Abstract
Enterococci are part of the normal intestinal flora in a large number of mammals, and these microbes are currently used as indicators of fecal contamination in water and food for human consumption. These organisms are considered one of the primary causes of nosocomial and environmental infections due to their ability to survive in the environment and to their intrinsic resistance to antimicrobials. The aims of this study were to determine the biochemical patterns and antimicrobial susceptibilities of Enterococcus faecalis and E. faecium isolates from clinical samples and from water (groundwater, water from the Xochimilco wetland, and treated water from the Mexico City Metropolitan Area) and to determine the genetic relationships among these isolates. A total of 121 enterococcus strains were studied; 31 and 90 strains were isolated from clinical samples and water (groundwater, water from the Xochimilco wetland, and water for agricultural irrigation), respectively. Identification to the species level was performed using a multiplex PCR assay, and antimicrobial profiles were obtained using a commercial kit. Twenty-eight strains were analyzed by pulsed-field gel electrophoresis (PFGE). E. faecium strains isolated from water showed an atypical biochemical pattern. The clinical isolates showed higher resistance to antibiotics than those from water. Both the enterococci isolated from humans, and those isolated from water showed high genetic diversity according to the PFGE analysis, although some strains seemed to be closely related. In conclusion, enterococci isolated from humans and water are genetically different. However, water represents a potential route of transmission to the community and a source of antimicrobial resistance genes that may be readily transmitted to other, different bacterial species.
Introduction
The genus Enterococcus is characterized by individual, paired, or short-chain gram-positive catalase-negative cocci. Enterococci emerged in the last decade of the twentieth century as one of the primary causes of hospital-acquired infections, although enterococci can also cause human infections in the community [1]. Enterococci can survive in a variety of environments, such as soil, water, food, plants, and animals [2], [3], [4]. In humans, as well as in other mammals and birds, these microbes are mainly found in the gastrointestinal tract as commensals but may become opportunistic pathogens in individuals with serious diseases whose immune systems are compromised and in patients who have been hospitalized for prolonged periods or who have received broad-spectrum antimicrobial therapy. Enterococci possess intrinsic or acquired resistance to several antimicrobials, such as glycopeptides, β-lactams, and fluoroquinolones, and can exhibit high levels of resistance to aminoglycosides (gentamicin and streptomycin), leading to drastically reduced therapeutic options for patients infected with enterococci, such that these bacteria are regarded as important pathogens with clinical relevance [5].
Enterococcus transmission occurs endogenously (through translocation from the gut to the bloodstream) and exogenously (in the hospital environment, e.g., via inanimate objects and the hands of health care workers and visitors) or through the consumption of contaminated food and water, the latter being the most common route of transmission, especially in developing countries [6], [7]. The Mexico City Metropolitan Area (MCMA) is a critical region from an environmental point of view, and the availability of clean water in this region is one of the most important issues for the maintenance and future development of the urban ecosystem. In the MCMA, the aquifer recharge and groundwater extraction and distribution systems are susceptible to contamination, and water quality varies seasonally. The presence of enterococci is predominantly reported during the dry season and is associated with fecal contamination [8].
The biochemical characteristics, antimicrobial susceptibility and molecular typing results of clinical and environmental E. faecalis and E. faecium isolates have not yet been reported for Mexico. The biochemical patterns and the antimicrobial susceptibility, as well as the genetic relationships between human and environmental E. faecalis and E. faecium isolates, will provide valuable information relevant to the epidemiology of infections caused by enterococci. Pulsed-field gel electrophoresis (PFGE) has been shown previously to be an excellent and highly reproducible method to identify clonal relationships among isolates [1], [9], [10], [11]. Therefore, the purpose of the present study was to determine the origins of E. faecalis and E. faecium strains isolated from hospitals, groundwater designated for human usage, water from the Xochimilco wetland, and treated wastewater used for agricultural irrigation in the MCMA.
Materials and Methods
Bacterial Strains
A total of 121 Enterococcus strains were studied: a) 31 (25.6%) strains were isolated as part of standard care (from blood, cerebrospinal fluid, eyes, livers, peripheral venous catheters, pleural fluid, sputum, urine, wounds, and the respiratory system) at five hospitals located in the southern part of the MCMA, and b) 90 (74.4%) strains were isolated from water; of these 90 strains, eight were isolated from groundwater for human use and consumption (wells), 72 were isolated from the Xochimilco wetland (created by a historical canal system), and 10 were isolated from treated wastewater from an important treatment plant. The 90 isolates from water were collected during both the rainy (45 strains) and dry (45 strains) seasons.
Presumptive Identification by Standard Biochemical Testing
The isolation of Enterococcus strains from clinical samples was performed using sheep blood agar as previously described [1]. The environmental strains were isolated using a standard membrane filtration method using K-F agar for streptococci according to procedures described by the American Public Health Association (APHA) [12]. In both cases, characteristic colonies of Enterococcus spp. were presumptively identified based on Gram staining, the catalase test, the hydrolysis of esculin in the presence of bile, and growth in brain-heart infusion broth containing 6.5% NaCl [1]. All isolates were kept in BHI broth with 15% glycerol at −70°C until further analysis.
Identification Using a Commercial Kit
Presumptive Enterococcus spp. were identified to the species level, and their antimicrobial susceptibility profiles were established using a semi-automated MicroScan system for the identification for gram-positive cocci (Positive Combo-12. Dade, Behring). The antibiotics tested by micro-dilution in broth were ampicillin (Am), ciprofloxacin (Cp), erythromycin (E), nitrofurantoin (Fd), imipenem (Imp), norfloxacin (Nxn), penicillin (P), rifampin (Rif), tetracycline (Te), and vancomycin (Va), with the additional detection of high-level resistance to gentamicin (HLR-G) and streptomycin (HLR-S). Glycopeptide resistance tests were performed according to the CLSI 2008 guidelines [13]; vancomycin resistance (VR) was confirmed using BHI agar plates containing 6 µg/mL of vancomycin, and a teicoplanin (TEC) resistance assessment was performed by agar dilution using Mueller-Hilton agar plates containing 4, 8, 16, and 32 µg/mL of the antimicrobial agent.
Identification by Multiplex PCR
Cell pellets of Enterococcus spp. were suspended in 180 µl of lysis buffer (20 µg/ml lysozyme, 20 mM Tris-HCl, 2 mM EDTA, 1.2% Triton X-100) and incubated at 37°C for 30 min, after which 20 µl of proteinase K was added to each reaction, and chromosomal DNA was extracted according to the manufacturer's instructions (QIAamp DNA Mini Kit, QIAGEN). For each strain, the DNA concentration was estimated using a NanoDrop spectrophotometer (ND-1000, Thermo Scientific), and DNA integrity was verified using a 0.8% agarose gel. A multiplex PCR assay was used to confirm the identification of E. faecalis and E. faecium strains as described previously by Layton et al. [14].
Pulsed-field Gel Electrophoresis
From among the clinical and environmental isolates identified as E. faecium and E. faecalis, 28 strains were selected randomly (considering origin, rainy or dry season, and antimicrobial susceptibility pattern). Nine of the selected strains came from three hospitals located in southern Mexico City, two strains came from groundwater (wells), 16 came from surface water in the Xochimilco wetland (Xochimilco canal), and one came from treated wastewater used for agricultural irrigation (treatment plant). These strains were collected during both the dry and rainy seasons. The PFGE patterns were obtained according to a protocol using the SmaI restriction enzyme as previously described [15]. The restriction pattern analyses were based on published criteria [16].
Statistical Analyses
The antimicrobial susceptibility patterns were analyzed for descriptive purposes based on relative frequencies. Comparisons of the antimicrobial susceptibility patterns of the clinical and environmental isolates were performed using the Fisher-Freeman-Halton exact test for contingency tables. All analyses were performed using Stata statistical software (Intercooled Stata 7.0 for Windows 98/95/NT Package, 2002) and StatXact (v. 4.0.1, Cytel Software Corp., 1999).
Results
When the identification of the Enterococcus spp. was performed to the species level using both a commercial kit and a multiplex PCR method (Figure 1), we found that 27 strains had results from the commercial kit and the multiplex PCR assay that were discordant. Table 1 shows that 13 strains identified by the commercial kit as E. faecium were identified as E. faecalis by multiplex PCR analysis. In addition, three strains identified by the commercial kit as E. faecalis were identified as E. faecium using the multiplex PCR analysis. Nine strains identified by the commercial kit as E. casseliflavus, E. durans/hirae, E. raffinosus and Enterococcus spp. were identified as E. faecium and E. faecalis by the multiplex PCR method. Finally, two strains identified as E. faecium gp and as E. faecalis by the commercial kit were classified as Enterococcus spp. by the multiplex PCR assay. Given the previously reported greater specificity of the multiplex PCR assay, the results obtained using this method were used for the subsequent analysis [14].
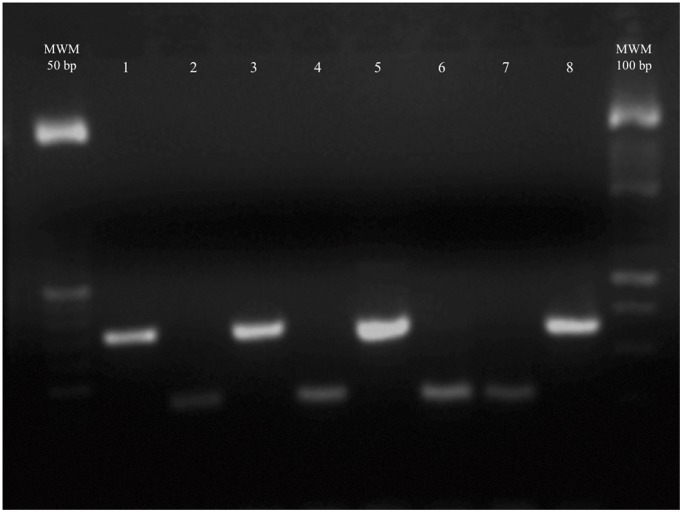
Figure 1. Multiplex PCR assay patterns for E. faecium and E. faecalis.
Lane 1) E. faecalis (other respiratory); lane 2) E. faecium (other respiratory); lane 3) E. faecalis (wetland, rainy season); lane 4) E. faecium (water treatment plant); lane 5) E. faecalis (water treatment plant); lane 6) E. faecium (wetland, rainy season); lane 7) E. faecium EF1 (positive control); and lane 8) E. faecalis ATCC 29212 (positive control).
Table 1. Strains with discordant results from the multiplex PCR assay and a commercial kit.
| Source | Identification | |
| Multiplex PCR | Commercial KIT | |
| Water treatment plant | E. faecalis | E. faecium |
| Groundwater (well) | E. faecalis | E. faecium |
| Groundwater (well) | E. faecalis | E. faecium |
| Wetland | E. faecalis | E. faecium |
| Wetland | E. faecalis | E. faecium |
| Wetland | E. faecalis | E. faecium |
| Wetland | E. faecalis | E. faecium |
| Wetland | E. faecalis | E. faecium gp |
| Wetland | E. faecalis | E. faecium gp |
| Wetland | E. faecalis | E. casseliflavus |
| Wetland | E. faecalis | E. faecium |
| Wetland | E. faecalis | E. durans/hirae |
| Wetland | E. faecalis | E. durans/hirae |
| Wetland | E. faecalis | E. raffinosus |
| Wetland | E. faecium | E. durans/hirae |
| Wetland | E. faecium | E. durans/hirae |
| Blood | E. faecalis | E. faecium |
| Blood | E. faecalis | E. faecium |
| Other respiratory | E. faecalis | E. faecium |
| Urine | E. faecalis | Enterococcus spp |
| Urine | E. faecium | E. faecalis |
| Cerebrospinal fluid | E. faecium | Enterococcus spp |
| Cerebrospinal fluid | E. faecium | Enterococcus spp |
| Catheter | E. faecium | E. faecalis |
| Eye | E. faecium | E. faecalis |
| Wetland | Enterococcus spp | E. faecium gp |
| Wetland | Enterococcus spp | E. faecalis |
Of the 121 enterococci studied by multiplex PCR, 102 (84.3%) were identified as E. faecium and E. faecalis; of these, 48.1% (49/102) were E. faecium and 51.9% (53/102) were E. faecalis. In total, 28.6% (14/49) of the E. faecium came from clinical samples and 28.6% (14/49) and 42.8% (21/49) were from water collected during the rainy and dry seasons, respectively. In total, 32.1% (17/53) of the E. faecalis isolates were clinical isolates, and 39.6% (21/53) and 28.3% (15/53) of the E. faecalis isolates were from water collected during the rainy and dry seasons, respectively.
Among the isolates, 19 (15.7%) strains from environmental water samples were identified using the Dade MicroScan system as E. durans/hirae (10/19, 52.6%), E. casseliflavus (4/19, 21%), E. gallinarum (2/19, 10.5%), E. raffinosus (1/19, 5.3%), E. faecium gp (1/19, 5.3%), or E. faecalis (1/19, 5.3%). These species were not included in the subsequent analysis because our study focused only on the clinically relevant species, i.e., E. faecium and E. faecalis.
The biochemical profiles of the E. faecium and E. faecalis strains are shown in Table 2. Some of these isolates performed atypical biochemical reactions, and this pattern was particularly common among the E. faecium and E. faecalis strains isolated from water. The atypical results for the water isolates included the utilization of sorbitol, arabinose, and raffinose; positive Voges-Proskauer reaction; growth in the presence of crystal violet; and alkaline phosphatase and pyrrolidonyl arylamidase activity. Interestingly, 15 (42.8%) of the E. faecium strains isolated from water samples were positive for the utilization of raffinose; in contrast, 6 (17.1%) E. faecium strains that were also isolated from water samples and four (28.6%) that were isolated from clinical samples were positive for the use of sorbitol (Table 2). Fewer of the E. faecalis strains isolated from water and clinical samples were able to use sorbitol.
Table 2. Biochemical profiles of E. faecium and E. faecalis strains from clinical and water samples.
| Characteristics | No. positive strainsa | |||
| E. faecium | E. faecalis | |||
| Clinicalb 14 (%) | Waterc 35 (%) | Clinical 17 (%) | Water 36 (%) | |
| Growth in crystal violet | 4 (28.6) | 17 (48.6) | 12 (70.6) | 30 (83.3) |
| Voges-Proskauer reaction | 12 (85.7) | 32 (91.4) | 17 (100) | 35 (97.2) |
| Alkaline phosphatase | 3 (21.4) | 2 (5.7) | 14 (82.4) | 22 (61.1) |
| Pyrrolidonyl arylamidase | 14 (100) | 32 (91.4) | 17 (100) | 32 (88.9) |
| Acid from: | ||||
| Mannitol | 14 (100) | 31 (88.6) | 16 (100) | 34 (94.4) |
| Sorbitol | 4 (28.6) | 6 (17.1) | 13 (76.5) | 15 (41.7) |
| Arabinose | 12 (85.7) | 29 (82.8) | 4 (23.5) | 12 (33.3) |
| Inulin | 0 (0) | 0 (0) | 0 (0) | 2 (5.6) |
| Raffinose | 0 (0) | 15 (42.8) | 1 (5.9) | 10 (27.8) |
| Pyruvate utilization | 3 (21.4) | 0 (0) | 12 (70.6) | 22 (61.1) |
All strains hydrolyzed esculin and grew in 40% bile and 6.5% NaCl.
Isolated strain from blood, cerebrospinal fluid, eyes, livers, peripheral venous catheters, pleural fluid, sputum, urine, wounds, and the respiratory system).
Isolated strains from groundwater for human use and consumption (wells), water from the Xochimilco wetland, and water from a water treatment plant (used for agricultural irrigation).
Of the strains identified as E. faecalis and E. faecium, 90.2% (92/102) were resistant to at least one antibiotic. The antimicrobial susceptibility tests showed that the clinical strains had, in general, a higher rate of resistance against the tested antimicrobials than did the isolates obtained from water samples (Table 3), particularly among the E. faecium strains (Table 4). We identified 34 different antimicrobial susceptibility patterns; furthermore, some specific resistance patterns were common among the clinical isolates and among the water isolates from both seasons (rainy and dry), (Table 4). Two E. faecium strains isolated from the Xochimilco wetland and two strains isolated from the water treatment plant during the dry season showed an intermediate level of resistance to vancomycin (Va) (8 µg/mL), as did one E. faecalis isolate from the Xochimilco wetland (rainy season) and one from the water treatment plant (dry season), as shown in Table 3. The clinical E. faecium (42.8% and 50%) and E. faecalis (29.4% and 41.2%) isolates showed high-level resistance to gentamicin and streptomycin, respectively (Table 3), but only two E. faecium strains and one E. faecalis strain isolated from water showed high-level resistance to streptomycin and gentamicin, respectively. All strains were susceptible to teicoplanin (data not shown). Comparing the antimicrobial patterns of the clinical and environmental isolates, we found four antibiotics that showed non-significant differences for E. faecium strains (Fd, Imp, Rif, and Va) and five for E. faecalis strains (Amp, Fd, Imp, Rif, and Va) (Table 3).
Table 3. Percentages of antibiotic resistance for the E. faecium and E. faecalis isolates from clinical and water samples.
| Species | Origin (n)b | Interpretivecriteria | Antibiotic (percentage)a | |||||||
| Amp | Fd | Imp | Rif | Te | Va | HLG | HLS | |||
| E. faecium | Clinical (14) | Susceptible | 42.8 | 92.8 | 100 | 42.8 | 14.3 | 100 | 57.2 | 50 |
| Intermediate | 0 | 7.1 | 0 | 14.3 | 7.1 | 0 | 0 | 0 | ||
| Resistant | 57.1 | 0 | 0 | 42.8 | 78.6 | 0 | 42.8 | 50 | ||
| Water (35) | Susceptible | 97.1 | 94.3 | 100 | 62.8 | 82.8 | 88.6 | 100 | 94.3 | |
| Intermediate | 0 | 2.8 | 0 | 2.8 | 0 | 11.4* | 0 | 0 | ||
| Resistant | 2.8 | 2.8 | 0 | 34.3 | 17.2 | 0 | 0 | 5.7 | ||
| §p | 0.0000 | 0.63 | NS | 0.21 | 0.0001 | 1.0 | 0.0001 | 0.0001 | ||
| E. faecalis | Clinical (17) | Susceptible | 88.2 | 100 | 94.1 | 58.8 | 35.3 | 100 | 70.6 | 58.8 |
| Intermediate | 0 | 0 | 0 | 11.8 | 0 | 0 | 0 | 0 | ||
| Resistant | 11.8 | 0 | 5.9 | 29.4 | 63.7 | 0 | 29.4 | 41.2 | ||
| Water (36) | Susceptible | 100 | 97.2 | 100 | 61.1 | 75 | 94.4 | 97.2 | 100 | |
| Intermediate | 0 | 2.8 | 0 | 13.9 | 0 | 5.6** | 0 | 0 | ||
| Resistant | 0 | 0 | 0 | 25 | 25 | 0 | 2.8 | 0 | ||
| §p | NS | NS | 0.41 | 1.0 | 0.0011 | NS | 0.0332 | 0.0076 | ||
Interpretive criteria are according to CLSI, 2008 (Clinical and Laboratory Standards Institute, 2008); Amp = ampicillin, Fd = nitrofurantoin, Imp = imipenem, Rif = rifampin, Te = tetracycline, Va = vancomycin, HLG = high-level resistance to gentamicin, and HLS = high-level resistance to streptomycin.
The numbers in parentheses are the numbers of strains tested.
Four and **two strains had intermediate resistance to vancomycin (8 µL/mL). Clinical samples (blood, cerebrospinal fluid, eye, liver, peripheral venous catheter, pleural fluid, sputum, urine, wound, and respiratory system). Water samples (groundwater for human use and consumption (wells), water from the Xochimilco wetland and water from a water treatment plant (water used for agricultural irrigation).
Fisher-Freeman-Halton exact test. NS = not significant.
Table 4. Antimicrobial susceptibility patterns of E. faecium and E. faecalis isolates from clinical and water samples.
| Antimicrobial susceptibility pattern | n a | E. faecium | E. faecalis | ||||||||||||
| Clin | Rainy season | Dry season | Clin | Rainy season | Dry season | ||||||||||
| WL | GW | WTP | WL | GW | WTP | WL | GW | WTP | WL | GW | WTP | ||||
| Amp, Cp, E, Nxn, P, Rif, Te | 3 | 3 | |||||||||||||
| Cp, E, Fd, Nxn, P, Rif, Te | 1 | 1 | |||||||||||||
| Amp, Cp, E, Nxn, P, Te | 3 | 2 | 1 | ||||||||||||
| Amp, E, Nxn, P, Rif, Te | 1 | 1 | |||||||||||||
| Amp, Fd, Nxn, P, Rif, Te | 1 | 1 | |||||||||||||
| Cp, E, Fd, Nxn, Rif, Te | 1 | 1 | |||||||||||||
| Cp, E, Fd, Nxn, Te | 1 | 1 | |||||||||||||
| Cp, E, Nxn, Rif, Te | 2 | 1 | 1 | ||||||||||||
| Amp, E, Nxn, Rif | 1 | 1 | |||||||||||||
| Amp, E, P, Rif | 1 | 1 | |||||||||||||
| Cp, E, Nxn, Te | 3 | 2 | 1 | ||||||||||||
| Cp, E, P, Te | 1 | 1 | |||||||||||||
| E, Imp, Nxn, Te | 1 | 1 | |||||||||||||
| E, Nxn, Rif, Te | 2 | 1 | 1 | ||||||||||||
| E, Nxn, Te, Va* | 1 | 1 | |||||||||||||
| E, Rif, Te, Va* | 1 | 1 | |||||||||||||
| Nxn, Rif, Te, Va* | 1 | 1 | |||||||||||||
| Amp, Nxn, Rif | 1 | 1 | |||||||||||||
| Cp, Nxn, Rif | 1 | 1 | |||||||||||||
| Cp, E, Nxn | 4 | 3 | 1 | ||||||||||||
| E, Nxn, Rif | 1 | 1 | |||||||||||||
| E, Nxn, Te | 1 | 1 | |||||||||||||
| E, Rif, Te | 5 | 2 | 1 | 2 | |||||||||||
| E, Rif, Va* | 2 | 1 | 1 | ||||||||||||
| Cp, Nxn | 4 | 2 | 2 | ||||||||||||
| E, P | 1 | 1 | |||||||||||||
| E, Rif | 11 | 2 | 1 | 1 | 2 | 1 | 4 | ||||||||
| E, Te | 6 | 1 | 3 | 1 | 1 | ||||||||||
| Rif, Te | 1 | 1 | |||||||||||||
| Te, Va* | 1 | 1 | |||||||||||||
| E | 20 | 1 | 5 | 1 | 1 | 9 | 3 | ||||||||
| Nxn | 2 | 2 | |||||||||||||
| Rif | 5 | 2 | 2 | 1 | |||||||||||
| Te | 1 | 1 | |||||||||||||
| TOTAL 34 | 92 | 14 | 6 | 2 | 1 | 14 | 1 | 4 | 17 | 18 | 1 | 0 | 12 | 1 | 1 |
Amp = ampicillin, Cp = ciprofloxacin, E = erythromycin, Fd = nitrofurantoin, Imp = imipenem, Nxn = norfloxacin, P = penicillin, Rif = rifampin, Te = tetracycline, and Va = vancomycin.
Numbers of strains. Clin = Clinical samples (blood, cerebrospinal fluid, eye, liver, peripheral venous catheter, pleural fluid, sputum, urine, wound, and respiratory system). WL = water from the Xochimilco wetland, GW = groundwater for human use and consumption (wells), WTP = water from a water treatment plant (used for agricultural irrigation).
Strains with intermediate vancomycin resistance.
From among the 102 strains identified as E. faecalis and E. faecium, 29 isolates were chosen randomly. Among these, nine (32%) corresponded to clinical strains (eight E. faecalis and one E. faecium) isolated from blood, pleural fluid, urine, wounds, and respiratory sites. Among the 19 (68%) environmental strains, six were E. faecium and 13 were E. faecalis isolated from wells, the wetland, and the water treatment plant. The comparison of the PFGE patterns with respect to the average level of diversity for both E. faecalis and E. faecium (Figures 2, 3 and 4).
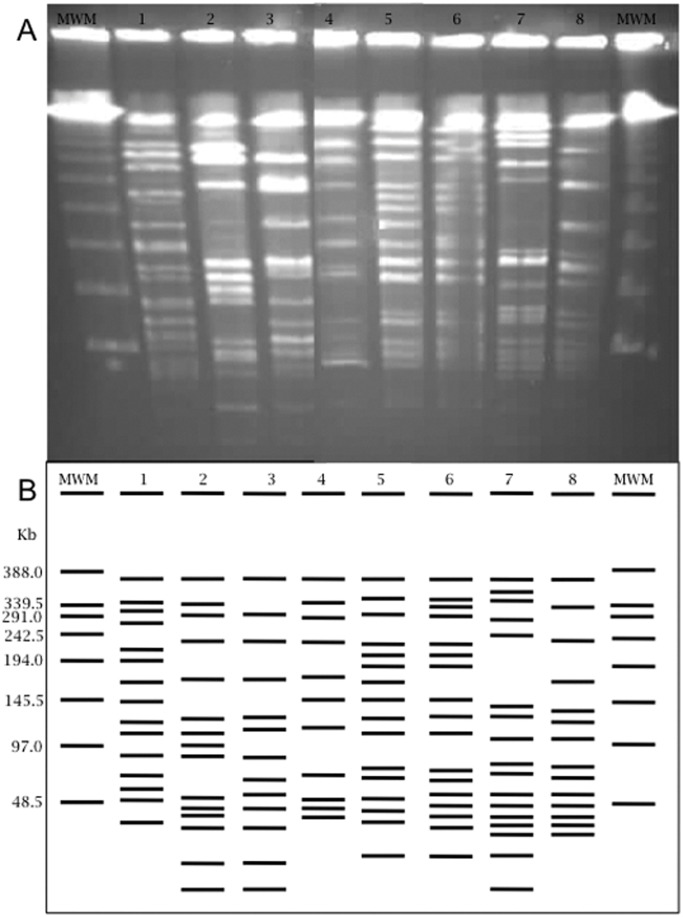
Figure 2. Electrophoretic PFGE patterns from E. faecium and E. faecalis.
A) Agarose gel electrophoresis showing the SmaI digestion patterns of enterococci. B) Graphical representation of the banding patterns. Lane 1) E. faecium (blood); lane 2) E. faecalis (pleural fluid); lane 3) E. faecalis (urine); lane 4) E. faecalis (wound); lane 5) E. faecalis (wetland, rainy season); lane 6) E. faecalis (wetland, dry season); lane 7) E. faecalis (wetland, dry season); and lane 8) E. faecium (wetland, dry season). MWM = Lambda Ladder PFG Marker (New England BioLabs).
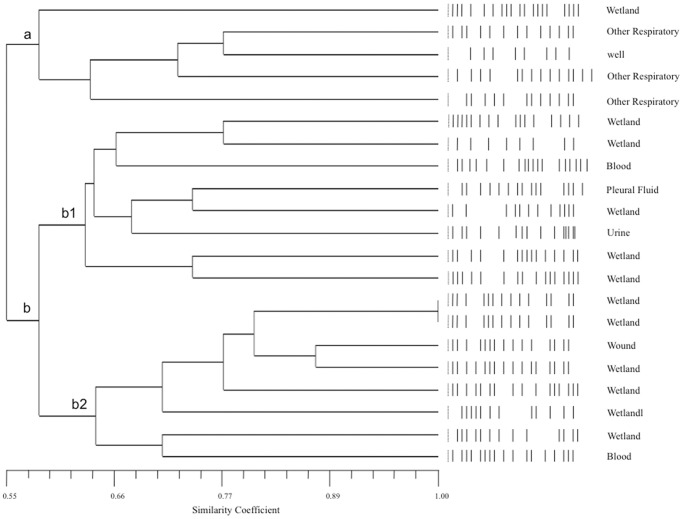
Figure 3. UPGMA dendrogram based on the PFGE patterns of E. faecalis isolates from clinical samples (blood, pleural fluid, urine, wounds, and other respiratory sites) and water samples (wells, wetland and water treatment plant).
The UPGMA dendrogram was constructed with the PFGE patterns of 21 strains using the NTSYS-pc program and Jacquard’s coefficient. Mantel test r = 0.75389, p = 1.0.
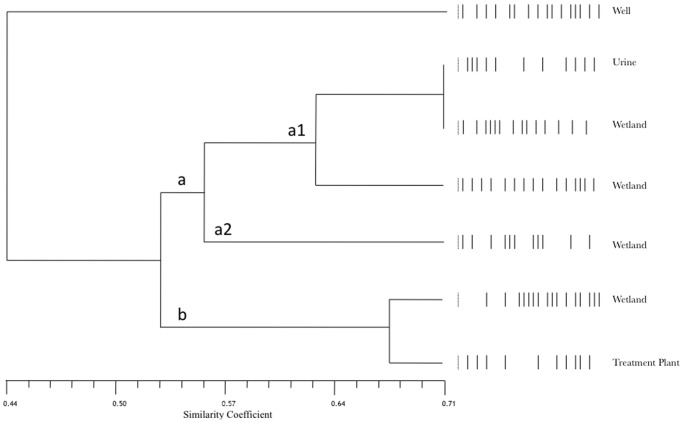
Figure 4. UPGMA dendrogram based on PFGE patterns of E. faecium isolates from clinical samples (urine) and water samples (wells, wetland and water treatment plant).
The UPGMA dendrogram was constructed with the PFGE patterns of 7 strains using the NTSYS-pc program and Jacquard’s coefficient. Mantel test r = 0.76432, p = 0.999.
Among the E. faecalis isolates, the clinical and water samples did not fall into a homogeneous group (Figure 3). Instead, the water isolates had a similarity coefficient greater than 0.50 and fell into two groups (a and b). Clinical isolates from respiratory samples formed the smallest group, which also included a wetland isolate and a water well isolate. The similarity of the clinical respiratory tract isolates (similarity coefficient, 0.77) to the well water isolates (for water for human use and consumption) suggests that there might be a closer association between clinical isolates and well water isolates than between clinical isolates and wetland water isolates.
The water isolate group b formed two distinct subgroups (b1 and b2) related by a similarity coefficient of 0.60. The b1 subgroup also contained clinical isolates from pleural fluid, urinary tract and blood samples, and subgroup b2 included wound and blood isolates. Two strains from the wetland water were shown to have identical PFGE patterns (coefficient of similarity = 1). The two strains with this PFGE pattern were grouped with a wound isolate by a similarity coefficient greater than 0.85, which was the closest association found between a clinical isolate and a water isolate, suggesting that these wetland water strains were related to the E. faecalis strain that caused a wound infection. Other isolates in the b2 subgroup were clustered with wetland water isolates with similarity coefficients above 0.70, indicating genetic proximity and suggesting that wetland water isolates could represent a genetic pool of E. faecalis strains with the potential to cause infections in hospitalized patients (Figure 3).
Among the E. faecium isolates, the isolate with the most distinct PFGE pattern was isolated from a water well and had similarity coefficients under 0.50 with all of the other strains (Figure 4). The rest of the isolates formed a group with PFGE patterns having similarities above 0.50 and consisting of two subgroups a and b. One urine isolated within a1 subgroup was grouped with two strains from the wetland and shared a similarity of 0.71 with one of them, which was the highest similarity coefficient obtained in this analysis (Figure 4). An isolate from the treatment plant water grouped with other isolates from the wetland water but formed a distinct b subgroup. It appears that one isolate among the wetland isolates is closely genetically related to the parental isolate of this treatment plant water isolate. In this study, no association was observed between the E. faecalis and E. faecium strains with respect to either their PFGE patterns or their antimicrobial susceptibility patterns.
Discussion
Enterococci identification was performed using the MicroScan gram-positive panel. This system has been previously evaluated using a broad distribution of enterococcal species and showed very good identification of the most common species of enterococci [17], [18], [19]. When d'Azevedo et al. evaluated an upgraded version of this system, they found that the system performed well for the identification of E. faecalis and typical E. faecium [20]. However, sometimes the identification of strains of E. faecium was problematic when trying to distinguish them from strains of E. gallinarum [21], and similarly, these authors reported errors when identifying strains of E. durans, E. avium, E. raffinosus, E. hirae, and E. mundtii [20]. However, this system has not been tested extensively with environmental isolates.
To overcome these limitations, methods based on the analysis of bacterial DNA have been successfully applied [22], [23], [24]. Identification to the species level using PCR with species-specific primers is a valuable method and can replace complex molecular clustering techniques and conventional microbiological tests that are otherwise necessary to identify species that are difficult to distinguish using phenotypic approaches [25], [26], [27], [28]. Multiplex PCR with specific primers is a simple molecular tool that allows the rapid and accurate identification of enterococci. This technique has been used successfully to identify vancomycin-resistant enterococci [25], [29], but it can also be used to identify all other enterococci [14], [30].
The microbial metabolic characteristics of the E. faecalis and E. faecium isolates were compared with the taxonomic system described by Teixeira et al. [1]. We found that only 41.7% (13/17, Table 2) of the E. faecalis strains isolated from the water samples produced acid from sorbitol, in contrast to 76.5% of the isolates from the clinical samples. According to data reported by Teixeira et al., it is expected that 80 to 90% of the isolates would be positive. However, the present findings are not surprising because most metabolic identification schemes are based on the biochemical characteristics of clinical isolates. The biochemical patterns of water isolates have not been studied extensively and may be different [1], [10]. We found sorbitol-positive E. faecium strains isolated from clinical and water samples, and it has been suggested that these strains are typically associated with dogs [31]. Our result may therefore indicate that dogs kept as pets play an important role in the transmission of E. faecium strains to humans and to the environment.
Regarding the E. faecium strains isolated from water samples that were able to utilize raffinose, it has been suggested that E. faecium isolates obtained from chicken or poultry can be phenotypically classified according to their raffinose utilization, a unique feature that is not present in this microorganism when isolated from other sources [31]. This observation may indicate that the origin of these environmental isolates may be linked to chicken husbandry in the MCMA.
We found no differences in the frequency of isolation of either E. faecalis or E. faecium between the clinical and water samples. It is important to note that the water samples exhibited greater diversity in Enterococcus species, most likely because the sources of contamination included a wide range of animal as well as human sources.
Enterococci isolated from clinical samples had a higher frequency of antimicrobial resistance than those obtained from water [1]. This result is not surprising because exposure to antibiotics is more common in the hospital setting than in the community (although antibiotics were still available over-the-counter in Mexico during the study period).
Because enterococci have been frequently found in water and because these bacteria are an important cause of infections, it is necessary to elucidate the association between human and water isolates. Therefore, 28 enterococci isolates selected based on their sites of origin, season of isolation (rainy or dry), and antimicrobial susceptibility patterns were genotyped by PFGE to determine their clonal relationships. This analysis demonstrated that the isolates of both clinical and environmental origin showed large genetic variabilities [16], presenting both related and unrelated patterns.
The antimicrobial susceptibility patterns and the PFGE patterns did not exhibit any association, in contrast to a previous report that found a clonal relationship between environmental and clinical isolates [32]. This result suggests that nosocomial infections may be of environmental origin or that these microorganisms may be transferred from hospitals to the environment, allowing their dissemination into the community.
The biochemical and antimicrobial susceptibility patterns of the Enterococcus isolates from the clinical and environmental samples showed wide variability, suggesting that the origin of water contamination comes from a variety of sources (including, most likely, animal husbandry). However, water could represent a potential route of transmission of multi-resistant bacteria in the community. Further studies are required to confirm these findings.
Acknowledgments
We would like to thank Maria Paulina Montañés Sentíes for her support in the experimental work.
Funding Statement
This research was funded by grant Consejo Nacional de Ciencia y Tecnología (CONACyT) 78787M, and Salud-2009-C01-112588 and the operative budget of the Facultad de Medicina, Universidad Nacional Autónoma de México. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Teixeira L, Carvalho M, Facklan R (2007) Enterococcus. In: Murray P, Baron E, Landry M, Jorgensen J, Pfaller M, editors. Manual of Clinical Microbiology. 9th ed. Washington, DC: ASM Press. 430–442.
- 2. Graves AK, Weaver RW (2010) Characterization of enterococci populations collected from a subsurface flow constructed wetland. J Appl Microbiol 108: 1226–1234. [DOI] [PubMed] [Google Scholar]
- 3. Franzetti L, Pompei M, Scarpellini M, Galli A (2004) Phenotypic and genotypic characterization of Enterococcus spp. of different origins. Curr Microbiol 49: 255–260. [DOI] [PubMed] [Google Scholar]
- 4. Devriese LA, Kerckhove AVD, Kilpper-Bälz R, Schleifer KH (1987) Characterization and Identification of Enterococcus Species Isolated from the Intestines of Animals. International Journal of Systematic Bacteriology 37: 257–259. [Google Scholar]
- 5. Arias CA, Contreras GA, Murray BE (2010) Management of multidrug-resistant enterococcal infections. Clin Microbiol Infect 16: 555–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Iversen A, Kuhn I, Rahman M, Franklin A, Burman LG, et al. (2004) Evidence for transmission between humans and the environment of a nosocomial strain of Enterococcus faecium . Environ Microbiol 6: 55–59. [DOI] [PubMed] [Google Scholar]
- 7. Silva J, Loyola P, Galleguillos J, Rodriguez Y, Colque-Navarro P, et al. (2005) [Prevalence of antibiotic resistant Enterococcus spp in waste waters in the north of Chile]. Rev Med Chil 133: 1201–1210. [DOI] [PubMed] [Google Scholar]
- 8.Mazari-Hiriart M, López-Vidal Y, Ponce de León S, Calva-Mercado J, Rojo-Callejas F, et al.. (2002) Calidad del agua para uso y consumo humano en la ciudad de México [Memoria in extenso]. Cancún, Quintana Roo, México: XXVIII Congreso Interamericano de Ingeniería Sanitaria y Ambiental.
- 9. Dicuonzo G, Gherardi G, Lorino G, Angeletti S, Battistoni F, et al. (2001) Antibiotic resistance and genotypic characterization by PFGE of clinical and environmental isolates of enterococci. FEMS Microbiol Lett 201: 205–211. [DOI] [PubMed] [Google Scholar]
- 10. Domig KJ, Mayer HK, Kneifel W (2003) Methods used for the isolation, enumeration, characterisation and identification of Enterococcus spp. 2. Pheno- and genotypic criteria. Int J Food Microbiol 88: 165–188. [DOI] [PubMed] [Google Scholar]
- 11. Arias CA, Reyes J, Zuniga M, Cortes L, Cruz C, et al. (2003) Multicentre surveillance of antimicrobial resistance in enterococci and staphylococci from Colombian hospitals, 2001–2002. J Antimicrob Chemother 51: 59–68. [DOI] [PubMed] [Google Scholar]
- 12.APHA (1998) American Public Health Association. Standard methods for the examination of water and wastewater; Association APH, editor. Washington, DC: American Public Health Association.
- 13.CLSI (2008) Clinical and Laboratory Standards Institute. Performance Stantards for Antimicrobial Susceptibility Testing; Eighteenth Informational Supplement. CLSI document M100-S18 (ISBN 1–56238–653–0). 940 West Valley Road, Suite 1400, Wayne, Pennsylvania 19087–1898 USA: Clinical and Laboratory Standards Institute.
- 14. Layton BA, Walters SP, Lam LH, Boehm AB (2010) Enterococcus species distribution among human and animal hosts using multiplex PCR. J Appl Microbiol 109: 539–547. [DOI] [PubMed] [Google Scholar]
- 15. Saeedi B, Hallgren A, Jonasson J, Nilsson LE, Hanberger H, et al. (2002) Modified pulsed-field gel electrophoresis protocol for typing of enterococci. APMIS 110: 869–874. [DOI] [PubMed] [Google Scholar]
- 16. Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, et al. (1995) Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol 33: 2233–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Iwen PC, Kelly DM, Linder J, Hinrichs SH (1996) Revised approach for identification and detection of ampicillin and vancomycin resistance in Enterococcus species by using MicroScan panels. J Clin Microbiol 34: 1779–1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Iwen PC, Rupp ME, Schreckenberger PC, Hinrichs SH (1999) Evaluation of the revised MicroScan dried overnight gram-positive identification panel to identify Enterococcus species. J Clin Microbiol 37: 3756–3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tritz DM, Iwen PC, Woods GL (1990) Evaluation of MicroScan for identification of Enterococcus species. J Clin Microbiol 28: 1477–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. d'Azevedo PA, Dias CA, Goncalves AL, Rowe F, Teixeira LM (2001) Evaluation of an automated system for the identification and antimicrobial susceptibility testing of enterococci. Diagn Microbiol Infect Dis 40: 157–161. [DOI] [PubMed] [Google Scholar]
- 21. Willey BM, Jones RN, McGeer A, Witte W, French G, et al. (1999) Practical approach to the identification of clinically relevant Enterococcus species. Diagn Microbiol Infect Dis 34: 165–171. [DOI] [PubMed] [Google Scholar]
- 22. Marino M, Maifreni M, Rondinini G (2003) Microbiological characterization of artisanal Montasio cheese: analysis of its indigenous lactic acid bacteria. FEMS Microbiol Lett 229: 133–140. [DOI] [PubMed] [Google Scholar]
- 23. Delgado S, Mayo B (2004) Phenotypic and genetic diversity of Lactococcus lactis and Enterococcus spp. strains isolated from Northern Spain starter-free farmhouse cheeses. Int J Food Microbiol 90: 309–319. [DOI] [PubMed] [Google Scholar]
- 24. Mannu L, Paba A (2002) Genetic diversity of lactococci and enterococci isolated from home-made Pecorino Sardo ewes' milk cheese. J Appl Microbiol 92: 55–62. [DOI] [PubMed] [Google Scholar]
- 25. Dutka-Malen S, Evers S, Courvalin P (1995) Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J Clin Microbiol 33: 24–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ke D, Picard FJ, Martineau F, Menard C, Roy PH, et al. (1999) Development of a PCR assay for rapid detection of enterococci. J Clin Microbiol 37: 3497–3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Knijff E, Dellaglio F, Lombardi A, Andrighetto C, Torriani S (2001) Rapid identification of Enterococcus durans and Enterococcus hirae by PCR with primers targeted to the ddl genes. J Microbiol Methods 47: 35–40. [DOI] [PubMed] [Google Scholar]
- 28. Jackson CR, Fedorka-Cray PJ, Barrett JB (2004) Use of a genus- and species-specific multiplex PCR for identification of enterococci. J Clin Microbiol 42: 3558–3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Free L, Sahm DF (1996) Detection of Enterococcal Vancomycin Resistance by Multiplex PCR. In: Persing DH, editor. PCR protocols for emerging infectious diseases: a supplement to Diagnostic Molecular Microbiology Principles and Applications. Washington, DC: ASM Press. 150.
- 30. Monstein HJ, Quednau M, Samuelsson A, Ahrne S, Isaksson B, et al. (1998) Division of the genus Enterococcus into species groups using PCR-based molecular typing methods. Microbiology 144 (Pt 5): 1171–1179. [DOI] [PubMed] [Google Scholar]
- 31. Devriese LA, Pot B, Van Damme L, Kersters K, Haesebrouck F (1995) Identification of Enterococcus species isolated from foods of animal origin. Int J Food Microbiol 26: 187–197. [DOI] [PubMed] [Google Scholar]
- 32. Grammenou P, Spiliopoulou I, Sazakli E, Papapetropoulou M (2006) PFGE analysis of enterococci isolates from recreational and drinking water in Greece. J Water Health 4: 263–269. [PubMed] [Google Scholar]