Abstract
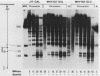
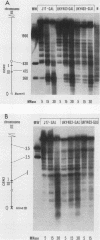
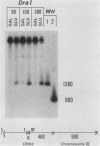
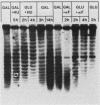
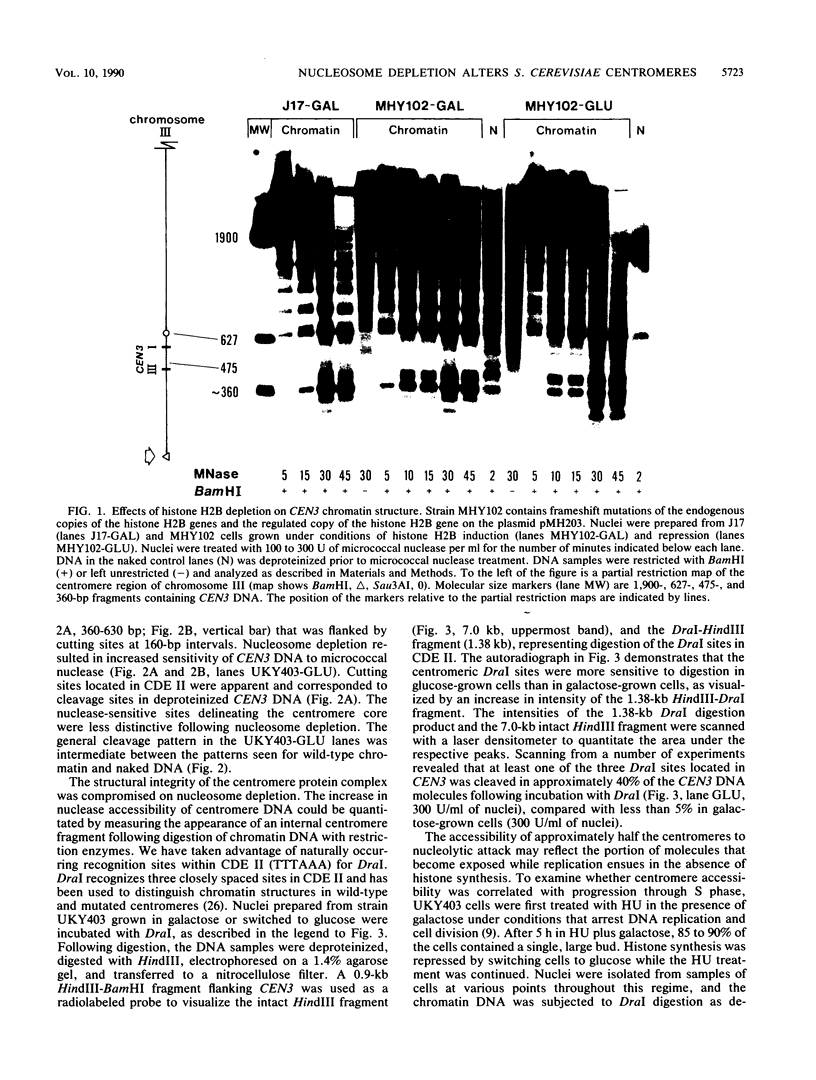
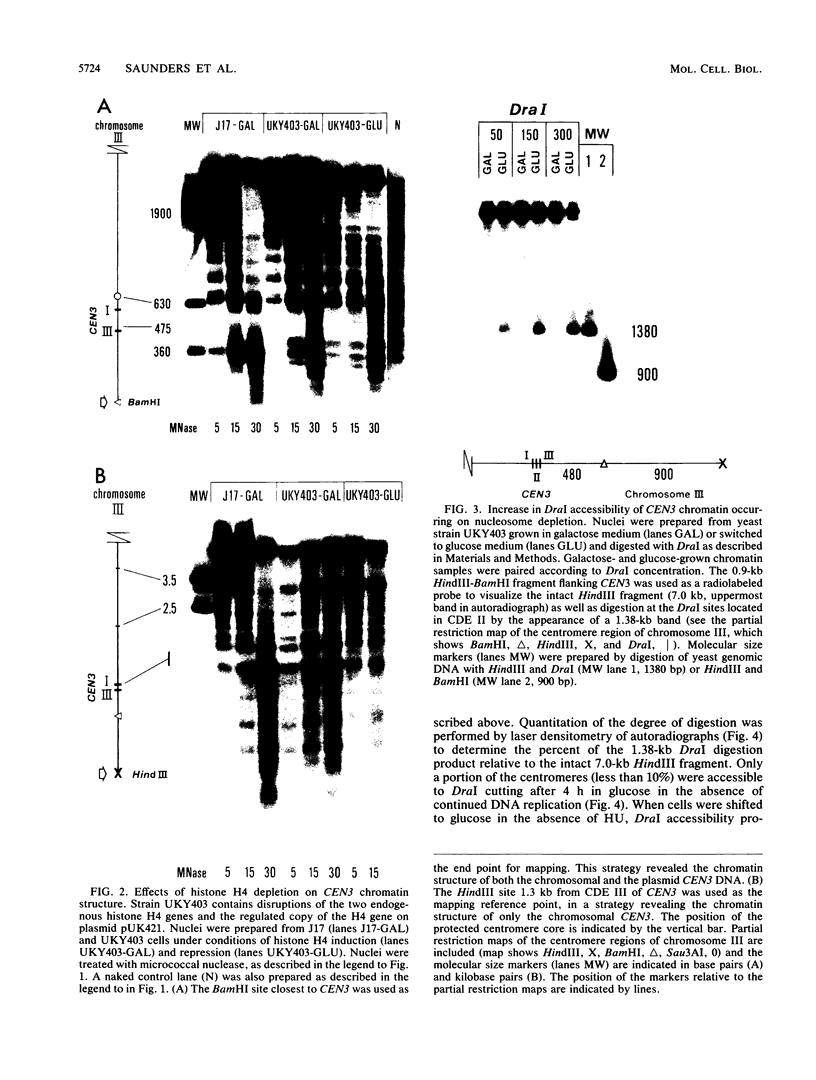
Saccharomyces cerevisiae centromeric DNA is packaged into a highly nuclease-resistant chromatin core of approximately 200 base pairs of DNA. The structure of the centromere in chromosome III is somewhat larger than a 160-base-pair nucleosomal core and encompasses the conserved centromere DNA elements (CDE I, II, and III). Extensive mutational analysis has revealed the sequence requirements for centromere function. Mutations affecting the segregation properties of centromeres also exhibit altered chromatin structures in vivo. Thus the structure, as delineated by nuclease digestion, correlated with functional centromeres. We have determined the contribution of histone proteins to this unique structural organization. Nucleosome depletion by repression of either histone H2B or H4 rendered the cell incapable of chromosome segregation. Histone repression resulted in increased nuclease sensitivity of centromere DNA, with up to 40% of CEN3 DNA molecules becoming accessible to nucleolytic attack. Nucleosome depletion also resulted in an alteration in the distribution of nuclease cutting sites in the DNA surrounding CEN3. These data provide the first indication that authentic nucleosomal subunits flank the centromere and suggest that nucleosomes may be the central core of the centromere itself.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bloom K. S., Amaya E., Carbon J., Clarke L., Hill A., Yeh E. Chromatin conformation of yeast centromeres. J Cell Biol. 1984 Nov;99(5):1559–1568. doi: 10.1083/jcb.99.5.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom K. S., Carbon J. Yeast centromere DNA is in a unique and highly ordered structure in chromosomes and small circular minichromosomes. Cell. 1982 Jun;29(2):305–317. doi: 10.1016/0092-8674(82)90147-7. [DOI] [PubMed] [Google Scholar]
- Bloom K., Kenna M., Saunders M. Cis- and trans-acting factors affecting the structure of yeast centromeres. J Cell Sci Suppl. 1989;12:231–242. doi: 10.1242/jcs.1989.supplement_12.19. [DOI] [PubMed] [Google Scholar]
- Clarke L., Carbon J. Isolation of a yeast centromere and construction of functional small circular chromosomes. Nature. 1980 Oct 9;287(5782):504–509. doi: 10.1038/287504a0. [DOI] [PubMed] [Google Scholar]
- Cumberledge S., Carbon J. Mutational analysis of meiotic and mitotic centromere function in Saccharomyces cerevisiae. Genetics. 1987 Oct;117(2):203–212. doi: 10.1093/genetics/117.2.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald-Hayes M., Clarke L., Carbon J. Nucleotide sequence comparisons and functional analysis of yeast centromere DNAs. Cell. 1982 May;29(1):235–244. doi: 10.1016/0092-8674(82)90108-8. [DOI] [PubMed] [Google Scholar]
- Funk M., Hegemann J. H., Philippsen P. Chromatin digestion with restriction endonucleases reveals 150-160 bp of protected DNA in the centromere of chromosome XIV in Saccharomyces cerevisiae. Mol Gen Genet. 1989 Oct;219(1-2):153–160. doi: 10.1007/BF00261171. [DOI] [PubMed] [Google Scholar]
- Gaudet A., Fitzgerald-Hayes M. Alterations in the adenine-plus-thymine-rich region of CEN3 affect centromere function in Saccharomyces cerevisiae. Mol Cell Biol. 1987 Jan;7(1):68–75. doi: 10.1128/mcb.7.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M., Chang M., Kim U. J., Grunstein M. Histone H2B repression causes cell-cycle-specific arrest in yeast: effects on chromosomal segregation, replication, and transcription. Cell. 1987 Feb 27;48(4):589–597. doi: 10.1016/0092-8674(87)90237-6. [DOI] [PubMed] [Google Scholar]
- Han M., Kim U. J., Kayne P., Grunstein M. Depletion of histone H4 and nucleosomes activates the PHO5 gene in Saccharomyces cerevisiae. EMBO J. 1988 Jul;7(7):2221–2228. doi: 10.1002/j.1460-2075.1988.tb03061.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hays T. S., Salmon E. D. Poleward force at the kinetochore in metaphase depends on the number of kinetochore microtubules. J Cell Biol. 1990 Feb;110(2):391–404. doi: 10.1083/jcb.110.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegemann J. H., Shero J. H., Cottarel G., Philippsen P., Hieter P. Mutational analysis of centromere DNA from chromosome VI of Saccharomyces cerevisiae. Mol Cell Biol. 1988 Jun;8(6):2523–2535. doi: 10.1128/mcb.8.6.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hieter P., Pridmore D., Hegemann J. H., Thomas M., Davis R. W., Philippsen P. Functional selection and analysis of yeast centromeric DNA. Cell. 1985 Oct;42(3):913–921. doi: 10.1016/0092-8674(85)90287-9. [DOI] [PubMed] [Google Scholar]
- Holm C., Stearns T., Botstein D. DNA topoisomerase II must act at mitosis to prevent nondisjunction and chromosome breakage. Mol Cell Biol. 1989 Jan;9(1):159–168. doi: 10.1128/mcb.9.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson V. In vivo studies on the dynamics of histone-DNA interaction: evidence for nucleosome dissolution during replication and transcription and a low level of dissolution independent of both. Biochemistry. 1990 Jan 23;29(3):719–731. doi: 10.1021/bi00455a019. [DOI] [PubMed] [Google Scholar]
- Kayne P. S., Kim U. J., Han M., Mullen J. R., Yoshizaki F., Grunstein M. Extremely conserved histone H4 N terminus is dispensable for growth but essential for repressing the silent mating loci in yeast. Cell. 1988 Oct 7;55(1):27–39. doi: 10.1016/0092-8674(88)90006-2. [DOI] [PubMed] [Google Scholar]
- Kenna M., Amaya E., Bloom K. Selective excision of the centromere chromatin complex from Saccharomyces cerevisiae. J Cell Biol. 1988 Jul;107(1):9–15. doi: 10.1083/jcb.107.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim U. J., Han M., Kayne P., Grunstein M. Effects of histone H4 depletion on the cell cycle and transcription of Saccharomyces cerevisiae. EMBO J. 1988 Jul;7(7):2211–2219. doi: 10.1002/j.1460-2075.1988.tb03060.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linxweller W., Hörz W. Reconstitution experiments show that sequence-specific histone-DNA interactions are the basis for nucleosome phasing on mouse satellite DNA. Cell. 1985 Aug;42(1):281–290. doi: 10.1016/s0092-8674(85)80123-9. [DOI] [PubMed] [Google Scholar]
- McGrew J., Diehl B., Fitzgerald-Hayes M. Single base-pair mutations in centromere element III cause aberrant chromosome segregation in Saccharomyces cerevisiae. Mol Cell Biol. 1986 Feb;6(2):530–538. doi: 10.1128/mcb.6.2.530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeill P. A., Berns M. W. Chromosome behavior after laser microirradiation of a single kinetochore in mitotic PtK2 cells. J Cell Biol. 1981 Mar;88(3):543–553. doi: 10.1083/jcb.88.3.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeks-Wagner D., Hartwell L. H. Normal stoichiometry of histone dimer sets is necessary for high fidelity of mitotic chromosome transmission. Cell. 1986 Jan 17;44(1):43–52. doi: 10.1016/0092-8674(86)90483-6. [DOI] [PubMed] [Google Scholar]
- Musich P. R., Brown F. L., Maio J. J. Nucleosome phasing and micrococcal nuclease cleavage of African green monkey component alpha DNA. Proc Natl Acad Sci U S A. 1982 Jan;79(1):118–122. doi: 10.1073/pnas.79.1.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes D. Nucleosome cores reconstituted from poly (dA-dT) and the octamer of histones. Nucleic Acids Res. 1979;6(5):1805–1816. doi: 10.1093/nar/6.5.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond T. J., Finch J. T., Rushton B., Rhodes D., Klug A. Structure of the nucleosome core particle at 7 A resolution. Nature. 1984 Oct 11;311(5986):532–537. doi: 10.1038/311532a0. [DOI] [PubMed] [Google Scholar]
- Saunders M., Fitzgerald-Hayes M., Bloom K. Chromatin structure of altered yeast centromeres. Proc Natl Acad Sci U S A. 1988 Jan;85(1):175–179. doi: 10.1073/pnas.85.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shure M., Pulleyblank D. E., Vinograd J. The problems of eukaryotic and prokaryotic DNA packaging and in vivo conformation posed by superhelix density heterogeneity. Nucleic Acids Res. 1977;4(5):1183–1205. doi: 10.1093/nar/4.5.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson R. T., Künzler P. Cromatin and core particles formed from the inner histones and synthetic polydeoxyribonucleotides of defined sequence. Nucleic Acids Res. 1979 Apr;6(4):1387–1415. doi: 10.1093/nar/6.4.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]