Abstract
Formate transport across the inner membrane is a critical step in anaerobic bacterial respiration. Members of the formate nitrite transport protein family function to shuttle substrate across the cytoplasmic membrane. In bacterial pathogens the nitrite transport protein is involved in protecting bacteria from peroxynitrite released by host cell macrophages. We have determined the 2.13 Å structure of the formate channel FocA from Vibrio cholerae, which reveals a pentamer, with each monomer possessing its own substrate translocation pore. Surprisingly, the fold of the FocA monomer resembles that found in water and glycerol channels. The selectivity filter in FocA consists of a cytoplasmic slit and a central constriction ring. A 2.5 Å high-formate structure shows two formate ions bound to the cytoplasmic slit via both hydrogen bonding and Van der Waals interactions, providing a structural basis for substrate selectivity of the channel.
Formate and nitrite are major metabolites formed during bacterial respiration under anaerobic conditions and, for certain bacterial pathogens, nitrite also plays a key role in overcoming immune responses of mammals. Up to one third of the glucose in fermenting Escherichia coli (E. coli) cells is converted to formate via the pyruvate formate lyase pathway 1,2. To prevent acidification of the bacterial cytoplasm, formate is exported from the cell across the inner membrane by the transport protein FocA. In the periplasm, formate is subsequently reduced by formate dehydrogenase into carbon dioxide. In contrast, the homologous nitrite transport protein NirC is employed by bacteria to import the solute into the cytoplasm, where it is reduced by nitrite reductase NirB into ammonia 3. The pathogen Salmonella typhimurium uses this transport system to defend itself against the attack of peroxynitrite produced by the inducible nitric oxide (NO) synthase of the host, by importing nitrite for neutralization 4–7. A higher NO output is observed from mouse macrophages infected with nirC-knockout Salmonella 7. Strikingly, the NirC mutant has exhibited a marked reduction in NO-dependent intracellular proliferation in both cultured macrophage cells and in liver, spleen and lymph tissues, highlighting NirC as a virulence protein in Salmonella.
Both the FocA and NirC proteins belong to the formate nitrite transporter family (FNT, TC 2A44) 8. A recent search in the InterPro database yielded ~1,500 members from bacteria, archae and eukaryotes. The organisms whose genome contain FocA or NirC orthologs include cadmium-resistant bacterium Euglena gracilis 9, pathogenic fungi such as Toxoplasma gondii, Candida albicans, Aspergillus fumigatus and A. flavus, the malaria parasite Plasmodium spp. and plants. However, no FNT member has been discovered in animals, making the NirC protein from human pathogens like Salmonella an attractive target for antimicrobial development.
The best-characterized FNT members thus far are the formate transport protein FocA (formate channel) 10,11 and the nitrite transport protein NirC 3,12, both from E. coli. Most FNT proteins are composed of ~280 amino acids (Supplementary Fig. 1), and the E. coli FocA has been shown by phoA- and lacZ-fusion to possess six transmembrane α-helices, with both the amino- and carboxyl-termini located in the cytoplasm 11. focA-knockouts displayed a 50 % reduction in formate export in E. coli, resulting in a cytoplasmic accumulation of formate up to 10 mM. NirC, on the other hand, mediates high-flux transport of nitrite across the E. coli inner membrane in both directions 12. However, the substrate transport mechanism by FNT proteins is unknown. It is even unclear whether they function as channels or transporters 11, which is reflected in the confusing nomenclature. While the founding member of the family is named as a channel (therefore the “c” in FocA) 11, the entire group of proteins is classified as a transporter family (therefore the “T” in FNT) 8. Importantly, if FNT proteins operate as transporters, one would need to know the substrate binding site, the driving force and the conformational changes that occur during substrate translocation. Alternatively, if the proteins function as channels, understanding of their transport mechanism would require identification of the substrate selectivity filter, characterization of the substrate-filter interaction, and suggestion of a possible gating mechanism.
RESULTS
FocA protein from Vibrio cholerae
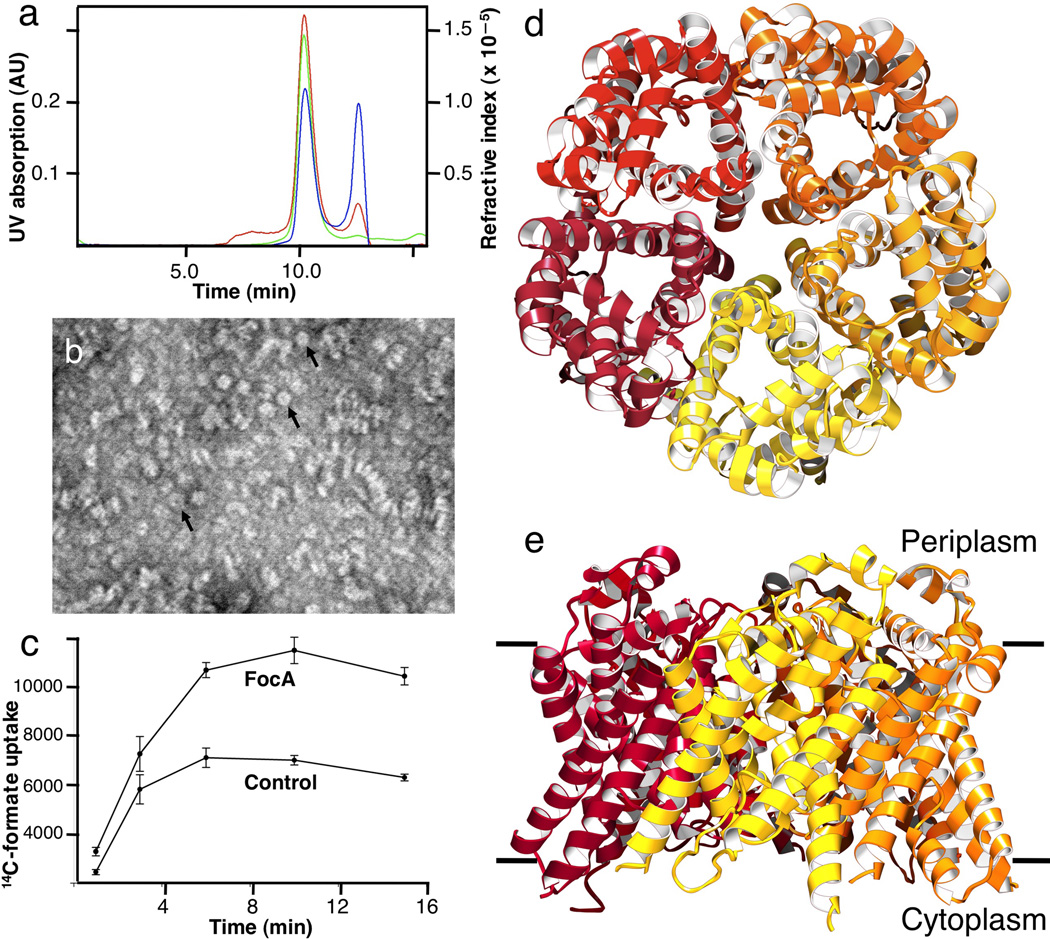
We set out to understand the transport mechanism of formate nitrite transport proteins by structural and biochemical approaches. We chose to characterize the FocA protein from Vibrio cholerae (V. cholerae). The protein was expressed in E. coli and purified in the detergent β-octylglucoside (β-OG) and 20 mM formate. Purified FocA exhibited high monodispersity and stability in a number of detergents, and thus appeared to be an ideal target for crystallization experiments. In octylglucoside solution, the protein has an apparent molecular weight of 303 kDa, which includes 142 kDa of bound detergent and phospholipid molecules, as measured by analytical size-exclusion chromatography coupled with static light scattering and refractive index techniques (Fig. 1a). The protein mass of 161 kDa agrees with a FocA pentamer of 155 kDa. Under the electron microscope, purified protein appears in various orientations on the grid, some as donut-like rings of 80–90 Å in diameter (Fig. 1b), which is consistent with a 155-kDa FocA pentamer. To investigate whether the FocA protein from V. cholerae transports formate across the membrane, we measured formate uptake into proteoliposomes using a concentrative uptake assay 13,14, which is suitable for measuring transport activities of both channels and transporters 15. Formate uptake was observed at pH 6.7 in proteoliposomes reconstituted with V. cholerae FocA (Fig. 1c), showing that the protein is able to transport formate. As no pH or additional electrochemical gradient existed across the proteoliposomes membrane other than the equal molar counterions (glutamate or acetate) used to balance the charges 14,15, the FocA protein probably functions either as a formate channel or uniporter 16.
Figure 1.
Characterization of the FocA protein from Vibrio Cholerae. (a) Measurements of protein mass in solution. The protein has an apparent molecular weight of 303 kDa, which includes 142 kDa of bound octylglucoside detergent and phospholipid, indicating a 161 kDa FocA pentamer. The green line indicates UV absorbance at 280 nm, the red line static light scattering, and the blue line differential refractive index measurements. (b) Electron micrograph of FocA showing purified protein in various orientations. A few ring-like particles with a central cavity, probably representing a pentamer, are indicated by arrowheads. The scale bar represents 500 Å. (b) Uptake of 14C-labeled formate into proteoliposomes reconstituted from purified FocA at pH 6.7. Liposomes devoid of protein (Control) showed marked permeability to formate, as expected for a monovalent substrate with a pKa of 3.8. (d) Pentameric structure of FocA from the 2.13 Å resolution crystal structure, viewed from the periplasm. (e) FocA pentamer viewed from within the membrane plane.
We grew three-dimensional crystals of V. cholerae FocA in the presence of 20 mM formate (designated low-formate crystal form) that diffracted X-rays to ~2 Å resolution. Its structure was determined at 2.13 Å using a platinum derivative by SIRAS phasing, and the map quality was improved markedly with non-crystallographic symmetry averaging (Table 1). The R-factor after refinement, upon the removal of the non-crystallographic symmetry constraints, is at 18.0%, and the Rfree is at 20.0%. As observed in solution and in the membrane, the FocA protein exists as a homo-pentamer in the crystal structure (Figs. 1d&e), with a diameter of 82 Å and a thickness of 58 Å. Whereas the construct used for crystallization contained the entire 280 amino acid sequence, the first 22 amino acids at the N-terminus and the last three at the C-terminus are disordered in the crystal. In addition to the protein, 16 octylglucoside detergent molecules and 346 water molecules are present in this low-formate crystal form (Table 1). All five FocA monomers in the pentamer, named A to E, have a similar overall structure (Figs. 1d&e). However, one particular loop adopts one configuration in Monomers A–C and a different configuration in Monomers D and E. We will first describe the structure of monomer A, the best ordered in the former group. This will be followed later by a description of the structure of the better-ordered monomer in the latter group, Monomer E.
Table 1.
Crystallography data and refinement statistics
| Crystal form: | Low-formate | High-formate | |
|---|---|---|---|
| Crystal parameters: | |||
| Space group | P212121 | P212121 | |
| Cell dimensions | |||
| a, b, c (Å) | 99.83, 100.90 , 192.34 | 99.78, 100.46 , 192.96 | |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 | |
| Data quality: | |||
| Resolution (Å) | 50 – 2.13 | 50 – 2.5 | |
| Rsym | 0.11 (0.72) | 0.17 (0.63) | |
| Mean redundancy | 7.2 | 7.3 | |
| Completeness | 94.60% | 98.60% | |
| Mean I/σ(I) | 12.43 (2.1) | 14.6 (2.6) | |
| Phasing Statistics: | |||
| Anomalous signal (Å) | 5.4 | ||
| Mean figure of merit | 0.52 | ||
| Refinement: | |||
| Resolution (Å) | 50 – 2.13 | 50 – 2.5 | |
| No. reflections | 102,406 | 62,189 | |
| Rwork/ Rfree | 0.18 / 0.21 | 0.17 / 0.22 | |
| Average B-factor (Å2) | |||
| Protein | 68.91 | 55.76 | |
| Formate, octylglucoside | 99.94 | 81.70 | |
| Water | 58.46 | 45.64 | |
| R.M.S deviations | |||
| Bond lengths (Å) | 0.008 | 0.008 | |
| Bond angles (°) | 0.995 | 1.026 | |
| Model Contents: | |||
| Monomer A: 256 | Monomer A: 257 | ||
| Monomer B: 254 | Monomer B: 255 | ||
| Protein residues | Monomer C: 259 | Monomer C: 254 | |
| Monomer D: 251 | Monomer D: 254 | ||
| Monomer E: 255 | Monomer E: 256 | ||
| Ligands | 16 β-OG | 13 β-OG, 51 formate * | |
| Water molecules | 346 | 227 | |
Notes:
Rsym = S|Ij – <Ij>|/SIj, where <Ij> is the averaged intensity for symmetry related reflections. Redundancy represents the ratio between the number of measurements and the number of unique reflections. R factor = S|F(obs) – F(cal)|/SF(obs); 5% of the data that were excluded from the refinement were to calculate Rfree. The average B factor was calculated for all non-hydrogen atoms. r.m.s.d. of bond is the root-mean-square deviation of the bond angle and length. Numbers in parentheses are statistics of the highest resolution shell.
While two formate ions are found at the selectivity filter of Monomer E, one is found at that of Monomer D. The rest of the formate ions are found on the protein surface.
FocA monomer
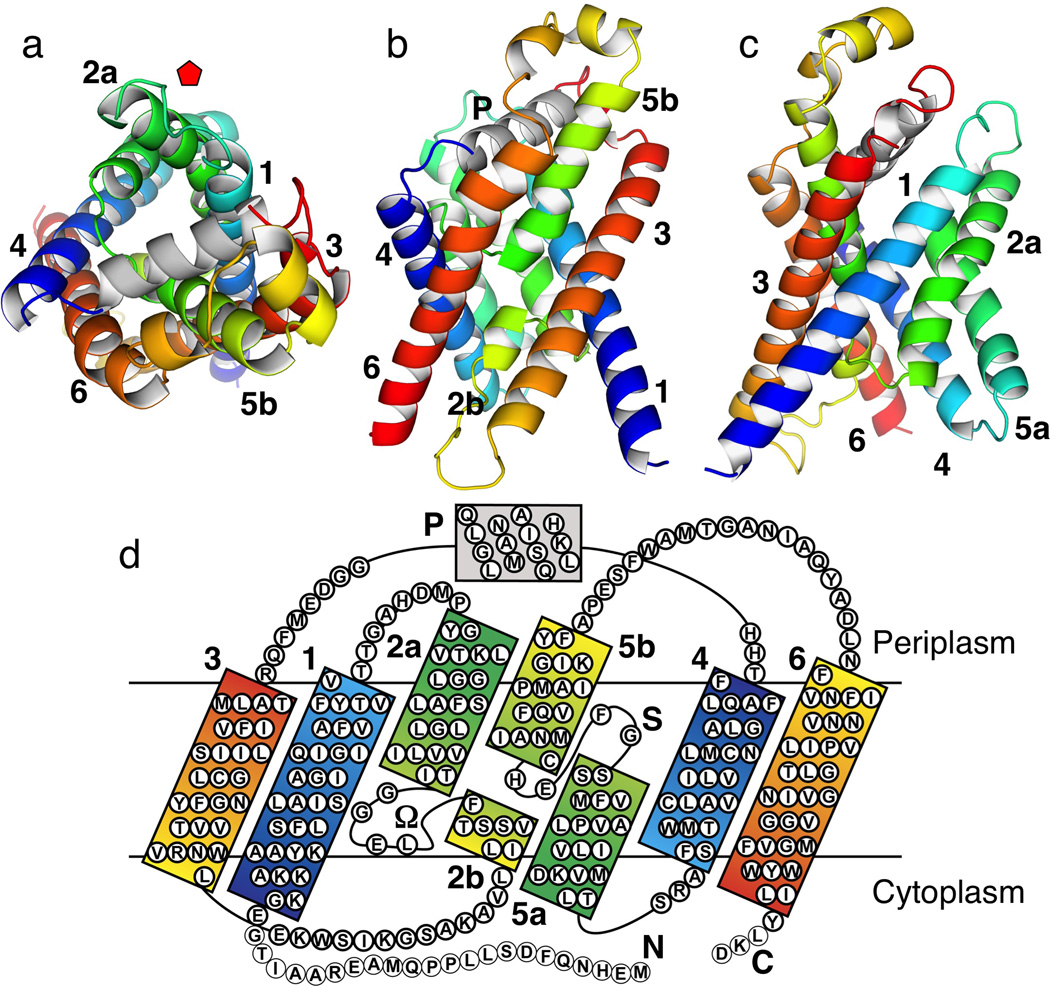
The FocA monomer, as depicted in Monomer A, consists of six transmembrane α-helices, TMs 1–6 (Fig. 2). The second and fifth helices are broken in the middle of the membrane. Viewed from the periplasmic side, the protein has the shape of a kite (Fig. 2a). The acute corner of the kite is located close to the fivefold axis of the pentamer and the angle at the corner is ~72° (Fig. 1d). The transmembrane helices form a right-handed twisted bundle with a pore in the middle of the monomer. Viewed from within the membrane, FocA resembles an hourglass, with most of the helices tilted from the membrane normal at a ~35° angle (Figs. 2b&c). The cytoplasmic surface of the protein, on which both the N- and C-termini reside, is relatively flat. On the periplasmic side, the L3-4 loop connecting TM3 and TM4 contains a 4-turn helix (Helix P), which lies parallel to the membrane surface in a groove formed by the tilted transmembrane helices.
Figure 2.
Overall structure and transmembrane topology of FocA monomer in the low-formate crystal form, represented by Monomer A. Transmembrane α-helices are numbered sequentially from TM1 to TM6. Helices in each half of the protein, TM1-3 and TM4-6, respectively, are colored from blue to green to red sequentially, highlighting the structurally related inverted repeats. (a) Top view from the periplasmic side. Position of the fivefold axis of the pentamer is indicated with a red pentagon. (b) Side view from within the membrane plane along the pseudo-twofold axis. (c) Side view from within the membrane plane as in (b) and rotated 90° counterclockwise along the membrane normal. (d) Topology diagram with sequence colored similarly as above. Helices TM2 and TM5 are broken in the middle of the membrane. The segment that connects TM2a and TM3 (residues Gly85—Glu108) is designated as the Ω loop. The short helix with loop L3-4 on the periplasmic side is designated as the P helix. Thin circles at the termini (residues Met1—Gly22; Leu278—Asp280) indicate disordered residues in the crystal structure.
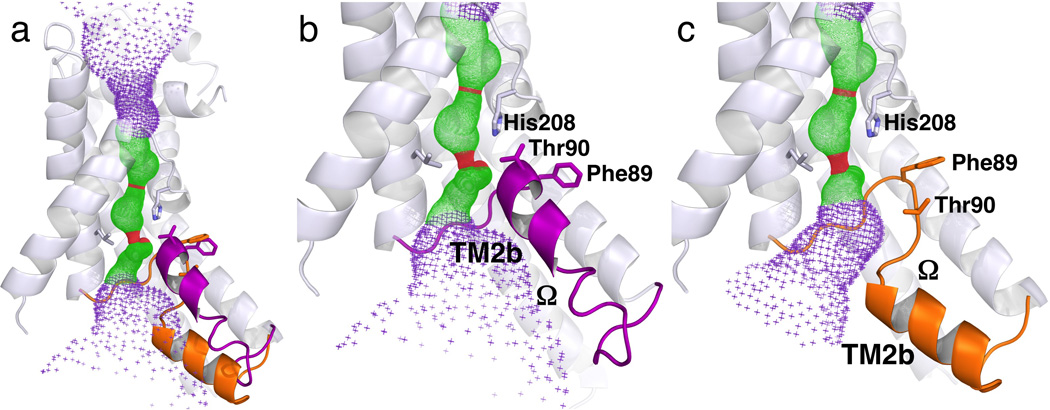
Strikingly, the first three transmembrane α-helices (TMs 1–3) and the second three (TMs 4–6) form inverted repeats in the membrane, with the two halves being related by pseudo-twofold symmetry around an axis that is parallel to the membrane plane (Figs. 2b&d). In the first half, TM1 is sandwiched between TM2 and TM3 (Fig. 2c). With its N-terminus in the cytoplasm, TM1 is a straight α-helix of 8.5 turns (residues Glu23–Val54). Following a short loop L1-2, the next transmembrane helix is broken into two. TM2a starts at Tyr63 and ends at T84 in the cytoplasmic half of the membrane, whereas TM2b begins at 16 Å away within the membrane at residues Phe90, and is tilted from the membrane surface at a small angle (Figs. 3a&b). The segment that connects TM2a and TM3, of which TM2b is a part, has the shape of the Greek letter “Ω.” TM3 (residues Leu107–Thr134) goes across the membrane at a 40° angle to the periplasmic side (Figs. 2a–c). Following the L3-4 loop which contains the P helix, the symmetry-related second half of the protein starts from the N-terminus of TM4 in the periplasm and ends with the C-terminus of TM6 in the cytoplasm, with TM5b broken into two halves and each penetrates through only halfway in the membrane (Figs. 2b–d). This pseudo-twofold symmetry between the two protein halves, however, is far from perfect. While the 4.5-turn TM5b penetrates halfway into the membrane, the symmetrically-related TM2b consists of only 1.5 turns and is tilted at a very different angle. The loop between TM5a and TM5b, with only four amino acids (residues Gly205–His208) and located in the middle of the membrane, is in an extended configuration with a shape similar to that of the letter “S” (Supplementary Fig. 2). The His208 reside from the S loop interacts with the Ω loop by forming a hydrogen bond to its Thr90 residue (Figs. 3b&4c).
Figure 3.
Different configurations of the Ω loop that connects TM2a and TM3 in different FocA monomers, both from the low-formate structure. (a) Structure of Monomer A overlaid with the Ω loop from Monomer E. The Ω loop is colored purple in Monomer A and orange in Monomer E. (b) Close up view of the Ω loop including TM2b in the UP position in Monomer A. (c) Identical view of the Ω loop in the DOWN position in Monomer E. The TM2b helix is at difference positions between the two monomers in both physical space and in sequence. The helices 6 and 4 are removed for clarity. The pore in the center of the monomer as computed by the program HOLE 47 is shown for reference. The structures of Monomers B and C are the same as Monomer A, whereas the structure of Monomer D is similar to that of Monomer E.
Figure 4.
Pore structure in the FocA monomer at low formate concentration. (a) Overall structure of Monomer A viewed from within the membrane, superimposed with the pore structure calculated by HOLE 47. Residues that form the cytoplasmic slit and the central constriction ring are shown as sticks. The helices 6 and 4 have been removed for clarity. (b) Electrostatic surface representation of the pore in cross section as colored by electrostatic properties. The surface of the cytoplasmic funnel is basic where the periplasmic funnel acidic. In the substrate selectivity filter, the cytoplasmic one third is basic, and the periplasmic two thirds is hydrophobic. The front of the monomer has been removed for clarity. (c) Detailed view of the selectivity filter in Monomer A. In this UP configuration, Thr90 of the Ω loop makes a hydrogen bond with His208 of the S loop. (d) Detailed view of the selectivity filter in Monomer E. In this DOWN configuration, the Ω loop moves in the cytoplasmic direction and the hydrogen bond between Thr90 and His208 is broken. As in Figures 3 and 5, residues are colored in purple in Monomer A and orange in Monomer E.
The structure of the FocA monomer (Figs. 2a–c) is reminiscent of the fold found in the water and glycerol channels of the AQP/GlpF family 17–19. Despite the lack of any detectable homology in amino acid sequence – the identity between the V. cholerae FocA to both human AQP1 and E. coli GlpF is merely 9–12% (Supplementary Fig. 1) – their protein folds share an unmistakable likeness (Supplementary Fig. 3). The similarity is particularly evident in the inverted twofold symmetry, the total number of transmembrane α-helices (six), their topology, the right-handed twist of the helix bundle, and even in the existence of a pore in the middle of the monomer. However, major differences are also obvious between the two families of transport proteins. First, AQP/GlpF channels form tetramers in the membrane and in detergent solution 20. All known crystal structures of such proteins, by now over ten of them have been solved from various species of bacteria, yeast, plant as well as mammals, are also uniformly tetrameric 17–19,21,22. The r.m.s.d between FocA and AQP1 is 7 Å for α-carbon. As expected from such difference in oligomeric state, the FocA monomer is more “squeezed” when viewed along the membrane normal (Supplementary Fig. 3). Within the transmembrane region, the structures of the two protein families also have significant differences, which may have mechanistic consequences. Helices TM2a and TM5a in FocA are notably shorter than their counterparts in AQP1 or GlpF 17–19, and therefore span only halfway in the membrane as opposed to the entire thickness. Two equal-length, short helices in AQP/GlpF, HA and HB, interact head-to-head via the conserved NPA motifs in the middle of the membrane, form essentially the seventh transmembrane α-helix in those proteins 17–19. In contrast, although TM5b of FocA is at a similar position as its counterpart HB in AQP1, the shorter TM2b helix is in a completely different position and tilted at a much smaller angle from the membrane plane (Figs. 2b&d). As expected, the signature NPA motifs of the AQP/GlpF family are missing in the FocA sequence (Supplementary Fig. 1). As a result of such structural differences, the geometry of the central channel in the monomer is also different between the two protein families (Supplementary Fig. 3).
FocA pentamer
FocA monomers assemble into a pentamer with the acute corner of each kite located near the fivefold axis (Fig. 1d). The fivefold symmetry is maintained for most parts of the protein, with an r.m.s.d. of 0.32 Å for the backbone Cα atoms and 0.62 Å for all non-hydrogen atoms, except for the Ω loop connecting TM2a and TM3, which varies among the five monomers in both its position and the degree of order in the crystal (Table 1). In Monomers B and C, the configuration of the Ω loop is similar to that found in Monomer A (Fig. 3b) (which we thereby named the UP position), except that the C-terminal end of the Ω loop is largely disordered. On the other hand, in Monomers D and E, although the beginning of the Ω loop (residues Gly85–Leu88) is in the same position as in Monomers A-C, the rest of the Ω loop moves in the direction of the cytoplasm by ~5 Å (Fig. 3c & Supplementary Figs. 4&5) (which we named the DOWN position for the loop). As a result, the hydrogen bond between Thr90 of the Ω loop and His208 of the S loop is broken and, interestingly, a water molecule occupies the original position of Thr90, forming a hydrogen bond with His208 (Supplementary Fig. 4). In addition, the 1.5-turn helix TM2b found in Monomers A–C completely unwinds, and a different segment in the sequence of the Ω loop (residues Val93–Ser101) forms a new TM2b (Figs. 3a&c, Supplementary Fig. 5). The C-terminal end of the Ω loop is ordered in Monomer E but disordered in Monomer D. Such a degree of variation in the configuration of the Ω loop in the crystal structure (Supplementary Table 1) probably reflects the intrinsic flexibility of the loop and therefore may have mechanistic consequences. Similar local structural variations between monomers have been observed for the E. coli water channel AqpZ 23.
The large, complementary interface between the monomers explains the high stability observed for the FocA pentamer (Figs. 1a&b). The interface between two monomers, formed between TMs1–3 of one monomer and TMs4–6 of the adjacent monomer, has an area of ~1,450 Å2. While there are no salt bridges and few hydrogen bonds between the neighboring monomers, the Van der Waals contacts are rather intimate, with a surface complementarity parameter of 0.68, comparable to that of a typical interface between an antibody with its protein antigen 24. As in the case of the water and glycerol channels 17–19, such large interface area and high degree of surface complementarity between the monomers give the FocA pentamer unusual stability (Figs. 1a&b).
The fivefold axis in the FocA pentamer prescribes a large central pore (Fig. 1d and Supplementary Fig. 6). Although with a diameter of only 2.8 Å at its cytoplasmic entrance, the central pore is wide open in the middle of the membrane, with a diameter of ~13 Å, and it narrows down to ~10 Å on the periplasmic side (Supplementary Fig. 6). This central pore is lined by the N-terminal half of TM5a on the cytoplasmic side and the N-terminal half of TM2a on the periplasmic side. The small diameter of its cytoplasmic entrance is mediated by the side chains of Met192 from each of the five TM5a helices. Five octylglucoside detergent molecules are found inside the central pore, suggesting that the pore is probably occupied by lipid molecules in the native membrane environment (Supplementary Fig. 6). As the residue Met192 is not conserved among FocA proteins (Supplementary Fig. 1) and also because the rest of the central pore is too wide to be the selectivity filter (for a channel) or a substrate binding site for an ion with the size of formate, the central pore is unlikely to facilitate formate transport across the membrane. This is consistent with the fact that in APQ1 or GlpF the conducting channel is not located at the fourfold axis of those tetramers 17–19.
Pore in the monomer
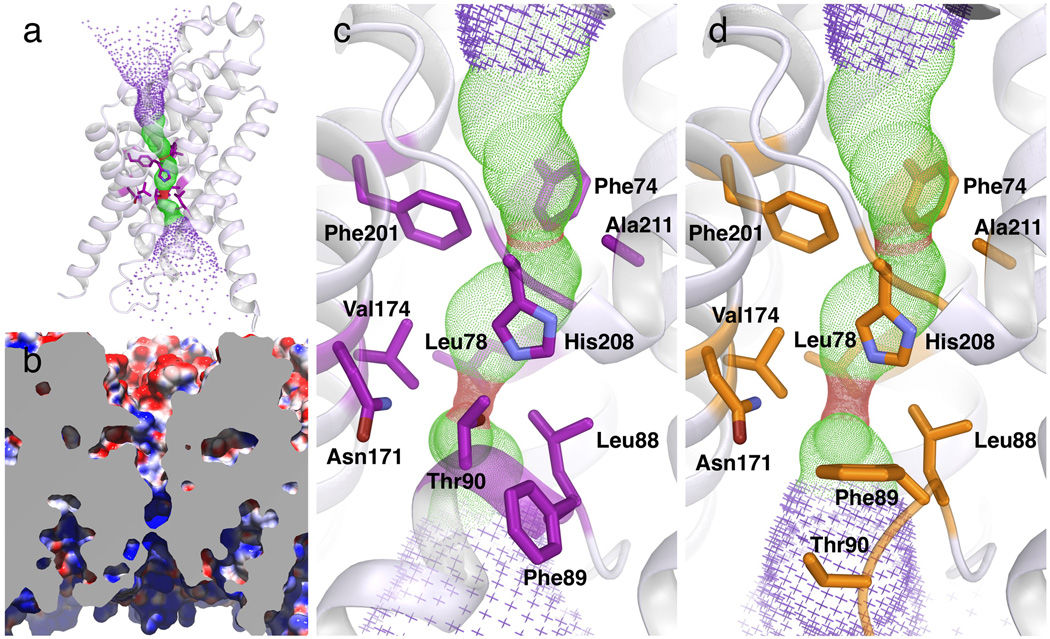
We then focus on the pore in the middle of the monomer, which is continuous and open to both sides of the membrane (Fig. 4a). The pore in the center of the FocA monomer can be divided into three parts along the membrane normal: the funnel or wide opening on the cytoplasmic side, the extended narrow pore in the middle and the funnel on the periplasmic side of the protein (Fig. 4b). The narrow pore is 18 Å long, whereas the cytoplasmic and periplasmic funnels are 16 Å and 20 Å long, respectively. The axis of the cytoplasmic funnel is tilted at ~35° from the membrane normal. The funnel has an opening on the cytoplasmic surface of the protein, of 30 Å by 15 Å, and it gradually narrows down into a slit of 2–3 Å at the inner end of the funnel. The funnel is lined by TM1, TM2b, the Ω-loop, TM4, the L4-5 loop, TM5 and TM6. The entire surface of the cytoplasmic funnel is very basic.
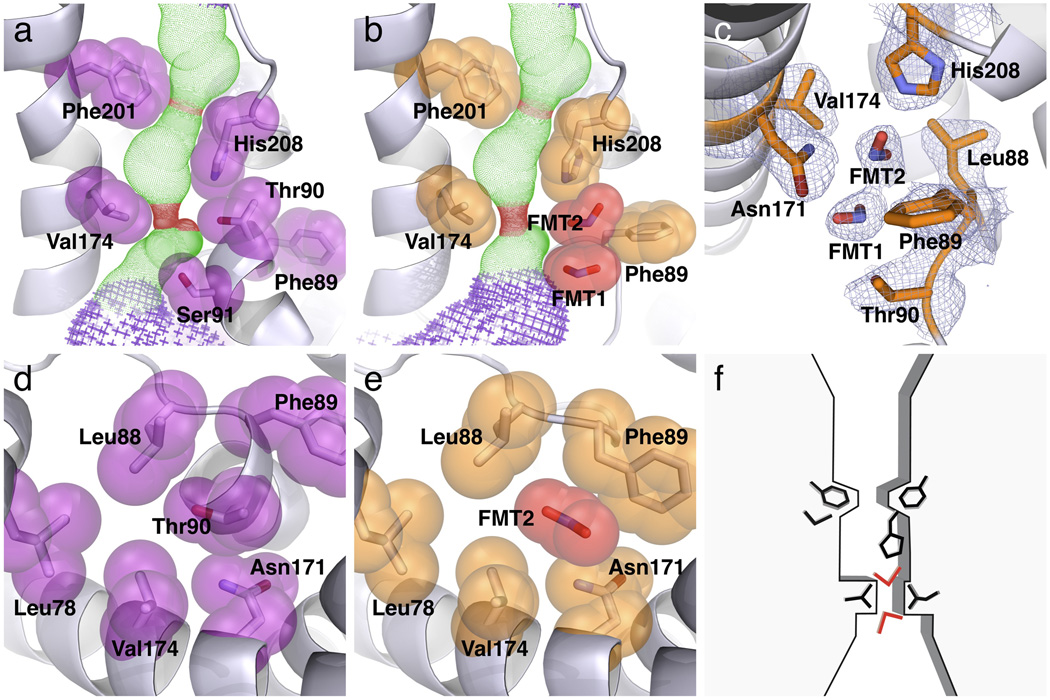
The cytoplasmic funnel becomes the extended narrow pore in the middle of the membrane (Figs. 4b&c). The cross section of the pore changes along the membrane normal in shape, size and surface property. Whereas the cytoplasmic half is elliptical and narrower, the periplasmic half is circular and wider, with the narrowest parts being a narrow slit and a central ring. The narrow slit, located at the cytoplasmic entrance of the pore, is 2.1 Å by 4.1 Å (Figs. 5a&d). It is formed by Leu78 from TM2a, Leu88 and Thr90 from the Ω loop, and Val174 from TM4. His208 from the S loop sits vertically on the periplasmic side of the slit, where the pore has an elliptical shape of 3.4 Å by 5.4 Å in cross section. Further in the periplasmic direction, the central constriction ring is 2.3 Å in diameter, which is formed by the side chains of Phe74 from TM2a, Phe201 from TM5a, the His208 Cβ atom from the S loop and Ala211 from TM5b (Figs. 4c&5a). The pore becomes ~6 Å wide on the periplasmic side of the central ring. While the cytoplasmic one third of the narrow pore is basic, the two thirds on the periplasmic side is completely hydrophobic. In Monomer E, however, with its Ω loop in the DOWN position and the Thr90 and Ser91 residues moved away, the cytoplasmic slit becomes 2.1 Å by 7.2 Å in cross section, and is surrounded by five residues (Leu78, Leu88, Phe89, Asn171 and Val174) instead of four as in Monomer A (Figs. 4d). The long edge of the slit is approximately parallel to the long axis of the FocA monomer kite. Importantly, all these residues both at the slit and the central ring are highly conserved among various FNT members including both FocA and NirC proteins (Supplementary Fig. 1).
Figure 5.
Selectivity structure and its interactions with formate. (a) Side view of the selectivity filter in the low-formate structure of Monomer A. Both the cytoplasmic slit and the central constriction ring are shown. The Ω loop is in the UP configuration, and Thr90 of the Ω loop makes a hydrogen bond with His208 of the S loop. (b) Side view of the selectivity filter in the high-formate structure of Monomer E, solved at 2.5 Å resolution from crystals grown in 120 mM formate. The Ω loop is in the DOWN configuration, and two formate ions (FMT1 and FMT2) are located in the cytoplasmic slit. (c) 2Fo-Fc electron density map showing the locations of the two bound formate ions in the selectivity filter (contoured at 1σ). Formate 2 makes hydrogen bonding with His208. (d) Top view of the cytoplasmic slit in the low-formate structure of Monomer A. Thr90 is surrounded by a ring of residues. (e) Top view of the cytoplasmic slit in the high-formate structure of Monomer E. Bound Formate 2 occupies the same position as the Thr90 side chain in Monomer A. (f) Schematic drawing showing the substrate selectivity filter in FocA.
Similar to the extended narrow pore, the periplasmic funnel is roughly normal to the membrane plane (Figs. 3a, 4a&b). The periplasmic funnel begins with a 7 Å long cylinder of 8 Å in diameter in the membrane before it opens up to ~30 Å in size at the periplasmic surface. The cylindrical part of the funnel is lined by TM1, TM2a, the S loop, the L3-4 loop including Helix P, and TM4, whereas the periplasmic opening of the funnel is lined by TM2a, the S loop and Helix P. Importantly, in contrast to the basic property of the cytoplasmic funnel, the surface of the periplasmic funnel is largely acidic.
As the central pore around the fivefold axis was unlikely to transport substrate, the pore structure in the middle of the FocA monomer immediately suggested a formate translocation pathway (Figs. 4a&b). This is again similar to the AQP/GlpF channels, for the water/glycerol pathway is located in the center of the monomer 17–19. Furthermore, no thin or thick gate is observed in FocA that blocks the transport pathway, as one would have expected for a membrane transporter 25,26. The observation of a narrow pore and absence of any gate-like structural features in the pore strongly imply that FocA operates via a channel mechanism instead of that of a transporter. It follows that the extended narrow pore constitutes its substrate selectivity filter. This notion that FNT proteins operate as channels also agrees with the following experimental evidence: (1) our observation that no gradient other than that of formate is required for its uptake into proteoliposomes by V. cholerae FocA (Fig. 1c); (2) the transport of nitrite across the E. coli inner membrane by NirC is bi-directional and its rate of transport is much faster than that of other nitrite transporters 12. Finally, the fact that we observed a water molecule in the selectivity filter of two of the FocA monomers (Supplementary Fig. 4 and Supplementary Table 1) suggests that the channel may be permeable to water.
Locations of formate ions in the pore
In order to characterize interaction between the substrate selectivity filter and formate in the FocA channel, we grew FocA crystals in the presence of 120 mM formate and determined its structure at 2.5 Å resolution (designated the high-formate structure, Table 1). The overall FocA structure does not change in the presence of high formate concentration, but variations among the monomers are again evident in the Ω loop (Supplementary Table 1). Like in the low-formate structure, the Ω loop in Monomers A, B and C is in the UP position, whereas the loop is in the DOWN position in Monomers D and E. Importantly, in the latter two monomers of the high-formate structure, instead of water molecules, two formate ions are found at the cytoplasmic slit in the selectivity filter (Figs. 5b,c&e). The observation that formate ions are visible in the structure in the presence of 120 mM but not 20 mM formate indicates that the affinity of the protein for its substrate is in the range of millimolar or even tens of millimolar. Such a concentration needed for substrate visualization in the selectivity filter is very similar to that for the voltage-gated K+-channel 27, further supporting the notion that FocA operates as a channel instead of a transporter 16.
In Monomers D and E, the first formate ion (named Formate 1) is located at the opening of the cytoplasmic slit, while the second formate (Formate 2) is 2.9 Å away on its periplasmic side (Figs. 5b,c&e). Both formate ions are dehydrated. Formate 2, the inner substrate, occupies the same position as the water molecule in the low-formate Monomer E structure (Supplementary Fig. 4). It is coordinated from the periplasmic side by His208, with both its oxygen atoms making a bidentate hydrogen bond with the ε2 nitrogen of the His208 imidazole ring (Supplementary Fig. 7 and Supplementary Table 2) 28,29. The proximity of His208 to the N-terminus of TM5b, due to the existence of the helix dipole moment, makes the histidine residue more positively charged than usual, probably resulting in a stronger hydrogen bond with the formate. Within the membrane plane, this formate ion sits within the cytoplasmic slit (Figs. 5b,c&e). In addition to Van der Waals contact with the side chains of Leu88, Phe89 and Asn171, the formate via its O2 atom also makes hydrogen bonding to the Nδ2 atom of Asn171. At the cytoplasmic side, Formate 2 makes hydrogen bonding and Van der Waals contact with the O2 of Formate 1. In the membrane plane, Formate 1 also makes contact with the side chains of Leu88, Thr90 and Asn171 (Supplementary Table 2). Thus, both formate substrates, particularly the inner one, make very specific interactions with the selectivity filter.
DISCUSSION
Structural basis of substrate selectivity
The specific interactions of two bound formate ions with the selectivity filter, as well as the cross-section size and electrostatic surface property of the selectivity filter (Figs. 5a–e), immediately suggests a structural basis for substrate selectivity of the formate channel (Fig. 5f). The positively-charged surface of the cytoplasmic funnel helps to concentrate formate ions — a pre-selection process — as observed in the case of nAChR, a ligand-gated ion channel 30. The selectivity filter of the formate channel begins at the cytoplasmic slit. A formate enters the slit as observed in Monomers D and E (Figs. 5c&f). At the inner site, both oxygen atoms of the formate form a hydrogen bond with the Nε2 atom of His208, and the formate makes Van der Waals contact with the slit, suggestive of a coin in a slot, except the coin cannot tilt or rotate due to the hydrogen bonds. The binding of the inner formate in turn facilitates the binding of the outer formate by making a hydrogen bond with the latter. Once the first formate passes the cytoplasmic slit, it goes through the remaining part of the selectivity filter fairly smoothly. There is probably little electrostatic interaction with the periplasmic half of the pore due to its hydrophobic surface; the size of the 2.3 Å central restriction ring fits with the dimensions of the formate. After the formate reaches the exit of the filter, the negatively-charged periplasmic funnel rapidly repels the formate into the extracellular space. The existence of the two deep funnels in the protein reduces the energetic penalty for ion access to the hydrophobic part of the membrane 31,32.
As expected from the similarities in protein fold, the overall shape of the FocA channel as well as the length of its selectivity filter is similar to those of AQP1 and GlpF 17–19. Compared to APQ/GlpF channels, the positions of the narrowest parts are more in the cytoplasmic half of the selectivity filter in FocA. The narrow dimension of constriction rings, at 2.1—2.3 Å, is in between of that of ~1.8 Å and ~2.5 Å, respectively, as found in the water and glycerol channels 17–19, consistent with the relative sizes of the three types of substrates. Interestingly, many key residues involved in the selectivity of FocA are from loops that connect broken α-helices in the membrane, as has been observed with other channels and transporters 32–34.
The position of the substrate translocation pathway in FocA is in stark contrast to that of other pentameric channels 30,35–37, or of those voltage- or ligand-gated ion channels 30,31,38, in which the substrate pathway is located at the center of the oligomers. Channels for larger ions and solutes often function as a monomer, which in turn forms a highly-stable oligomer each with a separate transport pathway 39–41. As the structural rigidity of the selectivity filter of a channel is crucial for achieving its required selectivity 31, the oligomerization of a channel protein with only six transmembrane helices obviously provides an efficient way to achieve that goal. Whether such oligomerization also implies cooperativity between the monomeric channels is unclear; we favor the opinion of lack of cooperativity as the rigidity of the monomer and of the large monomer-monomer interface make such cooperation hard to materialize 42.
Flexibility of the Ω loop
Given the well-ordered structure of the FocA protein both in the absence and presence of formate, the flexibility of the Ω loop observed in this work is striking (Supplementary Table 1). Whether such flexibility of the loop has any mechanistic significance is unclear. We, however, would hypothesize that the flexibility of the Ω loop, and in particular the position of the Thr90 residue of the loop, provides a gating mechanism for the formate channel. In the low-formate structure of Monomers A–C, the Ω loop is in the UP position with the Thr90 residue forming a hydrogen bond with the His208 residue of the S loop, thus preventing formate ions from entering the selectivity filter (Figs. 5a&d). In the high-formate structure of Monomers D and E, the Ω loop swings out to the DOWN position and the Thr90 and Ser91 residue moves away from the entrance of cytoplasmic slit, allowing formate ions to enter the selectivity filter (Figs. 5b,c&e). In short, we hypothesize that the competition of formate with the Thr90 residues for hydrogen bonding with His208 opens the channel—a substrate-gated mechanism for the formate channel. This resembles the voltage-dependent chloride-channel ClC-0, in which the channel opening is strongly facilitated by extracellular chloride 43,44. The movement of the Ω loop in such a gating mechanism, on the other hand, is similar to that observed for aquaporins from plants and yeast, in which the swinging in of a loop into the cytoplasmic funnel occludes the pore, without changing the structure of the selectivity filter 45,46. However, we have no experimental evidence that supports such a hypothesis of FocA gating. This hypothesis, along with other exciting mechanistic questions, such as whether FocA is permeable to water, whether it selects formate over nitrite, and if there is any cooperativity among monomers in the pentamer, await further biochemical, biophysical and structural investigations.
METHDOS
Protein purification and crystallization
We cloned FocA from Vibrio Cholerae into a pBAD vector with a C-terminal decahistidine tag and overexpressed in E. Coli (C43) cells 48–50. Following cell disruption and membrane solubilization with 1% (w/v) β-dodecylmaltocide (DDM) the supernatant was incubated with TALON Co2+ resin overnight. After washing, the protein was eluted from the resin using a buffer containing 1.1% (w/v) β-octylglucoside (OG), 20 mM sodium formate, 100 mM NaCl, 8% (v/v) glycerol and 10 mM Tris at pH 7.5. The eluate was concentrated and further purified via size-exclusion chromatography using a Sephadex 200 column on Akta FPLC (GE Healthcare). Peak fractions were collected and concentrated to approximately ~8 mg ml−1 before crystallization trials. FocA crystals were grown in two days using the vapor diffusion method and sitting drop trays (Emerald Biosciences) in a buffer containing 32% (v/v) PEG 550MME and 25 mM MgCl2. High-formate crystals were prepared similarly with the exception of the substitution of 120 mM sodium formate for NaCl in all buffers and the addition of 20 mM MOPS at pH 6.7 to the crystallization buffer.
Determination of protein oligomeric state
We measured the mass of the FocA protein in solution using a Wyatt miniDAWN TREOS 3 angle static light scattering detector, Wyatt Optilab rEX refractive index detector and Agilent VWD UV absorbance detector 51. Purified protein sample (5 µl) was injected onto a TSK-GEL SuperSW3000 4.6 mm ID by 30 cm silica gel size-exclusion column in buffer containing 0.016% of DDM at a rate of 0.25 ml min−1. A value for the differential refractive index (dn/dc) for DDM was calculated using the Wyatt Optilab rEX refractive index detector, to be 0.128 (ml g−1). The protein/detergent conjugate was de-convoluted using a previously-described method 52. Refractive index contributions due to bound lipid are not accounted for in the calculation.
Electron microscopy
Purified protein samples were applied to an electro microscopy grids coated with carbon film, stained with 2% (w/v) uranyl acetate, and taken to dryness 53. The grids were then checked in a CM12 electron microscope (Philips Electron Instruments, Eindhoven, Netherlands).
Transport activity assay
We measured the transport activity of FocA using a concentrative uptake assay 13,14. 10 mg ml−1 E. coli total phospholipid (Avanti Polar Lipids) was dissolved in chloroform and dried under nitrogen followed by high vacuum for 1 hour. Lipid was resuspended by vortexing and brief sonication (< 30 sec) into degassed, filtered buffer containing 150 mM sodium formate and 20 mM MOPS at pH 6.7. Following 10 cycles of freezing and thawing, vesicles were prepared via extrusion (Avanti Mini-Extruder) using a filter of 400 nm in pore size. Proteoliposomes were prepared by the addition of protein prior to extrusion in a protein to lipid ratio of 1:10,000 (w/w) and allowed to incubate on ice for 2 hours. Buffer exchange was performed immediately prior to assaying by application of 400 µl proteoliposomes to a fresh 1.8 ml G50 fine Sephadex (GE Healthcare) column equilibrated with 150 mM sodium glutamate and 20 mM MOPS at pH 6.7 and centrifuged at 735× g for 2 minutes. Transport assays were initiated by the addition of 450 µM 14C labeled sodium formate (American Radiolabeled Chemical) and terminated by the application of 20 µl from each time point to a G-50 Probequant™ Micro-column (GE Healthcare) and centrifuged for 2 min at 700× g. The resulting proteoliposomes were measured for activity using 1 ml scintillation fluid and counted on a Wallac 350 scintillation counter.
Structure determination
We collected X-ray diffraction data at beamlines X25 and X29 in the National Synchrotron Light Source of the Brookhaven National Laboratory. Images were processed and scaled using HKL2000 and SCALEPACK 54. For low-formate crystals, a heavy atom derivative was obtained by addition of 6 mM potassium tetrachloroplatinate(II) (Hampton Research) directly to the drop containing pre-formed crystals and allowed to incubate for 2 hours. Anomalous data was collected at the platinum L3 edge and phases were obtained by single isomorphous replacement with anomalous scattering (SIRAS) using the program Phenix Autosol 55. The high-formate crystal structure was solved using the low-formate structure as the initial model. Models were built using COOT 56 and refinement was carried out using Phenix Refine 55. Structure figures were generated using the programs PYMOL 57 and UCSF Chimera 58. Octylglucoside molecules were observed in some of the FocA monomers, as in the case of other membrane transport proteins 59,60.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to W.A. Hendrickson for support, to M. Punta and B. Rost for bioinformatics analysis of membrane transporters, to Brian Kloss for assistance in cloning, and to the staff at beamlines X25 and X29 of the National Synchrotron Light Source in the Brookhaven National Laboratory for assistance in X-ray diffraction experiments. We thank B. Czyzewski, Y. Fujiyoshi, N. K. Karpowich, C.J. Law, X.D. Li, J.J. Marden, R.L. Mancusso, H. Sui, J. Wu, R.M. Xu, N. Unwin, T. Walz, M. Zhou and Z. Zhou for participating synchrotron trips and helpful discussions. This work was financially supported by the Protein Structure Initiative II of the NIH (U54GM075026 to W.A. Hendrickson) and by NIDDK (R01-073973 to D.N.W). A.B.W. was partially supported by a predoctoral fellowship from the NIH-NYU Graduate Partnership Program in Structural Biology.
Footnotes
Accession codes. Protein Data Bank: Atomic coordinates and structure factors for low- and high-formate FocA have been deposited under accession codes 3KLY and 3KLZ.
Note: Supplementary information is available on the Nature Structural & Molecular Biology website.
AUTHOR CONTRIBUTIONS
A.B.W. expressed, purified, crystallized and solved the structure of FocA and functionally characterized the protein. J.L. provided the initial clone and measured the protein oligomeric state. D.N.W. performed electron microscopy. A.B.W. and D.N.W. wrote the manuscript.
References
- 1.Stokes JL. Fermentation of glucose by suspensions of Escherichia coli. J. Bacteriol. 1949;57:147–158. doi: 10.1128/jb.57.2.147-158.1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sawers G. The hydrogenases and formate dehydrogenases of Escherichia coli. Antonie Van Leeuwenhoek. 1994;66:57–88. doi: 10.1007/BF00871633. [DOI] [PubMed] [Google Scholar]
- 3.Clegg S, Yu F, Griffiths L, Cole JA. The roles of the polytopic membrane proteins NarK, NarU and NirC in Escherichia coli K-12: two nitrate and three nitrite transporters. Mol Microbiol. 2002;44:143–155. doi: 10.1046/j.1365-2958.2002.02858.x. [DOI] [PubMed] [Google Scholar]
- 4.De Groote MA, et al. Genetic and redox determinants of nitric oxide cytotoxicity in a Salmonella typhimurium model. Proc Natl Acad Sci U S A. 1995;92:6399–6403. doi: 10.1073/pnas.92.14.6399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shiloh MU, et al. Phenotype of mice and macrophages deficient in both phagocyte oxidase and inducible nitric oxide synthase. Immunity. 1999;10:29–38. doi: 10.1016/s1074-7613(00)80004-7. [DOI] [PubMed] [Google Scholar]
- 6.Chakravortty D, Hensel M. Inducible nitric oxide synthase and control of intracellular bacterial pathogens. Microbes Infect. 2003;5:621–627. doi: 10.1016/s1286-4579(03)00096-0. [DOI] [PubMed] [Google Scholar]
- 7.Das P, Lahiri A, Chakravortty D. Novel role of the nitrite transporter NirC in Salmonella pathogenesis: SPI2-dependent suppression of inducible nitric oxide synthase in activated macrophages. Microbiology. 2009;155:2476–2489. doi: 10.1099/mic.0.029611-0. [DOI] [PubMed] [Google Scholar]
- 8.Saier MH, Jr, et al. Phylogenetic characterization of novel transport protein families revealed by genome analyses. Biochim Biophys Acta. 1999;1422:1–56. doi: 10.1016/s0304-4157(98)00023-9. [DOI] [PubMed] [Google Scholar]
- 9.Delomenie C, et al. A new homolog of FocA transporters identified in cadmium-resistant Euglena gracilis. Biochem Biophys Res Commun. 2007;358:455–461. doi: 10.1016/j.bbrc.2007.04.145. [DOI] [PubMed] [Google Scholar]
- 10.Sawers G, Bock A. Novel transcriptional control of the pyruvate formate-lyase gene: upstream regulatory sequences and multiple promoters regulate anaerobic expression. J Bacteriol. 1989;171:2485–2498. doi: 10.1128/jb.171.5.2485-2498.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suppmann B, Sawers G. Isolation and characterization of hypophosphite--resistant mutants of Escherichia coli: identification of the FocA protein, encoded by the pfl operon, as a putative formate transporter. Mol Microbiol. 1994;11:965–982. doi: 10.1111/j.1365-2958.1994.tb00375.x. [DOI] [PubMed] [Google Scholar]
- 12.Jia W, Tovell N, Clegg S, Trimmer M, Cole J. A single channel for nitrate uptake, nitrite export and nitrite uptake by Escherichia coli NarU and a role for NirC in nitrite export and uptake. Biochem J. 2009;417:297–304. doi: 10.1042/BJ20080746. [DOI] [PubMed] [Google Scholar]
- 13.Garty H, Rudy B, Karlish SJ. A simple and sensitive procedure for measuring isotope fluxes through ion-specific channels in heterogenous populations of membrane vesicles. J Biol Chem. 1983;258:13094–13099. [PubMed] [Google Scholar]
- 14.Middleton RE, Pheasant DJ, Miller C. Purification, reconstitution, and subunit composition of a voltage-gated chloride channel from Torpedo electroplax. Biochemistry. 1994;33:13189–13198. doi: 10.1021/bi00249a005. [DOI] [PubMed] [Google Scholar]
- 15.Walden M, et al. Uncoupling and turnover in a Cl−/H+ exchange transporter. J Gen Physiol. 2007;129:317–329. doi: 10.1085/jgp.200709756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Law CJ, Maloney PC, Wang DN. Ins and outs of major facilitator superfamily antiporters. Annu Rev Microbiol. 2008;62:289–305. doi: 10.1146/annurev.micro.61.080706.093329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murata K, et al. Structural determinants of water permeation through aquaporin-1. Nature. 2000;407:599–605. doi: 10.1038/35036519. [DOI] [PubMed] [Google Scholar]
- 18.Fu D, et al. Structure of a glycerol-conducting channel and the basis for its selectivity. Science. 2000;290:481–486. doi: 10.1126/science.290.5491.481. [DOI] [PubMed] [Google Scholar]
- 19.Sui H, Han BG, Lee JK, Walian P, Jap BK. Structural basis of water-specific transport through the AQP1 water channel. Nature. 2001;414:872–878. doi: 10.1038/414872a. [DOI] [PubMed] [Google Scholar]
- 20.Walz T, Smith BL, Zeidel ML, Engel A, Agre P. Biologically active two-dimensional crystals of aquaporin CHIP. J. Biol. Chem. 1994;269:1583–1586. [PubMed] [Google Scholar]
- 21.Stroud RM, et al. Glycerol facilitator GlpF and the associated aquaporin family of channels. Curr Opin Struct Biol. 2003;13:424–431. doi: 10.1016/s0959-440x(03)00114-3. [DOI] [PubMed] [Google Scholar]
- 22.Walz T, Fujiyoshi Y, Engel A. The AQP structure and functional implications. Handb Exp Pharmacol. 2009;31:31–56. doi: 10.1007/978-3-540-79885-9_2. [DOI] [PubMed] [Google Scholar]
- 23.Jiang J, Daniels BV, Fu D. Crystal structure of AqpZ tetramer reveals two distinct Arg-189 conformations associated with water permeation through the narrowest constriction of the water-conducting channel. J Biol Chem. 2006;281:454–460. doi: 10.1074/jbc.M508926200. [DOI] [PubMed] [Google Scholar]
- 24.Lawrence MC, Colman PM. Shape complementarity at protein/protein interfaces. J Mol Biol. 1993;234:946–950. doi: 10.1006/jmbi.1993.1648. [DOI] [PubMed] [Google Scholar]
- 25.Karpowich NK, Wang DN. Structural biology. Symmetric transporters for asymmetric transport. Science. 2008;321:781–782. doi: 10.1126/science.1161495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ashcroft F, Gadsby D, Miller C. Introduction. The blurred boundary between channels and transporters. Philos Trans R Soc Lond B Biol Sci. 2009;364:145–147. doi: 10.1098/rstb.2008.0245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou Y, MacKinnon R. The occupancy of ions in the K+ selectivity filter: charge balance and coupling of ion binding to a protein conformational change underlie high conduction rates. J Mol Biol. 2003;333:965–975. doi: 10.1016/j.jmb.2003.09.022. [DOI] [PubMed] [Google Scholar]
- 28.Garrett TP, Clingeleffer DJ, Guss JM, Rogers SJ, Freeman HC. The crystal structure of poplar apoplastocyanin at 1.8-Å resolution. The geometry of the copper-binding site is created by the polypeptide. J Biol Chem. 1984;259:2822–2825. doi: 10.2210/pdb2pcy/pdb. [DOI] [PubMed] [Google Scholar]
- 29.Schmiedekamp A, Nanda V. Metal-activated histidine carbon donor hydrogen bonds contribute to metalloprotein folding and function. J Inorg Biochem. 2009;103:1054–1060. doi: 10.1016/j.jinorgbio.2009.04.017. [DOI] [PubMed] [Google Scholar]
- 30.Miyazawa A, Fujiyoshi Y, Unwin N. Structure and gating mechanism of the acetylcholine receptor pore. Nature. 2003;423:949–955. doi: 10.1038/nature01748. [DOI] [PubMed] [Google Scholar]
- 31.Doyle DA, et al. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 32.Yernool D, Boudker O, Jin Y, Gouaux E. Structure of a glutamate transporter homologue from Pyrococcus horikoshii. Nature. 2004;431:811–818. doi: 10.1038/nature03018. [DOI] [PubMed] [Google Scholar]
- 33.Dutzler R, Campbell EB, MacKinnon R. Gating the selectivity filter in ClC chloride channels. Science. 2003;300:108–112. doi: 10.1126/science.1082708. [DOI] [PubMed] [Google Scholar]
- 34.Hunte C, et al. Structure of a Na+/H+ antiporter and insights into mechanism of action and regulation by pH. Nature. 2005;435:1197–1202. doi: 10.1038/nature03692. [DOI] [PubMed] [Google Scholar]
- 35.Mosser G, Mallouh V, Brisson A. A 9 Å two-dimensional projected structure of cholera toxin B-subunit-GM1 complexes determined by electron crystallography. J. Mol. Biol. 1992;226:23–28. doi: 10.1016/0022-2836(92)90120-9. [DOI] [PubMed] [Google Scholar]
- 36.Chang G, Spencer RH, Lee AT, Barclay MT, Rees DC. Structure of the MscL homolog from Mycobacterium tubeculosis: A gated mechanosensitive ion channel. Science. 1998;282:2220–2226. doi: 10.1126/science.282.5397.2220. [DOI] [PubMed] [Google Scholar]
- 37.Lunin VV, et al. Crystal structure of the CorA Mg2+ transporter. Nature. 2006;440:833–837. doi: 10.1038/nature04642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Unwin N. The structure of ion channels in membranes of excitable cells. Neuron. 1989;3:665–676. doi: 10.1016/0896-6273(89)90235-3. [DOI] [PubMed] [Google Scholar]
- 39.Khademi S, et al. Mechanism of ammonia transport by Amt/MEP/Rh: structure of AmtB at 1.35 Å. Science. 2004;305:1587–1594. doi: 10.1126/science.1101952. [DOI] [PubMed] [Google Scholar]
- 40.Zheng L, Kostrewa D, Berneche S, Winkler FK, Li XD. The mechanism of ammonia transport based on the crystal structure of AmtB of Escherichia coli. Proc Natl Acad Sci U S A. 2004;101:17090–17095. doi: 10.1073/pnas.0406475101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Levin EJ, Quick M, Zhou M. Crystal structure of a bacterial homologue of the kidney urea transporter. Nature. 2009 doi: 10.1038/nature08558. Advance online publication 28 October |. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Creighton TE. Proteins: Structures and molecular properties. New York: W.H. Freeman and Company; 1983. pp. 335–396. [Google Scholar]
- 43.Pusch M, Ludewig U, Rehfeldt A, Jentsch TJ. Gating of the voltage-dependent chloride channel CIC-0 by the permeant anion. Nature. 1995;373:527–531. doi: 10.1038/373527a0. [DOI] [PubMed] [Google Scholar]
- 44.Chen TY, Miller C. Nonequilibrium gating and voltage dependence of the ClC-0 Cl− channel. J Gen Physiol. 1996;108:237–250. doi: 10.1085/jgp.108.4.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tornroth-Horsefield S, et al. Structural mechanism of plant aquaporin gating. Nature. 2006;439:688–694. doi: 10.1038/nature04316. [DOI] [PubMed] [Google Scholar]
- 46.Fischer G, et al. Crystal structure of a yeast aquaporin at 1.15 angstrom reveals a novel gating mechanism. PLoS Biol. 2009;7:e1000130. doi: 10.1371/journal.pbio.1000130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smart OS, Neduvelil JG, Wang X, Wallace BA, Sansom MS. HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J Mol Graph. 1996;14:354–360. 376. doi: 10.1016/s0263-7855(97)00009-x. [DOI] [PubMed] [Google Scholar]
- 48.Auer M, et al. High-yield expression and functional analysis of Escherichia coli glycerol-3-phosphate transporter. Biochemistry. 2001;40:6628–6635. doi: 10.1021/bi010138+. [DOI] [PubMed] [Google Scholar]
- 49.Ward A, et al. Expression of prokaryotic membrane transport proteins in Escherichia coli. Biochem Soc Trans. 1999;27:893–899. doi: 10.1042/bst0270893. [DOI] [PubMed] [Google Scholar]
- 50.Miroux B, Walker JE. Over-production of proteins in Escherichia coli : mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J Mol Biol. 1996;260:289–298. doi: 10.1006/jmbi.1996.0399. [DOI] [PubMed] [Google Scholar]
- 51.Slotboom DJ, Duurkens RH, Olieman K, Erkens GB. Static light scattering to characterize membrane proteins in detergent solution. Methods. 2008;46:73–82. doi: 10.1016/j.ymeth.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 52.Kendrick BS, Kerwin BA, Chang BS, Philo JS. Online size-exclusion high-performance liquid chromatography light scattering and differential refractometry methods to determine degree of polymer conjugation to proteins and protein-protein or protein-ligand association states. Anal Biochem. 2001;299:136–146. doi: 10.1006/abio.2001.5411. [DOI] [PubMed] [Google Scholar]
- 53.Wang DN, Lemieux MJ, Boulter JM. Purification and characterization of transporter proteins from human erythrocyte membrane. Methods Mol Biol. 2003;228:239–255. doi: 10.1385/1-59259-400-X:239. [DOI] [PubMed] [Google Scholar]
- 54.Otwinowski Z, Miror W. Processing of X-ray diffraction data collected in oscillation mode. Meth. Enzym. 1997;276(Part A):307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 55.Adams PD, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 56.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 57.DeLano WL. The PyMOL User's Manual. San Carlos, CA: DeLano Scientific; 2002. p. 1. [Google Scholar]
- 58.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 59.Huang Y, Lemieux MJ, Song J, Auer M, Wang DN. Structure and mechanism of the glycerol-3-phosphate transporter from Escherichia coli. Science. 2003;301:616–620. doi: 10.1126/science.1087619. [DOI] [PubMed] [Google Scholar]
- 60.Quick M, et al. Binding of an octylglucoside detergent molecule in the second substrate (S2) site of LeuT establishes an inhibitor-bound conformation. Proc Natl Acad Sci U S A. 2009;106:5563–5568. doi: 10.1073/pnas.0811322106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.