Abstract
Aims: To identify yeast mutants that show a strong redox dependence of the ability to respire, we systematically screened a yeast deletion library for mutants that require the presence of reductants for growth on nonfermentable carbon sources. Results: Respirative growth of 44 yeast mutants was significantly improved by the addition of dithiothreitol or glutathione. Two mutants that were strongly stimulated by reductants lacked the proteins Cmc1 and Coa4. Both proteins belong to the family of “twin Cx9C” proteins present in the intermembrane space of mitochondria. Deletion of CMC1 or COA4 leads to assembly defects of cytochrome c oxidase, in particular to the lack of Cox1 and rapid degradation of Cox2 and Cox3. Interestingly, the presence of the reductants does not suppress these assembly defects and the levels of cytochrome c oxidase remain reduced. Reductants and antioxidants such as ascorbic acid rather counteract the effects of hydrogen peroxide that is produced from partially assembled cytochrome c oxidase intermediates. Innovation: Here we show that oxidative stress generated by the accumulation of partially assembled respiratory chain complexes prevents growth on carbon sources that force cells to respire. Conclusion: Defects in the assembly of cytochrome c oxidase can lead to increased production of hydrogen peroxide, which is sensed in cells and blocks their proliferation. We propose that this redox-regulated feedback regulation specifically slows down the propagation of cells carrying respiratory chain mutations in order to select for cells of high mitochondrial fitness. Antioxid. Redox Signal. 18, 1597–1612.
Introduction
The human body produces about 60 kg of ATP from ADP every day and, for example, during a marathon run, up to 30 kg/h (8). By far most of this ATP production takes place at the inner membrane of mitochondria where the enzymes of the respiratory chain use reduction equivalents derived from catabolic metabolism to generate a proton gradient that is used by the ATP synthase to phosphorylate ADP. Three enzymes contribute to the proton gradient: complex I (NADH dehydrogenase), complex III (cytochrome c reductase), and complex IV (cytochrome c oxidase). All three complexes are large oligomeric structures that are embedded into the inner membrane. They are comprised of subunits of dual genetic origin. The membrane-embedded evolutionarily conserved core subunits that carry out the catalytic activity of the complexes are typically encoded by the mitochondrial genome. Nuclear encoded subunits are associated with these core components and contribute to the stability or the regulation of these enzymes.
Innovation.
Mitochondria are a major source of reactive oxygen species (ROS). Here we show that mutants lacking the mitochondrial proteins Cmc1 or Coa4 show an impaired assembly of cytochrome c oxidase, the terminal complex of the respiratory chain. Interestingly, the inability of these strains to grow under respiratory conditions is not due to their reduced respiratory competence per se, but rather due to their strongly increased production of hydrogen peroxide. Quenching of ROS completely suppresses their growth defect. We propose that assembly intermediates of respiratory chain complexes significantly contribute to mitochondrial ROS production, thereby preventing the propagation of cells that contain malfunctional mitochondria.
The assembly of respiratory chain complexes is an intricate process for several reasons: (i) The levels of mitochondrially encoded and nuclear encoded subunits need to be coordinated, which requires a regulatory cross talk from mitochondria to the nucleus that is only poorly understood (31); (ii) the complexes of the respiratory chain contain several reactive coenzymes, such as heme groups, iron-sulfur clusters, and copper or zinc ions, which need to be inserted during the assembly process; (iii) most mitochondrially encoded subunits consist of many transmembrane spans and belong to the most hydrophobic proteins of a eukaryotic cell; to prevent their unproductive aggregation, they need to be inserted into the inner membrane in a cotranslational fashion, which is achieved by physical tethering of the mitochondrial Oxa1 insertase to mitochondrial ribosomes (25, 27, 50); (iv) possibly the largest problem is the high reactivity of the reaction centers in respiratory chain complexes; assembly intermediates of these enzymes might lead to the direct transfer of single electrons to oxygen giving rise to the production of superoxide and other reactive oxygen species (ROS) (24, 29, 39). Although the potentially hazardous role of these intermediates was proposed a long time ago (46), experimental evidence for a physiological relevance of these intermediates was so far only described following exposure of cells to externally added hydrogen peroxide (26, 53).
Mainly by genetic studies carried out over the last three decades in the yeast Saccharomyces cerevisiae, a large number of assembly factors have been identified that coordinate the biogenesis of respiratory chain complexes (11, 52). Despite their early discovery and the relevance of respiratory chain assembly for human diseases, the function of many of these factors is still largely unclear. One ill-defined group of these assembly factors are the “twin Cx9C” proteins. They are characterized by two sequence motifs each containing two cysteine residues that are separated by nine amino acid residues. These proteins are located in the intermembrane space (IMS) of mitochondria. S. cerevisiae contains 14 members of this family, most of which are critical for the accumulation of normal amounts of cytochrome c oxidase and for growth on nonfermentable carbon sources (9, 32). The cysteine residues in these proteins are required for their targeting to mitochondria; during the import reaction, these proteins interact with the oxidoreductase Mia40 in the IMS, which converts the reduced thiols in the imported precursor into oxidized disulfides that are found in the mature forms of these proteins (3, 10, 36, 41).
In order to identify a role of redox regulation for the biogenesis of the respiratory chain, we performed an unbiased screen to identify yeast mutants in which the competence to respire is increased by the reductants dithiothreitol (DTT) and/or reduced glutathione (GSH). Thereby, we identified two mutants lacking “twin Cx9C” proteins whose respiratory deficiency was suppressed by DTT and GSH. Interestingly, the reductants did not cure the primary defects in respiratory chain assembly of these strains. However, they counteracted a toxic effect that is presumably due to increased oxidative stress resulting from the presence of assembly intermediates of cytochrome c oxidase. Our data suggest that defects in the function or biogenesis of mitochondrial enzymes lead to the respiration-induced production of hydrogen peroxide, which can impair cell growth. Based on the observations of this study we propose that an intracellular redox-dependent signaling pathway dampens the propagation of mutants that produce increased levels of mitochondria-derived oxidative stress.
Results
The growth defect on glycerol of many yeast mutants is suppressed by addition of reductants
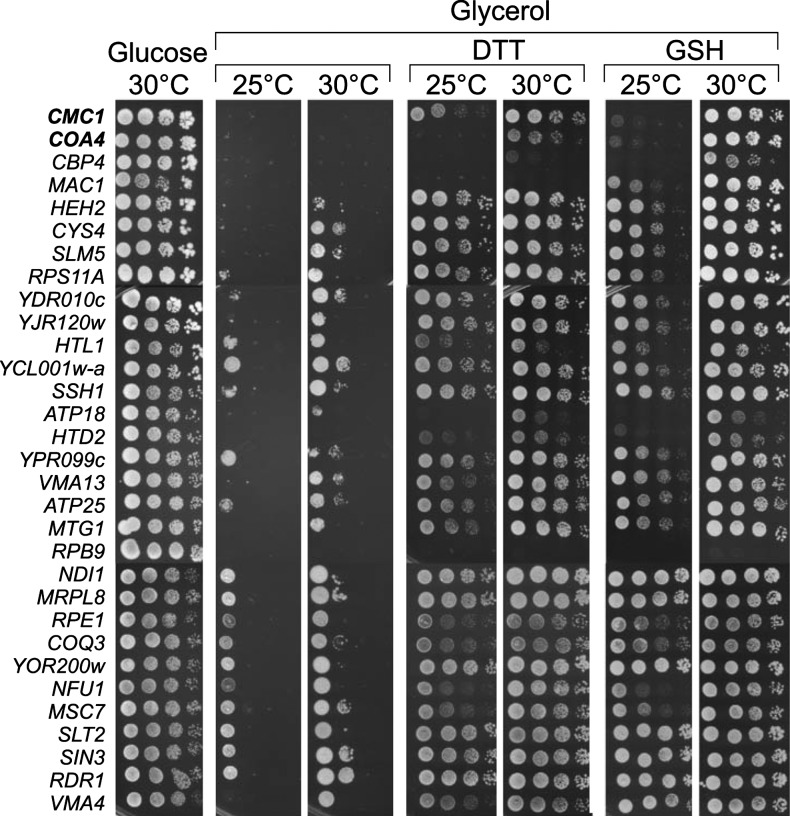
To screen for yeast mutants that depend on externally added reductants in order to grow on nonfermentable carbon sources, we performed a genome-wide unbiased growth analysis. To this end we used a subset of the yeast Matα deletion library which contained 386 deletion mutants that showed a reduced growth on the nonfermentable carbon source glycerol (35). These strains and a wild type for control were grown overnight in the presence of the fermentable carbon source glucose and then spotted with a pinning tool onto plates containing the nonfermentable carbon source glycerol in the absence or presence of 5 mM GSH or DTT. The plates were incubated at either 25°C or 30°C and the growth of the colonies was compared. The growth of 44 mutants was stronger on at least two of the four reductant-containing plates than on the control plates (Table 1). Of these strains 18 (40%) lacked mitochondrial proteins, most of which are components required for the biogenesis or function of the respiratory chain, including many factors required for mitochondrial protein synthesis. The second largest group of proteins (nine strains, i.e., 20%) comprises nuclear factors that influence transcription. The potential growth stimulation by GSH and DTT for the mutants that showed the strongest stimulation by reductants was further confirmed by drop dilution assays (Fig. 1). The minor differences between the growth phenotypes presented in Table 1 and Figure 1 are presumably due to the different conditions of the precultures from which the plates were prepared (glucose-based, largely stationary cultures for Table 1 and galactose-based exponentially growing cells for Fig. 1). In summary, we conclude that addition of reductants stimulates respiratory growth of many yeast mutants, in particular of those showing defects in gene expression.
Table 1.
Growth Stimulation of Yeast Mutants on YPG in the Presence of 5 μM Reduced Glutathione or Dithiothreitol
| ORF | Gene | YPG 25°C | YPG 30°C | GSH 25°C | GSH 30°C | DTT 25°C | DTT 30°C | Function |
|---|---|---|---|---|---|---|---|---|
| Respiratory chain function and assembly | ||||||||
| YLR218C | COA4 | Poor | Poor | Strong | Strong | Strong | Strong | Twin cx9c protein involved in cytochrome c oxidase assembly or stability |
| YKL137W | CMC1 | None | None | Strong | Strong | Strong | Strong | Twin cx9c protein involved in cytochrome c oxidase assembly or stability |
| YML120C | NDI1 | Poor | Reduced | Minor | Minor | Minor | Minor | Internal NADH-ubi oxidoreductase |
| YDL107W | MSS2 | Reduced | Reduced | Strong | None | Strong | None | Cytochrome c oxidase assembly |
| YER058W | PET117 | Reduced | Reduced | Strong | None | Minor | None | Cytochrome c oxidase assembly |
| YOL096C | COQ3 | Poor | Reduced | None | Minor | None | Minor | Ubiquinone biosynthesis |
| Mitochondrial protein synthesis | ||||||||
| YDR025W | RPS11A | Reduced | Reduced | Strong | Strong | Strong | Strong | Mitochondrial ribosome SSU |
| YJL063C | MRPL8 | Poor | Reduced | Strong | Strong | Minor | Strong | Mitochondrial ribosome LSU |
| YMR097C | MTG1 | Poor | Reduced | Minor | Minor | Minor | Minor | GTPase required for biogenesis of mitochondrial ribosomes |
| YMR098C | ATP25 | None | Reduced | Minor | Minor | Minor | Minor | Required for Atp9 synthesis |
| YOR200W | Poor | Reduced | Minor | Minor | Minor | Minor | Overlaps with MRM1, mitochondrial ribose rRNA methyltransferase | |
| YCR024C | SLM5 | Reduced | Reduced | Minor | Strong | None | Minor | Mitochondrial asparaginyl-tRNA synthetase |
| YJR113C | RSM7 | Reduced | Reduced | Strong | Minor | None | None | Mitochondrial ribosome SSU |
| Other metabolic enzymes | ||||||||
| YJL121C | RPE1 | Reduced | Reduced | Strong | Minor | Strong | Minor | D-ribulose-5-phosphate 3-epimerase |
| YIL155C | GUT2 | Reduced | Reduced | Strong | Minor | Strong | None | Mitochondrial glycerol-3-phosphate dehydrogenase |
| YGR155W | CYS4 | Poor | Poor | Strong | Strong | Minor | None | Cystathionine beta-synthase |
| YJL120W | Reduced | Reduced | Minor | None | Strong | None | Partially overlaps the verified gene YJL121C/RPE1 | |
| YDL114W | Reduced | Reduced | Strong | None | Strong | None | Similarity to acyl-carrier-protein reductases | |
| YHR067W | HTD2 | Poor | Poor | Strong | None | Minor | Mitochondrial 3-hydroxyacyl-thioester dehydratase | |
| YDR148C | KGD2 | None | Poor | Minor | None | Minor | None | Mitochondrial alpha-ketoglutarate dehydrogenase complex |
| YIL125W | KGD1 | None | Poor | Minor | None | Minor | None | Component of the mitochondrial alpha-ketoglutarate dehydrogenase complex |
| Nuclear gene expression | ||||||||
| YDR010C | Reduced | Reduced | Strong | Strong | Strong | Strong | Dubious, overlaps with Gal3 transcription factor | |
| YJL176C | SWI3 | Poor | Reduced | Strong | Minor | Minor | None | SWI remodeling complex |
| YCR020W-B | HTL1 | Reduced | Reduced | Minor | Minor | Minor | Minor | Component of the RSC chromatin remodeling complex |
| YOR380W | RDR1 | Reduced | Reduced | None | Minor | None | Strong | Transcriptional repressor nucleus |
| YOL051W | GAL11 | Reduced | Reduced | Strong | Minor | None | None | Transcription nucleus |
| YPL254W | HFI1 | Reduced | Reduced | Strong | None | Minor | None | SAGA complex nucleus |
| YGL066W | SGF73 | Reduced | Reduced | Minor | Minor | Minor | None | SAGA complex nucleus |
| YOL004W | SIN3 | Reduced | Reduced | None | Minor | None | Minor | Histone deacetylase |
| YGL070C | RPB9 | None | None | None | Minor | None | Minor | RNA polymerase II subunit B12.6 nucleus |
| Vacuolar transporters | ||||||||
| YDL128w | VCX1 | Reduced | Reduced | Strong | None | Strong | None | Vacuolar membrane antiporter with Ca2+/H+and K+/H+exchange activity |
| YOR332W | VMA4 | Reduced | Reduced | None | Strong | None | Minor | V-ATPase |
| YKL080W | VMA5 | Reduced | Reduced | Strong | None | None | Minor | V-ATPase |
| YPR036W | VMA13 | None | Reduced | None | Minor | None | Strong | V-ATPase |
| Other functions | ||||||||
| YBR283C | SSH1 | Reduced | Reduced | Strong | Strong | Strong | Strong | Sec61 homolog |
| YHR030C | SLT2 | Reduced | Reduced | Strong | Strong | Strong | Strong | MAP kinase, regulates autophagy |
| YIL153W | RRD1 | Reduced | Reduced | Strong | Minor | Strong | None | Peptidyl-prolyl cis/trans-isomerase |
| YKL040C | NFU1 | Poor | Reduced | Minor | Strong | None | None | Iron-sulfur cluster formation in mitochondria |
| YBL082C | ALG3 | Reduced | Reduced | Strong | None | Minor | None | Dolichol-P-Man dependent alpha(1–3) mannosyltransferase |
| YJL101C | GSH1 | Poor | Poor | Strong | None | Minor | None | Glutathion-synthase |
| Function unknown | ||||||||
| YCL001W-A | Reduced | Reduced | Strong | Strong | Strong | Strong | Function unknown | |
| YHR039C | MSC7 | Reduced | Reduced | Strong | None | Minor | Strong | Function unknown, ER-associated |
| YJR120W | Reduced | Reduced | Minor | Strong | None | Strong | Protein of unknown function; transporters Aus1p and Pdr11p | |
| YDR458C | HEH2 | Reduced | Reduced | None | Strong | Minor | None | Inner nuclear membrane (INM) protein, function unknown |
GSH, reduced glutathione.
FIG. 1.
Dithiothreitol (DTT) and reduced glutathione (GSH) improve respiratory growth of many yeast mutants. Strains showing a strong reductant-dependent growth stimulation in a systematic screen were grown overnight in galactose-containing medium and adjusted to optical density (OD) 1. Tenfold serial dilutions were spotted onto YPD (glucose) or YPG (glycerol) plates containing the indicated amounts of DTT and GSH. Plates were incubated for 2 (glucose) or 4 (glycerol) days at 25°C or 30°C.
Reductants suppress the growth defect of mutants lacking Cmc1 and Coa4 on nonfermentable carbon sources
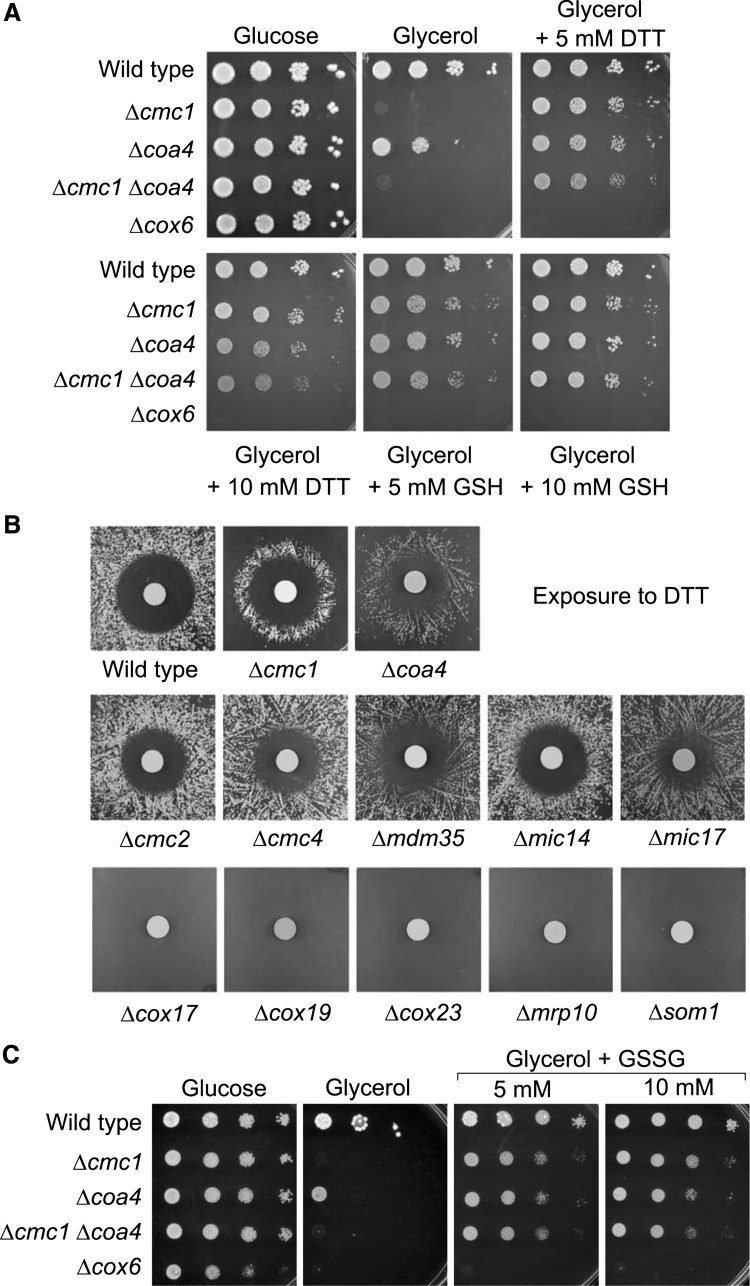
Two strains that showed a strong respiratory growth stimulation by GSH and DTT caught our attention because they lacked genes for the “twin Cx9C” proteins Cmc1 and Coa4, which were shown before to play a role in the biogenesis of cytochrome c oxidase (7, 22, 23, 32). These strains showed no (Δcmc1) or poor (Δcoa4) growth on glycerol but were strongly stimulated by either GSH or DTT. Even growth of a double mutant in which both Cmc1 and Coa4 are absent can be rescued by GSH and DTT (Fig. 2A). Reductant-dependent growth on glycerol was not observed for a mutant lacking the structural Cox6 subunit of cytochrome c oxidase.
FIG. 2.
Reductants stimulate the growth of Δcmc1 and Δcoa4 mutants on glycerol. (A) Tenfold serial dilutions were spotted onto YPD (glucose) or YPG (glycerol) plates containing the indicated amounts of DTT and GSH. (B) The strains indicated were grown overnight in YPGal and equally spread on YPG plates. A filter dish was placed on the plate to which 10 μl 3 M DTT was added. The plates were incubated for 4 days at 30°C. (C) Drop dilution experiment as shown in A except that here oxidized glutathione (GSSG) was added to the plates as indicated.
The positive effect of DTT on the growth of Δcmc1 and Δcoa4 cells was apparent when DTT-soaked filter papers were placed on a lawn of cells on YPG plates (Fig. 2B). The high DTT concentration directly around the filter prevented cell growth even of wild type leading to a colony-free halo. When Δcmc1 and Δcoa4 cells were used, a ring of colonies was observed that grew where the DTT concentration was neither too high nor too low. Ring-like halos were not observed with mutants lacking other “twin Cx9C” proteins (Fig. 2B).
The positive effect of the added reductants might be explained by a direct reduction of cellular thiols or, less directly, by an increase of the cellular capacity to buffer redox changes. To differentiate between both effects we next tested whether the addition of oxidized glutathione (GSSG) suppresses the growth defect of the mutants as well. As shown in Figure 2C, GSSG showed a strongly stimulating effect on respiratory growth of Δcmc1 and Δcoa4 cells. This suggests that it is not the direct reducing activity of the added reagent per se that is required as the uptake of GSSG, which can be reduced intracellularly to GSH by glutathione reductase in an NADPH-dependent process, obviously is sufficient to render the cells respiration-competent.
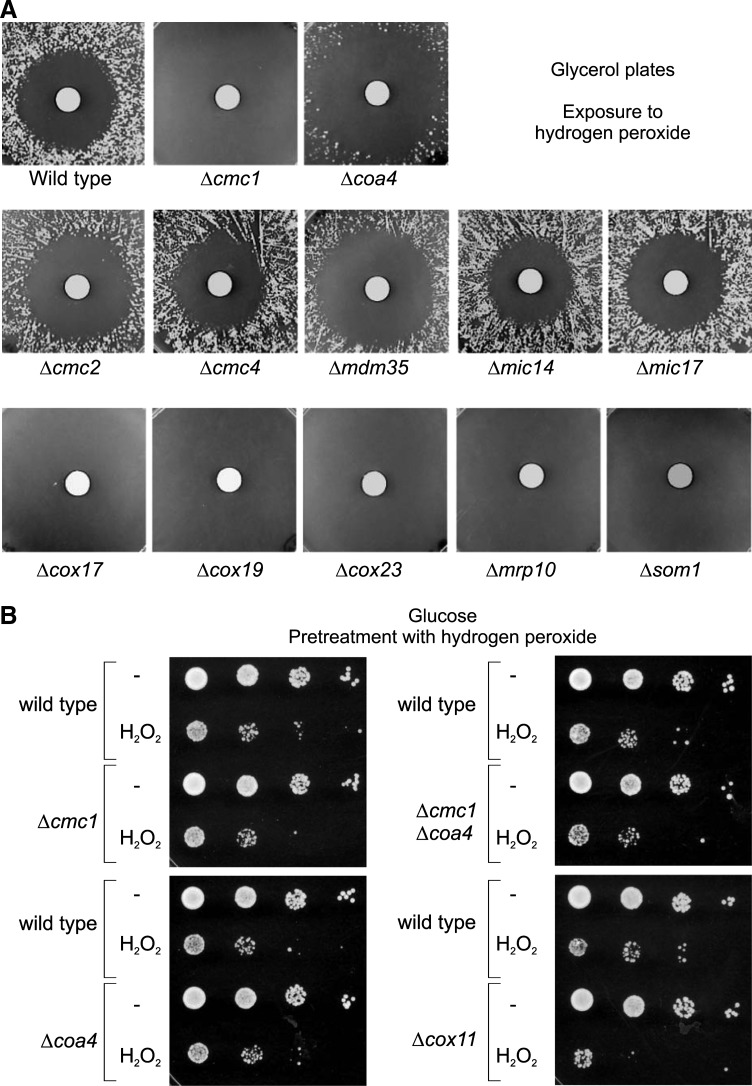
None of the yeast mutants lacking “twin Cx9C” proteins was suppressed upon exposure to the oxidizing reagent hydrogen peroxide (Fig. 3A). It was shown before that some mutants like Δcox11 that lead to defects in the assembly of cytochrome c oxidase show an increased toxicity upon incubation with hydrogen peroxide (26, 53). However, we did not find an increased hydrogen peroxide toxicity of Δcmc1 and Δcoa4 cells as long as they were cultured on glucose-containing medium (Fig. 3B).
FIG. 3.
Δcmc1 and Δcoa4 mutants show no strongly increased sensitivity to external hydrogen peroxide. (A) The strains indicated were grown overnight in YPGal and equally spread on YPG plates. A filter dish was placed on the plate to which 5 μl 30% hydrogen peroxide was added. The plates were incubated for 4 days at 30°C. The halos found in Δcoa4, Δcmc2, and Δmdm35 are somewhat larger than in wild-type cells, indicating that these mutants might be hypersensitive to oxidative stress. (B) Cells were pregrown on glucose-containing medium, adjusted to an OD of 0.5, and exposed to 5 mM hydrogen peroxide for 2 h. To assess the toxicity of hydrogen peroxide, serial dilutions of the samples were spotted on YPD plates.
Cmc1 or Coa4 exhibit specific nonredundant functions in mitochondria
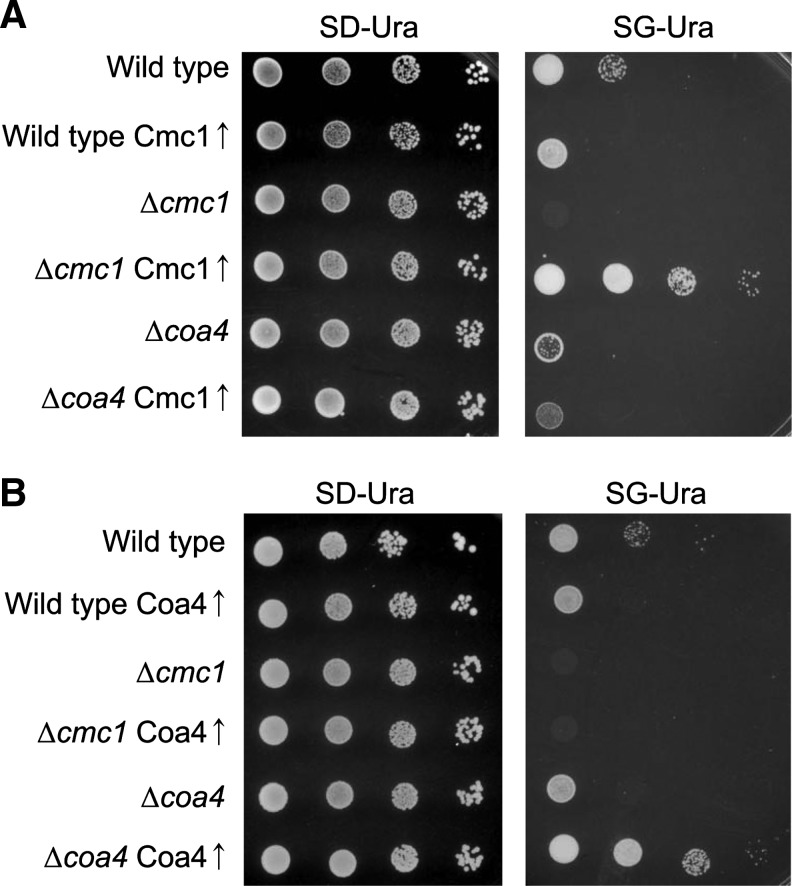
The similarities of Cmc1 and Coa4 in protein structure and in the growth phenotypes of the deletion mutants led us to test whether overexpression of one of the proteins can suppress the defects in the deletion mutants that lack the other protein (Fig. 4A, B). Whereas the overexpression plasmids carrying CMC1 and COA4 clearly suppressed the defects in Δcmc1 and Δcoa4 cells, respectively, they did not show any effect on the other mutant. Hence, we conclude that Cmc1 and Coa4 carry out nonredundant functions in mitochondria.
FIG. 4.
Cmc1 and Coa4 exhibit nonredundant functions in mitochondria. CMC1 (A) and COA4 (B) were cloned into a multi copy plasmid and expressed in the mutants indicated. Empty plasmids were transformed for control. Growth was tested by drop dilution assays as described in Figure 1.
Mutants lacking Cmc1 or Coa4 show reduced levels of cytochrome c oxidase
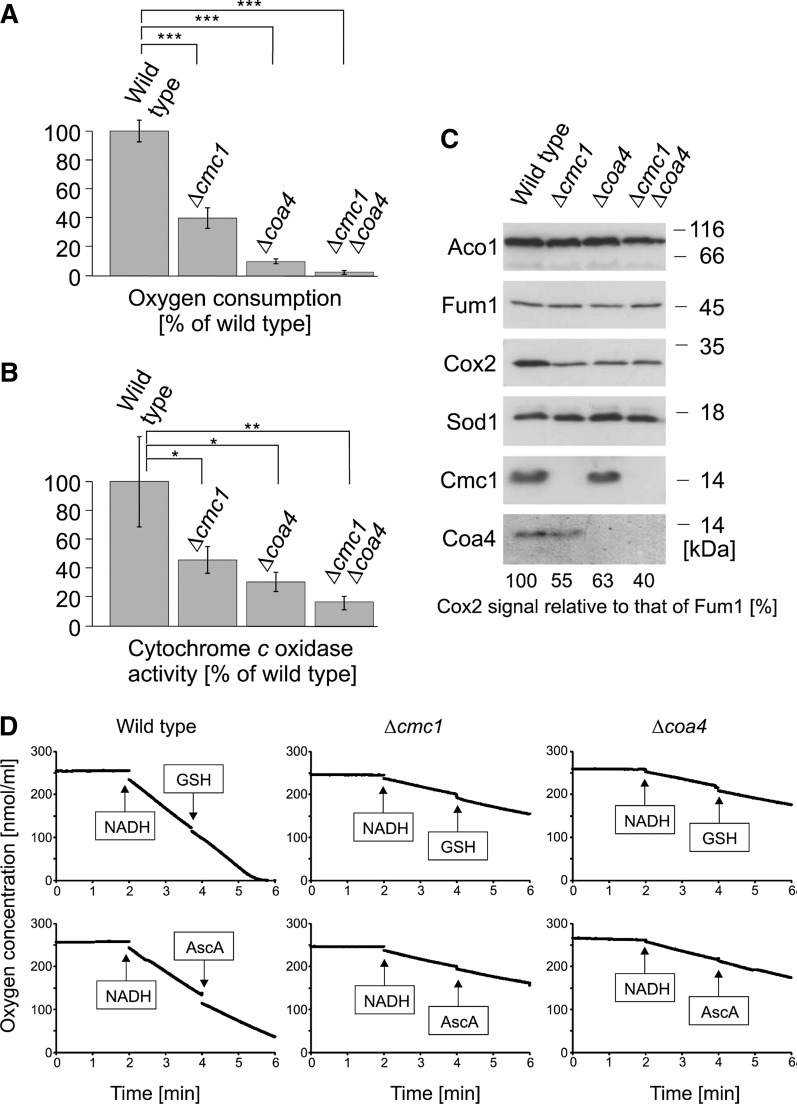
The growth defect of Δcmc1 and Δcoa4 mutants on nonfermentable carbon sources pointed to a defect in mitochondrial respiration in these strains. Indeed, oxygen consumption measurements with isolated mitochondria confirmed strongly reduced respiration levels in the mutants, particularly in Δcoa4 mitochondria (Fig. 5A).
FIG. 5.
Δcmc1 and Δcoa4 mutants show reduced levels of cytochrome c oxidase. (A) Mitochondria (100 μg) isolated from the indicated strains were incubated in 0.6 M sorbitol, 1 mM ethylenediaminetetraacetic acid (EDTA), 1 mM MgCl2, and 20 mM HEPES pH 7.4 in a reaction chamber containing a Clark electrode, and oxygen consumption was measured over time. Mean values and standard deviations of three independent measurements are shown. (B) Activity of cytochrome c oxidase was measured in mitochondrial extracts by following the oxidation of reduced cytochrome c over time. Mean values and standard deviations of three independent measurements are shown. (C) Mitochondrial proteins (100 μg) were analyzed by Western blotting using antibodies against the indicated proteins. The signals of Cox2 and Fum1 were quantified and the levels of Cox2, corrected for the levels of Fum1, are shown in the figure. (D) The addition of GSH or ascorbic acid (AscA) does not stimulate oxygen consumption of isolated mitochondria. Oxygen concentrations of mitochondria-containing solutions were recorded before and after the addition of NADH, GSH, and AscA. Shown are mean values of two measurements per sample. Asterisks indicate confidence levels as follows: *p<0.05, **p<0.01, and ***p<0.001.
It was previously demonstrated that Cmc1 and Coa4 play a direct or indirect role in the assembly of cytochrome c oxidase (7, 22, 23). We therefore analyzed the activity of cytochrome c oxidase in Δcmc1 and Δcoa4 mitochondria (Fig. 5B). This revealed that in the absence of Cmc1 about 40% of the cytochrome c oxidase activity of wild-type mitochondria is found, and in the absence of Coa4 only about 25%. This correlates with Western blot signals of Cox2, one of the mitochondrially encoded core subunits of cytochrome c oxidase, which was also diminished, but not absent, in Δcmc1 and Δcoa4 mitochondria (Fig. 5C). Oxygen consumption—thus, respiration—was not stimulated in mitochondria of these strains by addition of GSH or the antioxidant ascorbic acid (Fig. 5D), indicating that reductants had no direct effect on the performance of the respiratory chain in any of these mitochondria.
In summary, we find reduced levels of cytochrome c oxidase in both mutants, although it should be noted that cytochrome c oxidase is still present in these strains and still exhibits residual activity. Interestingly, respiratory activity was less affected in Δcmc1 than in Δcoa4 mitochondria although its growth defect on glycerol was much more pronounced. This indicated that the reduced activity of cytochrome c oxidase might not be exclusively responsible for the growth arrest.
Reductants do not increase the levels of cytochrome c oxidase in Δcmc1 and Δcoa4 mutants
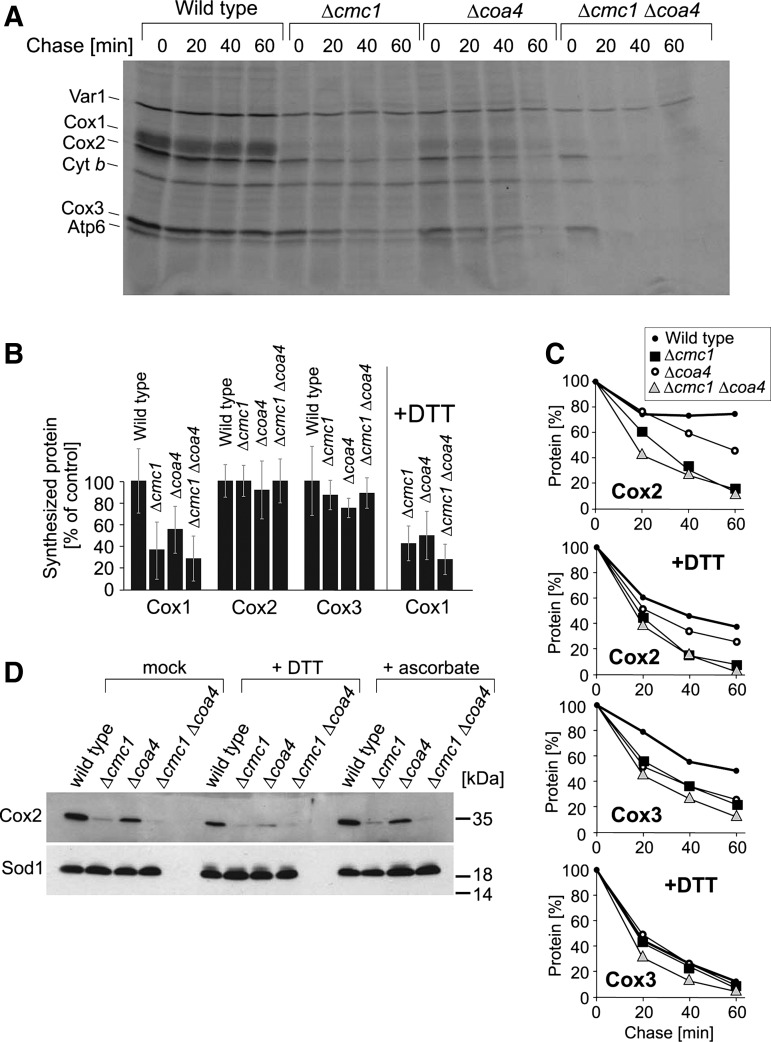
The suppression of the growth phenotype of the Δcmc1 and Δcoa4 mutants on nonfermentable media by DTT and GSH might suggest that reductants can restore normal cytochrome c oxidase levels in these strains. Diminished levels of cytochrome c oxidase might be due to a reduced production or an increased degradation of the enzyme. To monitor the synthesis of cytochrome c oxidase subunits as well as their stability, we radiolabeled mitochondrial translation products in whole yeast cells and followed their stability in subsequent chase reactions (Fig. 6A). In wild-type cells, all three mitochondria-encoded subunits of cytochrome c oxidase were efficiently synthesized and remained relatively stable during the 60 min of the postlabeling period. In contrast, Cox1 was synthesized only at reduced levels in the Δcmc1 and Δcoa4 cells (Fig. 6A, B). Diminished levels of Cox1 synthesis are characteristic for strains with defects in the assembly of cytochrome c oxidase (5, 21, 37, 44, 45). The levels of Cox1 were not increased in the presence of DTT, indicating that DTT did not suppress the assembly defect in these strains (Fig. 6B). Cox2 and Cox3, the two other subunits of cytochrome c oxidase, were synthesized at normal levels. However, their amounts rapidly declined during the chase reaction, most severely in the double mutant (Fig. 6A, C). The presence of DTT did not suppress the instability of Cox2 and Cox3, again indicating that reductants do not improve biogenesis of cytochrome c oxidase in these strains. This is further supported by Western blots (Fig. 6D), which revealed the presence of strongly reduced levels of Cox2 in the Δcmc1 and Δcoa4 mutants regardless of whether the cells were grown in the absence or the presence of DTT or ascorbic acid. In summary, although the presence of DTT suppressed the growth defects of Δcmc1 and Δcoa4 mutants, it did not alleviate the reduced biogenesis of cytochrome c oxidase in these strains. This suggests that the growth arrest is caused by side effects of these assembly mutants rather than directly by the reduction of the levels of cytochrome c oxidase.
FIG. 6.
The biogenesis of cytochrome c oxidase is impaired in Δcmc1 and Δcoa4 mutants. (A) Mitochondrial translation products were radiolabeled with [35S]-methionine for 15 min in cells after inhibition of cytosolic protein synthesis with cycloheximide. Labeling was stopped by washing of cells and addition of an excess of nonradioactive methionine. Cells were further incubated for the time periods indicated (“chase”), lysed, and analyzed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and autoradiography. (B, C) The experiment shown in (A) was repeated three times in the absence or presence of 1 mM DTT. The levels of newly synthesized Cox1, Cox2, and Cox3 normalized to the levels of the soluble matrix protein Var1 at chase time 0 were quantified (B). In (C), mean values of the intensities of Cox2 and Cox3 signals are shown relative to the values found at chase time 0. (D) Western blot signals of Cox2 and Sod1 of cells grown in the absence or presence of 5 mM DTT or ascorbic acid are shown. It should be noted that the addition of DTT or ascorbic acid did not increase the levels of Cox2 in the mutants.
Mitochondria of Δcmc1 and Δcoa4 mutants produce increased levels of hydrogen peroxide
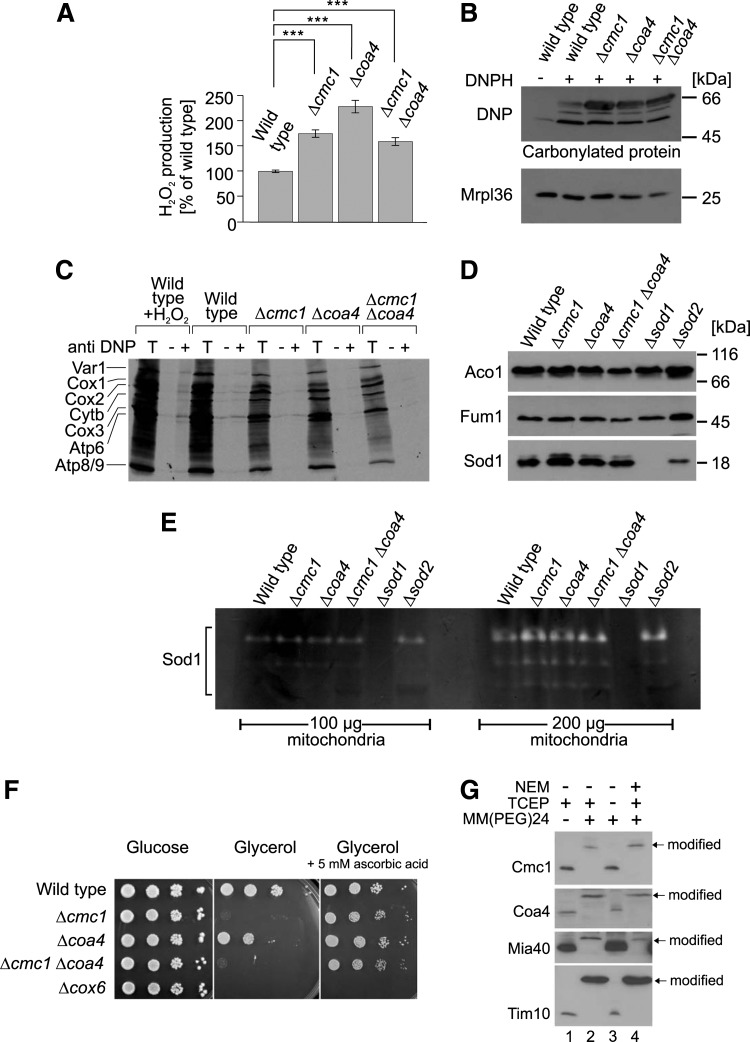
We speculated whether the problems in respiratory chain assembly in the Δcmc1 and Δcoa4 mutants result in an increased accumulation of ROS, which reduces their viability. To test this, we incubated mitochondria in the presence of succinate and measured the amount of hydrogen peroxide produced using a fluorescence-based Amplex red assay (Fig. 7A). Indeed we found significantly increased levels of hydrogen peroxide produced in these mutants. Moreover, we observed an increased carbonylation of mitochondrial proteins, which is indicative for oxidation-driven damage in these strains (42) (Fig. 7B). However, we do not regard it as likely that the rapid degradation of mitochondrial translation products that was observed in the Δcmc1 and Δcoa4 mitochondria was due to their oxidative damage since immunoprecipitation experiments with dinitrophenyl-specific antibodies after dinitrophenylhydrazone-modification did not show any elevated levels of carbonylated translation products in these mutants (Fig. 7C). Thus, we suspect that the oxidative damage is rather a consequence of the cytochrome c oxidase defect rather than its cause.
FIG. 7.
Δcmc1 and Δcoa4 mutants suffer from increased oxidative stress. (A) The production of hydrogen peroxide in the presence of succinate was measured with mitochondria isolated from the indicated strains. Mean values of eight independent measurements are shown. (B) Δcmc1 and Δcoa4 mitochondria show increased levels of protein carbonylation. Mitochondria were incubated with dinitrophenylhydrazine to modify carbonylated proteins which were then detected by Western blotting with a dinitrophenyl-specific antibody and quantified. (C) Translation products were radiolabeled in isolated mitochondria for 1 h. Mitochondria were either treated with 50 mM hydrogen peroxide or mock treated for 10 min. Mitochondria were reisolated and lysed, and proteins were treated with dinitrophenylhydrazine. Samples were either directly applied to the gel (5%) or after immunoprecipitation with dinitrophenyl-specific antibodies (DNP) or preimmune serum. (D) Western blot of mitochondria isolated from the strains indicated show that steady-state levels of Sod1 are not altered in Δcmc1 and Δcoa4 cells. (E) Superoxide dismutase activity is not considerably changed in Δcmc1 and Δcoa4 mitochondria. Activities of Sod1 and Sod2 were determined using an in gel activity assay that relied on a superoxide-dependent nitro blue tetrazolium staining. (F) Ascorbic acid suppresses growth defect of Δcmc1 and Δcoa4 mutants. Tenfold serial dilutions of yeast cultures were dropped on YP plates containing or lacking 5 mM ascorbic acid. (G) Cmc1 and Coa4 are oxidized at steady state. Wild-type cells were split into four samples and treated with N-ethylmaleimide (NEM) (to block free thiols), triscarboxyethyl phosphine (TCEP) (to reduce disulfide bonds) and MM(PEG)24 (adding about 1 kDa to each free thiol) in subsequent reactions as indicated. Samples were analyzed by Western blotting. It should be noted that Coa4 is slightly shifted by the addition of MM(PEG)24 (lane 3), which is explained by the presence of two reduced cysteine residues in addition to the oxidized twin Cx9C motif. ***p<0.001.
Mitochondria contain two superoxide dismutases that produce hydrogen peroxide from superoxide radicals: Sod1 in the IMS and Sod2 in the matrix. Increased levels of Sod1 in Δcmc1 mitochondria were reported before (22), but no changes in the protein levels of Sod1 or Sod2 were observed with the strains used in this study (Fig. 7D). Activity gels revealed comparable activities of superoxide dismutase activity in wild type, Δcmc1, and Δcoa4 mitochondria excluding that the different levels of hydrogen peroxide produced by the different mitochondria are due to different superoxide dismutase activities (Fig. 7E). Thus, it appears more likely that the increased ROS levels caused by the defects in the assembly of the respiratory chain complexes are responsible for the observed growth arrest. In order to test this hypothesis, we analyzed cell growth in the presence of the antioxidant ascorbic acid (Fig. 7F). Interestingly, ascorbic acid completely suppressed the growth defect of the mutants. Although ascorbic acid treatment can reduce sulfenylated thiols, which might prevent disulfide formation (38, 55), the strong suppression by ascorbic acid suggests that quenching of ROS rather than the reduction of disulfides allows growth of the Δcmc1 and Δcoa4 mutants on nonfermentable carbon sources.
It remained possible that Cmc1 and Coa4 serve as scavengers for hydrogen peroxide in the IMS since hydrogen peroxide might directly react with reduced cysteine residues in these proteins. We therefore determined the redox state of Cmc1 and Coa4 in wild-type mitochondria using the alkylating reagent MM(PEG)24. When mitochondrial proteins were denatured and reduced with the thiol-free reductant triscarboxyethyl phosphine (TCEP), treatment with MM(PEG)24 caused a large size shift of Cmc1 and Coa4 (Fig. 7G). However, in the absence of TCEP, both proteins were not shifted, indicating that their thiol residues were oxidized. In a “reverse shift” setup, that is, when samples were initially treated with N-ethylmaleimide (NEM) to block all accessible thiols, then reduced and treated with MM(PEG)24, Cmc1 and Coa4 shifted to a larger size. Thus, Cmc1 and Coa4, like Mia40 or Tim10, are present in an oxidized state in the IMS, making a direct function as quenchers of hydrogen peroxide unlikely.
The Δcoa4 mutant shows increased hydrogen peroxide levels in the cytosol
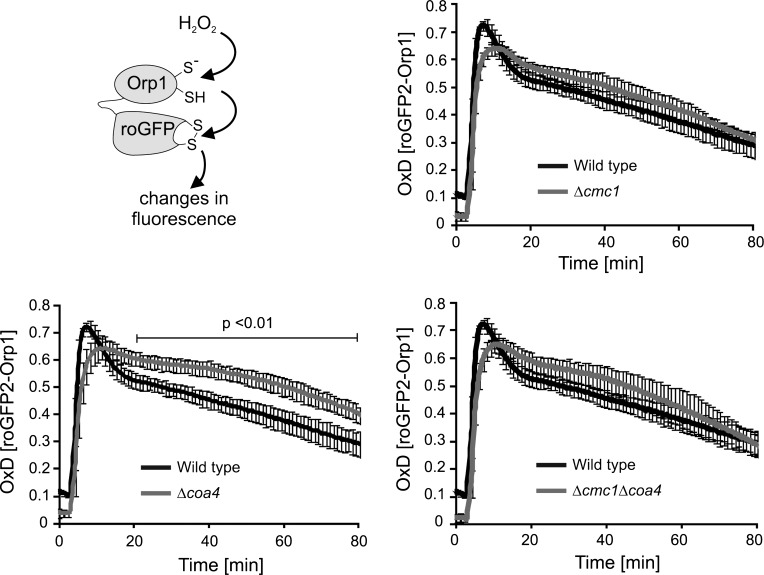
Next we tested whether the increased hydrogen peroxide levels produced in Δcmc1 and Δcoa4 mitochondria influence the redox milieu outside of mitochondria in these cells. To this end, we made use of a roGFP2-Orp1-sensor protein that was shown to serve as a tool to measure dynamic changes in cytosolic hydrogen peroxide levels (20, 40). We expressed this sensor protein in the cytosol of wild-type and mutant cells, which were then grown on galactose-based medium (i.e., fermentation conditions). Then, the cells were challenged by the addition of 5 mM hydrogen peroxide to the medium and the changes in the redox state of the sensor were followed by monitoring fluorescence in a plate-reader. In all strains, hydrogen peroxide treatment efficiently oxidized the sensor. However, there were considerable differences in the rate of sensor recovery between the strains. The sensor in the Δcoa4 cells recovered significantly slower compared to wild type, while in the Δcmc1 and Δcoa4Δcmc1 cells, probe recovery was slightly slower compared to wild type although not significantly so (Fig. 8).
FIG. 8.
Delayed recovery of a roGFP2-Orp1 probe in the absence of Coa4. The roGFP2-Orp1 probe was expressed in the cytosol of wt, Δcoa4, Δcmc1, and Δcoa4Δcmc1 cells. Probe oxidation was induced by the addition of 5 mM hydrogen peroxide. Significantly slower probe recovery was observed in Δcoa4 cells compared to wild-type cells.
While exact interpretation of the probe recovery data is complicated due to the oxidation state of the sensor being a product of both hydrogen peroxide-mediated oxidation and thioredoxin-mediated reduction, the data correlate well with both the differences in hydrogen peroxide production measured with the Amplex Red assay in Figure 7A and the growth phenotypes in Figure 9. Therefore, probably due to pre-existing increased endogeous hydrogen peroxide production, the capacity of the cells, particulary Δcoa4, to deal with additional exogenously added hydrogen peroxide is likely decreased.
FIG. 9.
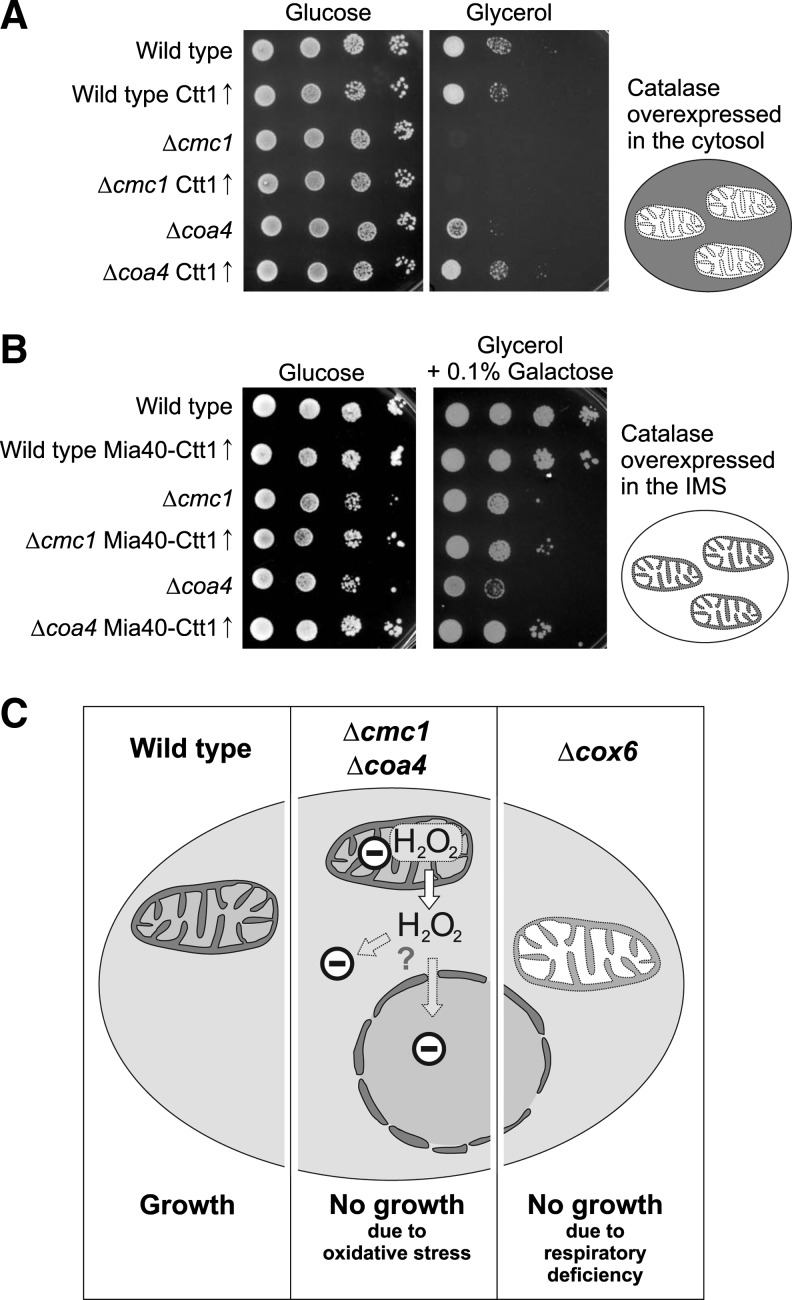
The accumulation of hydrogen peroxide in the cytosol contributes to the growth arrest of the Δcoa4 mutant. (A) The cytosolic catalase Ctt1 was expressed from a multi copy plasmid in the strains indicated. Note that overexpression of Ctt1 suppresses the defect observed in Δcoa4 but not in Δcmc1 cells. (B) Overexpression of Ctt1 in the intermembrane space (IMS) of mitochondria partially suppressed the growth defects of Δcmc1 and Δcoa4 mutants. (C) Model illustrating the growth arrest observed in Δcmc1 and Δcoa4 mutants. We propose that the increased production of hydrogen peroxide prevents growth of these strains under respiratory conditions. Hydrogen peroxide might be sensed outside the mitochondria, for example, to influence gene expression in the nucleus or Yca1-induced apoptosis in the cytoplasm.
It should also be noted that these measurements were carried out under fermentative conditions, thus when the respiratory chain was not fully active. The inability of the mutants to grow on glycerol prevented us from measuring the hydrogen peroxide levels under respirative conditions since nonproliferating cultures show strongly increased hydrogen peroxide levels at stationary phase (not shown).
Overexpression of cytosolic catalase suppresses growth defect of Δcoa4 mutants
Next we asked whether the increased hydrogen peroxide levels in the cytosol contribute to the growth defect observed in the Δcmc1 and Δcoa4 mutants. To reduce the levels of hydrogen peroxide specifically in the cytosol, we overexpressed the cytosolic catalase Ctt1 in these strains from a multi copy plasmid. This partially restored growth of the Δcoa4, but not of the Δcmc1 mutant (Fig. 9A). However, a slight improvement of respiratory growth of both mutants was observed when a fusion protein was expressed, which targeted Ctt1 to the IMS (Mia40-Ctt1, Fig. 9B). From this we conclude that the production of hydrogen peroxide in the absence of Cmc1 and Coa4 contributes to their inability to grow on nonfermentable carbon sources. At least in case of the Δcoa4 strain, the respiration-induced growth arrest is at least partially caused by cytosolic hydrogen peroxide, which was counteracted by overexpression of cytosolic catalase.
Discussion
The respiratory chain of mitochondria is a major source of ROS in eukaryotic cells (1, 13). Although there is still some debate about the degree of ROS production under physiological conditions, it is widely accepted that enzymes of the respiratory chain can transfer single electrons onto oxygen, giving rise to the production of superoxide, which is rapidly converted to hydrogen peroxide by superoxide dismutases. Hydrogen peroxide plays a physiological role as a spatially and temporally dynamic signaling molecule in the cell (2, 14). Under pathological conditions, the degree of mitochondrial ROS production can be significantly increased, as was reported, for example, for several neurodegenerative diseases, including Parkinson's disease (6), Alzheimer's disease (51), and amyotrophic lateral sclerosis (30).
To better understand the relevance of redox processes for mitochondrial activity, we performed a genome-wide mutant screen to identify yeast proteins that under respiring conditions are essential for cell viability unless reducing reagents like DTT or GSH are externally added. We thereby identified mutants that grow significantly better in the presence of reductants than in their absence. One group of mutants lacked nuclear components, in particular transcription factors and enzymes that modify or remodel histone complexes. For example, two components of the SAGA (Spt-Ada-Gcn5-Acyltransferase) complex were identified, which is critical for the metabolic adaptation to respiratory growth conditions (47). This suggests that an increased capacity of cellular redox buffer improves the ability of yeast cells to grow on nonfermentable conditions. The sensing of mitochondria-derived oxidative stress by a number of nuclear transcription factors or histone-modifying enzymes is well known; for example, histone deacetylases of the sirtuin family are strongly stimulated by low NADH levels to couple the redox state of cells to their metabolic activity (12, 15, 18).
The largest fraction of reductant-suppressed mutants lacked mitochondrial proteins (40%), two of which were specifically analyzed in this study, since the growth of respective mutants was strongly stimulated by DTT and GSH. Cmc1 and Coa4 are “twin Cx9C” proteins, which do not share any similarity at the level of their primary sequence except for the four cysteine residues of the Cx9C motifs. The data presented in this study confirm a—direct or indirect—role in the biogenesis of cytochrome c oxidase that had been proposed before (7, 22). Nevertheless, both mutants exhibit at steady-state levels about 20% to 50% of the cytochrome c oxidase activity found in wild-type cells. These reduced levels of complex IV are still high enough to enable cell growth under respiring conditions, at least in the presence of DTT, GSH, or ascorbic acid. Obviously, it is not the reduced activity of the respiratory chain, which prevents cell growth on nonfermentative carbon sources but rather the increased ROS production in these mutants (Fig. 9C). Oxidative stress was found to be toxic only under respiring conditions, and cell growth remained unaffected on glucose. It appears likely that assembly intermediates that accumulate in Δcmc1 and Δcoa4 mitochondria contribute to the growth defect (Fig. 9C). How this growth arrest is conveyed is unclear, but it is conceivable that the increased hydrogen peroxide levels are sensed and block cellular propagation via an intracellular signaling pathway, potentially including the nuclear components identified in this study. The observation that overexpression of cytosolic catalase partially rescued the growth of Δcoa4 cells on glycerol strongly suggests that the increased hydrogen peroxide levels in the cytosol are crucial for the observed growth arrest. It is conceivable that such an intracellular redox-dependent signaling pathway dampens the propagation of mutants that produce increased levels of mitochondria-derived oxidative stress in order to prevent the distribution of malfunctional mitochondria. A recent study showed that oxidative stress in mitochondria leads to cell death in yeast by inducing caspase-mediated apoptosis (19). The yeast metacaspase Yca1 is known to be activated by hydrogen peroxide although the details of this apoptotic signaling pathway in yeast are not entirely clear (33). It will have to be tested whether Yca1 is activated by mitochondria-derived oxidative stress to prevent cell growth of mutants in which the respiratory chain is not properly assembled.
A recent study systematically analyzed collections of yeast mutants to identify strains that are unable to grow on glycerol (35); it was found that in particular the growth of cytochrome c oxidase assembly mutants strongly depended on the strain background, which was interpreted by additional (epigenetic) cues that contribute to the growth phenotypes in these strains. This surprising observation might be explained by differences in the redox state or the redox sensitivity of the compared strains.
A cell-killing potential of assembly intermediates of cytochrome c oxidase was shown before, although only when cells were exposed to externally applied hydrogen peroxide (26, 53). In particular, assembly intermediates of Cox1 containing heme but no copper killed hydrogen peroxide-treated cytochrome c oxidase mutants on glucose medium. This cytotoxic potential might explain why mitochondrial Cox1 synthesis is under tight feedback control, which prevents the accumulation of assembly intermediates (5, 21, 37, 44, 45). At this stage it is not entirely clear where the ROS are produced. The strong genetic control of Cox1 and the large number of ROS-producing cytochrome c oxidase mutants suggest that the assembly intermediates of complex IV directly contribute to the generation of oxygen radicals. This cytotoxic effect is presumably not restricted to eukaryotes since it was shown before that bacteria lacking the cytochrome bd oxidase can be suppressed by addition of externally added glutathione (17). Nevertheless, it is also possible that the reduced levels of cytochrome c oxidase complexes lead to an accumulation of electrons at the level of complex III, which is known to have the potential to produce large amounts of superoxide (13). In vivo, ROS levels are presumably controlled by a number of mitochondrial reducing enzymes and by GSH, which diffuses across the outer membrane through porins (28, 34).
In summary, our study suggests that partially assembled cytochrome c oxidase complexes produce increased ROS levels under respirative conditions which trigger growth arrest of yeast cells. Presumably, these intermediates also form in wild-type cells but are rapidly converted into fully functional complexes and therefore produce ROS at much lower levels. Potentially, the significant levels of ROS that were reported to be produced by the respiratory chain are not exclusively byproducts of normal respiration but, at least in part, are generated by assembly intermediates of respiratory chain complexes. It will be interesting to monitor hydrogen peroxide production in mammalian cells that have cytochrome c oxidase assembly defects, for example, in cells from patients suffering from Leigh syndrome (43, 54). This might help to unravel the basic mechanisms of the ROS production by the respiratory chain under physiological and pathological conditions.
Materials and Methods
Yeast strains and media
All yeast strains were derived from the wild-type strain BY4742 (MATα; his3Δ1; leu2Δ0, lys2Δ0; ura3Δ0). The Δcmc1 Δcoa4 mutant was generated by replacement of the COA4 gene by a cassette containing the HIS5 gene of Schizosaccharomyces pombe in a Δcmc1 background.
The sequences of CMC1, COA4, and CTT1 from 300 bp upstream of the start codons to 150 bps downstream of the stop codons were amplified by PCR and cloned into the multi copy plasmid pRS426 (48). For the expression of Ctt1 in the IMS, the DNA sequence encoding for a fusion protein consisting of the N-terminal 75 amino acid residues of Mia40 and full-length Ctt1 was cloned into the expression vector pYX223 (Novagen) for expression under control of a GAL promoter. All strains were grown in YP (1% yeast extract, and 2% peptone) or synthetic medium with 2% glucose, galactose, or glycerol as carbon sources at 30°C. Mitochondria were prepared from galactose-grown cells as described (4).
Measurement of hydrogen peroxide production
Mitochondria (100 μg) were resolved in 600 mM sorbitol, 1 mM ethylenediaminetetraacetic acid (EDTA), 20 mM potassium phosphate (pH 7.4), 50 μM Amplex red, and 1 unit/ml horse radish peroxidase. By addition of 10 mM succinate respiration was induced. Hydrogen peroxide-induced formation of resorufin was measured in a fluorometer by excitation of 544 nm and emission of 590 nm for 10 min.
Measurements with the roGFP2-Orp1 probe
Yeast strains were grown for 24 h at 25°C in Hartwell's complete (HC) media containing 2% (w/v) glucose, followed by 1:50 dilution into HC + 2% (w/v) galactose and growth for a further 48 h. Following growth 4.5 OD600 units of cells were harvested by centrifugation and resuspended at a concentration of 7.5 OD600 units/ml in 100 mM Mes/Tris pH 6.0. The sample was divided into three equal aliquots which were either treated with 20 mM N,N,N′,N′-tetramethylazodicarboxamide (diamide) or 100 mM DTT for the fully oxidized and fully reduced controls respectively, or left untreated. Samples were loaded to a flat-bottomed 96-well plate (BD-Falcon; Product No. 353219) and centrifuged for 5 min at 20 g to form a loose cell pellet. Probe response in the untreated cells was induced by the addition of 5 mM hydrogen peroxide. Probe oxidation was followed by measuring fluorescence emission using a plate-reader system (FLUOstar Omega) with an emission filter of 510/20 nm and excitation filters of 390/10 nm and 480/10 nm. The degree of probe oxidation (OxDroGFP2) was determined according to Equation 1, where I is the fluorescence emission intensity at 510 nm, following excitation at either 390 or 480 nm, for the fully reduced (red) and fully oxidized (ox) controls, and the sample.
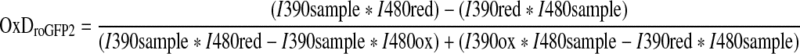 |
Protein carbonylation assay
Mitochondria (150 μg) were lysed by addition of 6% sodium dodecyl sulfate (SDS). The samples were incubated with 3.96 mg/ml 2,4-dinitrophenylhydrazine, 0.154 g/ml trifluoroacetic acid for 10 min at 25°C. After precipitation with trichloroacetic acid (TCA) the pellets were resuspended in SDS-loading buffer (60 mM Tris/HCl pH 6.8, 10% glycerine, 2% SDS, 0.01% bromophenol blue). The proteins were separated on SDS–polyacrylamide gels, transferred to nitrocellulose membranes, and analyzed by Western blotting with a 2,4-dinitrophenylhydrazone-specific antibody. For the analysis of carbonylated translation products, proteins were synthesized in isolated mitochondria as described (16) in the presence of [35S]-methionine. Then mitochondria were washed, lysed with SDS, treated as described above, and used for immunoprecipitation with the dinitrophenylhydrazone-specific antibody or preimmune serum for control.
Analysis of redox state of proteins
To determine the redox state of Cmc1 and Coa4, cells were grown to an optical density (OD) of 0.6 and harvested by centrifugation. Cell pellets were resuspended in 12% TCA. For the “inverse shift” sample, the cell pellet was incubated in 50 mM NEM for 15 min at 30°C before addition of TCA. Then, cells were broken by sonication and glass bead homogenization. TCA pellets were resuspended in 80 mM Tris pH 7.0, 10% glycerol, 2% SDS, and 0.05% bromocresol purple. For reduction of disulfides, samples were incubated with 10 mM TCEP for 20 min at 96°C followed by addition of 15 mM MM(PEG)24 (Thermo Scientific). After incubation for 1 h at 25°C in the dark and 1 min at 96°C samples were analyzed by Western blotting.
Oxygen consumption
Mitochondrial oxygen consumption was measured basically as described previously (26, 53) using a Clark electrode (Hansatech Instruments). Mitochondria (100 μg) were incubated in 0.6 M sorbitol, 1 mM MgCl2, and 20 mM HEPES, pH 7.4. Oxygen consumption was induced by addition of 5 mM NADH and measured for 2 min, and then 1 mM ascorbate or 1 mM GSH was added and the measurement was continued for 2 min. Each measurement was repeated twice.
Statistical analysis
T tests were performed to calculate p-values. Confidence levels are shown in figures as follows; *p<0.05, **p<0.01, and ***p<0.001.
Miscellaneous
In vivo labeling of translation products (16), enzyme activity assays (49), oxygen consumption, and generation of antibodies against Cmc1 and Coa4 (32) were performed as described.
Abbreviations Used
- DTT
dithiothreitol
- GSH
reduced glutathione
- GSSG
oxidized glutathione
- HC
Hartwell's complete
- IMS
intermembrane space
- NEM
N-ethylmaleimide
- OD
optical density at 578 nm
- ROS
reactive oxygen species
- SDS
sodium dodecyl sulfate
- TCA
trichloroacetic acid
- TCEP
triscarboxyethyl phosphine
Acknowledgments
We thank Sabine Knaus for experimental assistance, and Jan Riemer and Benedikt Westermann for discussion and advice. This work was supported by grants from the Landesschwerpunkt für Membrantransport and the Deutsche Forschungsgemeinschaft (IRTG1830 and He2803/4-1) to J.M.H. and by a Kekulé fellowship of the Fonds der Chemischen Industrie to K.B.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Adam-Vizi V. Chinopoulos C. Bioenergetics and the formation of mitochondrial reactive oxygen species. Trends Pharmacol Sci. 2006;27:639–645. doi: 10.1016/j.tips.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 2.Albrecht SC. Barata AG. Grosshans J. Teleman AA. Dick TP. In vivo mapping of hydrogen peroxide and oxidized glutathione reveals chemical and regional specificity of redox homeostasis. Cell Metab. 2011;14:819–829. doi: 10.1016/j.cmet.2011.10.010. [DOI] [PubMed] [Google Scholar]
- 3.Allen S. Balabanidou V. Sideris DP. Lisowsky T. Tokatlidis K. Erv1 mediates the Mia40-dependent protein import pathway and provides a functional link to the respiratory chain by shuttling electrons to cytochrome c. J Mol Biol. 2005;353:937–944. doi: 10.1016/j.jmb.2005.08.049. [DOI] [PubMed] [Google Scholar]
- 4.Altmann K. Dürr M. Westermann B. Saccharomyces cerevisiae as a model organism to study mitochondrial biology. In: Leister D, editor; Herrmann JM, editor. Mitochondria. Practical Protocols. Totowa, NJ: Humana Press; 2007. pp. 81–90. [DOI] [PubMed] [Google Scholar]
- 5.Barrientos A. Zambrano A. Tzagoloff A. Mss51p and Cox14p jointly regulate mitochondrial Cox1p expression in Saccharomyces cerevisiae. EMBO J. 2004;23:3472–3482. doi: 10.1038/sj.emboj.7600358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beal MF. Bioenergetic approaches for neuroprotection in Parkinson's disease. Ann Neurol. 2003;53(Suppl 3):S39–S47. doi: 10.1002/ana.10479. discussion S47–S48. [DOI] [PubMed] [Google Scholar]
- 7.Bestwick M. Jeong MY. Khalimonchuk O. Kim H. Winge DR. Analysis of Leigh syndrome mutations in the yeast SURF1 homolog reveals a new member of the cytochrome oxidase assembly factor family. Mol Cell Biol. 2010;30:4480–4491. doi: 10.1128/MCB.00228-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buono MJ. Kolkhorst FW. Submitting illuminations for review. Adv Physiol Educ. 2001;25:70–71. doi: 10.1152/advances.2001.25.2.45. [DOI] [PubMed] [Google Scholar]
- 9.Cavallaro G. Genome-wide analysis of eukaryotic twin CX9C proteins. Mol Biosyst. 2010;6:2459–2470. doi: 10.1039/c0mb00058b. [DOI] [PubMed] [Google Scholar]
- 10.Chacinska A. Pfannschmidt S. Wiedemann N. Kozjak V. Sanjuan Szklarz LK. Schulze-Specking A. Truscott KN. Guiard B. Meisinger C. Pfanner N. Essential role of Mia40 in import and assembly of mitochondrial intermembrane space proteins. EMBO J. 2004;23:3735–3746. doi: 10.1038/sj.emboj.7600389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Costanzo MC. Fox TD. Control of mitochondrial gene expression in Saccharomyces cerevisiae. Annu Rev Genet. 1990;24:91–113. doi: 10.1146/annurev.ge.24.120190.000515. [DOI] [PubMed] [Google Scholar]
- 12.Delaunay A. Pflieger D. Barrault MB. Vinh J. Toledano MB. A thiol peroxidase is an H2O2 receptor and redox-transducer in gene activation. Cell. 2002;111:471–481. doi: 10.1016/s0092-8674(02)01048-6. [DOI] [PubMed] [Google Scholar]
- 13.Dröse S. Brandt U. Molecular mechanisms of superoxide production by the mitochondrial respiratory chain. Adv Exp Med Biol. 2012;748:145–169. doi: 10.1007/978-1-4614-3573-0_6. [DOI] [PubMed] [Google Scholar]
- 14.Finkel T. Signal transduction by mitochondrial oxidants. J Biol Chem. 2012;287:4434–4440. doi: 10.1074/jbc.R111.271999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Finkel T. Deng CX. Mostoslavsky R. Recent progress in the biology and physiology of sirtuins. Nature. 2009;460:587–591. doi: 10.1038/nature08197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Funes S. Herrmann JM. Analysis of mitochondrial protein synthesis in yeast. Methods Mol Biol. 2007;372:255–263. doi: 10.1007/978-1-59745-365-3_18. [DOI] [PubMed] [Google Scholar]
- 17.Goldman BS. Gabbert KK. Kranz RG. The temperature-sensitive growth and survival phenotypes of Escherichia coli cydDC and cydAB strains are due to deficiencies in cytochrome bd and are corrected by exogenous catalase and reducing agents. J Bacteriol. 1996;178:6348–6351. doi: 10.1128/jb.178.21.6348-6351.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grant CM. Role of the glutathione/glutaredoxin and thioredoxin systems in yeast growth and response to stress conditions. Mol Microbiol. 2001;39:533–541. doi: 10.1046/j.1365-2958.2001.02283.x. [DOI] [PubMed] [Google Scholar]
- 19.Greetham D. Kritsiligkou P. Watkins RH. Carter Z. Parkin J. Grant CM. Oxidation of the yeast mitochondrial thioredoxin promotes cell death. Antioxid Redox Signal. 2012;18:376–385. doi: 10.1089/ars.2012.4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gutscher M. Sobotta MC. Wabnitz GH. Ballikaya S. Meyer AJ. Samstag Y. Dick TP. Proximity-based protein thiol oxidation by H2O2-scavenging peroxidases. J Biol Chem. 2009;284:31532–31540. doi: 10.1074/jbc.M109.059246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herrmann JM. Woellhaf MW. Bonnefoy N. Control of protein synthesis in yeast mitochondria: the concept of translational activators. Biochim Biophys Acta. 2013;1833:286–294. doi: 10.1016/j.bbamcr.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 22.Horn D. Al-Ali H. Barrientos A. Cmc1p is a conserved mitochondrial twin CX9C protein involved in cytochrome c oxidase biogenesis. Mol Cell Biol. 2008;28:4354–4364. doi: 10.1128/MCB.01920-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horn D. Zhou W. Trevisson E. Al-Ali H. Harris TK. Salviati L. Barrientos A. The conserved mitochondrial twin Cx9C protein Cmc2 Is a Cmc1 homologue essential for cytochrome c oxidase biogenesis. J Biol Chem. 2010;285:15088–15099. doi: 10.1074/jbc.M110.104786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iuso A. Scacco S. Piccoli C. Bellomo F. Petruzzella V. Trentadue R. Minuto M. Ripoli M. Capitanio N. Zeviani M. Papa S. Dysfunctions of cellular oxidative metabolism in patients with mutations in the NDUFS1 and NDUFS4 genes of complex I. J Biol Chem. 2006;281:10374–10380. doi: 10.1074/jbc.M513387200. [DOI] [PubMed] [Google Scholar]
- 25.Jia L. Dienhart M. Schramp M. McCauley M. Hell K. Stuart RA. Yeast Oxa1 interacts with mitochondrial ribosomes: the importance of the C-terminal hydrophilic region of Oxa1. EMBO J. 2003;22:6438–6447. doi: 10.1093/emboj/cdg624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khalimonchuk O. Bird A. Winge DR. Evidence for a pro-oxidant intermediate in the assembly of cytochrome oxidase. J Biol Chem. 2007;282:17442–17449. doi: 10.1074/jbc.M702379200. [DOI] [PubMed] [Google Scholar]
- 27.Kohler R. Boehringer D. Greber B. Bingel-Erlenmeyer R. Collinson I. Schaffitzel C. Ban N. YidC and Oxa1 form dimeric insertion pores on the translating ribosome. Mol Cell. 2009;34:344–353. doi: 10.1016/j.molcel.2009.04.019. [DOI] [PubMed] [Google Scholar]
- 28.Kojer K. Bien M. Gangel H. Morgan B. Dick TP. Riemer J. Glutathione redox potential in the mitochondrial intermembrane space is linked to the cytosol and impacts the Mia40 redox state. EMBO J. 2012;31:3169–3182. doi: 10.1038/emboj.2012.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leary SC. Sasarman F. Nishimura T. Shoubridge EA. Human SCO2 is required for the synthesis of CO II and as a thiol-disulphide oxidoreductase for SCO1. Hum Mol Genet. 2009;18:2230–2240. doi: 10.1093/hmg/ddp158. [DOI] [PubMed] [Google Scholar]
- 30.Liu J. Lillo C. Jonsson PA. Vande Velde C. Ward CM. Miller TM. Subramaniam JR. Rothstein JD. Marklund S. Andersen PM. Brannstrom T. Gredal O. Wong PC. Williams DS. Cleveland DW. Toxicity of familial ALS-linked SOD1 mutants from selective recruitment to spinal mitochondria. Neuron. 2004;43:5–17. doi: 10.1016/j.neuron.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 31.Liu Z. Butow RA. Mitochondrial retrograde signaling. Annu Rev Genet. 2006;40:159–185. doi: 10.1146/annurev.genet.40.110405.090613. [DOI] [PubMed] [Google Scholar]
- 32.Longen S. Bien M. Bihlmaier K. Kloeppel C. Kauff F. Hammermeister M. Westermann B. Herrmann JM. Riemer J. Systematic analysis of the twin cx9c protein family. J Mol Biol. 2009;393:356–368. doi: 10.1016/j.jmb.2009.08.041. [DOI] [PubMed] [Google Scholar]
- 33.Madeo F. Herker E. Maldener C. Wissing S. Lachelt S. Herlan M. Fehr M. Lauber K. Sigrist SJ. Wesselborg S. Frohlich KU. A caspase-related protease regulates apoptosis in yeast. Mol Cell. 2002;9:911–917. doi: 10.1016/s1097-2765(02)00501-4. [DOI] [PubMed] [Google Scholar]
- 34.Mari M. Morales A. Colell A. Garcia-Ruiz C. Kaplowitz N. Fernandez-Checa JC. Mitochondrial glutathione: features, regulation and role in disease. Biochim Biophys Acta. 2012:pii. doi: 10.1016/j.bbagen.2012.10.018. S0304-4165(12)00304-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Merz S. Westermann B. Genome-wide deletion mutant analysis reveals genes required for respiratory growth, mitochondrial genome maintenance and mitochondrial protein synthesis in Saccharomyces cerevisiae. Genome Biol. 2009;10:R95. doi: 10.1186/gb-2009-10-9-r95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mesecke N. Terziyska N. Kozany C. Baumann F. Neupert W. Hell K. Herrmann JM. A disulfide relay system in the intermembrane space of mitochondria that mediates protein import. Cell. 2005;121:1059–1069. doi: 10.1016/j.cell.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 37.Mick DU. Fox TD. Rehling P. Inventory control: cytochrome c oxidase assembly regulates mitochondrial translation. Nat Rev Mol Cell Biol. 2011;12:14–20. doi: 10.1038/nrm3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Monteiro G. Horta BB. Pimenta DC. Augusto O. Netto LE. Reduction of 1-Cys peroxiredoxins by ascorbate changes the thiol-specific antioxidant paradigm, revealing another function of vitamin C. Proc Natl Acad Sci U S A. 2007;104:4886–4891. doi: 10.1073/pnas.0700481104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moran M. Marin-Buera L. Gil-Borlado MC. Rivera H. Blazquez A. Seneca S. Vazquez-Lopez M. Arenas J. Martin MA. Ugalde C. Cellular pathophysiological consequences of BCS1L mutations in mitochondrial complex III enzyme deficiency. Hum Mutat. 2010;31:930–941. doi: 10.1002/humu.21294. [DOI] [PubMed] [Google Scholar]
- 40.Morgan B. Sobotta MC. Dick TP. Measuring E(GSH) and H2O2 with roGFP2-based redox probes. Free Radic Biol Med. 2011;51:1943–1951. doi: 10.1016/j.freeradbiomed.2011.08.035. [DOI] [PubMed] [Google Scholar]
- 41.Naoe M. Ohwa Y. Ishikawa D. Ohshima C. Nishikawa S. Yamamoto H. Endo T. Identification of Tim40 that mediates protein sorting to the mitochondrial intermembrane space. J Biol Chem. 2004;279:47815–47821. doi: 10.1074/jbc.M410272200. [DOI] [PubMed] [Google Scholar]
- 42.Nystrom T. Role of oxidative carbonylation in protein quality control and senescence. EMBO J. 2005;24:1311–1317. doi: 10.1038/sj.emboj.7600599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Papadopoulou LC. Sue CM. Davidson MM. Tanji K. Nishino I. Sadlock JE. Krishna S. Walker W. Selby J. Glerum DM. Coster RV. Lyon G. Scalais E. Lebel R. Kaplan P. Shanske S. De Vivo DC. Bonilla E. Hirano M. DiMauro S. Schon EA. Fatal infantile cardioencephalomyopathy with COX deficiency and mutations in SCO2, a COX assembly gene. Nat Genet. 1999;23:333–337. doi: 10.1038/15513. [DOI] [PubMed] [Google Scholar]
- 44.Perez-Martinez X. Broadley SA. Fox TD. Mss51p promotes mitochondrial Cox1p synthesis and interacts with newly synthesized Cox1p. EMBO J. 2003;22:5951–5961. doi: 10.1093/emboj/cdg566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pierrel F. Bestwick ML. Cobine PA. Khalimonchuk O. Cricco JA. Winge DR. Coa1 links the Mss51 post-translational function to Cox1 cofactor insertion in cytochrome c oxidase assembly. EMBO J. 2007;26:4335–4346. doi: 10.1038/sj.emboj.7601861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Poyton RO. Assembling a time bomb—cytochrome c oxidase and disease. Nat Genet. 1998;20:316–317. doi: 10.1038/3778. [DOI] [PubMed] [Google Scholar]
- 47.Roberts GG. Hudson AP. Transcriptome profiling of Saccharomyces cerevisiae during a transition from fermentative to glycerol-based respiratory growth reveals extensive metabolic and structural remodeling. Mol Genet Genomics. 2006;276:170–186. doi: 10.1007/s00438-006-0133-9. [DOI] [PubMed] [Google Scholar]
- 48.Sikorski RS. Hieter P. A system of shuttle vectors and host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stehling O. Smith PM. Biederbick A. Balk J. Lill R. Muhlenhoff U. Investigation of iron-sulfur protein maturation in eukaryotes. Methods Mol Biol. 2007;372:325–342. doi: 10.1007/978-1-59745-365-3_24. [DOI] [PubMed] [Google Scholar]
- 50.Szyrach G. Ott M. Bonnefoy N. Neupert W. Herrmann JM. Ribosome binding to the Oxa1 complex facilitates cotranslational protein insertion in mitochondria. EMBO J. 2003;22:6448–6457. doi: 10.1093/emboj/cdg623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tanaka S. Takehashi M. Matoh N. Iida S. Suzuki T. Futaki S. Hamada H. Masliah E. Sugiura Y. Ueda K. Generation of reactive oxygen species and activation of NF-kappaB by non-Abeta component of Alzheimer's disease amyloid. J Neurochem. 2002;82:305–315. doi: 10.1046/j.1471-4159.2002.00958.x. [DOI] [PubMed] [Google Scholar]
- 52.Tzagoloff A. Dieckmann CL. PET genes of Saccharomyces cerevisiae. Microbiol Rev. 1990;54:211–225. doi: 10.1128/mr.54.3.211-225.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Veniamin S. Sawatzky LG. Banting GS. Glerum DM. Characterization of the peroxide sensitivity of COX-deficient yeast strains reveals unexpected relationships between COX assembly proteins. Free Radic Biol Med. 2011;51:1589–1600. doi: 10.1016/j.freeradbiomed.2011.06.024. [DOI] [PubMed] [Google Scholar]
- 54.Zhu Z. Yao J. Johns T. Fu K. De Bie I. Macmillan C. Cuthbert AP. Newbold RF. Wang J. Chevrette M. Brown GK. Brown RM. Shoubridge EA. SURF1, encoding a factor involved in the biogenesis of cytochrome c oxidase, is mutated in Leigh syndrome. Nat Genet. 1998;20:337–343. doi: 10.1038/3804. [DOI] [PubMed] [Google Scholar]
- 55.Zito E. Hansen HG. Yeo GS. Fujii J. Ron D. Endoplasmic reticulum thiol oxidase deficiency leads to ascorbic acid depletion and noncanonical scurvy in mice. Mol Cell. 2012;48:39–51. doi: 10.1016/j.molcel.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]