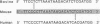Abstract
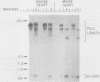
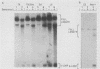
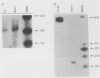
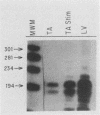
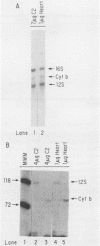
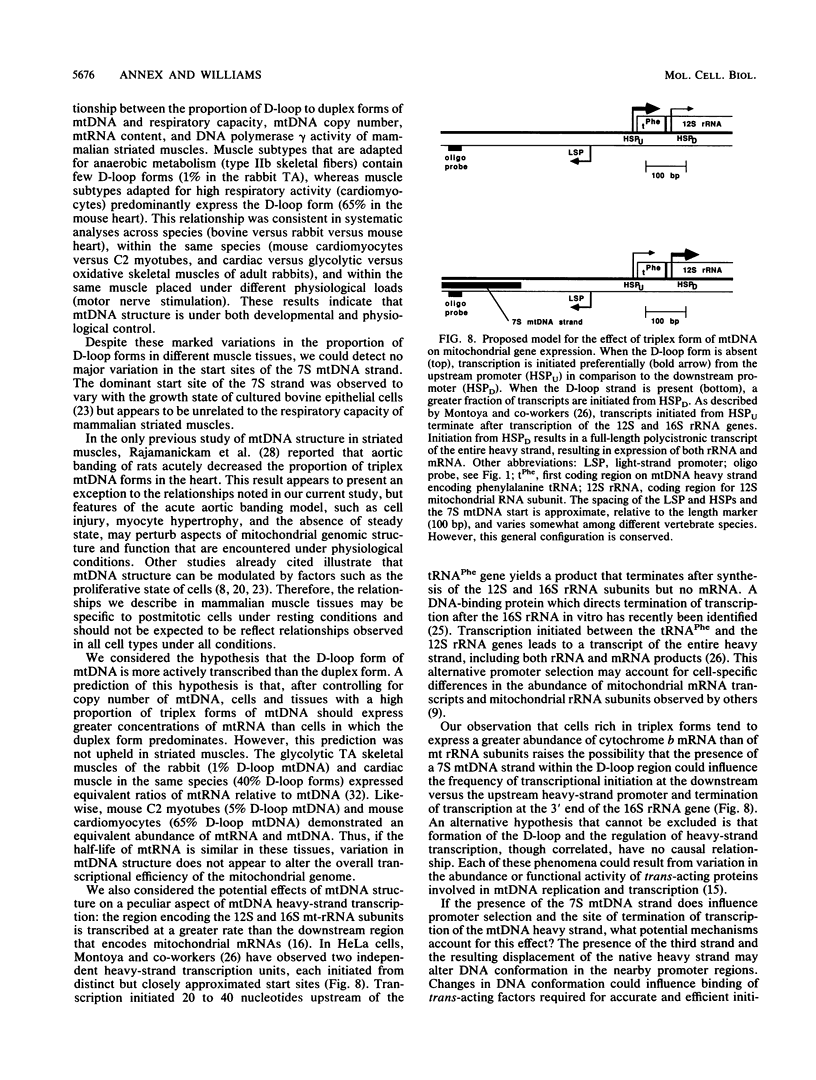
Mitochondrial DNA (mt DNA) in cells of vertebrate organisms can assume an unusual triplex DNA structure known as the displacement loop (D loop). This triplex DNA structure forms when a partially replicated heavy strand of mtDNA (7S mtDNA) remains annealed to the light strand, displacing the native heavy strand in this region. The D-loop region contains the promoters for both heavy- and light-strand transcription as well as the origin of heavy-strand replication. However, the distribution of triplex and duplex forms of mtDNA in relation to respiratory activity of mammalian tissues has not been systematically characterized, and the functional significance of the D-loop structure is unknown. In comparisons of specialized muscle subtypes within the same species and of the same muscle subtype in different species, the relative proportion of D-loop versus duplex forms of mtDNA in striated muscle tissues of several mammalian species demonstrated marked variation, ranging from 1% in glycolytic fast skeletal fibers of the rabbit to 65% in the mouse heart. There was a consistent and direct correlation between the ratio of triplex to duplex forms of mtDNA and the capacity of these tissues for oxidative metabolism. The proportion of D-loop forms likewise correlated directly with mtDNA copy number, mtRNA abundance, and the specific activity of the mtDNA (gamma) polymerase. The D-loop form of mtDNA does not appear to be transcribed at greater efficiency than the duplex form, since the ratio of mtDNA copy number to mtRNA was unrelated to the proportion of triplex mtDNA genomes. However, tissues with a preponderance of D-loop forms tended to express greater levels of cytochrome b mRNA relative to mitochondrial rRNA transcripts, suggesting that the triplex structure may be associated with variations in partial versus full-length transcription of the heavy strand.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson S., Bankier A. T., Barrell B. G., de Bruijn M. H., Coulson A. R., Drouin J., Eperon I. C., Nierlich D. P., Roe B. A., Sanger F. Sequence and organization of the human mitochondrial genome. Nature. 1981 Apr 9;290(5806):457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- Anderson S., de Bruijn M. H., Coulson A. R., Eperon I. C., Sanger F., Young I. G. Complete sequence of bovine mitochondrial DNA. Conserved features of the mammalian mitochondrial genome. J Mol Biol. 1982 Apr 25;156(4):683–717. doi: 10.1016/0022-2836(82)90137-1. [DOI] [PubMed] [Google Scholar]
- Barat-Gueride M., Dufresne C., Rickwood D. Effect of DNA conformation on the transcription of mitochondrial DNA. Eur J Biochem. 1989 Aug 1;183(2):297–302. doi: 10.1111/j.1432-1033.1989.tb14928.x. [DOI] [PubMed] [Google Scholar]
- Barat M., Mignotte B. A DNA binding protein from Xenopus laevis oocyte mitochondria. Chromosoma. 1981;82(4):583–593. doi: 10.1007/BF00295014. [DOI] [PubMed] [Google Scholar]
- Bibb M. J., Van Etten R. A., Wright C. T., Walberg M. W., Clayton D. A. Sequence and gene organization of mouse mitochondrial DNA. Cell. 1981 Oct;26(2 Pt 2):167–180. doi: 10.1016/0092-8674(81)90300-7. [DOI] [PubMed] [Google Scholar]
- Bogenhagen D., Clayton D. A. Mechanism of mitochondrial DNA replication in mouse L-cells: kinetics of synthesis and turnover of the initiation sequence. J Mol Biol. 1978 Feb 15;119(1):49–68. doi: 10.1016/0022-2836(78)90269-3. [DOI] [PubMed] [Google Scholar]
- Buzan J. M., Low R. L. Preference of human mitochondrial RNA polymerase for superhelical templates with mitochondrial promoters. Biochem Biophys Res Commun. 1988 Apr 15;152(1):22–29. doi: 10.1016/s0006-291x(88)80674-0. [DOI] [PubMed] [Google Scholar]
- Callen J. C., Tourte M., Dennebouy N., Mounolou J. C. Changes in D-loop frequency and superhelicity among the mitochondrial DNA molecules in relation to organelle biogenesis in oocytes of Xenopus laevis. Exp Cell Res. 1983 Jan;143(1):115–125. doi: 10.1016/0014-4827(83)90114-3. [DOI] [PubMed] [Google Scholar]
- Chang D. D., Fisher R. P., Clayton D. A. Roles for a promoter and RNA processing in the synthesis of mitochondrial displacement-loop strands. Biochim Biophys Acta. 1987 Jul 14;909(2):85–91. doi: 10.1016/0167-4781(87)90029-7. [DOI] [PubMed] [Google Scholar]
- Clayton D. A. Transcription of the mammalian mitochondrial genome. Annu Rev Biochem. 1984;53:573–594. doi: 10.1146/annurev.bi.53.070184.003041. [DOI] [PubMed] [Google Scholar]
- Devlin B. H., Wefald F. C., Kraus W. E., Bernard T. S., Williams R. S. Identification of a muscle-specific enhancer within the 5'-flanking region of the human myoglobin gene. J Biol Chem. 1989 Aug 15;264(23):13896–13901. [PubMed] [Google Scholar]
- Doda J. N., Wright C. T., Clayton D. A. Elongation of displacement-loop strands in human and mouse mitochondrial DNA is arrested near specific template sequences. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6116–6120. doi: 10.1073/pnas.78.10.6116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich R., Larousse A., Jacquet M. A., Marin M., Reiss C. In vitro transcription initiation from three different Escherichia coli promoters. Effect of supercoiling. Eur J Biochem. 1985 Apr 15;148(2):293–298. doi: 10.1111/j.1432-1033.1985.tb08838.x. [DOI] [PubMed] [Google Scholar]
- Fisher R. P., Topper J. N., Clayton D. A. Promoter selection in human mitochondria involves binding of a transcription factor to orientation-independent upstream regulatory elements. Cell. 1987 Jul 17;50(2):247–258. doi: 10.1016/0092-8674(87)90220-0. [DOI] [PubMed] [Google Scholar]
- Gelfand R., Attardi G. Synthesis and turnover of mitochondrial ribonucleic acid in HeLa cells: the mature ribosomal and messenger ribonucleic acid species are metabolically unstable. Mol Cell Biol. 1981 Jun;1(6):497–511. doi: 10.1128/mcb.1.6.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glisin V., Crkvenjakov R., Byus C. Ribonucleic acid isolated by cesium chloride centrifugation. Biochemistry. 1974 Jun 4;13(12):2633–2637. doi: 10.1021/bi00709a025. [DOI] [PubMed] [Google Scholar]
- Herendeen D. R., Kassavetis G. A., Barry J., Alberts B. M., Geiduschek E. P. Enhancement of bacteriophage T4 late transcription by components of the T4 DNA replication apparatus. Science. 1989 Sep 1;245(4921):952–958. doi: 10.1126/science.2672335. [DOI] [PubMed] [Google Scholar]
- Hutson S. M. Branched chain alpha-keto acid oxidative decarboxylation in skeletal muscle mitochondria. Effect of isolation procedure and mitochondrial delta pH. J Biol Chem. 1986 Apr 5;261(10):4420–4425. [PubMed] [Google Scholar]
- Kasamatsu H., Vinograd J. Replication of circular DNA in eukaryotic cells. Annu Rev Biochem. 1974;43(0):695–719. doi: 10.1146/annurev.bi.43.070174.003403. [DOI] [PubMed] [Google Scholar]
- King M. P., Attardi G. Injection of mitochondria into human cells leads to a rapid replacement of the endogenous mitochondrial DNA. Cell. 1988 Mar 25;52(6):811–819. doi: 10.1016/0092-8674(88)90423-0. [DOI] [PubMed] [Google Scholar]
- King T. C., Low R. L. Mapping of control elements in the displacement loop region of bovine mitochondrial DNA. J Biol Chem. 1987 May 5;262(13):6204–6213. [PubMed] [Google Scholar]
- King T. C., Low R. L. Mitochondrial DNA displacement loop structure depends on growth state in bovine cells. J Biol Chem. 1987 May 5;262(13):6214–6220. [PubMed] [Google Scholar]
- Kmiec E. B., Ryoji M., Worcel A. Gyration is required for 5S RNA transcription from a chromatin template. Proc Natl Acad Sci U S A. 1986 Mar;83(5):1305–1309. doi: 10.1073/pnas.83.5.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse B., Narasimhan N., Attardi G. Termination of transcription in human mitochondria: identification and purification of a DNA binding protein factor that promotes termination. Cell. 1989 Jul 28;58(2):391–397. doi: 10.1016/0092-8674(89)90853-2. [DOI] [PubMed] [Google Scholar]
- Montoya J., Christianson T., Levens D., Rabinowitz M., Attardi G. Identification of initiation sites for heavy-strand and light-strand transcription in human mitochondrial DNA. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7195–7199. doi: 10.1073/pnas.79.23.7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepe G., Gadaleta G., Palazzo G., Saccone C. Sequence-dependent DNA curvature: conformational signal present in the main regulatory region of the rat mitochondrial genome. Nucleic Acids Res. 1989 Nov 11;17(21):8803–8819. doi: 10.1093/nar/17.21.8803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajamanickam C., Merten S., Kwiatkowska-Patzer B., Chuang C. H., Zak R., Rabinowitz M. Changes in mitochondrial DNA in cardiac hypertrophy in the rat. Circ Res. 1979 Oct;45(4):505–515. doi: 10.1161/01.res.45.4.505. [DOI] [PubMed] [Google Scholar]
- Topper J. N., Clayton D. A. Identification of transcriptional regulatory elements in human mitochondrial DNA by linker substitution analysis. Mol Cell Biol. 1989 Mar;9(3):1200–1211. doi: 10.1128/mcb.9.3.1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams R. S., Garcia-Moll M., Mellor J., Salmons S., Harlan W. Adaptation of skeletal muscle to increased contractile activity. Expression nuclear genes encoding mitochondrial proteins. J Biol Chem. 1987 Feb 25;262(6):2764–2767. [PubMed] [Google Scholar]