Abstract
Mutations in LMNA, the gene that encodes A-type lamins, cause multiple diseases including dystrophies of the skeletal muscle and fat, dilated cardiomyopathy, and progeria-like syndromes (collectively termed laminopathies). Reduced A-type lamin function, however, is most commonly associated with skeletal muscle dystrophy and dilated cardiomyopathy rather than lipodystrophy or progeria. The mechanisms underlying these diseases are only beginning to be unraveled. We report that mice deficient in Lmna, which corresponds to the human gene LMNA, have enhanced mTORC1 (mammalian target of rapamycin complex 1) signaling specifically in tissues linked to pathology, namely, cardiac and skeletal muscle. Pharmacologic reversal of elevated mTORC1 signaling by rapamycin improves cardiac and skeletal muscle function and enhances survival in mice lacking A-type lamins. At the cellular level, rapamycin decreases the number of myocytes with abnormal desmin accumulation and decreases the amount of desmin in both muscle and cardiac tissue of Lmna–/– mice. In addition, inhibition of mTORC1 signaling with rapamycin improves defective autophagic-mediated degradation in Lmna–/– mice. Together, these findings point to aberrant mTORC1 signaling as a mechanistic component of laminopathies associated with reduced A-type lamin function and offer a potential therapeutic approach, namely, the use of rapamycin-related mTORC1 inhibitors.
INTRODUCTION
A-type lamins are type V intermediate filament proteins that form part of the nuclear lamina, providing structural integrity to the nucleus (1, 2) and regulating chromatin organization (3), transcription (4, 5), and DNA replication (6). In humans, mutations in LMNA result in several distinct diseases including autosomal dominant limb-girdle muscular dystrophy (LGMD1B) (7), Emery-Dreifuss muscular dystrophy (EDMD2/3) (8), dilated cardiomyopathy (DCM) and conduction-system disease (CMD1A) (9), familial partial lipodystrophy (10), Charcot-Marie-Tooth disease (11), and Hutchinson-Gilford progeria syndrome (HGPS) (12). Skeletal muscle dystrophy and dilated cardiomyopathy are commonly seen in people with LMNA missense mutations that impair the function of the protein (often in a dominant-negative fashion) (13–15). Stop codons, splice site variants, or insertions/deletions within the human lamin A gene can also reduce the amount of lamin A/C proteins (13–15). Lmna–/– mice also develop both DCM and skeletal muscle dystrophy, succumbing to complications resulting from cardiac dysfunction by 6 to 8 weeks of age (16, 17). Thus, Lmna–/– mice can serve as a clinically relevant model in which to investigate the molecular mechanisms of skeletal muscle dystrophy and DCM caused by reduced lamin A protein concentrations.
Cardiac and skeletal muscle tissues are highly adaptable and many signaling pathways regulate muscle remodeling. Muscle remodeling involves changes in the myofiber cytoarchitecture, gene expression, and protein composition in response to functional demands. One such path-way involves the mammalian target of rapamycin complex 1(mTORC1), which is a major regulator of protein synthesis. The mTORC1 complex consists of mTOR, regulatory associated protein of mTOR (Raptor) (18, 19), and mlst8/GβL (20). Activation of the mTORC1 signaling cascade, among other events, results in the phosphorylation of downstream substrates such as p70 S6 kinase and 4E-BP1 [eukaryotic translation initiation factor 4E (eIF4E)–binding protein 1] (21, 22), which in turn affect protein synthesis. Activation of S6 kinase by phosphorylation, for instance, results in the subsequent phosphorylation of ribosomal protein S6 (rpS6) and other components of the translational machinery, whereas phosphorylation of 4E-BP1 decreases its binding to eIF4E, freeing this initiation factor to promote cap-dependent translation (21). Enhanced mTOR-mediated phosphorylation of 4E-BP1 and S6 kinase is evident during normal cardiac and skeletal muscle remodeling as well as in the pathologic cardiac and skeletal remodeling that occurs in disease (23–27).
Another important function of the mTORC1 signaling pathway is the regulation of autophagy, a process in which the cell degrades damaged or excess cellular components—from individual proteins and protein aggregates to whole organelles—through the lysosomal machinery. When cells have sufficient nutrients, autophagy is inhibited by mTORC1; on the other hand, lack of nutrients inhibits the mTORC1 pathway and autophagy is initiated to reallocate nutrients from nonessential components to those that are vital to survival (28). Activation of mTORC1 in mammalian cells results in the phosphorylation of Unc-51–like kinase/human autophagy-related protein 1 (ULK1/hAtg1), disrupting its interaction with adenosine monophosphate–activated protein kinase (AMPK) and inhibiting AMPK-mediated ULK1 activation (29). Autophagy is important for protein homeostasis in cardiac and skeletal muscle; changes in autophagic markers in these tissues can be detected after nonpathological stimulation such as exercise, as well as in disease-induced situations such as dilated cardiomyopathy and muscular dystrophy (30–33).
Because of the importance of mTORC1 signaling in skeletal and cardiac tissue, we hypothesized that dysregulation of mTORC1 signaling may contribute to the cardiac and skeletal muscle pathology of Lmna–/– mice. Here, we examine mTORC1 signaling in Lmna–/– mice and test the effects of pharmacological manipulation of this pathway by rapamycin on Lmna–/– disease phenotypes.
RESULTS
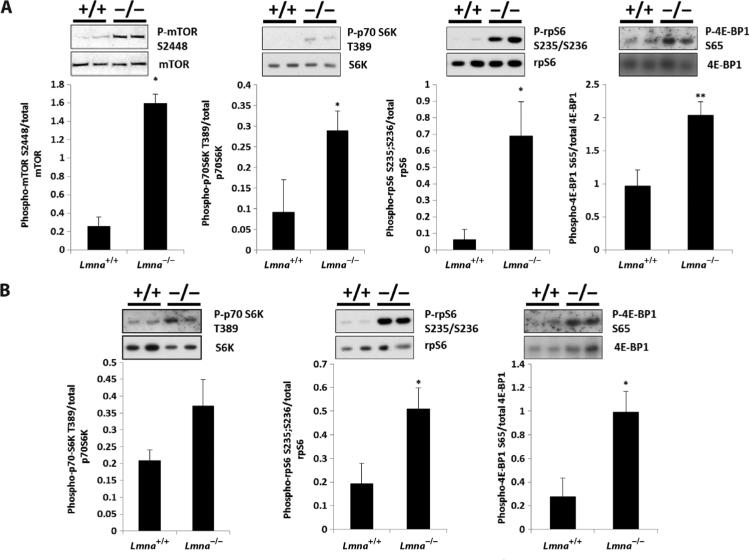
To test the hypothesis that dysregulation of mTORC1 signaling may contribute to the cardiac and skeletal muscle pathology of Lmna–/– mice, we looked at phosphorylation of downstream signaling components of the mTORC1 pathway in these mice by Western blot analysis. Consistent with this hypothesis, we observed significantly increased phosphorylation of mTOR(S2448) (P = 0.01), S6 kinase(T389) (P = 0.04), rpS6(S235/S236) (P = 0.02), and 4E-BP1(S65) (P = 0.005) in heart and increased phosphorylation of rpS6(S235/S236) (P = 0.03) and 4E-BP1(S65) (P = 0.01) in skeletal muscle of 4-week-old Lmna–/– animals compared to wild-type controls (Fig. 1, A and B). This hyperactivation of the mTORC1 pathway was not observed in nondiseased tissue such as liver (fig. S1C). Expression of mTORC1 components Raptor and GβL was unchanged in heart and skeletal muscle of Lmna–/– mice (fig. S1, A and B). The increased mTORC1 signaling in Lmna–/– mouse hearts was associated with early signs of left and right ventricular dilatation but not hypertrophy (fig. S1D). Indeed, other studies have noted the absence or attenuation of a hypertrophic response in both Lmna–/– and Lmna+/– mice with hearts subjected to pressure overload by transverse aortic constriction (17, 34). To further understand the impact of increased mTORC1 signaling in Lmna–/– mice, we performed polysome profile analysis on heart tissue using a protocol that was sufficiently sensitive to detect an angiotensin-induced increase in protein synthesis in wild-type heart tissue. Consistent with a lack of hypertrophy, hyperactivated mTORC1 signaling in Lmna–/– mice did not result in an alteration of global protein synthesis: Polysome profiles of heart tissue from Lmna–/– mice were not significantly different from those of Lmna+/+ mice (fig. S1, E and F).
Fig. 1.
Signaling through the mTORC1 pathway is increased in Lmna–/– mice. (A) Western blot analysis of Lmna+/+ (n = 7) and Lmna–/– (n = 8) mice heart tissue lysates. Phosphorylated mTOR(S2448), S6 kinase(T389), rpS6(S235/S236), and 4E-BP1(S65) are increased in Lmna–/– mice compared to Lmna+/+ mice. (B) Western blot analysis of Lmna+/+ (n = 7) and Lmna–/– (n = 8) mice skeletal muscle (quadriceps) tissue lysates. Phosphorylated rpS6(S235/S236) and 4E-BP1(S65) are increased in Lmna–/– mice compared to Lmna+/+ mice. However, phosphorylated S6 kinase(T389) is not significantly increased. *P < 0.05; **P < 0.01.
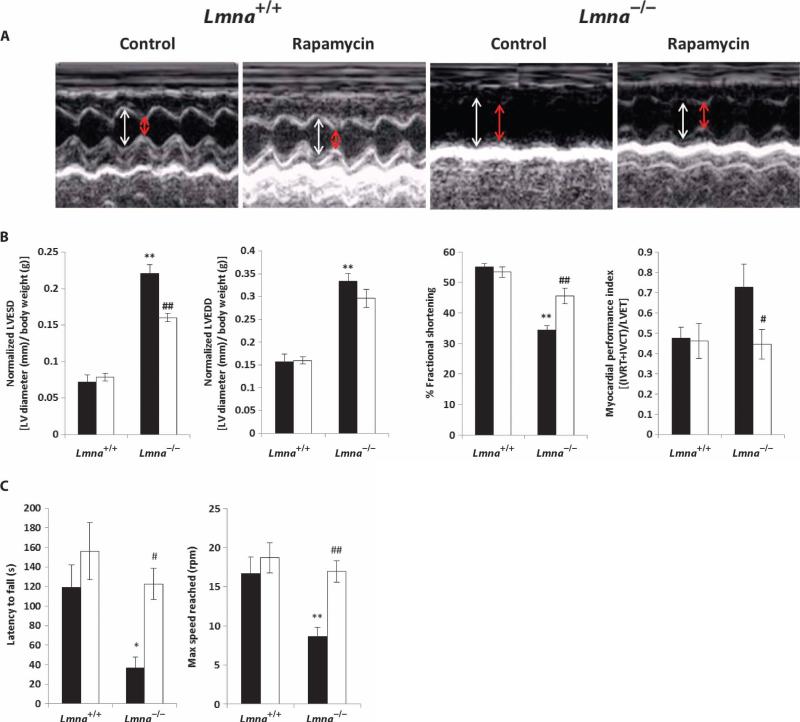
The small molecule rapamycin is a specific inhibitor of mTORC1, and derivatives are used clinically in cancer treatment, prevention of organ transplant rejection, and prevention of restenosis after angioplasty (35–38). Rapamycin also extends life span in genetically heterogeneous mice and in inbred C57BL/6 mice (39–41). To determine whether hyperactivation of the mTORC1 pathway contributes to the DCM and skeletal muscle dystrophy associated with loss of Lmna, we fed Lmna–/– mice chow containing encapsulated rapamycin beginning at an age of 3 to 4 weeks using the same protocol previously reported to extend life span (39). By 2 weeks of treatment, rapamycin was detectable in the blood of Lmna–/– mice (6.9 ± 0.45 ng/ml). We then used transthoracic echocardiography to determine the effect of dietary rapamycin on heart function in Lmna–/– mice. Echocardiographic analysis confirmed the results of previous studies that showed impaired cardiac function and left ventricular dilatation at 6 weeks of age in Lmna–/– mice compared to Lmna+/+ mice (17) (Fig. 2, A and B). This deficit is reflected in a significant increase in left ventricular end diastolic diameter (LVEDD) and left ventricular end systolic diameter (LVESD) (normalized to body weight) in Lmna–/– mice (both P < 0.001), which results in a significant decrease in fractional shortening (the fraction of diastolic diameter lost in systole) (P < 0.001) (Fig. 2B). Lmna–/– mice treated with rapamycin showed a significant decrease in LVESD (P < 0.001) relative to untreated Lmna–/– animals, as well as suppression of the defects in both fractional shortening (P < 0.001) and myocardial performance index (MPI) (P < 0.05). These data indicate that rapamycin improves cardiac function in mice lacking A-type lamins.
Fig. 2.
Treatment of Lmna–/– mice with rapamycin improves heart and skeletal muscle function. (A) Representative echocardiograms of Lmna+/+ and Lmna–/– mice on a control or rapamycin diet. White arrow, LVEDD; red arrow, LVESD. (B) Normalized LVESD (P < 0.001), normalized LVEDD, fractional shortening, and MPI. Values are significantly different in Lmna–/– mice fed control diet (n = 7) compared to Lmna+/+ mice fed control diet (n = 4). Rapamycin in the diet significantly improved LVESD, fractional shortening, and MPI in Lmna–/– mice (n = 7) compared to Lmna–/– mice fed the control diet (n = 7). Black bars, control-fed; white bars, rapamycin-fed. IVRT, isovolemic relaxation time; IVCT, isovolemic contraction; LVET, LV ejection time. (C) Analysis of muscle function by rotarod test. There was a significant decrease in latency to fall and maximum speed reached in Lmna–/– mice fed control diet (n = 5) compared to Lmna+/+ mice fed control diet (n = 5). Dietary rapamycin significantly increased latency to fall and maximum speed reached in Lmna–/– mice (n = 4) compared to Lmna–/– mice fed the control diet (n = 5). Black bars, control-fed; white bars, rapamycinfed. *P < 0.05, **P < 0.01, Lmna–/– control-fed versus Lmna+/+ control-fed; #P < 0.05, ##P < 0.01, Lmna–/– rapamycin-fed versus Lmna–/– control-fed.
We used the rotarod test of motor coordination to assess the effects of 2 weeks of dietary rapamycin on skeletal muscle function in Lmna–/– mice. This test is a complex motor task requiring muscle coordination and balance, as well as motor learning, and it has been used to determine the effectiveness of treatments for mouse muscular dystrophy models (42, 43). As expected, Lmna–/– mice were unable to stay on the rotating rod as long as Lmna+/+ mice (P < 0.05), and they did not reach the same maximum speed as Lmna+/+ mice given the same amount of time (P < 0.01) (Fig. 2C). Latency to fall off the rotating rod was significantly longer in Lmna–/– mice treated with rapamycin compared to control-fed Lmna–/– mice (P < 0.05). In addition, rapamycin-fed Lmna–/– mice were able to reach a significantly higher speed than control-fed Lmna–/– mice by the cutoff time of 5 min (P < 0.01) (Fig. 2C).
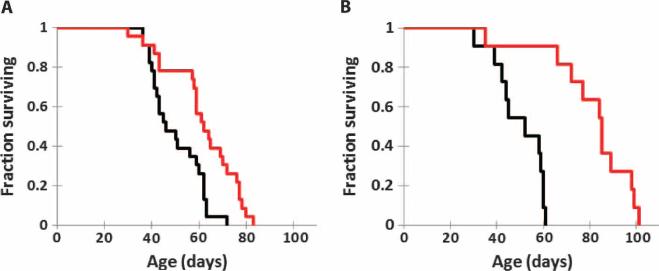
Because Lmna–/– mice treated with rapamycin show improved cardiac and muscle function, we examined the effect of rapamycin on the life span of these animals. Lmna–/– mice fed rapamycin chow from 3 to 4 weeks of age lived significantly longer than animals fed an identical diet without rapamycin (P = 0.0013, both groups n = 23) (Fig. 3A). Rapamycin feeding resulted in a 35% increase in median life span (62 days versus 46 days) and a 23% increase in mean life span (62 days versus 50 days). Maximum life span was also increased by rapamycin treatment: Of the Lmna–/– mice surviving to the 90th percentile, a significantly greater proportion of rapamycin-fed mice survived compared to mice fed the control diet (P < 0.05). Survival was significantly increased in both male Lmna–/– mice (P = 0.007) and female Lmna–/– mice fed dietary rapamycin (P = 0.04) (fig. S2, A and B). Analysis of weekly body weights of the mice during treatment revealed that male and female Lmna–/– mice fed rapamycin maintained their body weight over time better than did Lmna–/– mice fed the control diet (P < 0.05 and P = 0.006, respectively), although individual values at specific time points were not significantly different after a Bonferroni post hoc test (fig. S2, C and D). To determine whether a higher dose of rapamycin would result in a larger increase in survival, we administered rapamycin (8 mg/kg) by intraperitoneal injection every other day starting at 4 weeks of age. The higher dose of rapamycin also increased the survival of Lmna–/– mice (P < 0.0001, both groups n = 11) (Fig. 3B), causing a 57% increase in mean life span (81 days versus 51.5 days) and a 56% increase in median life span (85 days versus 54.5 days). Maximum life span of Lmna–/– mice at the 90th percentile revealed a significantly increased proportion of rapamycin-treated mice compared to control mice (P < 0.01).
Fig. 3.
Treatment of Lmna–/– mice with rapamycin increases survival. (A) Kaplan-Meier plot of Lmna–/– mice (mixed 129Sv-C57BL/6J genetic background) fed control (n = 23) or diet that included encapsulated rapamycin (n = 23). Survival was significantly increased in Lmna–/– mice fed dietary rapamycin (P = 0.0013). (B) Kaplan-Meier plot of Lmna–/– mice (mixed 129Sv-C57BL/6J background) injected with vehicle (n = 11) or a higher dose of rapamycin (8 mg/kg) (n = 11). Survival was significantly increased in Lmna–/– mice injected with rapamycin (P = 0.0002).
Given the potential of rapamycin as a therapeutic agent, we performed several other life span studies. Because rapamycin administration has been associated with side effects, we sought to determine whether administration of less rapamycin would have similar effects on longevity. First, we reduced the frequency of administration of rapamycin (8 mg/kg) to once weekly. Under these conditions, the mean and median life spans were increased by 52.7 and 43.1%, respectively (P < 0.0001) (fig. S3A). We also tested a brief 1-week administration (three doses every other day) of rapamycin starting at 4 weeks of age. Even this brief rapamycin treatment was sufficient to significantly extend life span (fig. S3B), albeit to a lesser degree. These studies indicate that the beneficial effects of rapamycin can be detected rapidly after drug administration and occur even with less frequent administration. Finally, we administered rapamycin (8 mg/kg) to Lmna–/– mice that were inbred with a C57BL/6J background, a strain in which rapamycin has been reported to extend the life span of Lmna+/+ mice (41). Analysis of blood from these rapamycin-injected Lmna–/– mice revealed levels of 85.7 ± 17.7 ng/ml after 1 week of injections (four doses), 24 hours after the last injection. Injection of rapamycin significantly increased the survival of C57BL/6J Lmna–/– mice (P = 0.0002, both groups n = 11) (fig. S3C), resulting in a 56% increase in mean life span (58 days versus 37 days) and a 60.5% increase in median life span (61 days versus 38 days). Although the maximum life span of Lmna–/– mice at the 90th percentile was not significantly different from those of controls (possibly as a result of low sample size), analysis of the mice at the 80th percentile revealed a significantly increased proportion of rapamycin-treated mice compared to control mice (P = 0.035).
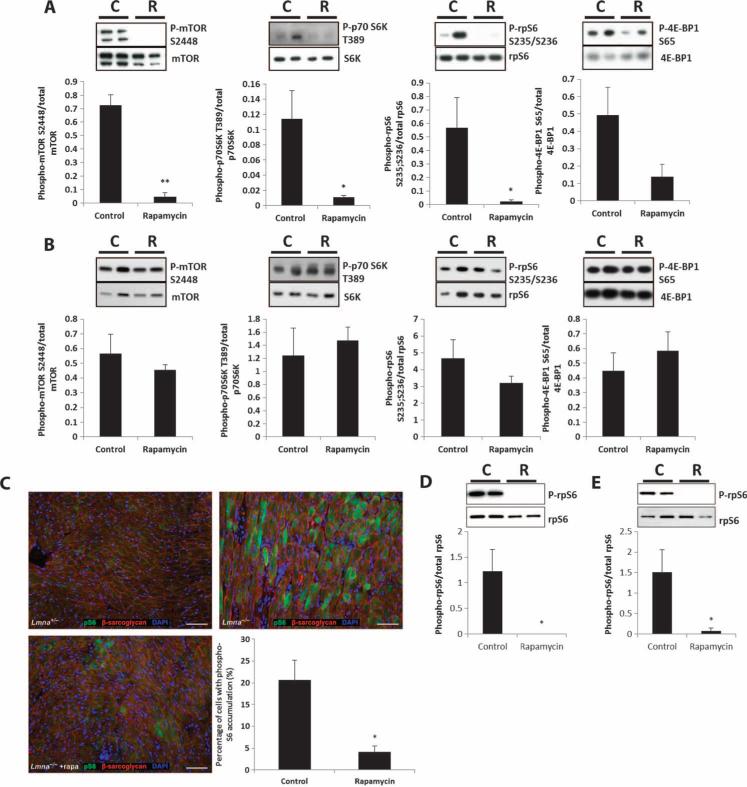
To further understand the effects of rapamycin on the heart and muscle of Lmna–/– mice, we examined molecular changes in heart and muscle (quadriceps) tissue. After 2 weeks of dietary rapamycin initiated at 3 to 4 weeks of age, inhibition of mTORC1 signaling was apparent in the heart tissue of rapamycin-treated animals (relative to controls) (Fig. 4A). Specifically, there was a significant decrease in phosphorylated mTOR(S2448) (P = 0.00001), S6 kinase(T389) (P = 0.02), and rpS6(S235/S236) (P = 0.03) in Lmna–/– mice fed rapamycin compared to Lmna–/– mice fed the control diet. However, phosphorylation of 4E-BP1(S65) was not significantly reduced by rapamycin treatment (P = 0.07), consistent with previous in vivo evidence suggesting that rapamycin has stronger effects on S6 kinase phosphorylation than on 4E-BP1 phosphorylation (44). Reduction of phosphorylated rpS6(S235/S236) in cardiomyocytes was confirmed by immunohistochemistry of heart tissue sections from Lmna–/– mice fed the rapamycin diet compared to those fed control diet (P = 0.02) (Fig. 4C). In contrast, dietary rapamycin did not significantly decrease mTORC1 signaling in skeletal muscle (Fig. 4B). Phosphorylation of mTOR(S2448), S6 kinase(T389), rpS6(S235/S236), and 4E-BP1(S65) was unchanged in skeletal muscle from rapamycin-fed Lmna–/– mice compared to controls (P = 0.4, 0.6, 0.2, and 0.5, respectively). However, after 1 week of rapamycin injections (8 mg/kg, every other day), phosphorylation of S6 was significantly inhibited in both heart and skeletal muscle, indicating that a higher dose is necessary for detection of a decrease in signaling through the mTORC1 pathway in skeletal muscle (Fig. 4, D and E).
Fig. 4.
Signaling through the mTORC1 pathway in heart and muscle of Lmna–/– mice is reduced by rapamycin treatment but is dependent on dosage. (A) Phosphorylated mTOR(S2448), S6 kinase(T389), and rpS6(S235/S236) were decreased in hearts from Lmna–/– mice fed the rapamycin diet (R) (n = 6) compared to Lmna–/– mice fed the control diet (C) (n = 6). Phosphorylation of 4E-BP1(S65) was not significantly reduced. (B) Phosphorylated mTOR(S2448), S6 kinase(T389), rpS6(S235/S236), and 4E-BP1(S65) were not significantly reduced in muscle from Lmna–/– mice fed the rapamycin diet (R) (n = 6) compared to Lmna–/– mice fed the control diet (C) (n = 5). (C) Immunohistochemistry of heart tissue sections from Lmna–/– mice fed the rapamycin diet (n = 3), which showed a significant decrease in the percentage of cardiomyocytes with accumulation of phosphorylated rpS6 compared to Lmna–/– mice fed the control diet (n = 3). (D and E) Phosphorylated rpS6(S235/S236) was decreased in heart and muscle from Lmna–/– mice injected with rapamycin (R) (8 mg/kg) (n = 4) compared to vehicle-injected Lmna–/– mice (C) (n = 4). Panel (D) is heart and panel (E) is muscle. *P < 0.05; **P < 0.01.
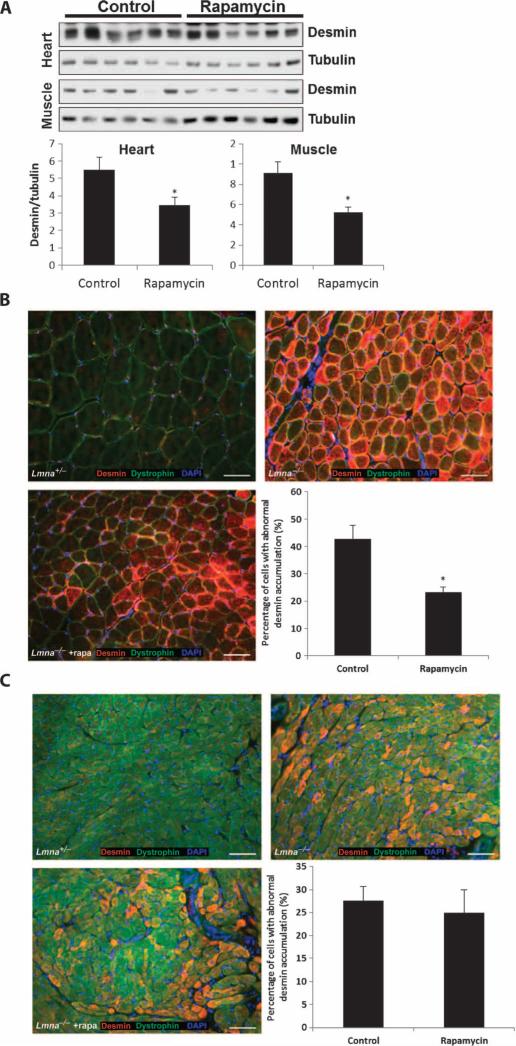
Lmna–/– mice have defects in the desmin filaments linking the cytoskeleton to the nucleus in both heart and muscle, resulting in accumulation of desmin aggregates in the cytoplasm and disruption of the Z-disc cross-striation pattern (17). We also observed these phenomena in our control-fed Lmna–/– mice; however, after 2 weeks of treatment with dietary rapamycin, we detected a significant reduction in the amount of desmin protein in both cardiac and skeletal muscle tissue as measured by Western blot (P = 0.042 and 0.012) (Fig. 5A). We also looked at desmin staining in tissue by immunohistochemistry and found that rapamycin diminished the number of myocytes containing abnormal desmin conglomerates in skeletal muscle (Fig. 5B) but not in the cardiac tissue of Lmna–/– mice (P = 0.025 and 0.68) (Fig. 5C). The differences in the effect of rapamycin in cardiac tissue by the two assays suggest that, although the number of cardiomyocytes containing desmin conglomerates did not change, the total amount of desmin present in the tissue was reduced. Last, we examined whether rapamycin rescued the aberrant cross-sectional area of Lmna–/– skeletal muscle fibers. Although there was a trend toward an increase in cross-sectional area of skeletal muscle fibers in rapamycin-fed Lmna–/– mice, it did not achieve statistical significance (P = 0.071) (fig. S4).
Fig. 5.
Rapamycin reduces abnormal desmin accumulation in Lmna–/– mice. (A) Desmin protein levels in heart and skeletal muscle of rapamycin-treated Lmna–/– mice (n = 6) compared to control-fed Lmna–/– mice (n = 6) as measured by Western blot analysis. (B) Accumulation of desmin in the myocytes of Lmna–/– mice compared to control Lmna+/– mice by immunohistochemistry of muscle sections. The percentage of cells with accumulation of desmin in the myocytes of Lmna–/– mice fed rapamycin (n = 4) was significantly reduced compared to Lmna–/– mice fed the control diet (n = 4). DAPI, 4′,6-diamidino-2-phenylindole. (C) Accumulation of desmin in the cardiomyocytes of Lmna–/– mice compared to control Lmna+/– mice by immunohistochemistry of heart sections. The percentage of cells with accumulation of desmin in the cardiomyocytes of Lmna–/– mice fed rapamycin (n = 4) was not different from that in Lmna–/– mice fed the control diet (n = 4). *P < 0.05.
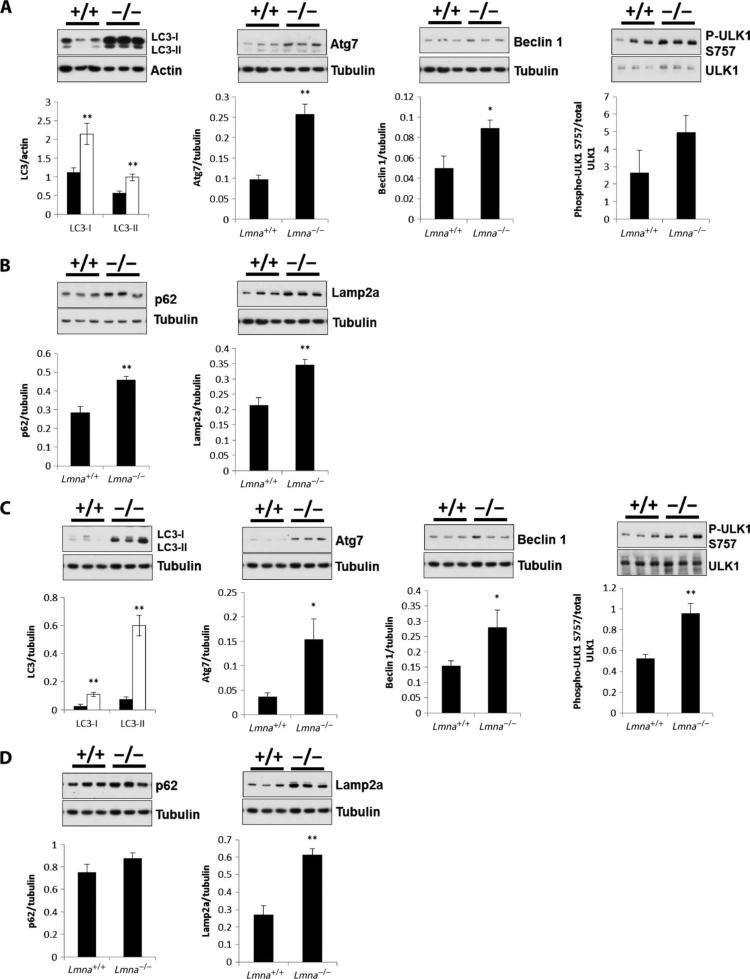
Because autophagy is implicated in mouse models of desmin-related cardiomyopathies (45) and muscular dystrophy (46) and because au tophagy is regulated by mTORC1 (47), we examined multiple autophagy markers in heart and skeletal muscle of Lmna+/+ and Lmna–/– mice at 3 to 4 weeks of age. Proteins involved in autophagosome formation, including microtubule-associated protein 1 light chain 3 (LC3), beclin 1, and autophagy-related protein 7 (Atg7), were significantly increased in both the heart and the muscle of Lmna–/– mice compared to Lmna+/+ mice (Fig. 6, A and B). In addition, the phosphorylation of the mTORC1 substrate and autophagy regulator, ULK1, was significantly increased in muscle (P = 0.0099) but not in heart (Fig. 6, A and B). Consistent with the increase in these autophagic markers, we also observed instances of the presence of autophagosomes by electron microscopy in heart sections from Lmna–/– mice (fig. S5). However, p62/sequestosome 1 (SQSTM1), a protein that is degraded during the autophagic process and used as a marker of autophagic flux, is also increased in the hearts (but not skeletal muscle) of Lmna–/– mice compared to controls (Fig. 6, B and D), indicating that in heart, whereas formation of autophagosomes is increased, degradation is inhibited. In addition, we found a significant increase in Lamp2a in both heart (P = 0.0051) and muscle (P = 0.00024) of Lmna–/– mice compared to Lmna+/+ mice (Fig. 6, B and D), indicative of an increase in chaperone-mediated autophagy, a process that can take place when autophagy is inhibited (48, 49).
Fig. 6.
Autophagy markers are increased in heart and skeletal muscle tissue of Lmna–/– mice. (A) LC3-I, LC3-II, Atg7, beclin 1, and phospho-ULK1 (P-ULK1) in heart. Western blot analysis of heart tissue lysates shows a significant increase in levels of LC3-I, LC3-II, Atg7, and beclin 1 protein in Lmna–/– mice compared to Lmna+/+ mice. Phospho-ULK1 levels are not significantly different. For LC3 blot: Lmna+/+ mice (n = 7) (black bars); Lmna–/– mice (n = 8) (white bars). For all other blots: Lmna+/+ mice (n = 5) and Lmna–/– mice (n = 4). (B) p62 and Lamp2a in heart. Western blot analysis of heart tissue shows a significant increase in p62 protein in Lmna–/– mice (n = 6) compared to Lmna+/+ mice (n = 7). Lamp2a protein levels are also significantly different in Lmna–/– mice (n = 4) compared to Lmna+/+ mice (n = 5). (C) LC3-I, LC3-II, Atg7, beclin 1, and phospho-ULK1 in muscle. Western blot analysis of muscle tissue lysates shows a significant increase in LC3-I, LC3-II, Atg7, and beclin 1 protein levels in Lmna–/– mice compared to Lmna+/+ mice. Phospho-ULK1 levels are also significantly different. For the LC3 blot: Lmna+/+ mice (n = 7) (black bars); Lmna–/– mice (n = 6) (white bars). For all other blots, Lmna+/+ mice (n = 7) and Lmna–/– mice (n = 6). (D) p62 and Lamp2a in muscle. Western blot analysis of muscle tissue shows a significant increase in Lamp2a protein levels in Lmna–/– mice (n = 6) compared to Lmna+/+ mice (n = 7) but not in p62 protein levels. *P < 0.05; **P < 0.01.
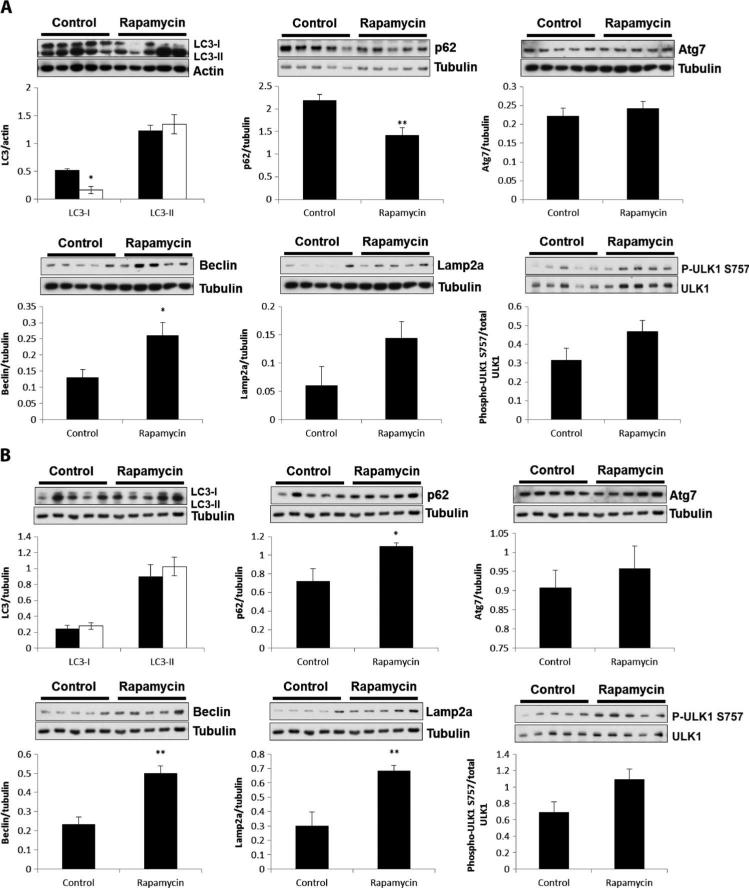
To test whether elevated mTOR signaling in Lmna–/– mice is responsible for the inhibition of autophagy-mediated degradation, we examined the effect on various autophagy markers of treating Lmna–/– mice with dietary rapamycin. In heart tissue, rapamycin caused a significant decrease in LC3-I (P = 0.029) but not LC3-II levels (P = 0.5), demonstrating a shift by heart cells to using LC3-II, which is indicative of autophagosome formation (Fig. 7A). In addition, we saw an increase in beclin 1 (P = 0.03). The significant decrease in p62 levels (P = 0.006) in rapamycin-treated Lmna–/– mice (Fig. 7A) indicated that autophagic-mediated degradation was increased when mTOR was inhibited. Unexpectedly, there was no change in phospho-ULK1 (Fig. 7A), a target of mTORC1 that has been shown to mediate its effect on autophagy (47). In muscle of Lmna–/– mice, rapamycin treatment did not change LC3-I or LC3-II levels (P = 0.6 and 0.5) (Fig. 7B) but did significantly increase beclin 1 (P = 0.001) (Fig. 7B). Unlike the heart, however, rapamycin did not decrease p62 expression, indicating that inhibition of mTOR did not activate autophagy in skeletal muscle. The differential effect of rapamycin on autophagy in heart and muscle of Lmna–/– mice in these experiments is likely a result of the different dosage requirements for effective inhibition of mTORC1 signaling in the two tissues (Fig. 4).
Fig. 7.
Rapamycin enhances autophagy-mediated degradation in hearts from Lmna–/– mice. (A) Effect of rapamycin on autophagy markers in heart. Western blot analysis of heart tissue shows a significant decrease in LC3-I (n = 6), p62 (n = 6), and beclin 1 (n = 6) protein levels [but no change in LC3-II (n = 6) protein levels] in Lmna–/– mice treated with rapamycin compared to control Lmna–/– mice (n = 5 to 6). Atg7, Lamp2a, and phospho-ULK1 are not significantly changed in Lmna–/– mice treated with rapamycin (n = 6) compared to control-treated Lmna–/– mice (n = 5). (B) Effect of rapamycin on autophagy markers in muscle. Western blot analysis of muscle tissue shows no significant change in LC3-I, LC3-II, Atg7, and phospho-ULK1 protein levels in Lmna–/– mice (n = 6) treated with rapamycin compared to control-treated Lmna–/– mice (n = 5). There was a significant increase in p62, beclin 1, and Lamp2a protein levels in Lmna–/– mice treated with rapamycin (n = 6) compared to control Lmna–/– mice (n = 5). *P < 0.05; **P < 0.01.
DISCUSSION
Our findings indicate that hyperactivation of the mTORC1 signaling pathway in Lmna–/– mice contributes to the heart-and muscle-specific defects in these mice and that inhibition of this pathway with rapamycin can significantly counteract this dysfunction and ultimately improve survival. Initially, activation of the mTORC1 pathway can be a beneficial response in muscle and cardiac tissue when hypertrophy is needed. In cardiac tissue, the hypertrophic response is activated by hemodynamic overload and provides temporary relief for heart function. Nevertheless, sustained activation of the hypertrophic response may not result in the long-term maintenance of heart function. For example, acute induction of hypertrophy by activation of Akt-1 specifically in the heart results in the preservation of heart function, whereas chronic activation leads to DCM (50). Moreover, treatment with rapamycin prevents the effects of both acute and chronic activation of Akt-1, suggesting that the mTORC1 pathway may be involved in both physiological responses (50). In addition, a recent report showed that mTORC1 signaling is elevated in a mouse model (LS/+ mice) of LEOPARD syndrome, an autosomal dominant disorder that manifests with congenital heart disease, among other symptoms (51). LS/+ mice initially develop hypertrophic hearts with sustained heart function; without treatment, however, this progresses to dilated cardiomyopathy and impaired heart function by 52 weeks of age (51). Treatment of these LS/+ mice with rapamycin reverses hypertrophic cardiomyopathy and rescues aberrant signaling through the Akt/mTOR pathway.
In Lmna–/– mice, activation of mTORC1 signaling and the hypertrophic response appear to be uncoupled because Lmna–/– mice do not show hypertrophy and instead develop only DCM and skeletal myopathy. It has been suggested that the lack of hypertrophy may be a result of the disorganization of the desmin filament network and accumulation of desmin ag gregates (17). Desmin, a major cytoskeletal protein in cardiac and muscle tissue, which connects the contractile apparatus to other organelles of the cell (including the mitochondria, lysosome, and nucleus), is necessary for the cytoskeletal rearrangements required for adaptive hypertrophy (52). Accordingly, DCM and skeletal myopathy are observed in humans with gene mutations (encoding desmin or desmin-interacting proteins like αB-crystallin) that lead to desmin disorganization and aggregation (53, 54), and desmin aggregates are often seen in human idiopathic DCM (55). Autophagy is up-regulated in response to desmin aggregation, and increases in autophagic markers such as autophagosomes, LC3-II levels, and p62 levels are seen in mouse models of desmin-related cardiomyopathies (45).
The increase that we observed in LC3-II levels, the lipidated form of LC3 that localizes to autophagosome membranes, in heart and skeletal muscle of Lmna–/– mice suggests that there may be an increase in the formation of autophagosomes because LC3-II is believed to be required for autophagosome closure (56). Increased levels of Atg7, an E1-like enzyme involved in the processing of LC3-I to LC3-II (57), and beclin 1, part of a multiprotein complex that participates in vesicle nucleation (47), in heart and skeletal muscle of Lmna–/– mice are also consistent with elevated initiation of autophagy. However, the increase in p62 protein levels in heart may indicate that there is a reduction in autophagic flux because p62, a protein that binds ubiquitinated proteins targeted for autophagic degradation, is itself degraded during the autophagic process (56). Consistently, the increase in Lamp2a expression in both heart and skeletal muscle of Lmna–/– mice can indicate an increase in chaperone-mediated autophagy (48), which has been shown to compensate when autophagy is blocked (49). Thus, although we noted increases in autophagic markers in Lmna–/– mice, because of the potentially antagonizing effect of aberrant mTORC1 signaling on autophagy, the efficiency of this process may be compromised. A related phenotype has been observed in Zmpste24-deficient mice. Zmpste24 is a metalloproteinase involved in the processing of pre– lamin A to mature lamin A, and deletion of this gene in mice results in progeria-like characteristics (58). In this model, mTORC1 signaling is decreased and autophagy markers indicate increased autophagic throughput (58). Furthermore, reduced pre–lamin A expression in Zmpste24-deficient mice reverses this phenotype (58).
We propose that inefficient autophagy contributes to the disease phenotypes observed in Lmna–/– mice and that treatment with rapamycin suppresses mTORC1 hyperactivation and thus may reduce mTORC1-mediated inhibitory signals to the autophagy pathway. In heart, this scenario is supported by a shift in LC3-I to LC3-II, decreased p62 levels, and increased beclin 1 levels after rapamycin treatment. In muscle, rapamycin had less effect, with LC3-I and LC3-II levels remaining unchanged, whereas p62 levels were increased. Nevertheless, rapamycin treatment increased beclin 1 and Lamp2a in muscle, suggesting that autophagy was affected. The differential effect of rapamycin on autophagy in heart and muscle of Lmna–/– mice may reflect the different dosage requirements for effective inhibition of mTORC1 signaling in the two tissues. In muscle, a greater dose was required to see changes in mTORC1 signaling, and this dose may be required to see further activation of autophagy. On the other hand, rapamycin may be acting through a different mechanism in muscle because signaling through Foxo3 may play a greater role in the regulation of autophagy in muscle (59). In addition, rapamycin improves the dystrophic phenotype of a mouse model of Duchenne muscular dystrophy by reducing the infiltration of CD4+ and CD8+ effector T cells while preserving Foxp3+ regulatory T cells (60). Although it is not known whether this mechanism applies to Lmna–/– mice, CD4+ infiltration and other inflammatory changes are seen in human infantile-onset LMNA myopathy (61). Nonetheless, we saw positive effects of rapamycin treatment on desmin in the heart and skeletal muscle of Lmna–/– mice, resulting in improvements in heart and skeletal muscle function and, ultimately, survival.
There are currently no effective treatments for conduction-system disease (CDM1A) and the two muscular dystrophies linked to LMNA mutation (EDMD2/3 and LGMD1B), with affected individuals succumbing to sudden cardiac failure in their fourth and fifth decade (62). Our findings and those in the paper by Choi et al. (63), in this issue of Science Translational Medicine, point to a potential clinical intervention that could benefit individuals who suffer from these diseases. Rapalogs, derivatives of rapamycin with better pharmacokinetics, are clinically approved for use in a range of human diseases. We propose that these agents may prove efficacious for improving cardiac function in patients with LMNA-associated CDM1A, EDMD2/3, and LGMD1B, as well as for other diseases with reduced A-type lamin function.
MATERIALS AND METHODS
Animals and experimental protocols
Lmna–/– mice [in which exons 8 through part of exon 11 of the Lmna gene is completely deleted (16)] and Lmna+/+ littermate controls were produced by mating Lmna+/– mice of a mixed 129Sv-C57BL/6J genetic background. For analysis of mTORC1 signaling in Lmna–/– mice and Lmna+/+ controls, tissues were dissected from mice aged 4 weeks and analyzed by Western blot. For behavioral and biochemical studies, Lmna–/– mice and Lmna+/+ littermate controls were fed either a control diet or a diet containing encapsulated rapamycin (39) (courtesy of R. Strong) ad libitum starting at 3 to 4 weeks of age for 2 weeks before analysis. For survival studies, Lmna–/– mice were fed either a control diet or a diet containing encapsulated rapamycin ad libitum starting at 3 to 4 weeks of age for the duration of their life. Mice were weighed weekly when their cage was changed, and food was added to the cage. Mice were examined daily to determine their health, particularly in terms of their cardiac defects. As determined by veterinary consultation, mice were examined for (i) pale feet (mice with good circulation should have pink feet, whereas ill mice may have white or pale feet), (ii) abnormal breathing (if the mice are experiencing heart failure, they have labored breathing in their abdomen), and (iii) reduced activity (mice are less active if they are experiencing heart failure). Mice exhibiting these signs were euthanized.
For studies with injections of rapamycin (8 mg/kg) (LC Laboratories), Lmna–/– mice and Lmna+/+ littermate controls were produced by mating Lmna+/– mice of a mixed 129Sv-C57BL/6J genetic background. For biochemical studies, Lmna–/– mice and Lmna+/+ littermate controls (4 weeks of age) were given intraperitoneal injections of rapamycin (8 mg/kg) or vehicle control every other day for 1 week (four injections). Twenty-four hours after the last injection, tissues were dissected from the mice and immediately frozen in liquid nitrogen. For survival studies, Lmna–/– mice (4 weeks of age) were given intraperitoneal injections of rapamycin (8 mg/kg) or vehicle control every other day for the duration of their life. The endpoint was determined as above. For studies with injections of rapamycin (8 mg/kg) (in C57BL/6J genetic background), Lmna–/– mice and Lmna+/+ littermate controls were produced by mating Lmna+/– mice of C57BL/6J genetic background. For survival studies, Lmna–/– mice (3 to 4 weeks of age) were given intraperitoneal injections of rapamycin (8 mg/kg) or vehicle control every other day for the duration of their life span. Endpoint was determined as stated above.
Polysome analysis
Heart polysome protocol was adapted from Zomzely et al. (64) Frozen hearts were homogenized with a Dounce homogenizer in 1.5 ml of homogenization buffer composed of 250 mM sucrose, 50 mM tris-HCl (pH 7.4), 100 mM KCl, 12 mM MgCl2, cycloheximide (0.1 mg/ml), and RiboLock (400 U/ml) (Fermentas). Samples were centrifuged at 10,000g for 10 min at 4°C. Deoxycholate was then added to super-natant to a final concentration of 1%. Samples were incubated for 10 min at 4°C and then recentrifuged at 16,000g. The supernatant was loaded on a 10.8-ml sucrose gradient with 50 mM tris-HCl, 100 mM KCl, and 15 mM MgCl2 with cycloheximide (100 μg/ml) and heparin (1 mg/ml). Samples were centrifuged in an SW41 Ti rotor (Beckman) at 39,000 rpm at 4°C for 2 hours, and fractions were assessed for optical density.
Western blot analysis
Mouse tissues were dissected and immediately frozen in liquid nitrogen. For tissue homogenization, frozen tissues were placed into the prechilled chamber of a cryogenic tissue pulverizer (RPI) and instantly turned into fine powder with the compression force of a pestle struck by a mallet. The finely ground tissue was transferred to a Dounce homogenizer containing ice-cold homogenization buffer [50 mM Hepes (pH 7.6), 150 mM NaCl, 20 mM sodium pyrophosphate, 20 mM β-glycero-phosphate, 10 mM NaF, 2 mM EDTA, 1% Igepal, 10% glycerol, 1 mM MgCl2, 1 mM CaCl2, 2 mM sodium orthovanadate, 2 mM phenylmethylsulfonyl fluoride, leupeptin (10 μg/ml), aprotinin (10 μg/ml), and 1:100 serine/threonine phosphatase inhibitor cocktail (Sigma)]. The tissues were homogenized on ice and then centrifuged at 13,000 rpm for 15 min at 4°C. The supernatants were collected and protein concentration was determined by BCA protein assay (Thermo Scientific 23227). Equal amounts of protein were resolved by SDS– polyacrylamide gel electrophoresis (4 to 12% gel), and Western blot analysis was performed with protein/phosphoprotein-specific antibodies. Anti-mTOR (2972, 1:1000), anti–phospho-mTOR(S2448) (5536, 1:1000), anti–p70 S6 kinase (2708, 1:1000), anti–phospho–p70 S6 kinase(T389) (9234, 1:500), anti-rpS6 (2217, 1:1000), anti–phospho-rpS6(S235/236) (2211, 1:1000), anti–4E-BP1 (9452, 1:1000), anti-tubulin (2125, 1:1000), anti–phospho-ULK1(S757) (6888, 1:1000), anti–beclin 1 (3495, 1:1000), anti-Atg7 (2631, 1:1000), anti-GβL (3274, 1:1000), anti-Raptor (2280, 1:1000), and anti–phospho–4E-BP1(S65) (9451, 1:500) were from Cell Signaling Technology. Anti-desmin (sc-23879, 1:1000) was from Santa Cruz Biotechnology. Anti-p62 (H00008878-M01, 1:1000) was from Abnova. Anti-LC3 (NB100-2331, 1:1000) was from Novus Biologicals. Anti-ULK1 was from Sigma (A7481, 1:1000). Anti-Lamp2a was from Abcam (18528, 1:1000). Anti-actin was from Chemicon (MAB1501R, 1:10,000).
Transthoracic echocardiography
Echocardiography examination was performed with a Siemens Acuson CV70. A mixture of 0.5% isoflurane and O2 was used to provide adequate sedation with minimal cardiac suppression during echocardiography. M-mode and Doppler imaging was performed to evaluate cardiac morphometry, systolic function, and MPI, which is calculated as the ratio of the sum of isovolemic contraction and relaxation time (IVCT + IVRT) to LV ejection time (LVET). An increase in MPI indicates that a larger fraction of systole is spent during isovolemic phases, which is the ineffective time fraction.
Rotarod
The day before the actual rotarod testing, the mice were placed on the rotarod set to a beginning speed of 5 rpm, with an acceleration rate of 0.1 rpm/s. The max speed was set at 25 rpm. They were allowed to practice the rotarod five times, one repetition every 5 min. If the animal did not fall off, each repetition would end at 5 min. The animals were tested 24 hours after the practice day, and the procedure was the same except scores were recorded. The score for each repetition was the time in seconds until the animal fell off the rotarod. The average of all five repetitions was used to score the sessions.
Tissue preparation and indirect immunofluorescence
Hearts and gastrocnemius muscles were rapidly excised and rinsed in phosphate-buffered saline (PBS) before mounting in Tissue-Tek optimal cutting temperature compound and subsequent freezing in liquid nitrogen–cooled isopentane. Special care was taken to prevent deformation of the hearts when dissecting with a plastic bulb pipette with the tip cut off to gently grasp the heart by suction rather than with forceps. Mounted samples were stored at –80°C until further processing. For analysis of ventricle wall thickness and ventricular dilatation, 10-μm-thick heart sections were mounted on Superfrost plus glass slides (Fisher) and then stained with hematoxylin and eosin. For immunohistochemistry, 8-μm-thick heart ventricle or gastrocnemius muscle sections were mounted on glass slides. Primary antibodies were desmin (Santa Cruz Biotechnology sc-23879; 1:100), dystrophin (a gift of J. S. Chamberlain) (1:600), phospho-rpS6(S240/244) (Cell Signaling Technology 2215; 1:100), and β-sarcoglycan (Leica NCL-b-SARC; 1:100). Sections were fixed in cold acetone for 20 min at –20°C followed by blocking in 5% bovine serum albumin in PBS with 0.3% Triton X-100. After staining was complete, tissues were mounted in Fluoromount (Southern Biotech). Samples were viewed on a Zeiss 200 M Axiovert, and images were acquired with Axiovision (Zeiss). Images were then scored for cross-sectional area in Axiovision with the “Outline Spline” tool to trace individual muscle fibers and calculate area. All fibers of an individual image were outlined until total fiber scored was >100 per mouse. Scoring of desmin and phospho-rpS6 accumulation was accomplished in a similar fashion with the cell counter tool in ImageJ to mark fibers with increased staining until >500 total fibers were scored per mouse. Increased desmin staining in muscle fibers was scored when there was any detectable cytoplasmic desmin or if there was significant desmin associated with the plasma membrane because these features were not detected in the control animals. Increased desmin staining in the heart was scored when increased accumulation of desmin was noted anywhere within the outline of the fiber.
Electron microscopy
Heart samples were fixed overnight in 2% paraformaldehyde, 2.5% glutaraldehyde, and 150 mM cacodylate buffer, osmicated in 2% osmium tetroxide plus 0.8% potassium ferrocyanide in 100 mM cacodylate buffer for 1 hour at room temperature, counterstained in 5% uranyl acetate in water for 30 min at room temperature, and then dehydrated in a precooled ethanol series, cleared in propylene oxide, and infiltrated and flat-embedded with Embed 812 (EM Sciences 22940-278). Ultrathin sections (50 to 70 nm) were collected from the blocks onto copper grids and imaged on an FEI Tecnai 12 transmission electron microscope.
Rapamycin analysis
High-performance liquid chromatography–tandem mass spectrometry on whole blood samples was performed by Rocky Mountain Labs.
Statistical analysis
A two-tailed Student's t test was used for comparisons between two groups. Two-way analysis of variance (ANOVA) with Bonferroni post hoc analysis was used for comparisons between more than two groups. Log-rank test was used for comparisons of survival curves. Quantile regression with a two-tailed Fisher's exact test was used for analysis of maximum life span. For all statistics, P < 0.05 was considered significant.
Supplementary Material
Acknowledgments
We thank R.Mangalindan, A. Safarli, J. Harrington, D. V. Touch, H. Hopkins, and other members of the Ladiges lab in the Department of Comparative Medicine, as well as V. Damian and R. Frock for their help and expertise in the maintenance of mouse colonies.
Funding: This study was supported by National Institute on Aging grants R01 AG024287 and R01 AG033373 (to B.K.K.) and U01 AG022307 (to R.S.). F.J.R. and M.G.G. are supported by NIH training grant T32AG000057. S.C.C. is supported by the cardiovascular and pathology training grant NIH T32 HL007312. M.K. is an Ellison Medical Foundation New Scholar in Aging.
Footnotes
Competing interests: Patent applications have been filed on discoveries related to this invention with B.K.K. listed as an inventor.
Data and materials availability: The rodent diet containing microencapsulated rapamycin is available under a Materials Transfer Agreement from R.S. at the University of Texas Health Science Center at San Antonio.
SUPPLEMENTARY MATERIALS
www.sciencetranslationalmedicine.org/cgi/content/full/4/144/144ra103/DC1
Fig. S1. Further characteristics of Lmna+/+ and Lmna−/− mice.
Fig. S2. Effect of rapamycin on survival of Lmna+/+ and Lmna−/− male and female mice.
Fig. S3. Effect of rapamycin at different dosing regimens on survival of Lmna+/+ and Lmna−/− mice.
Fig. S4. Effect of rapamycin on myofiber cross-sectional area in Lmna−/− mice.
Fig. S5. Autophagosome formation in Lmna−/− mice.
REFERENCES AND NOTES
- 1.Lopez-Soler RI, Moir RD, Spann TP, Stick R, Goldman RD. A role for nuclear lamins in nuclear envelope assembly. J. Cell Biol. 2001;154:61–70. doi: 10.1083/jcb.200101025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taimen P, Pfleghaar K, Shimi T, Möller D, Ben-Harush K, Erdos MR, Adam SA, Herrmann H, Medalia O, Collins FS, Goldman AE, Goldman RD. A progeria mutation reveals functions for lamin A in nuclear assembly, architecture, and chromosome organization. Proc. Natl. Acad. Sci. U.S.A. 2009;106:20788–20793. doi: 10.1073/pnas.0911895106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lattanzi G, Columbaro M, Mattioli E, Cenni V, Camozzi D, Wehnert M, Santi S, Riccio M, Del Coco R, Maraldi NM, Squarzoni S, Foisner R, Capanni C. Pre-lamin A processing is linked to heterochromatin organization. J. Cell. Biochem. 2007;102:1149–1159. doi: 10.1002/jcb.21467. [DOI] [PubMed] [Google Scholar]
- 4.Kumaran RI, Muralikrishna B, Parnaik VK. Lamin A/C speckles mediate spatial organization of splicing factor compartments and RNA polymerase II transcription. J. Cell Biol. 2002;159:783–793. doi: 10.1083/jcb.200204149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spann TP, Goldman AE, Wang C, Huang S, Goldman RD. Alteration of nuclear lamin organization inhibits RNA polymerase II–dependent transcription. J. Cell Biol. 2002;156:603–608. doi: 10.1083/jcb.200112047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moir RD, Spann TP, Herrmann H, Goldman RD. Disruption of nuclear lamin organization blocks the elongation phase of DNA replication. J. Cell Biol. 2000;149:1179–1192. doi: 10.1083/jcb.149.6.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muchir A, Bonne G, van der Kooi AJ, van Meegen M, Baas F, Bolhuis PA, de Visser M, Schwartz K. Identification of mutations in the gene encoding lamins A/C in autosomal dominant limb girdle muscular dystrophy with atrioventricular conduction disturbances (LGMD1B). Hum. Mol. Genet. 2000;9:1453–1459. doi: 10.1093/hmg/9.9.1453. [DOI] [PubMed] [Google Scholar]
- 8.Bonne G, Di Barletta MR, Varnous S, Bécane HM, Hammouda EH, Merlini L, Muntoni F, Greenberg CR, Gary F, Urtizberea JA, Duboc D, Fardeau M, Toniolo D, Schwartz K. Mutations in the gene encoding lamin A/C cause autosomal dominant Emery-Dreifuss muscular dystrophy. Nat. Genet. 1999;21:285–288. doi: 10.1038/6799. [DOI] [PubMed] [Google Scholar]
- 9.Fatkin D, MacRae C, Sasaki T, Wolff MR, Porcu M, Frenneaux M, Atherton J, Vidaillet HJ, Jr., Spudich S, De Girolami U, Seidman JG, Seidman C, Muntoni F, Müehle G, Johnson W, McDonough B. Missense mutations in the rod domain of the lamin A/C gene as causes of dilated cardiomyopathy and conduction-system disease. N. Engl. J. Med. 1999;341:1715–1724. doi: 10.1056/NEJM199912023412302. [DOI] [PubMed] [Google Scholar]
- 10.Cao H, Hegele RA. Nuclear lamin A/C R482Q mutation in Canadian kindreds with Dunnigan-type familial partial lipodystrophy. Hum. Mol. Genet. 2000;9:109–112. doi: 10.1093/hmg/9.1.109. [DOI] [PubMed] [Google Scholar]
- 11.De Sandre-Giovannoli A, Chaouch M, Kozlov S, Vallat JM, Tazir M, Kassouri N, Szepetowski P, Hammadouche T, Vandenberghe A, Stewart CL, Grid D, Lévy N. Homozygous defects in LMNA, encoding lamin A/C nuclear-envelope proteins, cause autosomal recessive axonal neuropathy in human (Charcot-Marie-Tooth disorder type 2) and mouse. Am. J. Hum. Genet. 2002;70:726–736. doi: 10.1086/339274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Sandre-Giovannoli A, Bernard R, Cau P, Navarro C, Amiel J, Boccaccio I, Lyonnet S, Stewart CL, Munnich A, Le Merrer M, Lévy N. Lamin A truncation in Hutchinson-Gilford progeria. Science. 2003;300:2055. doi: 10.1126/science.1084125. [DOI] [PubMed] [Google Scholar]
- 13.Pasotti M, Klersy C, Pilotto A, Marziliano N, Rapezzi C, Serio A, Mannarino S, Gambarin F, Favalli V, Grasso M, Agozzino M, Campana C, Gavazzi A, Febo O, Marini M, Landolina M, Mortara A, Piccolo G, Viganò M, Tavazzi L, Arbustini E. Long-term outcome and risk stratification in dilated cardiolaminopathies. J. Am. Coll. Cardiol. 2008;52:1250–1260. doi: 10.1016/j.jacc.2008.06.044. [DOI] [PubMed] [Google Scholar]
- 14.Chandar S, Yeo LS, Leimena C, Tan JC, Xiao XH, Nikolova-Krstevski V, Yasuoka Y, Gardiner-Garden M, Wu J, Kesteven S, Karlsdotter L, Natarajan S, Carlton A, Rainer S, Feneley MP, Fatkin D. Effects of mechanical stress and carvedilol in lamin A/C–deficient dilated cardiomyopathy. Circ. Res. 2010;106:573–582. doi: 10.1161/CIRCRESAHA.109.204388. [DOI] [PubMed] [Google Scholar]
- 15.Parks SB, Kushner JD, Nauman D, Burgess D, Ludwigsen S, Peterson A, Li D, Jakobs P, Litt M, Porter CB, Rahko PS, Hershberger RE. Lamin A/C mutation analysis in a cohort of 324 unrelated patients with idiopathic or familial dilated cardiomyopathy. Am. Heart J. 2008;156:161–169. doi: 10.1016/j.ahj.2008.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sullivan T, Escalante-Alcalde D, Bhatt H, Anver M, Bhat N, Nagashima K, Stewart CL, Burke B. Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J. Cell Biol. 1999;147:913–920. doi: 10.1083/jcb.147.5.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nikolova V, Leimena C, McMahon AC, Tan JC, Chandar S, Jogia D, Kesteven SH, Michalicek J, Otway R, Verheyen F, Rainer S, Stewart CL, Martin D, Feneley MP, Fatkin D. Defects in nuclear structure and function promote dilated cardiomyopathy in lamin A/C– deficient mice. J. Clin. Invest. 2004;113:357–369. doi: 10.1172/JCI19448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hara K, Maruki Y, Long X, Yoshino K, Oshiro N, Hidayat S, Tokunaga C, Avruch J, Yonezawa K. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell. 2002;110:177–189. doi: 10.1016/s0092-8674(02)00833-4. [DOI] [PubMed] [Google Scholar]
- 19.Kim DH, Sarbassov DD, Ali SM, King JE, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110:163–175. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- 20.Kim DH, Sarbassov DD, Ali SM, Latek RR, Guntur KV, Erdjument-Bromage H, Tempst P, Sabatini DM. GbL, a positive regulator of the rapamycin-sensitive pathway required for the nutrient-sensitive interaction between raptor and mTOR. Mol. Cell. 2003;11:895–904. doi: 10.1016/s1097-2765(03)00114-x. [DOI] [PubMed] [Google Scholar]
- 21.Hara K, Yonezawa K, Kozlowski MT, Sugimoto T, Andrabi K, Weng QP, Kasuga M, Nishimoto I, Avruch J. Regulation of eIF-4E BP1 phosphorylation by mTOR. J. Biol. Chem. 1997;272:26457–26463. doi: 10.1074/jbc.272.42.26457. [DOI] [PubMed] [Google Scholar]
- 22.Burnett PE, Barrow RK, Cohen NA, Snyder SH, Sabatini DM. RAFT1 phosphorylation of the translational regulators p70 S6 kinase and 4E-BP1. Proc. Natl. Acad. Sci. U.S.A. 1998;95:1432–1437. doi: 10.1073/pnas.95.4.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ono Y, Ito H, Tamamori M, Nozato T, Adachi S, Abe S, Marumo F, Hiroe M. Role and relation of p70 S6 and extracellular signal-regulated kinases in the phenotypic changes of hypertrophy of cardiac myocytes. Jpn. Circ. J. 2000;64:695–700. doi: 10.1253/jcj.64.695. [DOI] [PubMed] [Google Scholar]
- 24.Rommel C, Bodine SC, Clarke BA, Rossman R, Nunez L, Stitt TN, Yancopoulos GD, Glass DJ. Mediation of IGF-1-induced skeletal myotube hypertrophy by PI(3)K/Akt/mTOR and PI(3)K/Akt/GSK3 pathways. Nat. Cell Biol. 2001;3:1009–1013. doi: 10.1038/ncb1101-1009. [DOI] [PubMed] [Google Scholar]
- 25.Bodine SC, Stitt TN, Gonzalez M, Kline WO, Stover GL, Bauerlein R, Zlotchenko E, Scrimgeour A, Lawrence JC, Glass DJ, Yancopoulos GD. Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nat. Cell Biol. 2001;3:1014–1019. doi: 10.1038/ncb1101-1014. [DOI] [PubMed] [Google Scholar]
- 26.McMullen JR, Sherwood MC, Tarnavski O, Zhang L, Dorfman AL, Shioi T, Izumo S. Inhibition of mTOR signaling with rapamycin regresses established cardiac hypertrophy induced by pressure overload. Circulation. 2004;109:3050–3055. doi: 10.1161/01.CIR.0000130641.08705.45. [DOI] [PubMed] [Google Scholar]
- 27.Sasai N, Agata N, Inoue-Miyazu M, Kawakami K, Kobayashi K, Sokabe M, Hayakawa K. Involvement of PI3K/Akt/TOR pathway in stretch-induced hypertrophy of myotubes. Muscle Nerve. 2010;41:100–106. doi: 10.1002/mus.21473. [DOI] [PubMed] [Google Scholar]
- 28.Zoncu R, Efeyan A, Sabatini DM. mTOR: From growth signal integration to cancer, diabetes and ageing. Nat. Rev. Mol. Cell Biol. 2011;12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ogura Y, Iemitsu M, Naito H, Kakigi R, Kakehashi C, Maeda S, Akema T. Single bout of running exercise changes LC3-II expression in rat cardiac muscle. Biochem. Biophys. Res. Commun. 2011;414:756–760. doi: 10.1016/j.bbrc.2011.09.152. [DOI] [PubMed] [Google Scholar]
- 31.Grumati P, Coletto L, Sabatelli P, Cescon M, Angelin A, Bertaggia E, Blaauw B, Urciuolo A, Tiepolo T, Merlini L, Maraldi NM, Bernardi P, Sandri M, Bonaldo P. Autophagy is defective in collagen VI muscular dystrophies, and its reactivation rescues myofiber degeneration. Nat. Med. 2010;16:1313–1320. doi: 10.1038/nm.2247. [DOI] [PubMed] [Google Scholar]
- 32.Kassiotis C, Ballal K, Wellnitz K, Vela D, Gong M, Salazar R, Frazier OH, Taegtmeyer H. Markers of autophagy are downregulated in failing human heart after mechanical unloading. Circulation. 2009;120:S191–S197. doi: 10.1161/CIRCULATIONAHA.108.842252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim YA, Kim YS, Song W. Autophagic response to a single bout of moderate exercise in murine skeletal muscle. J. Physiol. Biochem. 2012;68:229–235. doi: 10.1007/s13105-011-0135-x. [DOI] [PubMed] [Google Scholar]
- 34.Cupesi M, Yoshioka J, Gannon J, Kudinova A, Stewart CL, Lammerding J. Attenuated hypertrophic response to pressure overload in a lamin A/C haploinsufficiency mouse. J. Mol. Cell. Cardiol. 2010;48:1290–1297. doi: 10.1016/j.yjmcc.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Giordano A. Molecular basis of different outcomes for drug-eluting stents that release sirolimus or tacrolimus. Curr. Opin. Drug Discov. Devel. 2010;13:159–168. [PubMed] [Google Scholar]
- 36.Borders EB, Bivona C, Medina PJ. Mammalian target of rapamycin: Biological function and target for novel anticancer agents. Am. J. Health Syst. Pharm. 2010;67:2095–2106. doi: 10.2146/ajhp100020. [DOI] [PubMed] [Google Scholar]
- 37.Stanfel MN, Shamieh LS, Kaeberlein M, Kennedy BK. The TOR pathway comes of age. Biochim. Biophys. Acta. 2009;1790:1067–1074. doi: 10.1016/j.bbagen.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weir MR, Diekmann F, Flechner SM, Lebranchu Y, Mandelbrot DA, Oberbauer R, Kahan BD. mTOR inhibition: The learning curve in kidney transplantation. Transpl. Int. 2010;23:447–460. doi: 10.1111/j.1432-2277.2010.01051.x. [DOI] [PubMed] [Google Scholar]
- 39.Harrison DE, Strong R, Sharp ZD, Nelson JF, Astle CM, Flurkey K, Nadon NL, Wilkinson JE, Frenkel K, Carter CS, Pahor M, Javors MA, Fernandez E, Miller RA. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460:392–395. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miller RA, Harrison DE, Astle CM, Baur JA, Boyd AR, de Cabo R, Fernandez E, Flurkey K, Javors MA, Nelson JF, Orihuela CJ, Pletcher S, Sharp ZD, Sinclair D, Starnes JW, Wilkinson JE, Nadon NL, Strong R. Rapamycin, but not resveratrol or simvastatin, extends life span of genetically heterogeneous mice. J. Gerontol. A Biol. Sci. Med. Sci. 2011;66:191–201. doi: 10.1093/gerona/glq178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen C, Liu Y, Zheng P. mTOR regulation and therapeutic rejuvenation of aging hematopoietic stem cells. Sci. Signal. 2009;2:ra75. doi: 10.1126/scisignal.2000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bogdanovich S, McNally EM, Khurana TS. Myostatin blockade improves function but not histopathology in a murine model of limb-girdle muscular dystrophy 2C. Muscle Nerve. 2008;37:308–316. doi: 10.1002/mus.20920. [DOI] [PubMed] [Google Scholar]
- 43.Nevo Y, Halevy O, Genin O, Moshe I, Turgeman T, Harel M, Biton E, Reif S, Pines M. Fibrosis inhibition and muscle histopathology improvement in laminin-a2-deficient mice. Muscle Nerve. 2010;42:218–229. doi: 10.1002/mus.21706. [DOI] [PubMed] [Google Scholar]
- 44.Feldman ME, Apsel B, Uotila A, Loewith R, Knight ZA, Ruggero D, Shokat KM. Active-site inhibitors of mTOR target rapamycin-resistant outputs of mTORC1 and mTORC2. PLoS Biol. 2009;7:e38. doi: 10.1371/journal.pbio.1000038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zheng Q, Su H, Ranek MJ, Wang X. Autophagy and p62 in cardiac proteinopathy. Circ. Res. 2011;109:296–308. doi: 10.1161/CIRCRESAHA.111.244707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grumati P, Coletto L, Sabatelli P, Cescon M, Angelin A, Bertaggia E, Blaauw B, Urciuolo A, Tiepolo T, Merlini L, Maraldi NM, Bernardi P, Sandri M, Bonaldo P. Autophagy is defective in collagen VI muscular dystrophies, and its reactivation rescues myofiber degeneration. Nat. Med. 2010;16:1313–1320. doi: 10.1038/nm.2247. [DOI] [PubMed] [Google Scholar]
- 47.Pattingre S, Espert L, Biard-Piechaczyk M, Codogno P. Regulation of macroautophagy by mTOR and Beclin 1 complexes. Biochimie. 2008;90:313–323. doi: 10.1016/j.biochi.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 48.Cuervo AM, Dice JF. Unique properties of lamp2a compared to other lamp2 isoforms. J. Cell Sci. 2000;113:4441–4450. doi: 10.1242/jcs.113.24.4441. [DOI] [PubMed] [Google Scholar]
- 49.Kaushik S, Massey AC, Mizushima N, Cuervo AM. Constitutive activation of chaperone-mediated autophagy in cells with impaired macroautophagy. Mol. Biol. Cell. 2008;19:2179–2192. doi: 10.1091/mbc.E07-11-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shiojima I, Sato K, Izumiya Y, Schiekofer S, Ito M, Liao R, Colucci WS, Walsh K. Disruption of coordinated cardiac hypertrophy and angiogenesis contributes to the transition to heart failure. J. Clin. Invest. 2005;115:2108–2118. doi: 10.1172/JCI24682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marin TM, Keith K, Davies B, Conner DA, Guha P, Kalaitzidis D, Wu X, Lauriol J, Wang B, Bauer M, Bronson R, Franchini KG, Neel BG, Kontaridis MI. Rapamycin reverses hyper-trophic cardiomyopathy in a mouse model of LEOPARD syndrome–associated PTPN11 mutation. J. Clin. Invest. 2011;121:1026–1043. doi: 10.1172/JCI44972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hein S, Kostin S, Heling A, Maeno Y, Schaper J. The role of the cytoskeleton in heart failure. Cardiovasc. Res. 2000;45:273–278. doi: 10.1016/s0008-6363(99)00268-0. [DOI] [PubMed] [Google Scholar]
- 53.McLendon PM, Robbins J. Desmin-related cardiomyopathy: An unfolding story. Am. J. Physiol. Heart Circ. Physiol. 2011;301:H1220–H1228. doi: 10.1152/ajpheart.00601.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Taylor MR, Slavov D, Ku L, Di Lenarda A, Sinagra G, Carniel E, Haubold K, Boucek MM, Ferguson D, Graw SL, Zhu X, Cavanaugh J, Sucharov CC, Long CS, Bristow MR, Lavori P, Mestroni L, Familial Cardiomyopathy Registry. BEST (Beta-Blocker Evaluation of Survival Trial) DNA Bank Prevalence of desmin mutations in dilated cardiomyopathy. Circulation. 2007;115:1244–1251. doi: 10.1161/CIRCULATIONAHA.106.646778. [DOI] [PubMed] [Google Scholar]
- 55.Di Somma S, Marotta M, Salvatore G, Cudemo G, Cuda G, De Vivo F, Di Benedetto MP, Ciaramella F, Caputo G, de Divitiis O. Changes in myocardial cytoskeletal intermediate filaments and myocyte contractile dysfunction in dilated cardiomyopathy: An in vivo study in humans. Heart. 2000;84:659–667. doi: 10.1136/heart.84.6.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, Baba M, Baehrecke EH, Bahr BA, Ballabio A, Bamber BA, Bassham DC, Bergamini E, Bi X, Biard-Piechaczyk M, Blum JS, Bredesen DE, Brodsky JL, Brumell JH, Brunk UT, Bursch W, Camougrand N, Cebollero E, Cecconi F, Chen Y, Chin LS, Choi A, Chu CT, Chung J, Clarke PG, Clark RS, Clarke SG, Clavé C, Cleveland JL, Codogno P, Colombo MI, Coto-Montes A, Cregg JM, Cuervo AM, Debnath J, Demarchi F, Dennis PB, Dennis PA, Deretic V, Devenish RJ, Di Sano F, Dice JF, Difiglia M, Dinesh-Kumar S, Distelhorst CW, Djavaheri-Mergny M, Dorsey FC, Dröge W, Dron M, Dunn WA, Jr., Duszenko M, Eissa NT, Elazar Z, Esclatine A, Eskelinen EL, Fésüs L, Finley KD, Fuentes JM, Fueyo J, Fujisaki K, Galliot B, Gao FB, Gewirtz DA, Gibson SB, Gohla A, Goldberg AL, Gonzalez R, González-Estévez C, Gorski S, Gottlieb RA, Häussinger D, He YW, Heidenreich K, Hill JA, Høyer-Hansen M, Hu X, Huang WP, Iwasaki A, Jäättelä M, Jackson WT, Jiang X, Jin S, Johansen T, Jung JU, Kadowaki M, Kang C, Kelekar A, Kessel DH, Kiel JA, Kim HP, Kimchi A, Kinsella TJ, Kiselyov K, Kitamoto K, Knecht E, Komatsu M, Kominami E, Kondo S, Kovács AL, Kroemer G, Kuan CY, Kumar R, Kundu M, Landry J, Laporte M, Le W, Lei HY, Lenardo MJ, Levine B, Lieberman A, Lim KL, Lin FC, Liou W, Liu LF, Lopez-Berestein G, López-Otín C, Lu B, Macleod KF, Malorni W, Martinet W, Matsuoka K, Mautner J, Meijer AJ, Meléndez A, Michels P, Miotto G, Mistiaen WP, Mizushima N, Mograbi B, Monastyrska I, Moore MN, Moreira PI, Moriyasu Y, Motyl T, Münz C, Murphy LO, Naqvi NI, Neufeld TP, Nishino I, Nixon RA, Noda T, Nürnberg B, Ogawa M, Oleinick NL, Olsen LJ, Ozpolat B, Paglin S, Palmer GE, Papassideri I, Parkes M, Perlmutter DH, Perry G, Piacentini M, Pinkas-Kramarski R, Prescott M, Proikas-Cezanne T, Raben N, Rami A, Reggiori F, Rohrer B, Rubinsztein DC, Ryan KM, Sadoshima J, Sakagami H, Sakai Y, Sandri M, Sasakawa C, Sass M, Schneider C, Seglen PO, Seleverstov O, Settleman J, Shacka JJ, Shapiro IM, Sibirny A, Silva-Zacarin EC, Simon HU, Simone C, Simonsen A, Smith MA, Spanel-Borowski K, Srinivas V, Steeves M, Stenmark H, Stromhaug PE, Subauste CS, Sugimoto S, Sulzer D, Suzuki T, Swanson MS, Tabas I, Takeshita F, Talbot NJ, Tallóczy Z, Tanaka K, Tanaka K, Tanida I, Taylor GS, Taylor JP, Terman A, Tettamanti G, Thompson CB, Thumm M, Tolkovsky AM, Tooze SA, Truant R, Tumanovska LV, Uchiyama Y, Ueno T, Uzcátegui NL, van der Klei I, Vaquero EC, Vellai T, Vogel MW, Wang HG, Webster P, Wiley JW, Xi Z, Xiao G, Yahalom J, Yang JM, Yap G, Yin XM, Yoshimori T, Yu L, Yue Z, Yuzaki M, Zabirnyk O, Zheng X, Zhu X, Deter RL. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y, Kominami E, Tanaka K, Chiba T. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J. Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mariño G, Ugalde AP, Salvador-Montoliu N, Varela I, Quirós PM, Cadiñanos J, van der Pluijm I, Freije JM, López-Otín C. Premature aging in mice activates a systemic metabolic response involving autophagy induction. Hum. Mol. Genet. 2008;17:2196–2211. doi: 10.1093/hmg/ddn120. [DOI] [PubMed] [Google Scholar]
- 59.Mammucari C, Milan G, Romanello V, Masiero E, Rudolf R, Del Piccolo P, Burden SJ, Di Lisi R, Sandri C, Zhao J, Goldberg AL, Schiaffino S, Sandri M. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 2007;6:458–471. doi: 10.1016/j.cmet.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 60.Eghtesad S, Jhunjhunwala S, Little SR, Clemens PR. Rapamycin ameliorates dystrophic phenotype in mdx mouse skeletal muscle. Mol. Med. 2011;17:917–924. doi: 10.2119/molmed.2010.00256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Komaki H, Hayashi YK, Tsuburaya R, Sugie K, Kato M, Nagai T, Imataka G, Suzuki S, Saitoh S, Asahina N, Honke K, Higuchi Y, Sakuma H, Saito Y, Nakagawa E, Sugai K, Sasaki M, Nonaka I, Nishino I. Inflammatory changes in infantile-onset LMNA-associated myopathy. Neuromuscul. Disord. 2011;21:563–568. doi: 10.1016/j.nmd.2011.04.010. [DOI] [PubMed] [Google Scholar]
- 62.Hermans MC, Pinto YM, Merkies IS, de Die-Smulders CE, Crijns HJ, Faber CG. Hereditary muscular dystrophies and the heart. Neuromuscul. Disord. 2010;20:479–492. doi: 10.1016/j.nmd.2010.04.008. [DOI] [PubMed] [Google Scholar]
- 63.Choi JC, Muchir A, Wu W, Iwata S, Homma S, Morrow JP, Worman HJ. Temsirolimus activates autophagy and ameliorates cardiomyopathy caused by lamin A/C gene mutation. Sci. Transl. Med. 2012;4:144ra102. doi: 10.1126/scitranslmed.3003875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zomzely CE, Roberts S, Gruber CP, Brown DM. Cerebral protein synthesis. II. Instability of cerebral messenger ribonucleic acid-ribosome complexes. J. Biol. Chem. 1968;243:5396–5409. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.