Abstract
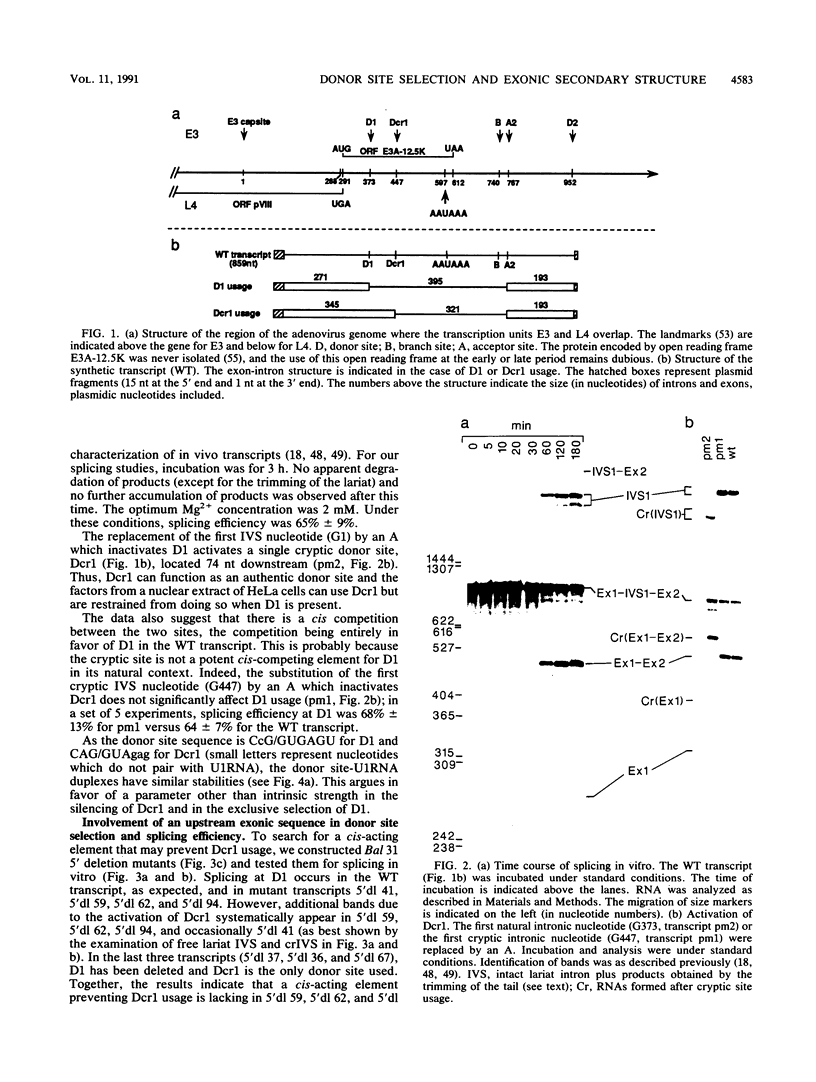
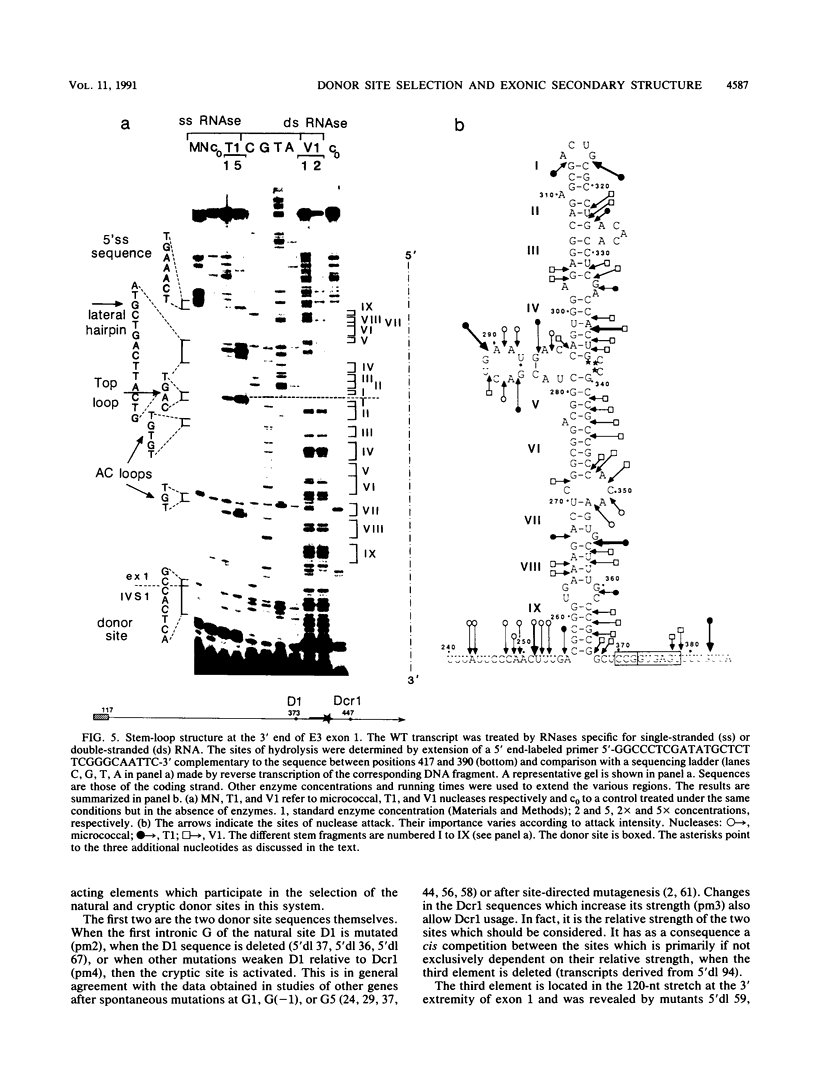
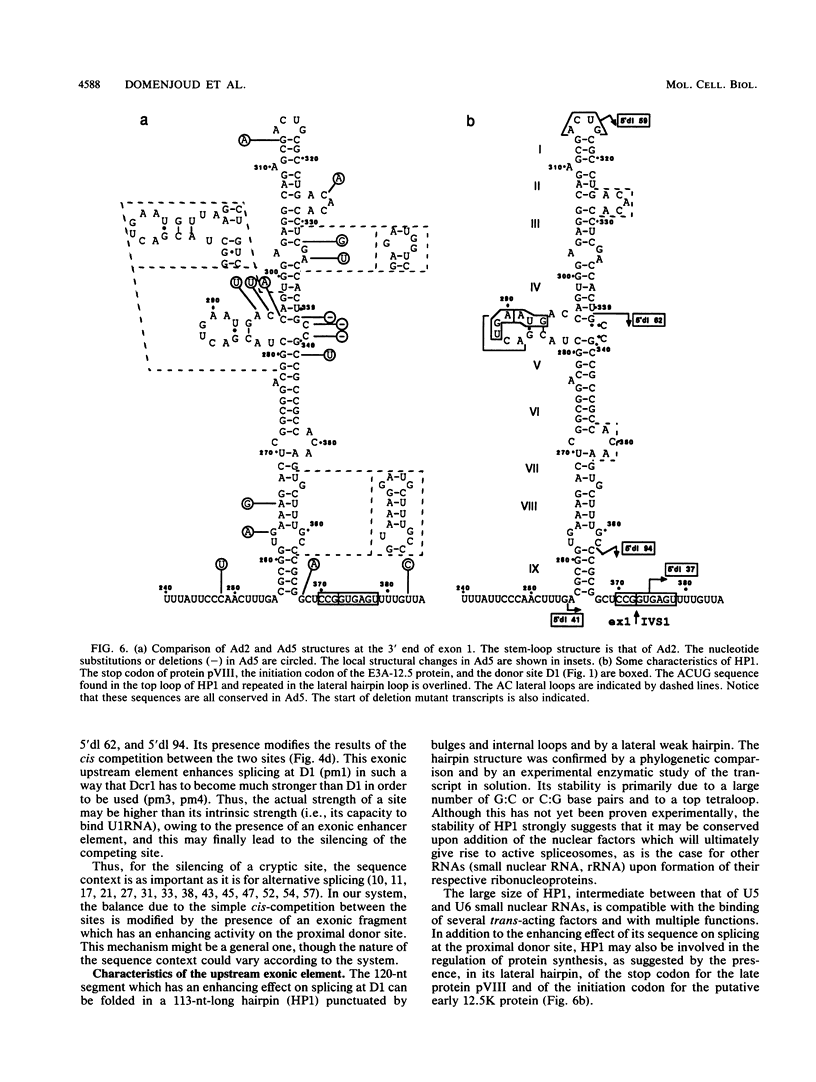
The first intron of the early region 3 from adenovirus type 2 contains a cryptic 5' splice site, Dcr1, 74 nucleotides downstream from the natural site D1. The cryptic site can be activated when the natural site is inactivated by mutagenesis. To investigate the basis for selection between a natural and a cryptic 5' splice site, we searched for cis-acting elements responsible for the exclusive selection of the natural site. We show that both the relative intrinsic strength of the sites and the sequence context affect the selection. A 120-nucleotide segment located at the 3' end of exon 1 enhances splicing at the proximal site D1; in its absence the two sites are used according to their strength. Thus, three cis-acting elements are involved in the silencing of the cryptic site: the sequence of D1, the sequence of Dcr1, and an upstream exonic sequence. We show that the exonic element folds, in solution, into a 113-nucleotide-long stem-loop structure. We propose that this potential stem-loop structure which is located 6 nucleotides upstream of the exon 1-intron junction is responsible for the preferential use of the natural 5' splice site.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adami G. R., Marlor C. W., Barrett N. L., Carmichael G. G. Leader-to-leader splicing is required for efficient production and accumulation of polyomavirus late mRNAs. J Virol. 1989 Jan;63(1):85–93. doi: 10.1128/jvi.63.1.85-93.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aebi M., Hornig H., Padgett R. A., Reiser J., Weissmann C. Sequence requirements for splicing of higher eukaryotic nuclear pre-mRNA. Cell. 1986 Nov 21;47(4):555–565. doi: 10.1016/0092-8674(86)90620-3. [DOI] [PubMed] [Google Scholar]
- Baudin F., Ehresmann C., Romby P., Mougel M., Colin J., Lempereur L., Bachellerie J. P., Ebel J. P., Ehresmann B. Higher-order structure of domain III in Escherichia coli 16S ribosomal RNA, 30S subunit and 70S ribosome. Biochimie. 1987 Oct;69(10):1081–1096. doi: 10.1016/0300-9084(87)90008-3. [DOI] [PubMed] [Google Scholar]
- Breathnach R., Chambon P. Organization and expression of eucaryotic split genes coding for proteins. Annu Rev Biochem. 1981;50:349–383. doi: 10.1146/annurev.bi.50.070181.002025. [DOI] [PubMed] [Google Scholar]
- Breitbart R. E., Andreadis A., Nadal-Ginard B. Alternative splicing: a ubiquitous mechanism for the generation of multiple protein isoforms from single genes. Annu Rev Biochem. 1987;56:467–495. doi: 10.1146/annurev.bi.56.070187.002343. [DOI] [PubMed] [Google Scholar]
- Chang D. D., Sharp P. A. Regulation by HIV Rev depends upon recognition of splice sites. Cell. 1989 Dec 1;59(5):789–795. doi: 10.1016/0092-8674(89)90602-8. [DOI] [PubMed] [Google Scholar]
- Chebli K., Gattoni R., Schmitt P., Hildwein G., Stevenin J. The 216-nucleotide intron of the E1A pre-mRNA contains a hairpin structure that permits utilization of unusually distant branch acceptors. Mol Cell Biol. 1989 Nov;9(11):4852–4861. doi: 10.1128/mcb.9.11.4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cladaras C., Wold W. S. DNA sequence of the early E3 transcription unit of adenovirus 5. Virology. 1985 Jan 15;140(1):28–43. doi: 10.1016/0042-6822(85)90443-x. [DOI] [PubMed] [Google Scholar]
- Cooper T. A., Cardone M. H., Ordahl C. P. Cis requirements for alternative splicing of the cardiac troponin T pre-mRNA. Nucleic Acids Res. 1988 Sep 12;16(17):8443–8465. doi: 10.1093/nar/16.17.8443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper T. A., Ordahl C. P. Nucleotide substitutions within the cardiac troponin T alternative exon disrupt pre-mRNA alternative splicing. Nucleic Acids Res. 1989 Oct 11;17(19):7905–7921. doi: 10.1093/nar/17.19.7905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam J. D., Lebovitz R. M., Roeder R. G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983 Mar 11;11(5):1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehresmann C., Baudin F., Mougel M., Romby P., Ebel J. P., Ehresmann B. Probing the structure of RNAs in solution. Nucleic Acids Res. 1987 Nov 25;15(22):9109–9128. doi: 10.1093/nar/15.22.9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eperon L. P., Estibeiro J. P., Eperon I. C. The role of nucleotide sequences in splice site selection in eukaryotic pre-messenger RNA. Nature. 1986 Nov 20;324(6094):280–282. doi: 10.1038/324280a0. [DOI] [PubMed] [Google Scholar]
- Eperon L. P., Graham I. R., Griffiths A. D., Eperon I. C. Effects of RNA secondary structure on alternative splicing of pre-mRNA: is folding limited to a region behind the transcribing RNA polymerase? Cell. 1988 Jul 29;54(3):393–401. doi: 10.1016/0092-8674(88)90202-4. [DOI] [PubMed] [Google Scholar]
- Freier S. M., Kierzek R., Jaeger J. A., Sugimoto N., Caruthers M. H., Neilson T., Turner D. H. Improved free-energy parameters for predictions of RNA duplex stability. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9373–9377. doi: 10.1073/pnas.83.24.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego M. E., Nadal-Ginard B. Myosin light-chain 1/3 gene alternative splicing: cis regulation is based upon a hierarchical compatibility between splice sites. Mol Cell Biol. 1990 May;10(5):2133–2144. doi: 10.1128/mcb.10.5.2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallinaro H., Vincendon P., Sittler A., Jacob M. Alternative use of a polyadenylation signal and of a downstream 3' splice site. Effect of 5,6-dichloro-1-beta-D-ribofuranosylbenzimidazole. J Mol Biol. 1988 Dec 20;204(4):1031–1040. doi: 10.1016/0022-2836(88)90059-9. [DOI] [PubMed] [Google Scholar]
- Green M. R., Maniatis T., Melton D. A. Human beta-globin pre-mRNA synthesized in vitro is accurately spliced in Xenopus oocyte nuclei. Cell. 1983 Mar;32(3):681–694. doi: 10.1016/0092-8674(83)90054-5. [DOI] [PubMed] [Google Scholar]
- Groebe D. R., Uhlenbeck O. C. Characterization of RNA hairpin loop stability. Nucleic Acids Res. 1988 Dec 23;16(24):11725–11735. doi: 10.1093/nar/16.24.11725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampson R. K., La Follette L., Rottman F. M. Alternative processing of bovine growth hormone mRNA is influenced by downstream exon sequences. Mol Cell Biol. 1989 Apr;9(4):1604–1610. doi: 10.1128/mcb.9.4.1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaphy S., Dingwall C., Ernberg I., Gait M. J., Green S. M., Karn J., Lowe A. D., Singh M., Skinner M. A. HIV-1 regulator of virion expression (Rev) protein binds to an RNA stem-loop structure located within the Rev response element region. Cell. 1990 Feb 23;60(4):685–693. doi: 10.1016/0092-8674(90)90671-z. [DOI] [PubMed] [Google Scholar]
- Hodges P. E., Rosenberg L. E. The spfash mouse: a missense mutation in the ornithine transcarbamylase gene also causes aberrant mRNA splicing. Proc Natl Acad Sci U S A. 1989 Jun;86(11):4142–4146. doi: 10.1073/pnas.86.11.4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hérissé J., Courtois G., Galibert F. Nucleotide sequence of the EcoRI D fragment of adenovirus 2 genome. Nucleic Acids Res. 1980 May 24;8(10):2173–2192. doi: 10.1093/nar/8.10.2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob M., Gallinaro H. The 5' splice site: phylogenetic evolution and variable geometry of association with U1RNA. Nucleic Acids Res. 1989 Mar 25;17(6):2159–2180. doi: 10.1093/nar/17.6.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakizuka A., Ingi T., Murai T., Nakanishi S. A set of U1 snRNA-complementary sequences involved in governing alternative RNA splicing of the kininogen genes. J Biol Chem. 1990 Jun 15;265(17):10102–10108. [PubMed] [Google Scholar]
- Krainer A. R., Maniatis T., Ruskin B., Green M. R. Normal and mutant human beta-globin pre-mRNAs are faithfully and efficiently spliced in vitro. Cell. 1984 Apr;36(4):993–1005. doi: 10.1016/0092-8674(84)90049-7. [DOI] [PubMed] [Google Scholar]
- Lapoumeroulie C., Acuto S., Rouabhi F., Labie D., Krishnamoorthy R., Bank A. Expression of a beta thalassemia gene with abnormal splicing. Nucleic Acids Res. 1987 Oct 26;15(20):8195–8204. doi: 10.1093/nar/15.20.8195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libri D., Goux-Pelletan M., Brody E., Fiszman M. Y. Exon as well as intron sequences are cis-regulating elements for the mutually exclusive alternative splicing of the beta tropomyosin gene. Mol Cell Biol. 1990 Oct;10(10):5036–5046. doi: 10.1128/mcb.10.10.5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malim M. H., Hauber J., Le S. Y., Maizel J. V., Cullen B. R. The HIV-1 rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature. 1989 Mar 16;338(6212):254–257. doi: 10.1038/338254a0. [DOI] [PubMed] [Google Scholar]
- Mardon H. J., Sebastio G., Baralle F. E. A role for exon sequences in alternative splicing of the human fibronectin gene. Nucleic Acids Res. 1987 Oct 12;15(19):7725–7733. doi: 10.1093/nar/15.19.7725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayeda A., Ohshima Y. Beta-globin transcripts carrying a single intron with three adjacent nucleotides of 5' exon are efficiently spliced in vitro irrespective of intron position or surrounding exon sequences. Nucleic Acids Res. 1990 Aug 25;18(16):4671–4676. doi: 10.1093/nar/18.16.4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mount S. M. A catalogue of splice junction sequences. Nucleic Acids Res. 1982 Jan 22;10(2):459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima H., Kono N., Yamasaki T., Hotta K., Kawachi M., Kuwajima M., Noguchi T., Tanaka T., Tarui S. Genetic defect in muscle phosphofructokinase deficiency. Abnormal splicing of the muscle phosphofructokinase gene due to a point mutation at the 5'-splice site. J Biol Chem. 1990 Jun 5;265(16):9392–9395. [PubMed] [Google Scholar]
- Nasim F. H., Spears P. A., Hoffmann H. M., Kuo H. C., Grabowski P. J. A Sequential splicing mechanism promotes selection of an optimal exon by repositioning a downstream 5' splice site in preprotachykinin pre-mRNA. Genes Dev. 1990 Jul;4(7):1172–1184. doi: 10.1101/gad.4.7.1172. [DOI] [PubMed] [Google Scholar]
- Nelson K. K., Green M. R. Mechanism for cryptic splice site activation during pre-mRNA splicing. Proc Natl Acad Sci U S A. 1990 Aug;87(16):6253–6257. doi: 10.1073/pnas.87.16.6253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen H. S., Nelbock P., Cochrane A. W., Rosen C. A. Secondary structure is the major determinant for interaction of HIV rev protein with RNA. Science. 1990 Feb 16;247(4944):845–848. doi: 10.1126/science.2406903. [DOI] [PubMed] [Google Scholar]
- Parent A., Zeitlin S., Efstratiadis A. Minimal exon sequence requirements for efficient in vitro splicing of mono-intronic nuclear pre-mRNA. J Biol Chem. 1987 Aug 15;262(23):11284–11291. [PubMed] [Google Scholar]
- Pavlakis G. N., Felber B. K. Regulation of expression of human immunodeficiency virus. New Biol. 1990 Jan;2(1):20–31. [PubMed] [Google Scholar]
- Reed R., Maniatis T. A role for exon sequences and splice-site proximity in splice-site selection. Cell. 1986 Aug 29;46(5):681–690. doi: 10.1016/0092-8674(86)90343-0. [DOI] [PubMed] [Google Scholar]
- Ris-Stalpers C., Kuiper G. G., Faber P. W., Schweikert H. U., van Rooij H. C., Zegers N. D., Hodgins M. B., Degenhart H. J., Trapman J., Brinkmann A. O. Aberrant splicing of androgen receptor mRNA results in synthesis of a nonfunctional receptor protein in a patient with androgen insensitivity. Proc Natl Acad Sci U S A. 1990 Oct;87(20):7866–7870. doi: 10.1073/pnas.87.20.7866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saga Y., Lee J. S., Saraiya C., Boyse E. A. Regulation of alternative splicing in the generation of isoforms of the mouse Ly-5 (CD45) glycoprotein. Proc Natl Acad Sci U S A. 1990 May;87(10):3728–3732. doi: 10.1073/pnas.87.10.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt P., Gattoni R., Keohavong P., Stévenin J. Alternative splicing of E1A transcripts of adenovirus requires appropriate ionic conditions in vitro. Cell. 1987 Jul 3;50(1):31–39. doi: 10.1016/0092-8674(87)90659-3. [DOI] [PubMed] [Google Scholar]
- Siebel C. W., Rio D. C. Regulated splicing of the Drosophila P transposable element third intron in vitro: somatic repression. Science. 1990 Jun 8;248(4960):1200–1208. doi: 10.1126/science.2161558. [DOI] [PubMed] [Google Scholar]
- Sittler A., Gallinaro H., Jacob M. In vivo splicing of the premRNAs from early region 3 of adenovirus-2: the products of cleavage at the 5' splice site of the common intron. Nucleic Acids Res. 1986 Feb 11;14(3):1187–1207. doi: 10.1093/nar/14.3.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sittler A., Gallinaro H., Kister L., Jacob M. In-vivo degradation pathway of an excised intervening sequence. J Mol Biol. 1987 Oct 20;197(4):737–741. doi: 10.1016/0022-2836(87)90480-3. [DOI] [PubMed] [Google Scholar]
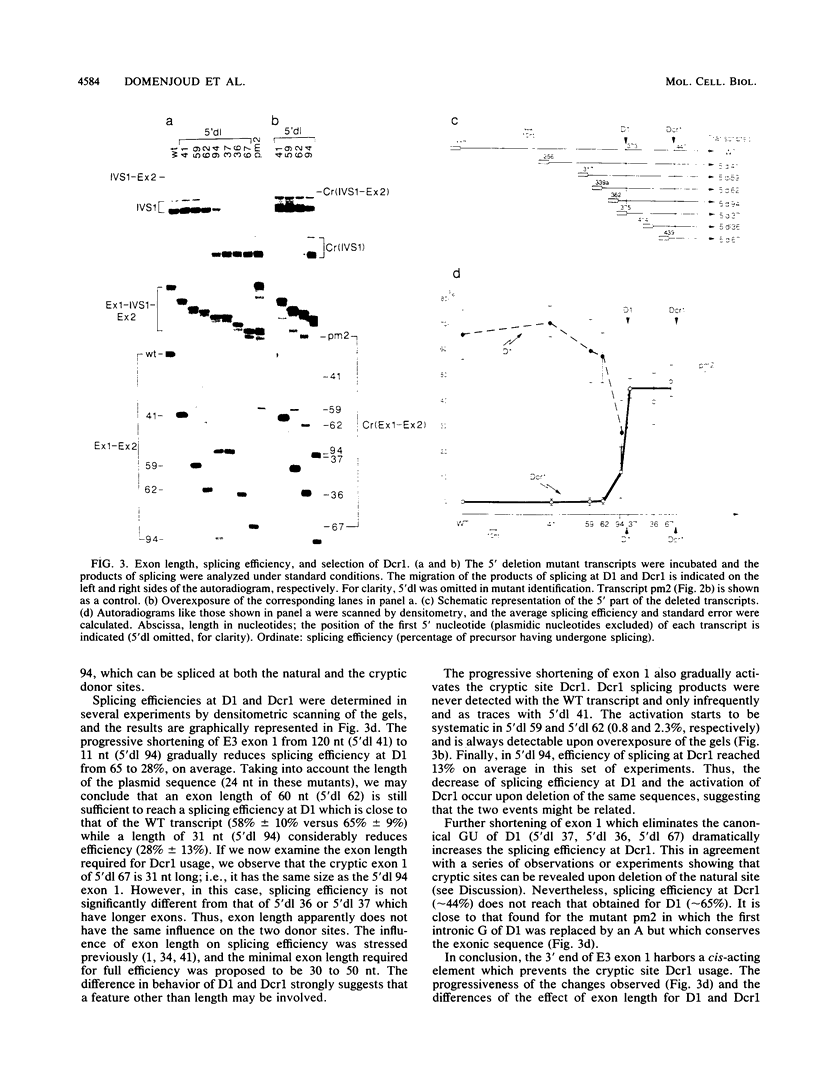
- Smith C. W., Patton J. G., Nadal-Ginard B. Alternative splicing in the control of gene expression. Annu Rev Genet. 1989;23:527–577. doi: 10.1146/annurev.ge.23.120189.002523. [DOI] [PubMed] [Google Scholar]
- Solnick D., Lee S. I. Amount of RNA secondary structure required to induce an alternative splice. Mol Cell Biol. 1987 Sep;7(9):3194–3198. doi: 10.1128/mcb.7.9.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somasekhar M. B., Mertz J. E. Exon mutations that affect the choice of splice sites used in processing the SV40 late transcripts. Nucleic Acids Res. 1985 Aug 12;13(15):5591–5609. doi: 10.1093/nar/13.15.5591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streuli M., Saito H. Regulation of tissue-specific alternative splicing: exon-specific cis-elements govern the splicing of leukocyte common antigen pre-mRNA. EMBO J. 1989 Mar;8(3):787–796. doi: 10.1002/j.1460-2075.1989.tb03439.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
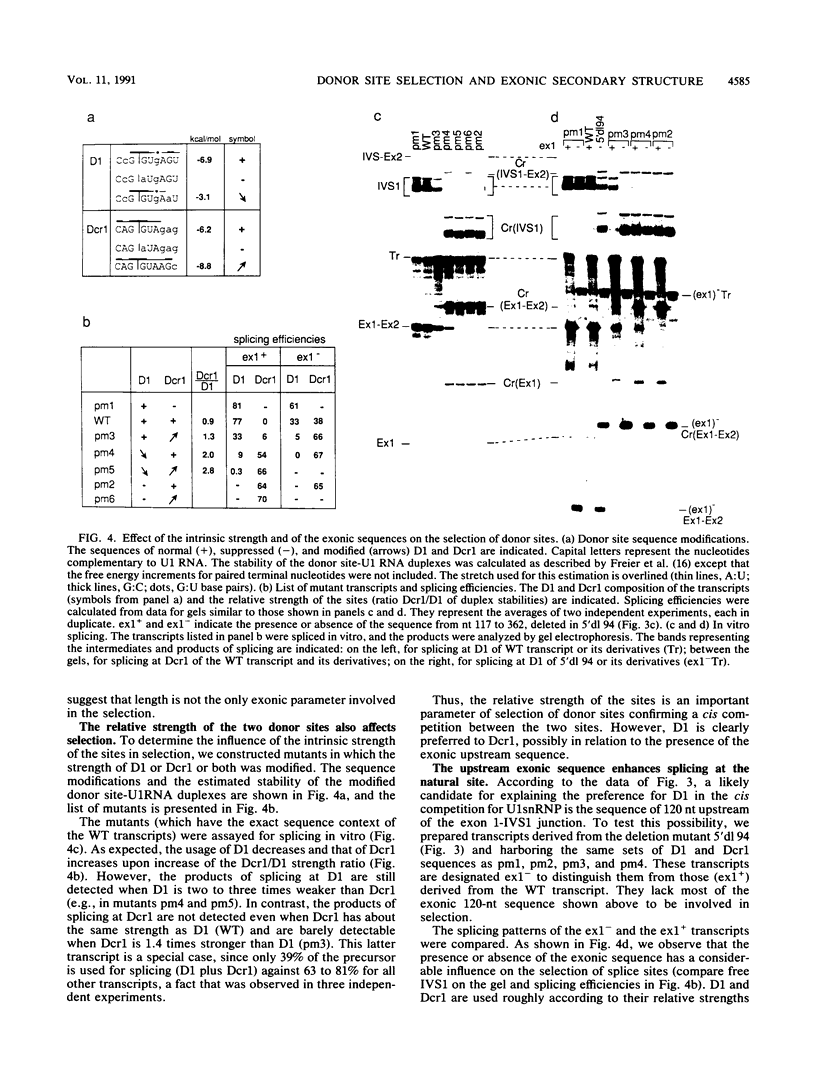
- Stålhandske P., Persson H., Perricaudet M., Philipson L., Pettersson U. Structure of three spliced mRNAs from region E3 of adenovirus type 2. Gene. 1983 May-Jun;22(2-3):157–165. doi: 10.1016/0378-1119(83)90099-9. [DOI] [PubMed] [Google Scholar]
- Tollefson A. E., Krajcsi P., Yei S. P., Carlin C. R., Wold W. S. A 10,400-molecular-weight membrane protein is coded by region E3 of adenovirus. J Virol. 1990 Feb;64(2):794–801. doi: 10.1128/jvi.64.2.794-801.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treisman R., Orkin S. H., Maniatis T. Specific transcription and RNA splicing defects in five cloned beta-thalassaemia genes. Nature. 1983 Apr 14;302(5909):591–596. doi: 10.1038/302591a0. [DOI] [PubMed] [Google Scholar]
- Tsai A. Y., Streuli M., Saito H. Integrity of the exon 6 sequence is essential for tissue-specific alternative splicing of human leukocyte common antigen pre-mRNA. Mol Cell Biol. 1989 Oct;9(10):4550–4555. doi: 10.1128/mcb.9.10.4550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidaud M., Gattoni R., Stevenin J., Vidaud D., Amselem S., Chibani J., Rosa J., Goossens M. A 5' splice-region G----C mutation in exon 1 of the human beta-globin gene inhibits pre-mRNA splicing: a mechanism for beta+-thalassemia. Proc Natl Acad Sci U S A. 1989 Feb;86(3):1041–1045. doi: 10.1073/pnas.86.3.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watakabe A., Inoue K., Sakamoto H., Shimura Y. A secondary structure at the 3' splice site affects the in vitro splicing reaction of mouse immunoglobulin mu chain pre-mRNAs. Nucleic Acids Res. 1989 Oct 25;17(20):8159–8169. doi: 10.1093/nar/17.20.8159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber S., Aebi M. In vitro splicing of mRNA precursors: 5' cleavage site can be predicted from the interaction between the 5' splice region and the 5' terminus of U1 snRNA. Nucleic Acids Res. 1988 Jan 25;16(2):471–486. doi: 10.1093/nar/16.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieringa B., Meyer F., Reiser J., Weissmann C. Unusual splice sites revealed by mutagenic inactivation of an authentic splice site of the rabbit beta-globin gene. Nature. 1983 Jan 6;301(5895):38–43. doi: 10.1038/301038a0. [DOI] [PubMed] [Google Scholar]
- Winship P. R. An improved method for directly sequencing PCR amplified material using dimethyl sulphoxide. Nucleic Acids Res. 1989 Feb 11;17(3):1266–1266. doi: 10.1093/nar/17.3.1266. [DOI] [PMC free article] [PubMed] [Google Scholar]