Abstract
Objective
To investigate whether hypoxia regulates Notch signaling, and whether Notch plays a role in intervertebral disc cell proliferation.
Methods
Reverse transcription–polymerase chain reaction and Western blotting were used to measure expression of Notch signaling components in intervertebral disc tissue from mature rats and from human discs. Transfections were performed to determine the effects of hypoxia and Notch on target gene activity.
Results
Cells of the nucleus pulposus and annulus fibrosus of rat disc tissue expressed components of the Notch signaling pathway. Expression of Notch-2 was higher than that of the other Notch receptors in both the nucleus pulposus and annulus fibrosus. In both tissues, hypoxia increased Notch1 and Notch4 messenger RNA (mRNA) expression. In the annulus fibrosus, mRNA expression of the Notch ligand Jagged1 was induced by hypoxia, while Jagged2 mRNA expression was highly sensitive to hypoxia in both tissues. A Notch signaling inhibitor, L685458, blocked hypoxic induction of the activity of the Notch-responsive luciferase reporters 12xCSL and CBF1. Expression of the Notch target gene Hes1 was induced by hypoxia, while coexpression with the Notch–intracellular domain increased Hes1 promoter activity. Moreover, inhibition of Notch signaling blocked disc cell proliferation. Analysis of human disc tissue showed that there was increased expression of Notch signaling proteins in degenerated discs.
Conclusion
In intervertebral disc cells, hypoxia promotes expression of Notch signaling proteins. Notch signaling is an important process in the maintenance of disc cell proliferation, and thus offers a therapeutic target for the restoration of cell numbers during degenerative disc disease.
The intervertebral disc is a specialized tissue that permits rotation, as well as flexure and extension, of the spine. One overriding aspect of disc cell biology is that cells of the nucleus pulposus and cells residing in the inner annulus are removed from the blood supply (1). For example, blood vessels originating in the vertebral body traverse the superficial region of the end plates; none of these vessels infiltrate the nucleus pulposus. With respect to the annulus, this tissue is considered to be avascular, except for small discrete capillary beds present in the dorsal and ventral surfaces; in no case does the annulus vasculature enter the nucleus pulposus (2–4). Modeling studies by Bartels and colleagues indicate that the PO2 levels within the disc are low (5); related to this observation, cells in the transitional zone between the inner annulus and the nucleus, as well as in the nucleus itself, display a robust hypoxic signal (6). In line with the avascular nature of this tissue, there is a robust and constitutive expression of both hypoxia-inducible factor 1α (HIF-1α) and HIF-2α, confirming that the inner annulus and nucleus pulposus cells reside in a hypoxic environment (7–9).
Consistent with the characteristics of most types of connective tissue, cell turnover within the disc is slow. Moreover, progenitor cells are present in the disc, which, on activation, could replace resident cells (10). Thus, tissue renewal in the intervertebral disc is dependent on the ability of progenitor cells to commit to the annulus fibrosus/nucleus pulposus lineage and undergo terminal differentiation. These events become even more important during disc degeneration and aging, since the number of cells in the annulus and nucleus pulposus decrease as a result of apoptosis and senescence (11–13).
Proliferation of progenitor cells is regulated by the hypoxia-sensitive Notch signaling pathway (14). Interaction between HIF-1α and the intracellular domain of the Notch protein (Notch-ICD) inhibits differentiation of myogenic and neural precursor cells (15). In skeletal tissue, disruption of Notch signaling markedly increases trabecular bone mass; indeed, mouse studies have shown that, with aging, osteopenia develops, due to a sharp reduction in mesenchymal progenitor cell populations in the mice (16,17). In articular cartilage, Notch signaling maintains the proliferation of chondrocytes, while during cell differentiation, there is a decrease in Notch signaling as well as a decrease in the expression of the target gene Hes5 (18). Hypoxia also increases the expression of known Notch target genes, such as Hes1 and Hey1 (15). Accordingly, in the nucleus, HIF-1α may directly interact with the Notch-ICD and direct cell fate.
The major objective of the present investigation was to examine the hypothesis that hypoxia regulates Notch signaling activity in intervertebral disc cells, and that this pathway is critical for the maintenance of cell proliferation. Our results show, for the first time, that in disc cells, hypoxia serves as a positive modulator of Notch signaling and Hes1 expression.
MATERIALS AND METHODS
Reagents and plasmids
Plasmids were kindly provided by Dr. Urban Lendahl (Karolinska Institute, Stockholm, Sweden) (plasmids 12xCSL-Luc and Hes1-Luc [−194/+160 bp]) (15) and Dr. Diane Hayward (Johns Hopkins University, Baltimore, MD) (plasmids 4xwtCBF1-Luc and 4xmtCBF1-Luc). Notch1-ICD, developed by Dr. Conie Cepko (Harvard Medical School, Department of Genetics, Boston, MA), was obtained from Addgene (no. 15131) (19). As an internal transfection control, vector pRL-TK (Promega), containing the Renilla reniformis luciferase gene, was used. The amount of transfected plasmid, the pretransfection period after seeding, and the posttransfection period before harvesting have been optimized for rat nucleus pulposus cells using pSV β-galactosidase plasmid (Promega) (9). The γ-secretase/Notch signaling inhibitor L685458 was from Sigma.
Isolation of rat intervertebral disc cells, and culture of cells in a state of hypoxia
Nucleus pulposus and inner annulus fibrosus cells were isolated from rat lumbar discs (obtained from 10–12-week-old rats), using a method previously described by Risbud et al (9). Cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) and 10% fetal bovine serum (FBS) supplemented with antibiotics. Both inner annulus fibrosus cells and nucleus pulposus cells were cultured in a Hypoxia Work Station (Invivo2 300) with a mixture of 1% O2, 5% CO2, and 94% N2 for 24–72 hours. The concentration of oxygen chosen for this study was based on our previous observations from in vitro studies, as well as on information generated on the oxemic status of the disc in vivo.
Collection and grading of human tissue
Lumbar disc tissue samples were collected as surgical waste from individuals undergoing elective spinal surgical procedures (mean age 54 years, range 38–82 years). Control samples from cadaver donors were obtained from regional tissue banks. In accordance with the guidelines of the Thomas Jefferson University Institutional Review Board, informed consent for sample collection was obtained from each patient. Assessment of intervertebral disc disease was performed using the modified Thompson grading system (20).
Immunohistologic studies
Freshly isolated rat spines were immediately fixed in 4% paraformaldehyde in phosphate buffered saline (PBS) and then embedded in paraffin. Transverse sections, 6–8 μm in thickness, were deparaffinized in xylene and rehydrated through graded ethanol. For localization of Notch-4 protein in the tissue, sections were incubated with anti–Notch-4 antibodies (1:150) in 2% bovine serum albumin in PBS, at a dilution of 1:100, at 4°C overnight. After thoroughly washing the sections, the bound primary antibody was incubated with a biotinylated universal secondary antibody (Vector), at a dilution of 1:20, for 10 minutes at room temperature. Sections were incubated with a streptavidin–peroxidase complex for 5 minutes and washed with PBS. The color reaction was developed using 3,3′-diaminobenzidine (Vectastain Universal Quick Kit; Vector).
Real-time reverse transcription–polymerase chain reaction (RT-PCR) analysis
Total RNA was extracted from rat annulus fibrosus and nucleus pulposus cells or tissue sections using RNeasy Mini columns (Qiagen). Before elution from the column, RNA was treated with RNase-free DNase I (Qiagen). In human tissue samples, total RNA was isolated from 100–300 mg of disc tissue. The tissue was homogenized in Trizol (Invitrogen) on ice, using an Omni TH Homogenizer (Omni International). Following extraction with TRIzol, RNA was passed through the RNeasy Mini columns. The purified, DNA-free RNA was converted to complementary DNA (cDNA) using Superscript III Reverse Transcriptase (Invitrogen).
Reactions were set up in triplicate in 96-well plates, using 1 μl cDNA with SYBR Green PCR Master Mix (Applied Biosystems), to which gene-specific forward and reverse PCR primers were added (synthesized by Integrated DNA Technologies). PCR reactions were performed in a StepOnePlus real-time PCR system (Applied Biosystems), in accordance with the manufacturer’s instructions; expression values were normalized to those of 18S cDNA and GAPDH. Melting curves were analyzed to verify the specificity of the RT-PCR and the absence of primer dimer formation.
Protein extraction and Western blotting
Rat disc cells were placed on ice immediately following treatment, and then washed with ice-cold Hanks’ balanced salt solution. All of the wash buffers and the final resuspension buffer included 1X protease inhibitor cocktail (Roche), NaF (5 mM), and Na3VO4 (200 μM). Nuclear or total cell proteins were resolved on 8–12% sodium dodecyl sulfate–polyacrylamide gels and transferred by electroblotting to PVDF membranes (Bio-Rad). The membranes were blocked with 5% nonfat dry milk in TBST (50 mM Tris, pH 7.6, 150 mM NaCl, 0.1% Tween 20) and incubated overnight at 4°C in 3% nonfat dry milk in TBST with anti–Notch-4 antibodies (1:600; Santa Cruz Biotechnology), anti–Jagged-1 antibodies (1:1,000; Cell Signaling Technology), anti-Hes1 antibodies (1:500; Santa Cruz Biotechnology), or anti–β-tubulin antibodies (1:2,000; Developmental Studies Hybridoma Bank). Immunolabeling was detected using an enhanced chemiluminescence reagent (Amersham Biosciences).
Immunofluorescence microscopy
Rat disc cells were plated in flat-bottomed 96-well plates (5 × 103/well) and cultured under hypoxic conditions for 24 hours. After incubation, cells were fixed with 4% paraformaldehyde, permeabilized with 0.2% Triton X-100 in PBS for 10 minutes, blocked with PBS containing 5% FBS, and incubated with antibodies against cleaved Notch1 (1:200; Cell Signaling Technology), Notch3 (1:200), Notch4 (1:100), or Hes1 (1:100) (the latter 3 from Neuromics), at 4°C overnight. As a negative control, cells were reacted with isotype IgG under similar conditions. After washing, the cells were incubated with an Alexa Fluor 488–conjugated anti-mouse secondary antibody (Invitrogen), at a dilution of 1:50, for 1 hour at room temperature. Cells were imaged using a laser scanning confocal microscope (Olympus Fluoview).
Transfections and dual luciferase assay
Rat disc cells were transferred, one day before transfection, to 24-well plates at a density of 4 × 104 cells/well. To investigate the effect of Notch-ICD overexpression on CBF1 and Hes1 promoter activity, cells were cotransfected with Notch-ICD (100–300 ng), or appropriate backbone vector, along with 300–350 ng CBF1 or Hes1 reporter and 300–350 ng pRL-TK plasmid. To measure the effect of hypoxia on Notch signaling, cells were transfected with 500 ng of 12xCSL or CBF1 reporter plasmids along with 500 ng pRL-TK plasmid; in some experiments, cells were treated with the Notch inhibitor L685458 (4 μM; Sigma). Lipofectamine 2000 (Invitrogen) was used as a transfection reagent. For each transfection, plasmids were premixed with the transfection reagent.
Within 48–72 hours after transfection, the cells were harvested and a dual luciferase reporter assay system (Promega) was used for sequential measurements of firefly and Renilla luciferase activities. Quantification of the luciferase activities and calculation of relative expression ratios were carried out using a luminometer (TD-20/20; Turner Designs). At least 3 independent transfections were performed, and all analyses were carried out in triplicate.
Assessment of cell proliferation by MTT assay
To measure disc cell proliferation, an MTT assay was carried out as described previously (5). Briefly, after treatment of the rat disc cells, MTT diluted in serum-free DMEM was added to the culture medium, to a final concentration of 0.5 mg/ml. At the end of the incubation period (2 hours at 37°C), the medium was removed, and the precipitated formazan crystals were solubilized in dimethyl sulfoxide. Product formation was measured by reading the absorbance at 560 nm, using a microplate reader (Tecan; Spectra Flour Plus).
Analysis of cell cycle
Following treatment of the rat annulus fibrosus cells, a single-cell suspension was prepared from cell cultures and fixed in ice-cold 70% ethanol for 1 hour. Cells were washed and resuspended in PBS with 5% FBS, and then incubated with 50 μM propidium iodide for 30 minutes at 37°C. Analysis of the cell cycle was conducted using a Coulter Epics XL-MCL system, with results analyzed using the XL System II software.
Statistical analysis
All measurements were performed in triplicate. Results are presented as the mean ± SEM. Differences between groups were assessed by analysis of variance. P values less than 0.05 were considered statistically significant.
RESULTS
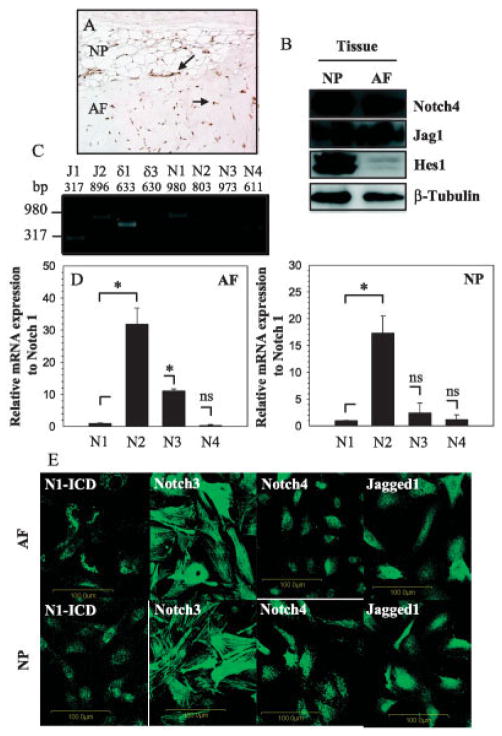
Transverse sections of skeletally mature rat discs were stained with antibodies to Notch-4 (Figure 1A). Notch-4 was observed to be present in cells of the annulus fibrosus and nucleus pulposus. Staining was evident in the plasma membrane and cytosol, as well as nucleus (Figure 1A).
Figure 1.
Expression of Notch receptors and ligands, as well as Notch target gene Hes1, in rat intervertebral discs. A, Transverse sections of disc tissue from mature rats were treated with anti–Notch-4 antibodies, and expression of Notch-4 was detected in cells of the nucleus pulposus (NP) and annulus fibrosus (AF) (arrows). Original magnification × 20. B, Expression of Notch-4, Jagged-1, and Hes1 was assessed in NP and AF tissue by Western blotting; anti–β-tubulin was used as a positive control. C, Expression of select Notch ligands (Jagged1 [J1], Jagged2 [J2], Delta1, and Delta3) and Notch receptors (N1–N4) was assessed in AF tissue by reverse transcription–polymerase chain reaction (RT-PCR). D, Messenger RNA expression of the 4 Notch receptors was assessed in primary cells of the AF (left) and NP (right) by real-time RT-PCR. Values for Notch2–Notch4 are expressed relative to that of Notch1 and are the mean ± SEM results from 3 independent experiments. * = P < 0.05. NS = not significant. E, AF and NP cells were assessed by immunofluorescence analysis for the expression of Notch3, Notch4, and Jagged1, as well as cleaved Notch1 (Notch1–intracellular domain [N1-ICD]). Original magnification × 20.
Expression of select Notch pathway proteins in the rat disc tissue was studied using Western blot analysis. As shown in Figure 1B, the rat nucleus pulposus and annulus fibrosus tissue expressed Notch-4, Jagged-1, and Notch target gene Hes1. Expression of Hes1 was most prominent in the nucleus pulposus (Figure 1B), while that of Jagged-1 was higher in the annulus fibrosus.
To further examine the expression of different Notch receptors and ligands in the disc tissue, messenger RNA (mRNA) expression in the annulus fibrosus tissue was analyzed by RT-PCR (Figure 1C). We observed expression of mRNA for the Notch ligands Jagged1, Jagged2, and Delta1 and Notch receptors 1–4. However, expression of Delta3 was undetectable. Nucleus pulposus tissue elicited a similar pattern of ligand and receptor mRNA expression (results not shown).
In parallel, we measured the relative mRNA expression of Notch receptors in primary cells of the annulus fibrosus and nucleus pulposus. As shown in Figures 1D and E, a raised expression of Notch2, when compared with the other receptors, was observed in both cell types. Moreover, in annulus fibrosus cells, expression of Notch3 was higher than that of Notch1 and Notch4. Immunofluorescence analysis confirmed that Notch1, Notch3, and Notch4, as well as Jagged1, were expressed in annulus fibrosus and nucleus pulposus cells (Figure 1F).
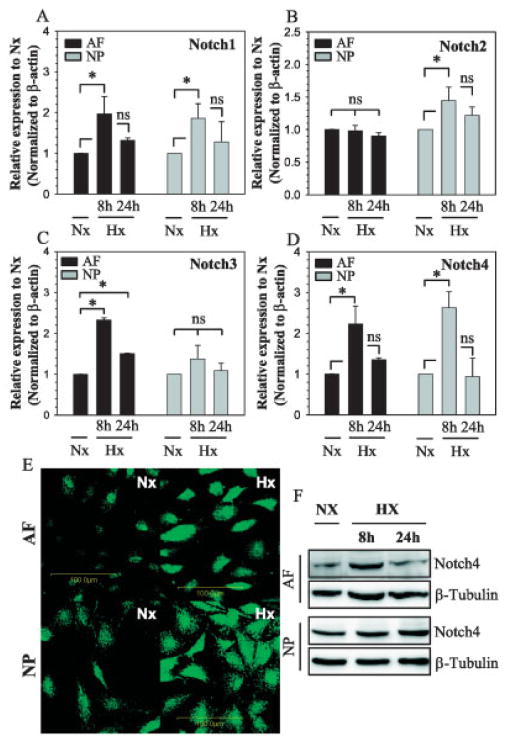
We also examined the relationship between expression of Notch receptors in disc cells and the oxemic status of the cells. As shown in Figure 2A, when annulus fibrosus and nucleus pulposus cells were cultured under conditions of hypoxia for 8 hours, Notch1 mRNA was induced. In contrast, Notch2 mRNA expression was elevated with hypoxia in nucleus pulposus cells only (Figure 2B). With regard to Notch3 mRNA, expression was induced by hypoxia in annulus fibrosus cells but not in nucleus pulposus cells (Figure 2C). Finally, expression of Notch4 mRNA was induced by hypoxia in both cell types (Figure 2D).
Figure 2.
Hypoxic regulation of Notch receptor expression in rat disc cells. A–D, AF and NP cells were cultured under conditions of hypoxia (Hx) for 8 or 24 hours, or under normoxic (Nx) conditions, and the response to hypoxia was assessed by real-time reverse transcription–polymerase chain reaction analysis of mRNA expression for Notch receptors Notch1 (A), Notch2 (B), Notch3 (C), and Notch4 (D). Values are the mean ± SEM results from 3 independent experiments. * = P < 0.05. E and F, Notch-4 protein expression under conditions of hypoxia, or under normoxic conditions, was assessed in AF and NP cells by immunofluorescence analysis (after 24 hours of hypoxia) (E) and Western blotting (after 8 and 24 hours of hypoxia) (F). Anti–β-tubulin was used as a positive control in Western blots. Original magnification × 20. See Figure 1 for other definitions.
To determine whether there was a concomitant elevation in Notch-4 protein expression associated with the oxemic status, the cells were evaluated by immunofluorescence analysis and Western blotting. In both cell types, there was an increase in Notch-4 expression in the hypoxic cultures, as indicated in Figures 2E and F. Protein expression was maximally elevated at 8 hours and returned to baseline levels by 24 hours in annulus fibrosus cells, while in nucleus pulposus cells, protein expression remained elevated up to 24 hours.
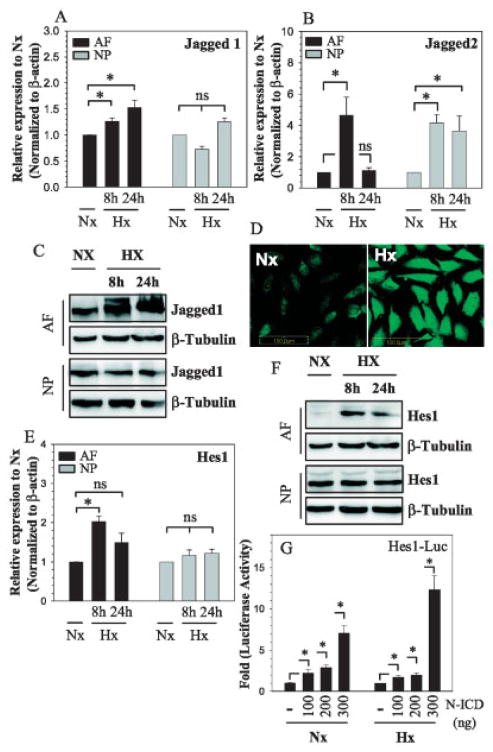
We then examined the effect of hypoxia on mRNA expression of the Notch ligands Jagged1 and Jagged2 in disc cells. As shown in Figure 3A, under conditions of hypoxia, there was induction of Jagged1 mRNA in annulus cells but not in nucleus pulposus cells. In contrast, in both annulus fibrosus and nucleus pulposus cells, Jagged2 mRNA expression was highly sensitive to hypoxia (Figure 3B). In both cell types, mRNA expression remained elevated for the first 8 hours; in annulus fibrosus cells, mRNA expression returned to baseline levels by 24 hours. In addition, the effect of hypoxia on Jagged-1 protein expression was examined in annulus fibrosus cells by Western blotting (Figure 3C) and immunofluorescence microscopic analysis (Figure 3D). In the hypoxic environment, a significant increase in the levels of Jagged-1 protein was seen (Figures 3C and D).
Figure 3.
Hypoxic regulation of Notch ligand expression in rat disc cells. A and B, AF and NP cells were cultured under conditions of hypoxia (Hx) for 8 or 24 hours, or under normoxic conditions (Nx), and the response to hypoxia was assessed by real-time RT-PCR analysis of Jagged1 (A) and Jagged2 (B) mRNA expression. C and D, Jagged-1 protein expression under conditions of hypoxia, or under normoxic conditions, was assessed in AF and NP cells by Western blotting (after 8 and 24 hours) (C) and immunofluorescence analysis (after 24 hours) (D). Anti–β-tubulin was used as a positive control in Western blots. Original magnification × 20. E and F, The effect of hypoxia on Hes1 gene expression and promoter activity was assessed in AF and NP cells cultured for 8 or 24 hours under conditions of hypoxia, or under normoxic conditions, using real-time RT-PCR analysis of Hes1 mRNA expression (E) and Western blotting of Hes-1 protein expression, with anti–β-tubulin used as a positive control (F). G, AF cells were cotransfected with a Hes1 reporter (−194/+160 bp) along with increasing doses of Notch1–intracellular domain (N-ICD), and Hes1 reporter activity was measured under normoxic and hypoxic conditions. Values in A, B, E, and G are the mean ± SEM results from 3 independent experiments. * = P < 0.05. See Figure 1 for other definitions.
To further evaluate the relationship between Notch signaling and oxygen tension, we measured the expression of Notch target gene Hes1 in the rat disc cells under normoxic and hypoxic conditions. In annulus fibrosus cells, expression of Hes1 mRNA was induced after 8 hours of hypoxia and returned to baseline levels by 24 hours (Figure 3E). In contrast, in nucleus pulposus cells, Hes1 mRNA expression was insensitive to hypoxia. Western blot analysis showed that Hes-1 protein expression was also elevated in hypoxia, and stayed high in annulus fibrosus cells under hypoxic conditions for 24 hours (Figure 3F). As expected, Hes-1 protein expression in nucleus pulposus cells remained unchanged (Figure 3F).
To investigate whether Hes1 responds to Notch signaling, we cotransfected annulus fibrosus cells with a Notch-ICD expression plasmid, and then measured Hes1 promoter activity. Induced overexpression of Notch resulted in increased Hes1 promoter activity under conditions of normoxia, and led to an even greater level of Hes1 promoter activity in the hypoxic state (Figure 3G).
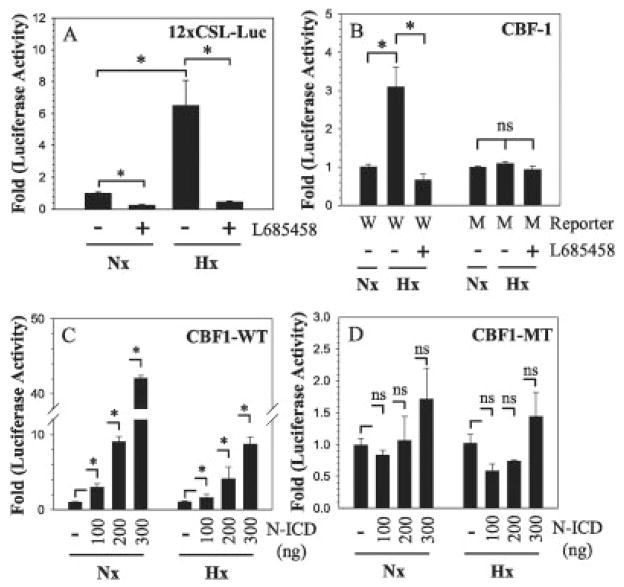
We next evaluated the effect of hypoxia on the activity of the Notch-responsive reporter 12xCSL and Notch target gene CBF1 in annulus fibrosus cells (Figures 4A and B). Hypoxia significantly induced 12xCSL reporter activity. Moreover, induction of reporter activity was completely blocked by the κ-secretase inhibitor L685458. Similarly, hypoxia increased CBF1 promoter activity, and this was also significantly blocked by inhibitor treatment.
Figure 4.
Effect of hypoxia on Notch signaling activity. A and B, The 12xCSL reporter (A) or CBF1 reporter containing a wild-type (W) or mutant (M) RBPJκ motif (B) was transfected into rat annulus fibrosus cells along with pRL-TK vector. Cells were cultured for 24 hours under normoxic conditions (21% O2) (Nx) or hypoxic conditions (1% O2) (Hx), and some cells were treated with γ-secretase inhibitor L685458 (4 μM), and luciferase reporter activity was measured. C and D, Rat annulus fibrosus cells were cotransfected with wild-type (WT) (C) or mutant (MT) (D) CBF1 reporters along with increasing doses of Notch1-ICD, and reporter activity was measured under normoxic and hypoxic conditions. Values are the mean ± SEM results from 3 independent experiments. * = P < 0.05. See Figure 1 for other definitions.
A CBF1 promoter harboring a mutation in the Notch-interacting RBPJκ site is insensitive to both hypoxia and L685458 treatment. To further validate that CBF1 promoter activity is responsive to Notch signaling, we cotransfected annulus fibrosus cells with a plasmid encoding the Notch1-ICD. This plasmid significantly increased wild-type CBF1 promoter activity, under both normoxic and hypoxic conditions (Figure 4C). The stimulatory effect of Notch-ICD on CBF1 promoter activity was evident in cultures with 100 ng of plasmid, and the effect was further enhanced when the concentration of plasmid was increased to 300 ng (Figure 4C). Moreover, the inductive effect was more pronounced in a state of normoxia. As expected, mutant CBF1 promoter activity was unresponsive to Notch coexpression (Figure 4D).
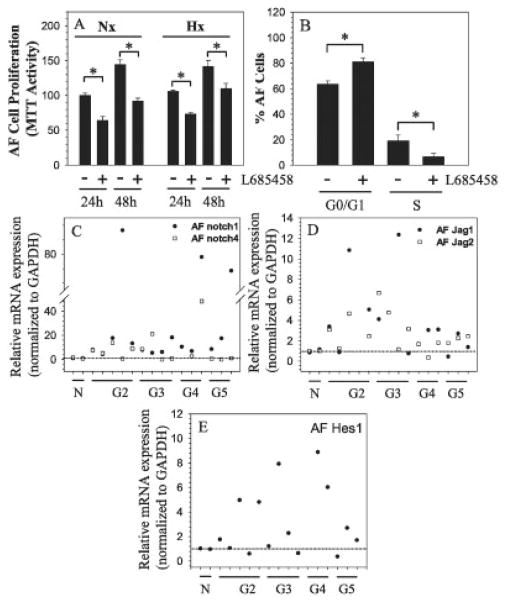
We also examined the relationship between Notch signaling and proliferation of annulus fibrosus cells. Treatment with γ-secretase inhibitor L685458 resulted in a significant suppression of annulus fibrosus cell proliferation under both normoxic and hypoxic conditions (Figure 5A). With the use of flow cytometry, we determined whether the cell cycle status was affected by L685458 treatment. The number of annulus fibrosus cells in S phase was significantly decreased after treatment with the inhibitor (mean ± SEM 6.7 ± 2.8%) when compared to the number of untreated control cells in S phase (mean ± SEM 19.25 ± 4.4%) (Figure 5B). In contrast, there was a greater accumulation of cells in G0/G1 phase after treatment (mean ± SEM 81.10 ± 3.3%) as compared to control cells left untreated (mean ± SEM 63.4 ± 2.9% in G0/G1 phase).
Figure 5.
Role of Notch signaling in controlling AF cell proliferation. A, AF cells were cultured under normoxia (Nx) or hypoxia (Hx), with or without γ-secretase inhibitor L685458 (4 μM), for 24–48 hours, and cell proliferation was measured by MTT assay. B, The cell cycle of AF cells treated with or without L685458 was determined by flow cytometry, with results expressed as the percentage of AF cells in G0/G1 phase or S phase. Values in A and B are the mean ± SEM results from 3 independent experiments. * = P < 0.05. C–E, Messenger RNA expression of Notch receptors Notch1 and Notch4 (C), Notch ligands Jagged1 and Jagged2 (D), and Notch target gene Hes1 (E) was assessed by real-time RT-PCR in multiple human AF tissue samples in varying degrees of degeneration (Thompson grades 2–5 [G2–G5]) compared with normal controls (N) (controls n = 2, G2 n = 5, G3 n = 4, G4 n = 3, G5 n = 3). The mean expression of mRNA in normal control samples was set at 1.0 (horizontal line). See Figure 1 for other definitions.
Finally, we evaluated the expression of select Notch receptors, ligands, and the target gene Hes1 in Thompson-graded degenerated human annulus fibrosus tissue (Figures 5C–E). Results of real-time RT-PCR analysis showed that there was a trend toward an increase in expression of Notch1 and Notch4 mRNA (Figure 5C) in degenerated human disc tissue when compared to normal controls. Similarly, both Jagged1 and Jagged2 mRNA exhibited a similar trend in expression in degenerated disc tissue (Figure 5D). Hes1 mRNA expression was also responsive to degeneration, as indicated by a higher mRNA expression level at the earlier Thompson stages (grade 2 and grade 3) (Figure 5E). Not surprisingly, there was a considerable between-patient variation within the same grade.
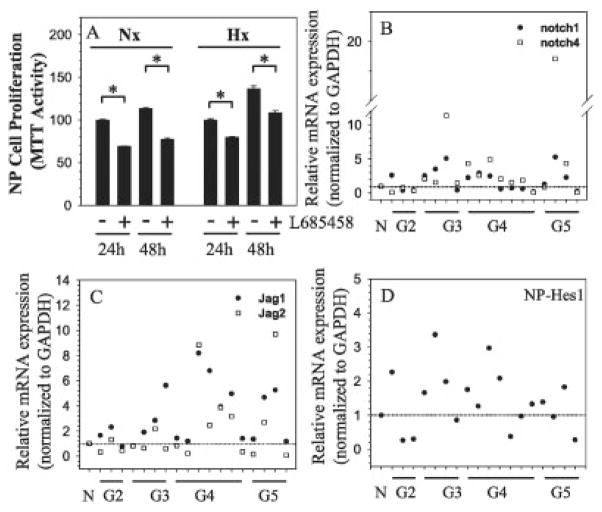
We also examined the role of Notch signaling in regulating proliferation of nucleus pulposus cells. Similar to the effects on annulus fibrosus cells and irrespective of the oxemic tension, L685458 treatment resulted in a significant inhibition of the proliferation of nucleus pulposus cells (Figure 6A).
Figure 6.
Role of Notch signaling in controlling NP cell proliferation. A, NP cells were cultured under conditions of normoxia (Nx) or hypoxia (Hx), with or without γ-secretase inhibitor L685458 (4 μM), for 24–48 hours, and cell proliferation was measured by MTT assay. Values are the mean ± SEM results from 3 independent experiments. * = P < 0.05. B–D, Messenger RNA expression of Notch receptors Notch1 and Notch4 (B), Notch ligands Jagged1 and Jagged2 (C), and Notch target gene Hes1 (D) was assessed by real-time RT-PCR in multiple human NP tissue samples in varying degrees of degeneration (Thompson grades 2–5 [G2–G5]) compared with normal controls (N) (controls n = 1, G2 n = 3, G3 n = 4, G4 n = 7, G5 n = 4). The mean expression of mRNA in normal control samples was set at 1.0 (horizontal line). See Figure 1 for other definitions.
Finally, we measured the expression of Notch receptors, ligands, and Hes1 in graded degenerated human disc tissue from the nucleus pulposus (Figures 6B–D). Results of real-time RT-PCR analysis showed that there was a trend toward an increase in mRNA expression of Notch1 and Notch4 (Figure 6B) and Jagged1 and Jagged2 (Figure 6C) in degenerated human disc tissue when compared to normal controls. Similarly, there was a trend toward an increase in the expression of Hes1 mRNA in the degenerated nucleus pulposus (Figure 6D). As in the degenerated tissue of the annulus fibrosus, a considerable between-patient variation in mRNA expression was observed in the degenerated nucleus pulposus, among tissue of the same Thompson grade.
DISCUSSION
The basis for the studies described herein is that progenitor cell populations are present in the hypoxic tissue of the intervertebral disc (10). Accordingly, it would not be unreasonable to assume that the commitment of these cells into functioning cells of the annulus fibrosus and nucleus pulposus is dependent on the highly conserved, hypoxia-sensitive Notch signaling pathway (14,15,21). The present study revealed that there is a hypoxia-dependent increase in expression of specific Notch ligands and receptors and a concomitant increase in Notch signaling activity. In addition, we found that hypoxia induces expression of Hes1, the Notch target gene that is associated with maintenance of the undifferentiated state and determination of cell fate (22,23).
Accordingly, as has also been observed in articular cartilage, the hypoxic status of the disc may serve to conserve cell number and maintain the differentiated state of the cells by regulating Notch signaling activity (18). Our results showed that there is an increase in levels of Notch signaling proteins in degenerated human disc tissue; however, the response is probably insufficient to maintain cell numbers and preserve the differentiated state of the cells. This observation could explain why there is a gradual age-dependent loss of cells and a decrease in the ability of resident cells to mount a reparative response.
In this study, we examined the expression and localization of Notch receptors and ligands in the intervertebral disc of skeletally mature animals. Both annulus fibrosus and nucleus pulposus tissue expressed Notch receptors (Notch-1–Notch-4) and ligands (Jagged-1, Jagged-2, and Delta-1). The observation that there was robust expression of the Notch target gene, Hes1, indicated the presence of an active Notch signaling pathway in the disc, a notion supported by the plethora of receptors and ligands that were present in both tissue types; a similar spectrum of receptors and ligands has been observed in cartilage (24). Consistent with this observation, although the study focused on defining components of the signaling pathway in specific tissues of the intervertebral disc, it would not be surprising to find that ligands from the inner annulus are associated with Notch receptors in the nucleus pulposus; likewise, ligands in the nucleus would serve to activate Notch signaling pathways in the annulus fibrosus. This interaction would be consistent with the notion that the disc itself represents an environmental niche that serves to integrate the functional activities of each of the component tissues (1).
There were marked differences in the pattern of expression of Notch receptors and ligands in the disc. While Notch2 mRNA transcript levels were elevated in both the annulus and the nucleus pulposus, Jagged-1 protein levels were significantly greater in the annulus than in the nucleus pulposus. In both tissues, Notch1 and Notch4 reporter expression was responsive to the level of PO2. In nucleus cells, Notch reporter expression was increased with hypoxia.
To examine the premise that hypoxia influences Notch signaling activity, the Notch-responsive reporters 12xCSL and CBF1 were utilized. Our results showed that there was increased activity of both reporters under hypoxic conditions. Moreover, reporter activity was completely inhibited when the hypoxic cells were treated with the γ-secretase inhibitor L685458. We confirmed the involvement of the signaling pathway using a reporter with a mutation in the CSL-binding site. In this case, hypoxic induction of Notch reporter activity was completely blocked. Results of these experiments lend direct support to the hypothesis that hypoxia promotes Notch signaling in disc cells. This result is consistent with observations of activation of Notch signaling in stem cells and other cell types (16,17) and confirms previous findings showing that the Notch signaling pathway is responsive to hypoxia (14,15,21). Of note, since disc cells function in vivo in a low oxemic environment, it is likely that hypoxia is a key factor regulating the activity of this pathway (14,15,21,25).
In addition to the effects of hypoxia on Notch ligands and receptors, we noted that there was a hypoxia-dependent increase in Notch target gene activity. This finding suggests that in disc cells, Hes1 expression is regulated in a hypoxia-dependent manner. It should be noted that in many types of tissue, Hes1 is considered to be a key regulator of differentiation (23,26). Hes1 is expressed by almost all undifferentiated cells and is required for the maintenance of this pool of cells. The observation that a progenitor cell pool is present in the disc, even in degenerated disc tissue (10), adds credence to the view that the hypoxic state enhances the activity of this powerful repressor of differentiation (23,26). Not surprisingly, the response of disc cells to HIF is markedly different from that of myogenic C2C12 cells (7,9,27). In stem cells and many cancer cells, HIF-1 interacts with the Notch coactivator Mastermindlike-1 to increase Hes-1 promoter activity (25). Possibly, in the disc, HIF-1α may promote recruitment of the Notch-ICD to the CSL-binding motifs in the Hes1 promoter (15). While the mechanism by which this type of regulation is achieved is still under examination, functionally, it is more than likely a reflection of the normative environment of hypoxia in the disc.
Although regulation of Notch activity may be tissue specific, the signaling pathway has been shown to modulate cell proliferation and differentiation, as well as self renewal (18,22,23,26). To determine whether the Notch pathway regulates disc cell number, we evaluated nucleus pulposus and annulus fibrosus cell proliferation under normoxic and hypoxic conditions. In both cases, when treated with a γ-secretase inhibitor, a marked decrease in proliferation was observed. We reasoned that this suppressive effect would account for the decrease in cell number that characterizes degenerated human disc tissue (11–13).
Based on this observation, we evaluated major components of the Notch signaling pathway in cells of human discs in varying degrees of degeneration. While the number of control tissue samples was very limited (due to practical difficulties in acquiring normal human discs), measurement of mRNA expression levels suggested that as the tissue became degenerated, there was a rise in expression of Jagged1 and Jagged2 and possibly Notch4 and Hes1. Our results are consistent with those of a recent study that showed increased expression of Notch1, Jagged1, and Hes5 in osteoarthritic cartilage (28). One explanation for the increase is that it possibly represents a compensatory response by resident cells to replace the lost or nonfunctional cells. Indeed, in the disc, as well as cartilage, a clonal proliferative response is a hallmark of degeneration (29–31). In both tissues, aside from the effects of hypoxia, other regulatory elements, including elevated signaling through the transforming growth factor β/Smad3 and Wnt/β-catenin pathways, are likely to modulate Notch pathway activity (32–35). Thus, although some increase in the levels of Notch signaling proteins may occur, the dysregulated response is insufficient to mount a complete functional recovery and restoration of cell numbers in the tissue.
In terms of pursuing therapeutic strategies, it should be possible to promote tissue repair by 3 different approaches. First, specific ligands and receptors of the Notch signaling pathway may be manipulated to reactivate the surviving endogenous progenitor cells to promote their proliferation and subsequent commitment to the annulus fibrosus and nucleus pulposus cell phenotype. Second, molecules, such as the numb protein, that antagonize Notch signaling activity may be targeted in disc progenitor cells (36). Finally, the oxemic status of the intervertebral niche may be restored using redox-sensitive agents.
Acknowledgments
Supported by the NIH (grants R01-AR-050087 to Drs. Shapiro and Risbaud and R01-AR-055655 to Dr. Risbud).
Footnotes
AUTHOR CONTRIBUTIONS
All authors were involved in drafting the article or revising it critically for important intellectual content, and all authors approved the final version to be published. Dr. Risbud had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study conception and design. Anderson, Sakai, Mochida, Albert, Shapiro, Risbud.
Acquisition of data. Hiyama, Skubutyte, Markova, Anderson, Yadla.
Analysis and interpretation of data. Hiyama, Skubutyte, Markova, Anderson, Yadla, Shapiro, Risbud.
References
- 1.Risbud MV, Schipani E, Shapiro IM. Hypoxic regulation of nucleus pulposus cell survival: from niche to notch. Am J Pathol. 2010;176:1577–83. doi: 10.2353/ajpath.2010.090734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gruber HE, Ashraf N, Kilburn J, Williams C, Norton HJ, Gordon BE, et al. Vertebral endplate architecture and vascularization: application of micro-computerized tomography, a vascular tracer, and immunocytochemistry in analyses of disc degeneration in the aging sand rat. Spine. 2005;30:2593–600. doi: 10.1097/01.brs.0000187877.30149.83. [DOI] [PubMed] [Google Scholar]
- 3.Hassler O. The human intervertebral disc: a micro-angiographical study on its vascular supply at various ages. Acta Orthop Scand. 1969;40:765–72. doi: 10.3109/17453676908989540. [DOI] [PubMed] [Google Scholar]
- 4.Rudert M, Tillmann B. Lymph and blood supply of the human intervertebral disc: cadaver study of correlations to discitis. Acta Orthop Scand. 1993;64:37–40. doi: 10.3109/17453679308994524. [DOI] [PubMed] [Google Scholar]
- 5.Bartels EM, Fairbank JC, Winlove CP, Urban JP. Oxygen and lactate concentrations measured in vivo in the intervertebral discs of patients with scoliosis and back pain. Spine. 1998;23:1–7. doi: 10.1097/00007632-199801010-00001. [DOI] [PubMed] [Google Scholar]
- 6.Lee DC, Adams CS, Albert TJ, Shapiro IM, Evans SM, Koch CJ. In situ oxygen utilization in the rat intervertebral disc. J Anat. 2007;210:294–303. doi: 10.1111/j.1469-7580.2007.00692.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rajpurohit R, Risbud MV, Ducheyne P, Vresilovic EJ, Shapiro IM. Phenotypic characteristics of the nucleus pulposus: expression of hypoxia inducing factor-1, glucose transporter-1 and MMP-2. Cell Tissue Res. 2002;308:401–7. doi: 10.1007/s00441-002-0563-6. [DOI] [PubMed] [Google Scholar]
- 8.Agrawal A, Gajghate S, Smith H, Anderson DG, Albert TJ, Shapiro IM, et al. Cited2 modulates hypoxia-inducible factor–dependent expression of vascular endothelial growth factor in nucleus pulposus cells of the rat intervertebral disc. Arthritis Rheum. 2008;58:3798–808. doi: 10.1002/art.24073. [DOI] [PubMed] [Google Scholar]
- 9.Risbud MV, Guttapalli A, Stokes DG, Hawkins D, Danielson KG, Schaer TP, et al. Nucleus pulposus cells express HIF-1α under normoxic culture conditions: a metabolic adaptation to the intervertebral disc microenvironment. J Cell Biochem. 2006;98:152–9. doi: 10.1002/jcb.20765. [DOI] [PubMed] [Google Scholar]
- 10.Risbud MV, Guttapalli A, Tsai TT, Lee JY, Danielson KG, Vaccaro AR, et al. Evidence for skeletal progenitor cells in the degenerate human intervertebral disc. Spine. 2007;32:2537–44. doi: 10.1097/BRS.0b013e318158dea6. [DOI] [PubMed] [Google Scholar]
- 11.Bertram H, Nerlich A, Omlor G, Geiger F, Zimmermann G, Fellenberg J. Expression of TRAIL and the death receptors DR4 and DR5 correlates with progression of degeneration in human intervertebral disks. Mod Pathol. 2009;22:895–905. doi: 10.1038/modpathol.2009.39. [DOI] [PubMed] [Google Scholar]
- 12.Tschoeke SK, Hellmuth M, Hostmann A, Robinson Y, Ertel W, Oberholzer A, et al. Apoptosis of human intervertebral discs after trauma compares to degenerated discs involving both receptor-mediated and mitochondrial-dependent pathways. J Orthop Res. 2008;26:999–1006. doi: 10.1002/jor.20601. [DOI] [PubMed] [Google Scholar]
- 13.Gruber HE, Ingram JA, Davis DE, Hanley EN., Jr Increased cell senescence is associated with decreased cell proliferation in vivo in the degenerating human annulus. Spine J. 2009;9:210–5. doi: 10.1016/j.spinee.2008.01.012. [DOI] [PubMed] [Google Scholar]
- 14.Silvan U, Diez-Torre A, Arluzea J, Andrade R, Silio M, Arechaga J. Hypoxia and pluripotency in embryonic and embryonal carcinoma stem cell biology. Differentiation. 2009;78:159–68. doi: 10.1016/j.diff.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 15.Gustafsson MV, Zheng X, Pereira T, Gradin K, Jin S, Lundkvist J, et al. Hypoxia requires Notch signaling to maintain the undifferentiated cell state. Dev Cell. 2005;9:617–28. doi: 10.1016/j.devcel.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 16.Engin F, Yao Z, Yang T, Zhou G, Bertin T, Jiang MM, et al. Dimorphic effects of Notch signaling in bone homeostasis. Nat Med. 2008;14:299–305. doi: 10.1038/nm1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hilton MJ, Tu X, Wu X, Bai S, Zhao H, Kobayashi T, et al. Notch signaling maintains bone marrow mesenchymal progenitors by suppressing osteoblast differentiation. Nat Med. 2008;14:306–14. doi: 10.1038/nm1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karlsson C, Jonsson M, Asp J, Brantsing C, Kageyama R, Lindahl A. Notch and HES5 are regulated during human cartilage differentiation. Cell Tissue Res. 2007;327:539–51. doi: 10.1007/s00441-006-0307-0. [DOI] [PubMed] [Google Scholar]
- 19.Bao ZZ, Cepko CL. The expression and function of Notch pathway genes in the developing rat eye. J Neurosci. 1997;17:1425–34. doi: 10.1523/JNEUROSCI.17-04-01425.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hilibrand AS, Carlson GD, Palumbo MA, Jones PK, Bohlman HH. Radiculopathy and myelopathy at segments adjacent to the site of a previous anterior cervical arthrodesis. J Bone Joint Surg Am. 1999;81:519–28. doi: 10.2106/00004623-199904000-00009. [DOI] [PubMed] [Google Scholar]
- 21.Diez H, Fischer A, Winkler A, Hu CJ, Hatzopoulos AK, Breier G, et al. Hypoxia-mediated activation of Dll4-Notch-Hey2 signaling in endothelial progenitor cells and adoption of arterial cell fate. Exp Cell Res. 2007;313:1–9. doi: 10.1016/j.yexcr.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 22.Kobayashi T, Mizuno H, Imayoshi I, Furusawa C, Shirahige K, Kageyama R. The cyclic gene Hes1 contributes to diverse differentiation responses of embryonic stem cells. Genes Dev. 2009;23:1870–5. doi: 10.1101/gad.1823109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grogan SP, Olee T, Hiraoka K, Lotz MK. Repression of chondrogenesis through binding of Notch signaling proteins HES-1 and HEY-1 to N-box domains in the COL2A1 enhancer site. Arthritis Rheum. 2008;58:2754–63. doi: 10.1002/art.23730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Williams R, Nelson L, Dowthwaite GP, Evans DJ, Archer CW. Notch receptor and Notch ligand expression in developing avian cartilage. J Anat. 2009;215:159–69. doi: 10.1111/j.1469-7580.2009.01089.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen J, Imanaka N, Chen J, Griffin JD. Hypoxia potentiates Notch signaling in breast cancer leading to decreased E-cadherin expression and increased cell migration and invasion. Br J Cancer. 2010;102:351–60. doi: 10.1038/sj.bjc.6605486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Akimoto M, Kameda Y, Arai Y, Miura M, Nishimaki T, Takeda A, et al. Hes1 is required for the development of craniofacial structures derived from ectomesenchymal neural crest cells. J Craniofac Surg. 2010;21:1443–9. doi: 10.1097/SCS.0b013e3181ebd1a0. [DOI] [PubMed] [Google Scholar]
- 27.Agrawal A, Guttapalli A, Narayan S, Albert TJ, Shapiro IM, Risbud MV. Normoxic stabilization of HIF-1α drives glycolytic metabolism and regulates aggrecan gene expression in nucleus pulposus cells of the rat intervertebral disk. Am J Physiol Cell Physiol. 2007;293:C621–31. doi: 10.1152/ajpcell.00538.2006. [DOI] [PubMed] [Google Scholar]
- 28.Karlsson C, Brantsing C, Egell S, Lindahl A. Notch1, Jagged1, and HES5 are abundantly expressed in osteoarthritis. Cells Tissues Organs. 2008;188:287–98. doi: 10.1159/000121610. [DOI] [PubMed] [Google Scholar]
- 29.Johnson WE, Eisenstein SM, Roberts S. Cell cluster formation in degenerate lumbar intervertebral discs is associated with increased disc cell proliferation. Connect Tissue Res. 2001;42:197–207. doi: 10.3109/03008200109005650. [DOI] [PubMed] [Google Scholar]
- 30.Sharp CA, Roberts S, Evans H, Brown SJ. Disc cell clusters in pathological human intervertebral discs are associated with increased stress protein immunostaining. Eur Spine J. 2009;18:1587–94. doi: 10.1007/s00586-009-1053-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lotz MK, Otsuki S, Grogan SP, Sah R, Terkeltaub R, D’Lima D. Cartilage cell clusters [review] Arthritis Rheum. 2010;62:2206–18. doi: 10.1002/art.27528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tran CM, Markova D, Smith HE, Susarla B, Ponnappan RK, Anderson DG, et al. Regulation of CCN2/connective tissue growth factor expression in the nucleus pulposus of the intervertebral disc: role of Smad and activator protein 1 signaling. Arthritis Rheum. 2010;62:1983–92. doi: 10.1002/art.27445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hiyama A, Sakai D, Risbud MV, Tanaka M, Arai F, Abe K, et al. Enhancement of intervertebral disc cell senescence by Wnt/β-catenin signaling–induced matrix metalloproteinase expression. Arthritis Rheum. 2010;62:3036–47. doi: 10.1002/art.27599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fu Y, Chang A, Chang L, Niessen K, Eapen S, Setiadi A, et al. Differential regulation of transforming growth factor β signaling pathways by Notch in human endothelial cells. J Biol Chem. 2009;284:19452–62. doi: 10.1074/jbc.M109.011833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Corada M, Nyqvist D, Orsenigo F, Caprini A, Giampietro C, Taketo MM, et al. The Wnt/β-catenin pathway modulates vascular remodeling and specification by upregulating Dll4/Notch signaling. Dev Cell. 2010;18:938–49. doi: 10.1016/j.devcel.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McGill MA, McGlade CJ. Mammalian numb proteins promote Notch1 receptor ubiquitination and degradation of the Notch1 intracellular domain. J Biol Chem. 2003;278:23196–203. doi: 10.1074/jbc.M302827200. [DOI] [PubMed] [Google Scholar]