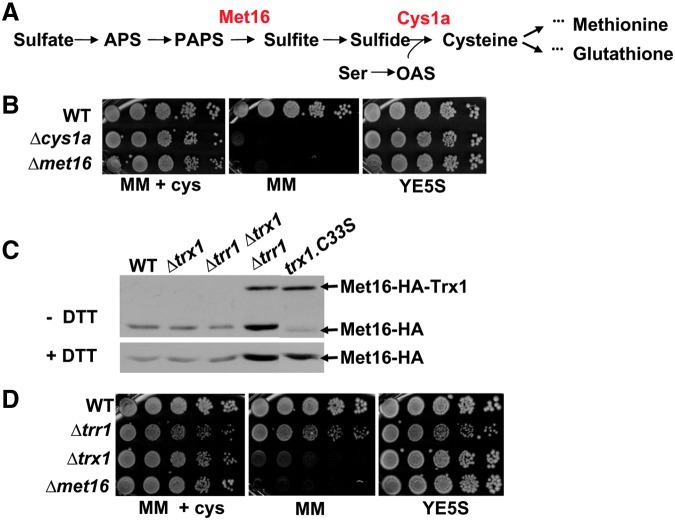
Cysteine-containing peptides of classical Trx substrates such as PAPS reductase-Met16 appear at high rates in both Δtrr1 and Δtrx1 extracts (Table 1). We analyzed the nonessential Met16 to determine the effect of the absence of Trx1 or Trr1 on protein activity. Regarding PAPS reductase enzymatic activity, Trx is used as the electron donor for the reduction of PAPS to phospho-adenosine-phosphate (PAP) and sulfite in the biosynthesis of all sulfur-containing metabolites (8). Thus, deletion of met16 renders cells auxotrophic for cysteine, to the same extend as deletion of the biosynthetic gene cys1a does (Fig. 4A, B). Even though cells lacking Trr1 display enhanced levels of oxidized Met16 (Table 1), this is due to the accumulation of a mixed disulfide with Trx1, as demonstrated with nonreducing electrophoresis of acidic protein extracts (Fig. 4C). Consistently, cells lacking Trx1 are fully auxotrophic for cysteine but cells lacking Trr1 are not (Fig. 4D). This suggests that there is sufficient turnover of oxidized Trx1 reduction in Δtrr1 cells and therefore Met16 can perform its catalytic cycle in this strain background.
FIG. 4.
In the absence of its reductase, Trx1 is able to recycle substrates at the expense of an alternative electron donor. (A) Cysteine biosynthesis pathway in Schizosaccharomyces pombe. The enzymatic roles of Met16 and Cys1a are indicated. (B) Exponentially growing 972 (WT), SG178 (Δcys1a), and SG171 (Δmet16) strains were serially diluted and spotted on minimal medium (MM), MM containing 0.66 mM cysteine (MM+cys), and YE5S plates. (C) Mixed disulfide of Met16 with Trx1. Immunodetection of Met16-HA from TCA extracts of strains SG54 (WT), SG71 (Δtrx1), SG78 (Δtrr1 Δtrx1), SG59 (Δtrr1), and SG181 (trx1.C33S), under nonreducing (−DTT, upper panel) and reducing (+DTT, lower panel) electrophoresis. Since cysteine 33 in Trx1 resolves the mixed disulfides with its substrates, the Trx1.C33S mutant allows in vivo trapping of an intermolecular disulfide of Trx1 with Met16. Arrows indicate Met16-HA and Met16-HA covalently linked to Trx1. (D) S. pombe cells lacking Trx1 are auxotrophic for cysteine, whereas S. pombe cells lacking Trr1 are not. Exponentially growing cultures of strains 972 (WT), SG167 (Δtrr1), MJ16 (Δtrx1), and SG171 (Δmet16) were serially diluted and spotted on MM, MM containing 0.66 mM cysteine (MM+cys), and YE5S plates. (To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars.)
Concluding Remarks and Future Directions
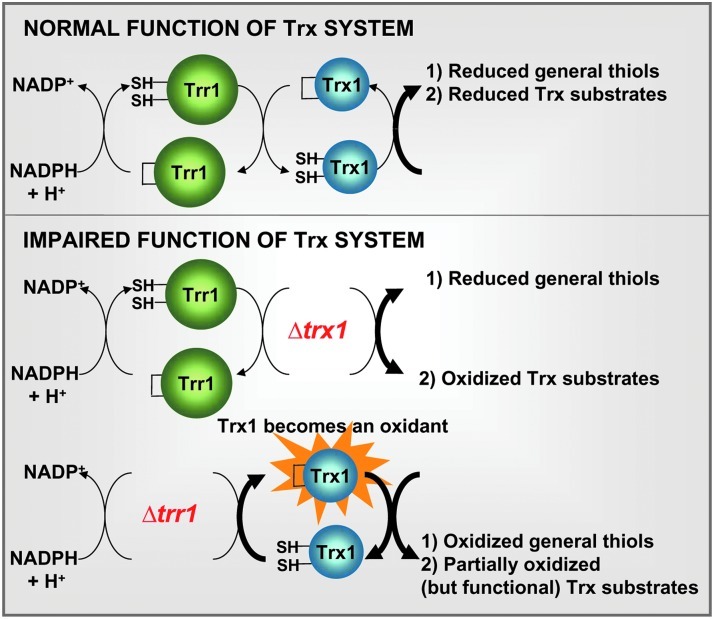
Using a proteomic approach, we have provided an inventory of sensitive thiols upon both peroxide treatment (6) and lack of the Trx system. An important conclusion of our study is that a massive thiol oxidation stress cannot be attributed to the absence of the Trx system, but rather to the accumulation of potent cytoplasmic oxidants when Trr is eliminated. We confirm with our work that the main role of Trxs is to recycle proteins that accumulate disulfides as part of their catalytic cycles (Fig. 5). However, the absence of Trr1 inverts the natural role of these electron donors to convert them in potent oxidants. One Trx substrate, Tpx1, also contributes to the formation of oxidized thiols in a strain lacking Trr1 and Trx1.
FIG. 5.
Proposed model for the participation of the thioredoxin system in general thiol homeostasis and in the recycling of specific Trx substrates. Thioredoxins specifically recycle proteins that form disulfides as part of their catalytic activities (upper panel). In the absence of Trx (lower panel, Δtrx1), Trx substrates are not recycled and appear as oxidized, but other thiols in proteins remain reduced. Only when Trr1 is absent (lower panel, Δtrr1), thioredoxins are converted into potent oxidants leading to massive reversible thiol oxidation; Trx substrates are partially recycled in this background (see text for details). (To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars.)
Lack of Trr1 impairs general fitness as a result of the oxidized Trx-dependent thiol oxidation stress. However, more detrimental to the cell can be the absence of Trx1 alone, since substrates that rely only on this electron donor for recycling are fully inactivated. In cells lacking Trr1, we suspect that the GSH pool, a major contributor to thiol homeostasis in the cell, can be an alternative electron donor for Trx1, since the ratio of oxidized GSH is dramatically increased (4-5-fold) in Δtrr1 but not in Δtrr1 Δtrx1 cells (our own unpublished data). This idea would be in concordance with a recent report that suggests an essential role for GSH in iron-sulfur cluster assembly, and a secondary role in redox control only when the Trx system is impaired (7).
Notes
Strains and growth conditions
The names and genotypes of the strains used in this study are as follows: 972 (h−), NG25 (h+ caf4/trr1::ura4+ura4-D18), MJ15 (h+ trx1::kanMX6), SG167 (h− caf4/trr1::natMX6), IC2 (h− pap1 ura4-D18 leu1-32), IC71 (h− caf4/trr1::natMX6 ura4-D18 leu1-32), MJ2 (h− trx1::kanMX6 ura4-D18 leu1-32), IC1 (h− pap1::ura4+ura4-D18 leu1-32), MJ8 (h− caf4/trr1-HA::kanMX6), SG202 (h+ trx1::natMX6 caf4/trr1-HA::kanMX6), SB50 (h+ srx1-HA::natMX6), SB69 (h− caf4/trr1::ura4+ura4-D18 srx1-HA::natMX6), SB68 (h− trx1::kanMX6 srx1-HA::natMX6), JF17 (h+ ctt1-HA::natMX6), SG200 (h− caf4/trr1::kanMX6 ctt1-HA::natMX6), SG198 (h+ trx1::ura4+ura4-D18 ctt1-HA::natMX6), SG18 (h− zwf1-HA::kanMX6), SB32 (h− caf4/trr1::ura4+ura4-D18 zwf1-HA::kanMX6), SG37 (h+ trx1::natMX6 zwf1-HA::natMX6), SG189 (h− trx3::hgh), IC76 (h+ trx1::natMX6 trx3::kanMX6 ura4-D18 leu1-32), SG170 (h+ tpx1::natMX6 trr1::kanMX6 trx1::ura4+ura4-D18 ade6-M210 leu1-32), SG178 (h− cys1a::ura4+ura4-D18), SG171 (h+ met16::kanMX6), SG54 (h− met16-HA::natMX6), SG71 (h− trx1::kanMX6 met16-HA::natMX6), SG78 (h− trx1::kanMX6 caf4/trr1::ura4+ura4-D18 met16-HA::natMX6), SG59 (h− caf4/trr1::ura4+ura4-D18 met16-HA::kanMX6), SG181 (h− trx1.C33S ura4-D18 met16-HA::kanMX6 leu1-32), PG22 (h? caf4/trr1::natMX6 trx1::kanMX6), SG185 (h− trx1::natMX6 trx3::kanMX6 trr1::phleo), SG164 (h−caf4/trr1::kanMX6 tpx1::natMX6). Cells were grown in standard rich medium (YE5S) or synthetic minimal medium (MM) as previously described (6).
Labeling of reversibly oxidized thiols for 1D electrophoresis analysis
To quantify levels of reversible cysteine oxidation, protein extracts of exponentially growing S. pombe cells (OD600∼0.5) were obtained and labeled with a fluorescently labeled iodoacetamide derivative as previously described (6). Image Quant software (GE Healthcare) was used to quantify fluorescently labeled thiols in Typhoon 8600 (GE Healthcare) scanned gels, using an excitation wavelength of 532 nm and an emission filter of 526 nm with a short-pass filter. ImageJ software (NIH) was used to quantify protein concentrations in silver stained protein gels.
RNA analysis
Total RNA from exponentially growing S. pombe cells was extracted, processed, and transferred to a membrane as previously reported (2). Membranes were hybridized with [α-32P] dCTP-labeled trr1, tpx1, srx1, ctt1, zwf1, caf5, p25, and SPCC663.08c probes. Ribosomal RNA was used as loading control.
S. pombe trichloroacetic extracts and immunoblot analysis
For the in vivo redox analysis of Pap1, trichloroacetic acid (TCA) protein extracts were prepared as previously reported (6) and analyzed by nonreducing sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Specific polyclonal Pap1 antibodies (6) were used for immunodetection. A similar protocol, but without alkaline phosphatase treatment, was followed to detect the the in vivo redox state of Met16-HA. Since cysteine 33 in Trx1 resolves the mixed disulfides with its substrates, the Trx1.C33S mutant allows in vivo trapping of an intermolecular disulfide of Trx1 with Met16. In this case, immunobloting was performed using house-made monoclonal anti-HA antiserum (12CA5). TCA protein extracts and reducing SDS-PAGE (sample buffer containing dithiothreitol) were used to detect differences in protein expression. Immunodetection was carried out using monoclonal anti-HA antiserum, or anti-Tpx1 and anti-Trx1 polyclonal antibodies (6).
Labeling of oxidized cysteine residues by ICAT
Protein extracts of 150 ml cultures (OD600∼0.5) were obtained and labeled as previously described (6) for the following pairs of samples: wild-type treated with 0.2 mM of H2O2 for 30–60 s versus wild-type untreated (6); untreated Δtrr1 versus untreated wild-type cells; and untreated Δtrx1 versus untreated wild-type cells. Briefly, the ICAT-based methodology consists of first blocking the thiol-redox status by adding TCA to each one of the two cultures/biological samples to be compared. Protein extracts were then obtained, and reduced thiols were alkylated with iodoacetamide. Originally oxidized cysteines were then reduced with (2-carboxyethyl) phosphine hydrochloride. At this point, a small fraction of the protein extracts were withdrawn for dimethyl labeling (heavy and light isotopes) and protein quantification, while the rest was treated with light (for one of the samples) or heavy (for the other sample) ICAT reagents (Light and Heavy Cleavable ICAT™ Reagent; AB Sciex). The extracts were then pooled, diluted, and trypsinized. Samples were processed and analyzed by LC-MS/MS (6). For the pair wild-type treated with H2O2 versus wild-type untreated, three biological replicates with two, two and three technical replicates respectively, were obtained. For the pair untreated Δtrr1 versus untreated wild-type cells, three biological replicates, with two, three, and four technical replicates respectively, were obtained. In one of these three biological replicates, ICAT labeling was swapped. For the pair untreated Δtrx1 versus untreated wild-type cells, two biological replicates, with two technical replicates each, were obtained; again, ICAT reagents were swapped.
Liquid chromatography, mass spectrometry of ICAT labeled peptides and ICAT data analysis
Peptides were analyzed by LC-MS/MS; MS/MS spectra were extracted and searched against the S. pombe GeneDB database as described (6).
ICAT data analysis: quantification analysis
Peptides were quantified with MSQuant 2.0b7 (www.msquant.sourceforge.net) as previously described (6). For each peptide, a ratio of cysteine oxidation was obtained by comparing its abundance in each biological sample (wild-type treated with H2O2, untreated Δtrr1, and untreated Δtrx1 cells) with the levels of the same peptide in untreated wild-type cells. An oxidation average ratio was calculated for those peptides having values>1.5-fold in 2 out of 3 biological replicates (for wild-type treated with H2O2 and untreated Δtrr1 samples), or>1.5-fold in 2 out of 2 biological replicates (for Δtrx1 samples). Bias due to differential protein loading was corrected by dividing each cysteine oxidation ratio by the median values of protein quantification (see section below Analysis of dimethyl labeled proteins).
Quantification and analysis of proteins by dimethyl labeling
One of the biological replicates of each ICAT pair was subjected to protein quantification by dimethyl labeling as reported (6). For each peptide, a ratio of protein level was obtained by comparing its abundance in each biological sample (either from wild-type treated with H2O2, or from untreated Δtrr1 or Δtrx1 cells) with the levels of the same peptide in untreated wild-type cells. Ratios of protein levels of peptides belonging to the same protein were used to calculate an average ratio for each protein. The overall median of protein levels obtained for each protein was used to normalize both ICAT and dimethyl data.
Cysteine auxotrophy assay
For survival on solid plates, S. pombe strains were grown in liquid YE5S media to an OD600 of 0.5. Cells were then washed in MM, and 105 cells in 2 μl were first spotted and then serially diluted (each dilution 1/10 from the previous concentration) onto agar plates of YE5S, MM, or MM with 0.66 mM cysteine. Plates were incubated at 30°C for 3–4 days.
Abbreviations Used
- APS
5′-adenylylsulfate
- DTT
dithiothreitol
- Grx
glutaredoxin
- GSH
reduced glutathione
- H2O2
hydrogen peroxide
- IAM
iodoacetamide
- ICAT
isotope-coded affinity tag
- LC-MS/MS
liquid chromatography and analyzed by mass spectrometry
- MM
minimal medium
- PAPS
3′-phosphoadenosine-5′-phosphosulfate
- SDS-PAGE
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- TCA
trichloroacetic acid
- Trr
thioredoxin reductase
- Trx
thioredoxin
Acknowledgments
This work was supported by the Spanish Ministry of Science and Innovation (BFU2009-06933 and BFU2012-32045), PLAN E and FEDER, by the Spanish program Consolider-Ingenio 2010 Grant CSD 2007-0020, and by SGR2009-196 from Generalitat de Catalunya (Spain) to E.H. E. H. and J.A. are recipients of ICREA Academia Awards (Generalitat de Catalunya).
References
- 1.Bulleid NJ. Ellgaard L. Multiple ways to make disulfides. Trends Biochem Sci. 2011;36:485–492. doi: 10.1016/j.tibs.2011.05.004. [DOI] [PubMed] [Google Scholar]
- 2.Castillo EA. Ayte J. Chiva C. Moldon A. Carrascal M. Abian J. Jones N. Hidalgo E. Diethylmaleate activates the transcription factor Pap1 by covalent modification of critical cysteine residues. Mol Microbiol. 2002;45:243–254. doi: 10.1046/j.1365-2958.2002.03020.x. [DOI] [PubMed] [Google Scholar]
- 3.Chen D. Wilkinson CR. Watt S. Penkett CJ. Toone WM. Jones N. Bahler J. Multiple pathways differentially regulate global oxidative stress responses in fission yeast. Mol Biol Cell. 2008;19:308–317. doi: 10.1091/mbc.E07-08-0735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chiappetta G. Ndiaye S. Igbaria A. Kumar C. Vinh J. Toledano MB. Proteome screens for Cys residues oxidation: the redoxome. Methods Enzymol. 2010;473:199–216. doi: 10.1016/S0076-6879(10)73010-X. [DOI] [PubMed] [Google Scholar]
- 5.Depuydt M. Messens J. Collet JF. How proteins form disulfide bonds. Antioxid Redox Signal. 2011;15:49–66. doi: 10.1089/ars.2010.3575. [DOI] [PubMed] [Google Scholar]
- 6.Garcia-Santamarina S. Boronat S. Espadas G. Ayte J. Molina H. Hidalgo E. The oxidized thiol proteome in fission yeast—optimization of an ICAT-based method to identify H2O2-oxidized proteins. J Proteomics. 2011;74:2476–2486. doi: 10.1016/j.jprot.2011.05.030. [DOI] [PubMed] [Google Scholar]
- 7.Kumar C. Igbaria A. D'Autreaux B. Planson AG. Junot C. Godat E. Bachhawat AK. Delaunay-Moisan A. Toledano MB. Glutathione revisited: a vital function in iron metabolism and ancillary role in thiol-redox control. Embo J. 2011;30:2044–2056. doi: 10.1038/emboj.2011.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meyer Y. Buchanan BB. Vignols F. Reichheld JP. Thioredoxins and glutaredoxins: unifying elements in redox biology. Annu Rev Genet. 2009;43:335–367. doi: 10.1146/annurev-genet-102108-134201. [DOI] [PubMed] [Google Scholar]
- 9.Stewart EJ. Aslund F. Beckwith J. Disulfide bond formation in the Escherichia coli cytoplasm: an in vivo role reversal for the thioredoxins. Embo J. 1998;17:5543–5550. doi: 10.1093/emboj/17.19.5543. [DOI] [PMC free article] [PubMed] [Google Scholar]