Abstract
Three peroxiredoxins (Prxs) were identified in Thermotoga maritima, which possesses neither glutathione nor typical thioredoxins: one of the Prx6 class; one 2-Cys PrxBCP; and a unique hybrid protein containing an N-terminal 1-Cys PrxBCP domain fused to a flavin mononucleotide-containing nitroreductase (Ntr) domain. No peroxidase activity was detected for Prx6, whereas both bacterioferritin comigratory proteins (BCPs) were regenerated by a NADH/thioredoxin reductase/glutaredoxin (Grx)-like system, constituting a unique peroxide removal system. Only two of the three Grx-like proteins were able to support peroxidase activity. The inability of TmGrx1 to regenerate oxidized 2-Cys PrxBCP probably results from the thermodynamically unfavorable difference in their disulfide/dithiol Em values, −150 and −315 mV, respectively. Mutagenesis of the Prx-Ntr fusion, combined with kinetic and structural analyses, indicated that electrons are not transferred between its two domains. However, their separate activities could function in a complementary manner, with peroxide originating from the chromate reductase activity of the Ntr domain reduced by the Prx domain. Antioxid. Redox Signal. 18, 1613–1622.
Introduction
Organisms have developed diverse detoxification systems, including catalases, ascorbate-peroxidases, NADH-peroxidases, and thiol-peroxidases (Tpxs) such as glutathione-peroxidases and peroxiredoxins (Prxs) to prevent the damages caused by hydroperoxides (8). Current classifications distinguish six Prx classes: Prx1, Prx5, Prx6, Tpx, AhpE, and BCP (bacterioferritin comigratory protein)/PrxQ (4). During hydroperoxide reduction, their peroxidatic cysteine (CysP) is oxidized to a sulfenic acid. The regeneration of CysP proceeds either via direct reduction of the sulfenic acid (1-Cys mechanism) or via the formation of intra- or intermolecular disulfide bonds with a resolving cysteine (CysR) (2-Cys mechanism) (8).
In prokaryotes, several Tpx systems were identified. Besides NADH-peroxidases, Prxs of the AhpC, Tpx, or BCP type are recycled via AhpF, AhpD, NADH oxidase (Nox)/dehydrogenase or through glutaredoxin (Grx) or thioredoxin (Trx)-dependent systems (4, 8). Consistently, a Prx gene is often adjacent to Nox, AhpF, or AhpD genes in bacteria (6). The biochemical and structural properties of Thermotoga maritima Prxs are described, with emphasis on the reducing systems that are required for their regeneration. Three Prxs {TmPrx6 (TM0807), TmPrxBCP (TM0780), and TmPrxNtr (TM0386)} were identified (Fig. 1A). TmPrxBCP is a 2-Cys BCP, whereas TmPrxNtr, which is restricted to Thermotogaceae, uniquely contains an N-terminal 1-Cys BCP domain that is fused to a C-terminal nitroreductase (Ntr) module. In addition, three putative disulfide reductases of the Trx/Grx/NrdH family, all containing Grx-like active sites [TmGrx1 (TM0996), TmGrx2 (TM1031), and TmGrx3 (TM0868)], were identified (Fig. 1B). In most thermotogales, TmGrx3 gene is adjacent to a thioredoxin reductase (TR) gene, TmTR (TM0869) (9).
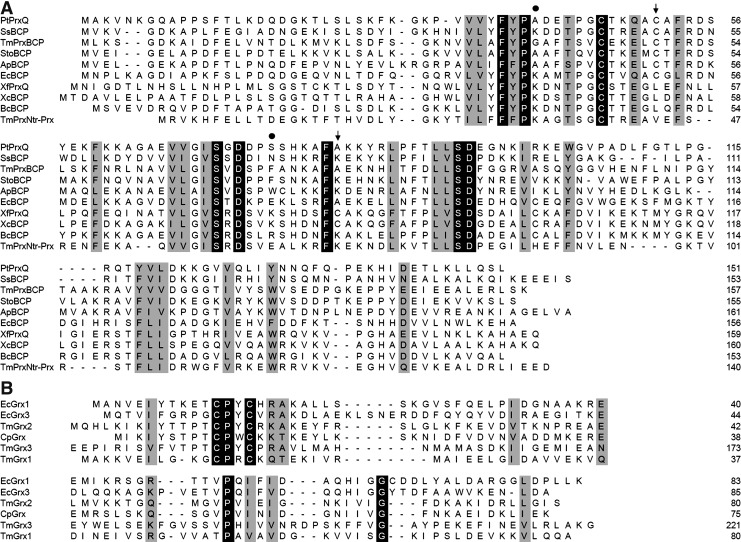
FIG. 1.
Amino acid sequence alignments of BCP/Q-type Prxs and Grx-like proteins. The alignments were done with ClustalW. In white on black: strictly conserved amino acids. In black on gray: functional amino acid conservation. (A) Sequence comparison of biochemically or structurally characterized BCPs. Accession numbers are as follows: Populus trichocarpa: PtPrxQ POPTR_0006s13980, Bulkholderia cenocepacia: BcBCP YP_002231064, Escherichia coli: EcBCP AAB88562, Xylella fastidiosa: XfPrxQ AAF83771, Sulfolobus solfataricus: SsBCP1 NP_343463, Aeropyrum pernix: ApBCP NP_148402, TmPrxNtr-Prx EHA59032, TmPrxBCP NP_228589, Xanthomonas campestris: XcBCP AEL0686, Sulfolobus tokadaii: StoBCP NP_377766. Only the BCP domain of TmPrxNtr is shown here. While the peroxidatic cysteine is strictly conserved, the position of the resolving cysteine, indicated by an arrow, can vary. In addition, some residues that are likely important for the formation of the A-type interface in dimeric BCP are indicated by a black circle. (B) Sequence comparison of Grx. Accession numbers are as follows: EcGrx3 NP_418067, EcGrx1, NP_308956, Clostridium pasteurianum CpGrx AAA23277. Only the C-terminal domain of TmGrx3 has been used for alignment. BCP, bacterioferritin comigratory protein; Grx, glutaredoxin; Ntr, nitroreductase; Prx, peroxiredoxin.
Innovation.
Peroxide removal is important for all living organisms. In the anaerobic thermophilic bacterium Thermotoga maritima, which possesses neither glutathione nor typical thioredoxins, two different bacterioferritin comigratory proteins, including a hybrid protein unique to Thermotogales, were reduced by two glutaredoxin-like proteins that were themselves regenerated by an NADH-thioredoxin reductase. Another original property of the fusion protein is its ability to couple both chromate and quinone reductase activities with peroxidase activities.
Reductase Activity of Grx-Like Proteins
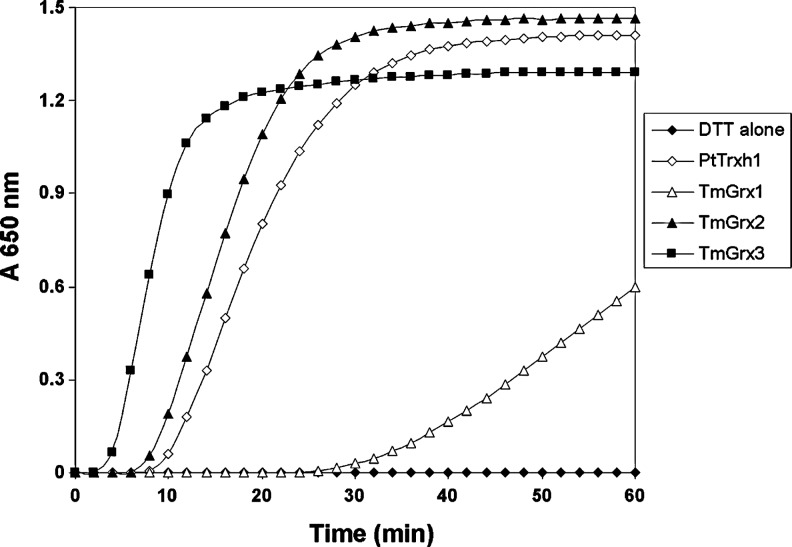
The disulfide reductase activity of T. maritima Grx-like proteins was tested using the reduction of insulin disulfide bridges (Fig. 2). TmGrx2 and TmGrx3 were more efficient than TmGrx1. The regeneration of these Grxs by TmTR was next evaluated by measuring the reduction of 5,5′-dithio-bis-2-nitrobenzoic acid (DTNB) by the NADH/TR/Grx system. The catalytic efficiencies (kcat/KGrx) were 6.8×105 M−1 s−1 for TmGrx1, 1.4×106 M−1 s−1 for TmGrx2, and 5.8×104 M−1 s−1 for TmGrx3 (Table 1). Oxidation-reduction midpoint potentials (Em) were measured to determine whether differences in activity might arise from differences in Em. The analysis of redox titrations for TmTR and the newly identified TmGrx1 and TmGrx2 was simple, because all three contain only one redox-active disulfide. TmGrx3, which contains two disulfides, was not titrated. The disulfides in TmTR and TmGrx2 had similar Em values of −295±5 mV and −270±5 mV, respectively, but the −150±5 mV value of TmGrx1 is significantly less negative.
FIG. 2.
Insulin reduction by Grx-like proteins. Insulin reduction was measured in the presence of DTT and 2 μM poplar Trxh1 (used a control), TmGrx2 and TmGrx3 and 5 μM TmGrx1, by measuring the turbidity at 650 nm caused by the precipitation of reduced insulin. Each trace is a representative experiment, selected from two to three replicates. DTT, dithiothreitol; Trx, thioredoxin.
Table 1.
Kinetic Parameters of the Recombinant Proteins
| Substrates | Enzymes | Km (μM) | kcat (s−1) | kcat/Km (M−1·s−1) |
|---|---|---|---|---|
| DTNB | TmGrx1 | 5.15±0.80 | 3.50±0.24 | 6.8×105 |
| TmGrx2 | 0.84±0.08 | 1.21±0.04 | 1.4×106 | |
| TmGrx3 | 11.39±1.55 | 0.66±0.04 | 5.8×104 | |
| Benzoquinone | TmPrxNtr | ≤20 | 52.80±7.20 | ≥9.4×106 |
| TmPrxNtrC40S | ≤20 | 44.10±5.45 | ≥6.3×106 | |
| TmPrxNtr-Ntr2 | ≤20 | 66.80±6.70 | ≥1.2×107 | |
| Chromate | TmPrxNtr | ≤20 | 21±1.34 | ≥3.7×106 |
| TmPrxNtrC40S | ≤20 | 19.80±1.05 | ≥2.8×106 | |
| TmPrxNtr-Ntr2 | ≤20 | 25±1.52 | ≥4.6×106 | |
| H2O2 | TmPrxNtr-Prx | 13.3±4.1 | 0.27±0.01 | 2.0×104 |
| TmPrxBCP | 39.6±4.5 | 0.12±0.01 | 3.1×103 |
All kinetic parameters have been obtained under steady-state conditions. For DTNB and H2O2 reducing activities, the results are expressed as nmol substrates reduced by nmole enzymes per second. For DTNB assay, the Km values represent the affinity of TmTR for the various Grxs-like. Hence, the catalytic efficiency corresponds here to a kcat/KGrx. For benzoquinone and chromate reductase activities, the affinity is so high that the Km cannot be accurately determined with this spectrophotometric assay. The turnover numbers have been expressed as nmol substrate reduced by nmol FMN per second, owing to the fact that the recombinant FMN-containing enzymes (TmPrxNtr, TmPrxNtrC40S, and TmPrxNtr-Ntr2) only contained 0.28, 0.35, and 0.27 mole FMN/mole monomer, respectively.
DTNB, 5,5′-dithio-bis-2-nitrobenzoic acid; FMN, flavin mononucleotide; Grx, glutaredoxin; Nrx, nitroreductase; Prx, peroxiredoxin.
Catalytic and Redox Properties of Prxs
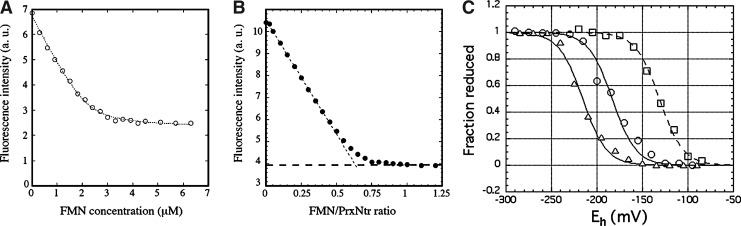
TmPrxNtr is the only TmPrx with a flavin-containing C-terminal Ntr domain (identified as flavin mononucleotide [FMN] after acid treatment and by mass spectrometry). FMN quantitation gave average values of 0.28, 0.35, and 0.27 mol FMN/mol enzyme for TmPrxNtr, TmPrxNtrC40S, and TmPrxNtr-Ntr2, respectively, due to the loss of flavin during protein purification. The FMN was reduced by both NADPH and NADH. The affinity of TmPrxNtr for FMN was measured using tryptophan fluorescence to monitor FMN binding (Fig. 3A, B). Fitting the data gave dissociation constants of 100±58 nM for TmPrxNtr and 1.4±0.2 μM for TmPrxNtr-Ntr2, with FMN binding levels about two-thirds that of the protein amount. Since 0.3 mol FMN/mol protein was initially present, the result indicates a single FMN binding site per TmPrxNtr monomer. Redox titrations of FMN in TmPrxNtr, TmPrxNtrC40S, and TmPrxNtr-Ntr2 gave Em values of −215±5 mV for TmPrxNtr-Ntr2, −185±5 mV for TmPrxNtr, and −130±5 mV for TmPrxNtrC40S (Fig. 3C). The excellent fits of the titrations to the Nernst equation for a single two-electron redox couple are consistent with the detection of only fully oxidized FMN and fully reduced flavin mononucleotide reduced (FMNH2) at all Eh values, with no appreciable levels of FMN semiquinone.
FIG. 3.
Stoichiometry of FMN binding, FMN affinity, and titration in TmPrxNtr. (A) Steady-state intrinsic tryptophan fluorescence intensity of TmPrxNtr as a function of FMN concentration. A 5 μM TmPrxNtr solution in 50 mM Tris pH 7.6 was excited at 290 nm, and the fluorescence emitted between 330 and 350 nm was integrated. The dashed curve corresponds to the least-square fit of the experimental values using the following equation: F=Finitial – Ffinal×((Kd + At + X) − ((Kd + At + X)2 – 4AtX)1/2)/2At, where At is the FMN binding site concentration, X is the concentration of added FMN, Finitial is the fluorescence of free TmPrxNtr, and Ffinal is the fluorescence at saturation. The fitted parameters are Kd, Finitial, Ffinal, and At; the results of the fit were for At: 2.98 μM and for Kd: 68 nM (R=0.999). (B) Steady-state intrinsic tryptophan fluorescence intensity of TmPrxNtr as a function of FMN/TmPrxNtr ratio. The TmPrxNtr concentration was 20 μM in 50 mM Tris pH 7.6. (C) Flavin titration. The reaction mixture contained 30 mM Tris HCl pH 8.0, 1 mM EDTA, either 380 μM TmPrxNtr (open circles) or 150 μM TmPrxNtrC40S (open squares) or 630 μM TmPrxNtr-Ntr2 (open triangles) and the following redox mediators, all at concentrations of 5 μM: 1,4-naphtoquinone, anthraquinone-2-sulfonate and safranin T (TmPrxNtr and TmPrxNtrC40S) or benzyl viologen (TmPrxNtr-Ntr2). The ambient potential, Eh, was adjusted using a potentiostat as described in the “Notes” section. Absorbance changes are relative, with the absorbance of the fully oxidized protein taken as 1 and that of the fully reduced protein taken as 0. FMN, flavin mononucleotide.
The possible H2O2 reducing activity of TmPrxNtr was investigated using an NADPH-coupled assay. A kcat value of 0.45 s−1 was measured at 50°C. Similar values were obtained using TmPrxNtrC40S and TmPrxNtr-Ntr2, suggesting that NADPH oxidation may involve a non-enzymatic reaction between FMNH2 and H2O2. Since all TmGrx-like proteins are reduced by TmTR, their capacity to reduce Prxs could be evaluated using a steady-state bi-substrate assay in which initial rates of peroxide reduction are monitored by a coupled NADH oxidation. No H2O2 reducing activity was detected using TmPrx6 in the presence of any of the TmGrxs raising questions about its peroxidase activity and regeneration mechanism. Of the three reductants tested, only TmGrx1 was unable to support the activity of both TmPrxBCP and TmPrxNtr-Prx (used to avoid possible artefacts arising from the FMN present in TmPrxNtr). All other kinetics gave good fits to the Michaelis–Menten equation, allowing determination of kcat and apparent Km values for the different substrates. The catalytic efficiency (kcat/KH2O2) of TmPrxNtr-Prx, 2.0×104 M−1 s−1, was higher than that of TmPrxBCP (3.1×103 M−1 s−1), and both values are similar to those of EcBCP (1.3×104 M−1 s−1) and poplar PrxQ (8×103 M−1 s−1) (Table 1) (5, 7). TmPrxNtr-Prx displayed comparable Km values for TmGrx2 and TmGrx3 (14±4 μM and 18±4 μM, respectively). TmPrxBCP exhibited a lower Km value for TmGrx3 than for TmGrx2 (11±0.8 μM and 30±4 μM, respectively). Consistent with the close evolutionary relationship between thermotogales and clostridia, the TR/Grx system used by T. maritima BCPs is similar to that used by Clostridium pasteurianum AhpC (6).
The reactivity and regeneration of TmPrxs are partially governed by thermodynamic properties such as the pKa of CysP and the Em values of disulfide/dithiol couples. The apparent pKa values for the single cysteine thiol groups in TmPrxNtr-Prx and TmPrxBCPC50S were 5.9±0.2 and 7.0±0.1, respectively. Thus, for both proteins, the thiolate ion, and not the protonated thiol, is the dominant species at pH values above 7.0. The best fit for redox titrations of TmPrxBCP were obtained for a model with a single disulfide/dithiol redox couple in each monomer, with Em=−315±5 mV. This value, which is similar to poplar PrxQ (−325 mV) but very different from EcBCP (−146 mV) (5, 7), may explain why TmGrx1 cannot reduce oxidized TmPrxBCP.
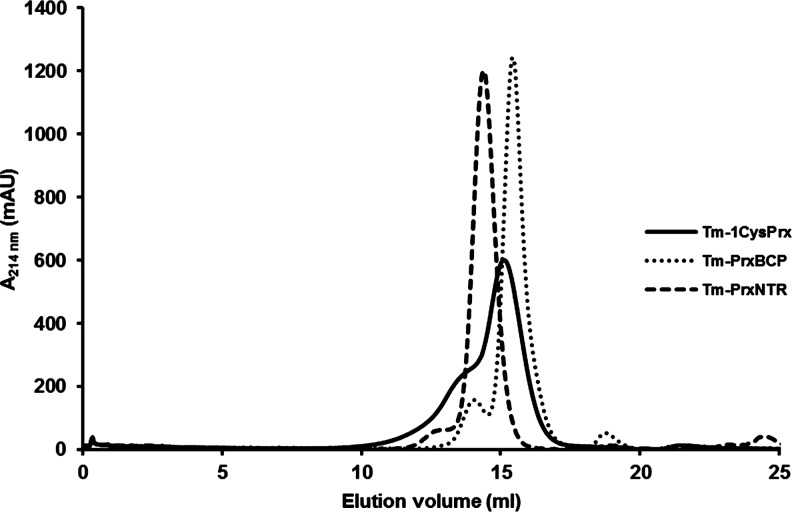
While the regeneration of 2-Cys BCP proceeds through the Trx-dependent reduction of an intramolecular disulfide, the mechanism for 1-Cys BCP is less clear. In Bulkholderia cenocepacia BCP, the sulfenic acid can be directly reduced either by Trx or by the GSH/Grx couple through the formation of a glutathione adduct (3). Alternatively, it is possible that an intermolecular disulfide is formed, as occurs with a CysR mutant of EcBCP (2). Both mechanisms might operate in TmPrxNtr. However, both analytical gel filtration and dynamic light scattering (DLS) measurements suggest the presence of covalent decamers involving CysP (Fig. 4), making the latter mechanism more likely. In contrast, the dimeric forms observed for TmPrx6 and TmPrxBCP during gel filtration do not involve disulfide linkages (Fig. 4). Only a few BCP members were shown to form non-covalent dimers. Consistent with previous predictions, a small hydrophobic (Gly39) and an aromatic residue (Phe75) may be important for the dimerization of TmPrxBCP (Fig. 1A) (5).
FIG. 4.
Oligomerization state of Thermotoga maritima Prxs. Analytical gel filtration of purified recombinant proteins was performed on a Superdex S200 column, pre-equilibrated with 30 mM Tris HCl pH 8.0, 200 mM NaCl. 250 μl samples of reduced proteins, at a concentration of 0.4 μg/μl, were loaded, and elution was carried out at a flow rate of 0.5 ml/min. TmPrx6 (theoretical mass of ca. 24 kDa) separated into two peaks: a major one with an estimated molecular mass of 41 kDa and a minor one of 80 kDa, suggesting that enzyme preparation contains both dimeric and tetramer forms. TmPrxBCP eluted as a major peak corresponding to a molecular mass of 35 kDa, which is consistent with a dimer, a monomer having a predicted molecular mass of 17.3 kDa. A minor peak at 70 kDa likely corresponding to a small fraction of tetramer is also visible. For as-isolated TmPrxNtr (theoretical monomeric mass of 37.2 kDa), an apparent molecular weight of 386 kDa, which is best interpreted as indicating a decamer, was found using DLS performed under non-reducing conditions. Pre-reducing TmPrxNtr decreased the apparent molecular mass to a value between 62 kDa, estimated by gel filtration, and 70 kDa, estimated by DLS, which is consistent with a dimeric form. In contrast, TmPrxNtrC40S displayed an apparent molecular mass of 74 kDa both in the presence and absence of reductants. DLS, dynamic light scattering.
Structure of TmPrxNtrC40S
Diffraction-quality crystals, with a homodimer in the asymmetric unit, were obtained for TmPrxNtrC40S. The Prx and Ntr domains of each monomer are connected by a short loop (Fig. 5). In the dimer, the A and B polypeptide chains are related by a non-crystallographic two-fold symmetry axis (0.51 Å rmsd for 311 α-carbons). The Ntr domain of one subunit is in contact with the Prx and Ntr domains of the other, but no contact was observed between Prx domains (Fig. 6A).
FIG. 5.
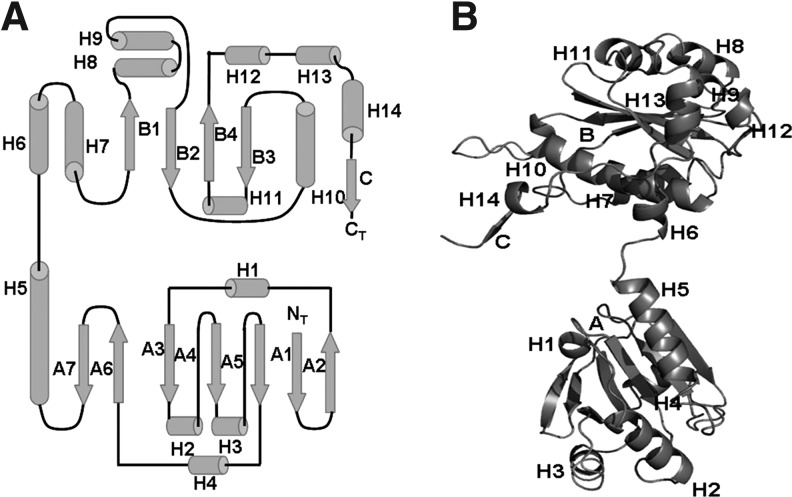
Monomer of TmNtrPrxC40S. (A) Topology diagram. Helices: In Prx domain: H1, H19-L22; H2, S38-R49; H3, V65-N75; H4, I88-F93; H5, H126-E140. In Ntr domain: H6, K146-W150; H7, R164-L176; H8, E194-A203; H9, Y209-N213; H10, Y236-M255; H11, P267-L274; H12, E294-E296; H13, E299-R306; H14, L314-I317. Sheets: In Prx domain: A, A1, E8-T11; A2, G14-T18; A3, Y26-F31; A4, A55-S61; A5, T80-S83; A6, S104-I108; A7, R115-R119. In Ntr domain: B, B1, R187-V191; B2, A216-S222; B3, V258-V262; B4, V281-G289; C, V318-W320. The C strand completes the B sheet of the other monomer. (B) Overall fold of the monomer. The Prx and Ntr domains are connected by a loop formed by three residues, Ser143, Leu144, and Asn145.
FIG. 6.
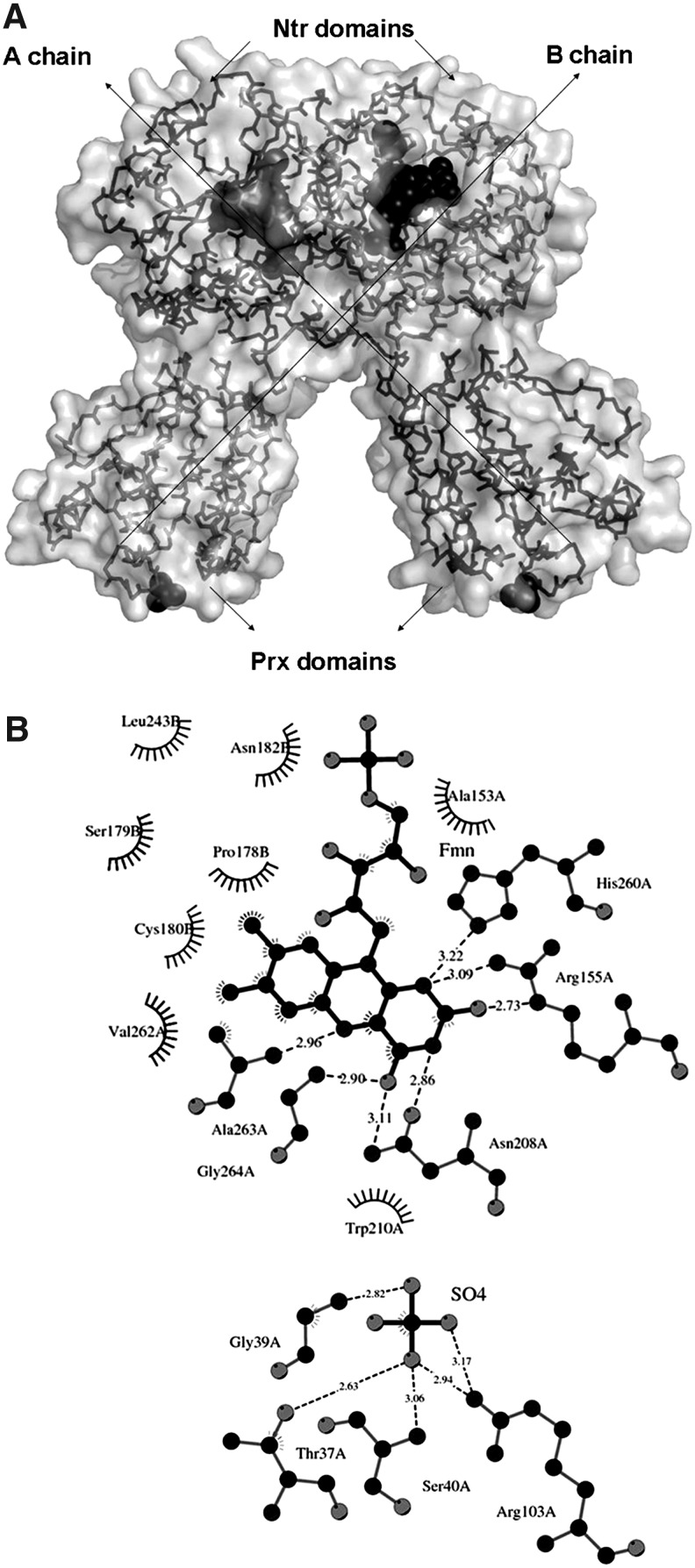
Active sites of TmNtrPrxC40S. (A) Transparent accessible surface representation of TmNtrPrxC40S. Main chain of subunits A (dark gray) and B (light gray) are represented in sticks, whereas FMN, SO4 molecules, and S atoms of disulfide bridges are represented as black spheres. (B) Schematic diagram generated using LIGPLOT showing the coordination of FMN and of SO4 binding in the active site of Prx module of chain A. In both chains, FMN and SO4 molecules have the same coordination.
The core of the Prx domains possesses a Trx-fold composed of a four-stranded β-sheet (A4, A6, A3, and A7) with three flanking α-helices (H2, H4, and H5). This core is associated with three additional β-strands (A1, A2, and A5) and two more helices (the 310-helices H1 and H3), forming a seven-stranded mixed β-sheet surrounded by five helices. A β-hairpin, formed by two small anti-parallel strands (residues 95–97 and 100–102), is present as a part of the loop connecting H4-helix with A6-strand. Ser40, which replaces the native CysP, is located at the N-terminus of the H2-helix. A sulfate ion is positioned in the fully folded catalytic site in both Prx domains, mimicking an enzyme reduced form. This ion is stabilized by hydrogen bond and salt bridge interactions with residues Thr37, Gly39, Ser40, Arg103, and active-site water molecules (Figs. 6B and 7A). The conserved Arg103 forms hydrogen bonds with Ser40, the sulfate ion, and a water molecule. This is different from other monomeric BCPs, where it forms hydrogen bonds with backbone carbonyls of the loop between A7-strand and H5-helix (4).
FIG. 7.
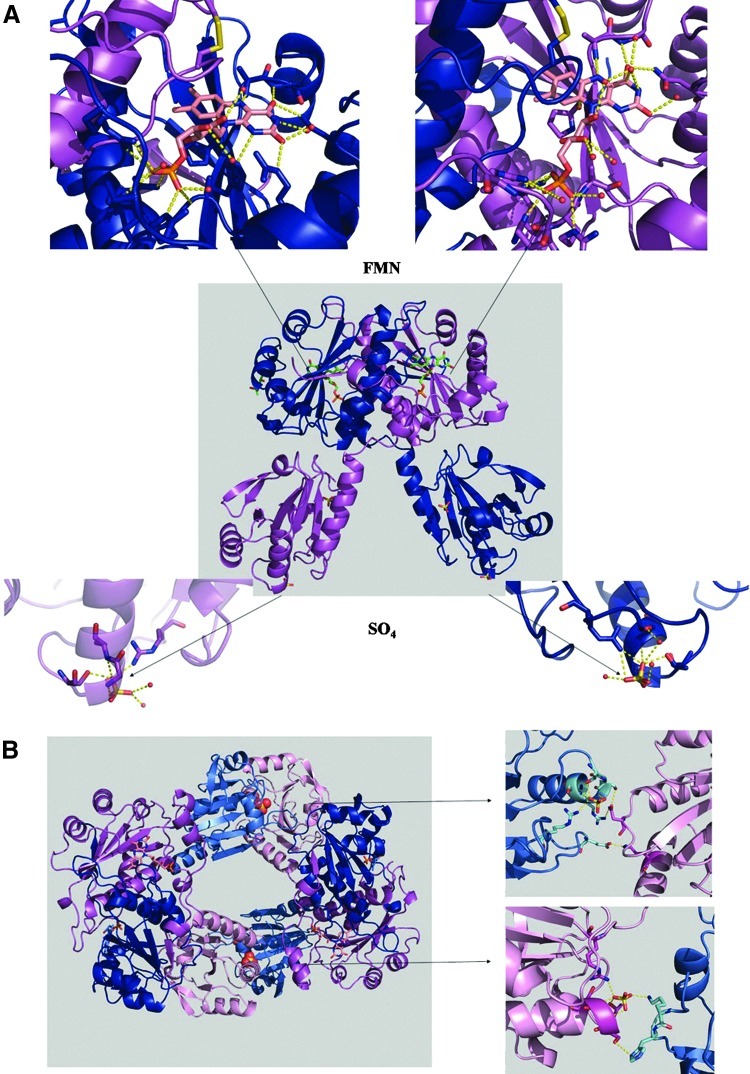
Cartoon diagram of TmNtrPrxC40S structure. (A) TmNtrPrxC40S dimer is shown in the middle with FMN, SO4 molecules into the active sites and into the cavities formed by the absence of the first amino acid, and MPD solvent molecule shown as stick models. In blue subunit A, magenta subunit B, yellow disulfide bonds. Around, zoom of the four active sites of the protein. Red spheres represent the water molecules located in these sites. (B) Crystal packing and intermolecular interactions. Prx modules are colored in marine and pink, and Ntr modules are colored in blue and magenta, in the A and B chains, respectively. On the left, view along the a-axis of two TmNtrPrxC40S dimer symmetrically related, linked via the Prx module. These bricks are piled along the a-axis forming channels. On the right, the two different networks of contact between Prx modules that promote the formation of these channels. At top: Ser38 A/Asn51 Bsym (3.0 Å), Arg42 A/Asn51 Bsym (3.1 Å), Glu124 A/Lys131 Bsym (2.6 Å). At bottom: Ser38 B/His6 Asym (3.4 Å), SO4 B/Lys5 Asym (2.7 Å). (To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars.)
The Ntr domains possess a three-layer α−β−α sandwich fold. Their C-terminal regions, consisting of the short H14-helix and the C-strand, overlap the surface of a neighboring monomer, acting as a fifth strand to the β-sheet. Each Ntr domain binds one FMN non-covalently (Fig. 6A). The cap covering the cofactor site is formed by the H12-helix and the loop between H7-helix and B1-strand of the other subunit, a region containing the Cys180-Cys228 disulfide. DTNB titrations of TmPrxNtr-Ntr2 thiols indicated that this disulfide is not reduced by electrons transferred from NAD(P)H via FMN, suggesting that it plays a structural, rather than catalytic, role. The re-face of FMN is solvent accessible, whereas the si face is buried. FMN binding involves hydrogen bonds to the residues of one subunit and hydrophobic contacts with both subunits (Figs. 6B and 7A). The oxygen of the phosphate group of FMN makes hydrogen bonds with several Arg residues (151, 152, 309, and 311). The main chain amide of Ala263 interacts with N5 of the isoalloxazine moiety, the possible acceptor of hydride ion, while N1–C2=O2 interacts with Arg155. The main chain carbonyl oxygen of Pro261 is near the π system of the FMN isoalloxazine (3.2 Å to C9A). Several water molecules located in this region also interact with FMN.
In TmPrxNtrC40S crystals, the dimers are perpendicular and interact in an inverted head-to-head orientation (Fig. 7B). These piled structures form channels along the a-axis. Since TmPrxNtrC40S only forms dimers in solution, this higher-order organization might arise from crystal packing between TmPrxNtrC40S molecules.
What Is the Physiological Interest of Fusing a Peroxidase to a Ntr Domain?
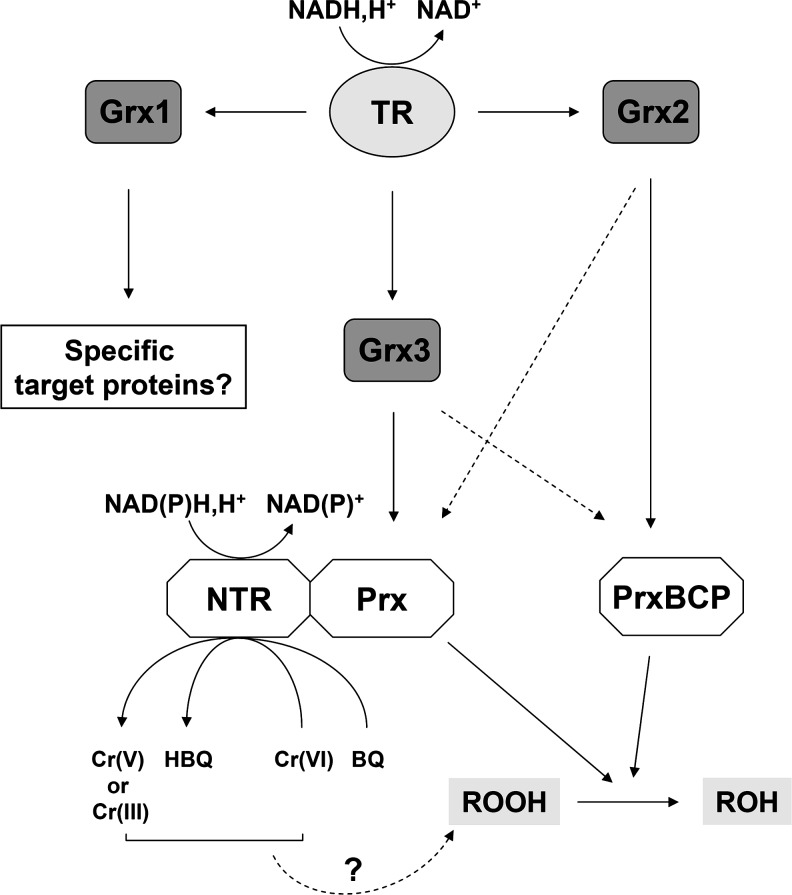
The fusion of genes is usually indicative of a functional link between their products. Although FMN redox potential and affinity are modified in TmPrxNtrC40S or TmPrxNtr-Ntr2 and the crystal structure of the TmPrxNtrC40S variant indicates that both domains can interact, electron transfer between the two modules of TmPrxNtr is unlikely, because the Prx active site is distant from the Ntr domain disulfide and as the disulfide is not reduced by FMN. We have demonstrated that the Ntr domain of TmPrxNtr, similar to several oxidoreductases (1), can reduce chromate and quinones with catalytic efficiencies between 106 and 107 M−1 s−1. Thus, a possible explanation for the linkage of the two domains is that the Prx module might rapidly remove peroxides arising from reduction of chromate-containing compounds by the Ntr module (Fig. 8).
FIG. 8.
Proposed regeneration mechanism for T. maritima PrxBCP-containing proteins. In TmPrxNtr, the Ntr domain, which exhibits quinone and chromate reductase activities, cannot provide electrons to the BCP domain, which could instead be regenerated by an NADH/TR/Grx-like system. Using the kcat/KGrx kinetic parameter, a slight preference of TmPrxNtr-Prx for TmGrx3 and of TmPrxBCP for TmGrx2 was obtained. Our current model with regard to the existence of this fusion enzyme in Thermotogales is that the Prx domain will scavenge the peroxide formed from the functioning of the Ntr module. TmPrx6 has not been represented here, as we did not detect any activity. BQ, benzoquinone; HBQ, hydrobenzoquinone; TR, thioredoxin reductase.
Notes
Cloning, expression, and purification of full-length, truncated, and mutated recombinant proteins
The open-reading frames of TmTR, TmGrx1 to 3, TmPrx6, TmPrxBCP, and TMPrxNtr were amplified by polymerase chain reaction from genomic DNA of T. maritima and cloned into pET-3d using primers listed in Table 2. For TmGrx2, TmPrxBCP, TmPrx6, and TmPrxNtr, owing to the use of the NcoI restriction, a codon for an alanine was added in the primer to keep the sequence in frame. For TmPrxNtr, the Prx and Ntr domains were expressed separately, and TmPrxNtr-Prx was cloned using TmPrxNtr for and TmPrxNtr-Prx rev primers; whereas TmPrxNtr-Ntr1 and TmPrxNtr-Ntr2 were cloned using TmPrxNtr rev and TmPrxNtr-Ntr for TmPrxNtr-Ntr, respectively. TmPrxNtr-Ntr2 is 13 amino acids longer at the N-terminus compared with TmPrxNtr-Ntr1. The substitution of CysP (Cys40) of the Prx module by serine, in TmPrxNtr or TmPrxNtr-Prx, and of CysR (Cys50) of TmPrxBCP, was accomplished by site-directed mutagenesis using two complementary mutagenic primers (Table 2). The residue numbering used to designate these cysteines differs by one amino acid compared with the theoretical sequence, due to the presence of the alanine after the initial methionine in these proteins.
Table 2.
List of Primers Used in This Study
| Name | Nucleotide sequence | N-ter or C-ter protein sequence |
|---|---|---|
| TmPrxNtr for | 5′ CCCCCCATGGCTAGGGTGAAGCACTTTGAA 3′ | MARVKHFE |
| TmPrxNtr rev | 5′ CCCCGGATCCCTAAAGGTTCCACCTGAC 3′ | VRWNL |
| TmPrxNtr C40S for | 5′ GCAGGAACGAGCGGTAGCACGAGAGAAGCCGTC 3′ | |
| TmPrxNtr C40S rev | 5′ GACGGCTTCTCTCGTGCTACCGCTCGTTCCTGC3′ | |
| TmPrxNtr-Prx rev | 5′ CCCCGGATCCTTAGTCTTCTTCTATCAGTCG 3′ | RLIEED |
| TmPrxNtr-Ntr for | 5′ CCCCCCATGGCTTTGAAAAAAGACAGAGTACCG 3′ | MALKKDRVP |
| TmPrxNtr-Ntr for2 | 5′ CCCCCCATGGCTCTGAACAAGCACATAGAG 3′ | MALNKHIE |
| TmTR for | 5′ CCCCCCATGGTTTTCTTTGATACTGGATCC 3′ | MVFFDTGS |
| TmTR rev | 5′ CCCCCCATGGTCAGAAGTAATGTTTTGCGGC 3′ | AAKHYF |
| TmGrx1 for | 5′ CCCCCCATGGCTAAAAAGGTGGAGATACTA 3′ | MAKKVEIL |
| TmGrx1 rev | 5′ CCCCGGATCCTTAAGCCTGCTGAAGGACCTT 3′ | KVLQQA |
| TmGrx2 for | 5′ CCCCCCATGGCTCAGCACCTGAAAATCAAA 3′ | MAQHLKIK |
| TmGrx2 rev | 5′ CCCCGGATCCTTACGATATTCCAAGGAG 3′ | LLGIS |
| TmGrx3 for | 5′ CCCCCCATGGGTATTTTGTCGGAC 3′ | MGILSD |
| TmGrx3 rev | 5′ CCCCGGATCCTCACCCCTTTGCGAGTCT 3′ | RLAKG |
| TmPrxBCP for | 5′ CCCCCCATGGCTTTGAAATCAGGCGATAAG 3′ | MALKSGDK |
| TmPrxBCP rev | 5′ CCCCGGATCCTTATTTTGAAAGACGCTCGAG 3′ | LERLSK |
| TmPrxBCP C50S for | 5′ TGTGAGAAAGAACTCTCCACGTTCAGGGATTCT 3′ | |
| TmPrxBCP C50S rev | 5′ AGAATCCCTGAACGTGGAGAGTTCTTTCTCACA 3′ | |
| TmPrx6 for | 5′ CCCCCCATGGCTGAAGGAAGAATTCCTCTC 3′ | MAEGRIPL |
| TmPrx6 rev | 5′ CCCCGGATCCTTATACCTTTTTGTAACA 3′ | CYKKV |
The NcoI and BamHI restriction sites are underlined, and the mutagenic codons as well as the GCT codon that are added in some sequences to keep them in frame are in bold characters.
The Escherichia. coli expression strain BL21(DE3), containing the pSBET plasmid, was transformed with each recombinant plasmid. Luria Bertani cultures grown at 37°C were progressively increased to 2 l and induced at the exponential phase by the addition of 100 μM isopropyl-ß-D-thiogalactopyranoside. The bacteria were pelleted by centrifugation for 15 min at 4400 g and resuspended in a 30 mM Tris-HCl pH 8.0, 1 mM ethylenediaminetetraaceticacid (EDTA), and 200 mM NaCl buffer. Subsequent purification steps, that is, cell lysis, ammonium sulfate precipitation, gel filtration (ACA44), and anion exchange (diethylaminoethyl [DEAE] Sephacel), were performed at 4°C. For TmPrxNtr, TmPrxNtrC40S, and TmPrxNtr-Ntr2, an important amount of free flavin was visibly released and very lately eluted. TmPrxNtr-Ntr1, which did not incorporate FMN, was not characterized further. With regard to the DEAE chromatography, TmTR, TmGrx3, TmPrx1, TmPrxBCP wt and C50S, TmPrxNtr wt and C40S, TmPrxNtr-Prx wt and C40S, and TmPrxNtr-Ntr2 were retained on the column; whereas TmGrx1 and TmGrx2 passed through. Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS PAGE) was used to check protein homogeneity, and mass spectrometry analyses were performed to unambiguously confirm the identity of the protein.
Determination of FMN content
After protein precipitation of TmPrxNtr, TmPrxNtrC40S, and TmPrxNtr-Ntr2 by the addition of trichloroacetic acid (to a final concentration of 10%), the pellet was resuspended in buffer 30 mM Tris-HCl pH 8.0, 1 mM EDTA, and 2% SDS and used to measure protein concentration at 280 nm. The FMN-containing supernatant was neutralized with NaHCO3 (a final concentration of 330 mM). FMN concentration was calculated from the absorption spectra using an extinction coefficient of 12,500 M−1 cm−1 at 446 nm for free FMN.
Determination of flavin stoichiometry and binding affinity constants
Intrinsic tryptophan-emission fluorescence measurements were performed using Bowman 2 Amincon or Fluoromax Spex spectrometers. Measurements were recorded at room temperature in a 1×0.4 cm cuvette under continuous stirring. The excitation wavelength was set to 290 nm. Emitted light was detected between 310 and 380 nm. Stoichiometry determination assays were performed using 20 μM TmPrxNtr or TmPrxNtr-Ntr2 in 50 mM Tris-HCl pH 7.6. For Kd determination, concentrations of TmPrxNtr or TmPrxNtr-Ntr2 in the range 1–5 μM were used, and aliquots from 0.5 to 1.45 mM FMN stock solutions were added stepwise to obtain a final concentration of twice the protein concentration. Fluorescence intensity signals at 340 nm were first corrected from the dilution due to the addition of flavin solutions, and then from the decrease of signal induced by the absorption of the incident light at 290 nm by the flavin species.
Determination of the oligomerization state
The oligomerization state of purified Prxs was analyzed by size-exclusion chromatography. Samples containing 100 μg of protein were loaded onto Superdex S200 10/300 columns equilibrated in 30 mM Tris-HCl pH 8.0, 200 mM NaCl and connected to an Akta purifier system (GE Healthcare). The flow rate was fixed at 0.5 ml min–1, and detection was recorded at 214 and 280 nm. The columns were calibrated using the 29–700 kDa molecular weight standards (Sigma). DLS measurements were performed at 20°C using a DynaPro-MS800 molecular-sizing instrument (Wyatt Technology). The samples in 100 mM Tris-HCl buffer pH 8.5, 0.1 mM NaCl, were loaded into 45 μl quartz cuvette. The hydrodynamic radii and molecular masses were determined from 50 DLS measurements (each measurement is a sum of 5 s photon acquisition), analyzed using Dynamics 6.9.2.11 software.
Determination of midpoint redox potentials
For disulfide-containing proteins, oxidation-reduction titrations were carried out at pH 7.0, as in (7). Oxidation-reduction titrations of FMN in TmPrxNtr proteins were carried out electrochemically under an argon atmosphere using an anaerobic titration vessel with a 0.3 mm optical pathlength and a BAS Voltammograph potentiostat to control potentials. Complete visible spectra were recorded at each ambient potential (Eh) value, using a Shimadzu UV-2401PC spectrophotometer. The reaction mixtures contained buffer (pH 8.0) and the following oxidation-reduction mediators, all at a concentration of 5 μM: anthraquinone-2-sulfonate; 2-hydroxy-1,4-naphthoquinone and safranin O for TmPrxNtr and TmPrxNtrC40S and anthraquinone-2-sulfonate; 2-hydroxy-1,4-naphthoquinone and benzyl viologen for TmPrxNtr-Ntr2. The change in absorbance at 465/462/460 nm minus 402 nm (TmPrxNtr/TmPrxNtrC40S/TmPrxNtr-Ntr2), as a function of Eh, was fitted to the Nernst Equation for a two-electron carrier, and the best fit was used to calculate the midpoint potential (Em) values.
pKa determination of Prx CysP
The determination of CysP pKa values of TmPrxNtr-Prx and TmPrxBCPC50S was performed using (2-pyridyl)dithiobimane at 50°C. Reactions were started by the addition of PDT-bimane to a final concentration of 25 μM to 10 μM reduced proteins in 500 μl of sodium citrate or phosphate buffer ranging from pH 3.0 to 8.0. The absorbance at 343 nm was recorded over 120 min with a Cary 50 spectrophotometer (Agilent). Absorbance data were fitted directly to the Michaelis- Menten equation with the GraphPad Prism 5 program and the t1/2 (the time to reach half-maximal reactivity as monitored by half-maximal release of pyridyl-2-thione) at each pH was determined. Those values were plotted against pH using sigmoidal curve fit and GraphPad Prism 5 (GraphPad software).
Insulin and DTNB reduction
The insulin reduction assay was carried out at 25°C in a 500 μl reaction mixture containing 100 mM phosphate pH 7.0, 2 mM EDTA, 0.75 mg/ml bovine insulin, and either with 330 μM dithiothreitol (DTT) and 2 μM TmGrx1, 2 μM TmGrx2 or 5 μM TmGrx3. The reduction of insulin was monitored as the increase in turbidity at 650 nm due to insulin precipitation. Non-enzymatic reduction of insulin by DTT was monitored in the absence of TmGrxs.
The ability of TmGrx1-3 to catalyze the reduction of DTNB in the presence of TmTR was measured at 25°C by monitoring the increase in absorbance at 412 nm caused by the release of TNB−. The reaction medium contained 30 mM Tris-HCl pH 8.0, 200 μM NADH, 950 nM TmTR, 100 μM DTNB, and varying concentrations of TmGrxs ranging from 0.13 to 25 μM. Control experiments were performed in the same conditions in the absence of TmGrx. Activity was expressed as nmol of TNB− released per nmol TmTR per second using a molar extinction coefficient of 13,600 M−1 cm−1 at 412 nm for TNB−. The apparent Km values were calculated by a non-linear regression using the program GraphPad Prism 4. DTNB reduction by TmPrxNtr (1.25 μM) was also assayed in the same conditions but at 50°C.
Peroxidase, chromate, and quinone reductase activities
The peroxidase activity of TmPrx6, TmPrxBCP, TmPrxNtr-Prx wt, and C40S and the activities of the flavoproteins (TmPrxNtr, TmPrxNtrC40S, and TmPrxNtr-Ntr2) versus benzoquinone, chromate (K2CrO4), and H2O2 were determined at 50°C under steady-state conditions by monitoring absorbance changes at 340 nm, arising from NADH or NADPH oxidation respectively, using a Cary 50 spectrophotometer (Agilent). For peroxidase activity measurements, the 500 μl reaction contained 30 mM Tris-HCl pH 8.0, 1 mM EDTA, 210 μM NADH, and 300 nM TmTR, varying concentrations of TmGrxs (from 0 to 35 μM) and of H2O2 (from 0 to 500 μM) and either 0.48 μM TmPrxBCP or 1.5 μM TmPrxNtr-Prx or 8 μM TmPrx6, unless otherwise indicated. The peroxidase activity was expressed as nmol NADH oxidized per nmol Prx per second using a molar extinction coefficient of 6220 M−1 cm−1 at 340 nm for NADH. For the other activities, a 500 μl reaction mixture contained 180 μM NADPH, 6.25, 25, or 900 nM enzymes with benzoquinone, K2CrO4, and H2O2 respectively and various amounts of substrates ranging from 0 to 200 μM (benzoquinone, K2CrO4) or to 2.5 mM (H2O2). The activities were expressed as nmol NADPH oxidized per nmol FMN per second using a molar extinction coefficient of 6220 M−1 cm−1 at 340 nm for NADPH. Steady-state kinetic parameters were obtained by varying concentrations of one substrate at a fixed saturating concentration of the other substrate. The apparent Km values were calculated by a non-linear regression using the program GraphPad Prism 4.
Crystallization, X-ray data collection, structure solution, and refinement of TmPrxNtrC40S
Initial identification of crystallization conditions was carried out using the vapour diffusion method in Cartesian technology workstation. Four hundred nanoliter of sitting drops of 1:1 mixture of TmPrxNtrC40S and a solution of crystallization (672 different conditions, commercially available) equilibrating against 150 μl reservoir in Greiner plate were used. The protein crystallizes only in the condition C4 from JBSscreen Classic 7 from JenaBioscience. The best crystals were obtained manually in limbro plates by the hanging-drop method at 18°C by mixing 1.5 μl of protein solution at 24.5 mg/ml with 1.5 μl of reservoir solution containing 30% (v/v) 2-methyl-2,4-pentanediol (MPD), 5% (w/v) PEG4000, and 0.1 M HEPES-Na pH 7.5. Pale yellow crystals of dimensions of up to 0.1×0.2×0.4 mm and that crystallized in space group P212121-unit cell parameters (a=60.5, b=113.18, c=121.83 Å) appeared within 2 weeks. The crystallization solution was used as a cryoprotectant for flash-frozing crystals in liquid nitrogen.
X-ray diffraction data up to 1.65 Å resolution were collected at 110 nK temperature on PROXIMA-1 beamline at Synchrotron SOLEIL, using wavelength 0.82656 Å and Dectris Pilatus 6M detector. The data were indexed and integrated with XDS, scaled, and merged with SCALA from the CCP4 package, giving a data set ∼99.9% complete with an overall Rmerge of 6.3% on intensities. The TmPrxNtrC40S crystal structure was determined by molecular replacement with the AMoRe program using data from 15 to 3 Å resolution. The Sulfolobus solfataricus Bcp1 (PDB ID: 3DRN) and a homology model built using the 322 residues of TmPrxNtrC40S with I-TASSER provided the peroxidase and Ntr models. The solution was found by searching four independent molecules with two sets of coordinates associated to the peroxidase and Ntr modules, respectively. The orientations of the molecules associated to the same model were constrained to satisfy the non-crystallographic two-fold axis resulting from the self-rotation function.
The model was adjusted manually using the COOT program. REFMAC 5.0 program was then used for refinement to a final Rcryst of 17.8% and Rfree of 20.7%. Most residues correspond to well-defined electron densities, except the first N-terminal residue of both monomers that did not appear, pointing to their cleavage. The cavities formed by the loss of the first amino acids are partially occupied by sulfate ions. A MPD molecule and 612 water molecules were placed. Last refinement and model validation (98.9% Ramachandran favored, 0% outliers) have been undertaken with the PHENIX and MolProbity programs. All structural representations were generated by the PyMOL program. Atomic coordinates and structure factors were deposited in the Protein Data Bank (4EO3 accession number).
Abbreviations Used
- BCP
bacterioferritin comigratory protein
- BQ
benzoquinone
- CysP
peroxidatic cysteine
- CysR
resolving cysteine
- DEAE
diethylaminoethyl
- DLS
dynamic light scattering
- DTNB
5,5′-dithio-bis-2-nitrobenzoic acid
- DTT
dithiothreitol
- EDTA
ethylenediaminetetraaceticacid
- FMN
flavin mononucleotide
- FMNH2
flavin mononucleotide reduced
- Grx
glutaredoxin
- HBQ
hydrobenzoquinone
- MPD
2-methyl-2,4-pentanediol
- Nox
NADH oxidase
- Ntr
nitroreductase
- PAGE
polyacrylamide gel electrophoresis
- Prx
peroxiredoxin
- SDS
sodium dodecylsulfate
- Tpxs
thiol-peroxidases
- TR
thioredoxin reductase
- Trx
thioredoxin
Acknowledgments
The oxidation-reduction titrations were carried out using support from the Division of Chemical Sciences Geosciences and Biosciences, Office of Basic Energy Sciences of the U.S. Department of Energy through Grant DE-FG03-99ER20346 (to D.B.K.). The authors acknowledge the Synchrotron Soleil (Proxima1) for provision of synchrotron radiation facilities and their staff for their helpful assistance. This work was also supported by Institut Universitaire de France grants to Jean Pierre Jacquot and Nicolas Rouhier.
References
- 1.Ackerley DF. Gonzalez CF. Keyhan M. Blake R., 2nd Matin A. Mechanism of chromate reduction by the Escherichia coli protein, NfsA, and the role of different chromate reductases in minimizing oxidative stress during chromate reduction. Environ Microbiol. 2004;6:851–860. doi: 10.1111/j.1462-2920.2004.00639.x. [DOI] [PubMed] [Google Scholar]
- 2.Clarke DJ. Mackay CL. Campopiano DJ. Langridge-Smith P. Brown AR. Interrogating the molecular details of the peroxiredoxin activity of the Escherichia coli bacterioferritin comigratory protein using high-resolution mass spectrometry. Biochemistry. 2009;48:3904–3914. doi: 10.1021/bi900189e. [DOI] [PubMed] [Google Scholar]
- 3.Clarke DJ. Ortega XP. Mackay CL. Valvano MA. Govan JR. Campopiano DJ. Langridge-Smith P. Brown AR. Subdivision of the bacterioferritin comigratory protein family of bacterial peroxiredoxins based on catalytic activity. Biochemistry. 2010;49:1319–1330. doi: 10.1021/bi901703m. [DOI] [PubMed] [Google Scholar]
- 4.Hall A. Nelson K. Poole LB. Karplus PA. Structure-based insights into the catalytic power and conformational dexterity of peroxiredoxins. Antioxid Redox Signal. 2011;15:795–815. doi: 10.1089/ars.2010.3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reeves SA. Parsonage D. Nelson KJ. Poole LB. Kinetic and thermodynamic features reveal that Escherichia coli BCP is an unusually versatile peroxiredoxin. Biochemistry. 2011;50:8970–8981. doi: 10.1021/bi200935d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reynolds CM. Meyer J. Poole LB. An NADH-dependent bacterial thioredoxin reductase-like protein in conjunction with a glutaredoxin homologue form a unique peroxiredoxin (AhpC) reducing system in Clostridium pasteurianum. Biochemistry. 2002;41:1990–2001. doi: 10.1021/bi011802p. [DOI] [PubMed] [Google Scholar]
- 7.Rouhier N. Gelhaye E. Gualberto JM. Jordy MN. De Fay E. Hirasawa M. Duplessis S. Lemaire SD. Frey P. Martin F. Manieri W. Knaff DB. Jacquot JP. Poplar peroxiredoxin Q. A thioredoxin-linked chloroplast antioxidant functional in pathogen defense. Plant Physiol. 2004;134:1027–1038. doi: 10.1104/pp.103.035865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rouhier N. Jacquot JP. The plant multigenic family of thiol peroxidases. Free Radic Biol Med. 2005;38:1413–1421. doi: 10.1016/j.freeradbiomed.2004.07.037. [DOI] [PubMed] [Google Scholar]
- 9.Yang X. Ma K. Characterization of a thioredoxin-thioredoxin reductase system from the hyperthermophilic bacterium Thermotoga maritima. J Bacteriol. 2010;192:1370–1376. doi: 10.1128/JB.01035-09. [DOI] [PMC free article] [PubMed] [Google Scholar]