Abstract
During breast carcinoma progression, the three-dimensional (3D) microenvironment is continuously remodeled, and changes in the composition of the extracellular matrix (ECM) occur. High throughput screening platforms have been used to decipher the complexity of the microenvironment and to identify ECM components responsible for cancer progression. However, traditional screening platforms are typically limited to two-dimensional (2D) cultures, and often exclude the influence of ECM and stromal components. In this work, a system that integrates 3-dimensional cell culture techniques with an automated microfluidic platform was used to create a new ECM screening platform that cultures cells in more physiologically relevant 3D in vitro microenvironments containing stromal cells and different ECM molecules. This new ECM screening platform was used to culture T47D breast carcinoma cells in mono- and co-culture with human mammary fibroblasts (HMF) with seven combinations of three different ECM proteins (collagen, fibronectin, laminin). Differences in the morphology of T47D clusters, and the proliferation of T47D cells were found in ECM compositions rich in fibronectin or laminin. In addition, an MMP enzyme activity inhibition screening showed the capabilities of the platform for small molecule screening. The platform presented in this work enables screening for the effects of matrix and stromal compositions and show promises for providing new insights in the identification of key ECM components involved in breast cancer.
INTRODUCTION
The mammary gland is a dynamic tissue in which cells in the mammary epithelium continuously interact with cells in the surrounding microenvironment. When the microenvironment receives signals from cells in the mammary epithelium, it sends back cues that help to maintain normal mammary tissue functions. If these interactions are disturbed, changes in the morphology, differentiation, proliferation, and migration of cells occur that can ultimately lead to the formation of a tumor and its progression to malignancy. It is believed that the major contributors to these changes are genetic alterations within the epithelial cells. However, evidence shows that the extracellular matrix (ECM) composition can also influence these interactions.
The ECM is composed of different molecules with specialized properties that not only provide a physico-mechanical and geometrical scaffolding to cells, but also influence cell behavior1. Some of the major ECM proteins found in the mammary gland include collagens, fibronectin (FN), and laminin (LN). Type-I collagen (CN) is the major fibrillar component in the mammary gland and serves as a backbone that provides structural integrity to the mammary gland, whereas FN and LN regulate cell adhesion to the ECM. Therefore, the interactions between these ECM components and mammary epithelial cells are important for maintaining normal mammary gland tissue functions. In fact, previous studies have shown that luminal epithelial cells polarize, resemble acini structures similar to those seen in vivo, and express milk proteins in response to lactogenic hormones when cultured in a three-dimensional (3D), LN-rich ECM gel2. However, if luminal epithelial cells are cultured using traditional, 2-dimensional (2D) surfaces or CN gels lacking LN, the cells lose their polarity and their mammary-specific gene expression patterns change2. These results demonstrate that both the 3D microenvironment and the ECM composition play a critical role in guiding normal mammary tissue function.
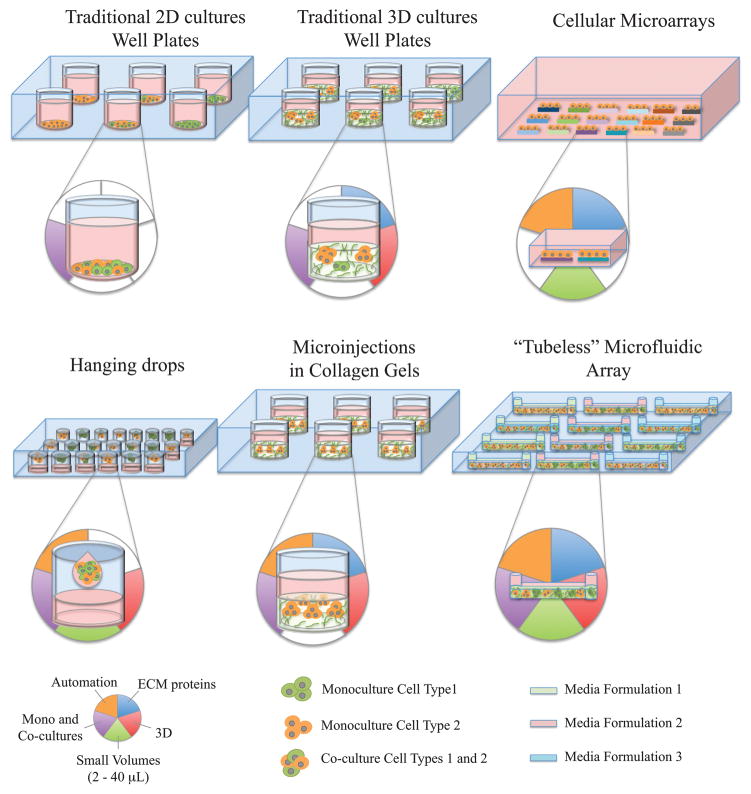
During breast cancer progression, the composition of the surrounding 3D microenvironment is continuously changed and represents a major challenge for identifying specific components and/or mechanisms. Traditional 96- and 384-well plates have shown to be useful for performing high-throughput screening (HTS) toxicology assays in cancer3–4. However, traditional well-plate screening platforms are typically limited to the 2D culture of cells and often exclude the influence of stromal cells and ECM molecules in modulating cellular behavior of cancer cells. Figure 1 shows some of the platforms that have been developed to address the limitations of the traditional 2D culture system. For example, three-dimensional cultures of cells in ECM proteins5–6 have shown to be valuable tools for providing cells with a more structurally appropriate context. However, the relatively large volumes of reagents required in these assays make them more expensive and limit their throughput capabilities. Cellular microarrays have been developed to increase the throughput capacity by depositing small spots of ECM molecules on a flat surface and growing cells on the ECM spots. Cellular microarrays have shown to be useful for studying the effect of the ECM composition in the maintenance of primary rat hepatocyte phenotype, and the differentiation of mouse embryonic stem cells7 and human mammary progenitor cells8. Multiple soluble formulations have also been included within cellular microarrays to examine the effect of growth factors in the growth and differentiation of embryonic stem cells9. However, multiple ECM spots are exposed to the same media formulation, and potential cross talk between spots can complicate the interpretation of results. Moreover, cellular microarrays are typically limited to the 2D culture of cells on top of ECM patterns, and do not represent the 3D microenvironment that is observed in vivo. Another approach that has been used for screening 3D cultures utilizes the hanging drop method, where spherical aggregates of cells are obtained in static or stirred suspension cultures10–11. The spherical aggregates have been used for testing anti-cancer drugs12, studying tumor cell biology13, and growing tumor cells and fibroblasts in co-cultures14. More recently, cell-polymer suspensions microinjected in collagen gels have been used to form 3D cell spheroids and visualize the distinct 3D migration of cells15. Although the hanging drop and microinjection methods have shown to be useful for screening monocultures and co-cultures in 3D, media is typically shared across the arrays such that results are confounded by soluble factor cross talk between array locations. Moreover, little work has been done to develop systems that include co-cultures with stromal cells as part of the 3D microenvironment. This is particularly important in breast cancer research since stromal fibroblasts play important roles in cancer development by modulating carcinoma cell proliferation both in vivo16 and in vitro17. Therefore, there is a need for more biologically relevant screening platforms that provide cancer cells with 3D microenvironments rich in ECM molecules and stromal cells, while providing independent experimental conditions.
Figure 1.
Currently available screening platforms and their capabilities to include ECM proteins (blue panel), perform 3D culture (red panel), utilize small volumes of cells and reagents (green panel), include monocultures and co-cultures within a single array (purple panel), and provide a fully automated loading (orange panel). Media formulations represent the different types of soluble formulations that can be added inside separate microchannels in a microchannel array.
In this study, an automated “tubeless” microfluidic screening platform previously developed for 3D cell culture18 was adapted to culture T47D breast carcinoma cells and human mammary fibroblasts (HMF) in 3D microenvironments and expanded to include different ECM molecules (CN, FN, LN) in the culture. The major advancements of the ECM automated microfluidic platform18 over the previously reported 3D microfluidic platform and traditional screening platforms include the ability to culture breast carcinoma cells in 3D microenvironments of different ECM compositions, the capacity of culturing monocultures and/or co-cultures, the ability of treating cells in separate microchannels with different soluble formulations, and the potential for performing small-molecule screenings. The platform screened for ECM compositions that affect: 1) the morphology of T47D breast carcinoma clusters, 2) the proliferation of T47D breast carcinoma cells, and 3) the enzyme activity inhibition of several matrix metalloproteinases (MMP). Applying the concepts presented in this work to higher throughput screening platforms will be useful for studying cell-ECM interactions in more physiologically relevant 3D in vitro microenvironments, identifying specific ECM proteins, and providing new insights on key mechanisms involved in breast cancer biology.
MATERIALS AND METHODS
Cell culture and ECM gels preparation
Human T47D breast carcinoma cells were cultured in flasks with low-glucose DMEM (1.0mg/mL, Gibco, Grand Island, NY), supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin (Invitrogen, Grand Island, NY). Human mammary fibroblasts immortalized with telomerase (HMFs) were cultured in high glucose DMEM (4.5mg/mL, Gibco), supplemented with 10% calf serum (CS) and 1% penicillin/streptomycin. Both cell lines were cultured in separate flasks inside a humidified incubator at 37°C and 5% CO2 before mixing with ECM gels and seeding in microchannels.
Extracellular matrix gels were prepared by mixing CN with FN or LN to get a total of seven different ECM compositions. FN (1mg/mL, human; BD Biosciences, Bedford, MA) and LN (1.88mg/mL, mouse; BD Biosciences) were reconstituted as specified by the manufacturer. A stock solution of CN (3.64 mg/mL, rat tail; BD Biosciences) was neutralized with a solution of 100mM HEPES buffer in 2X PBS in a 1:1 ratio, and incubated inside a bucket with ice for 10 minutes. Cells were re-suspended in serum-free DMEM (SF-DMEM). FN or LN were mixed with the neutralized CN, 1.5% (v/v) calf serum, and SF-DMEM to get final ECM concentrations of 1.3 mg/mL of CN with 0, 10, 50 or 100 μg/mL of FN or LN and a final cell density of 6 × 105 cells/mL (approximately 700 cells/microchannel). In co-culture experiments, HMF and T47D cells were added to the ECM gels in a 1:2 (HMF: T47D) ratio. 2D culture experiments were performed by coating microchannels with ECM proteins (prepared the same way as in 3D experiments) and incubating microchannels at 4°C for 2 hours. Microchannels were rinsed with PBS three times, cells were added in the microchannels, and incubated at 37°C afterwards. MMP activity was inhibited using the broad-spectrum inhibitor GM6001 (2.5mM, Millipore, Billerica, MA). GM6001 was diluted in culture media and added to the gels until a final concentration of 500nM was obtained. GM6001 was also added to cell culture media (500nM), and cultures were replaced every other day with fresh GM6001.
Tubeless microfluidic device fabrication and automated loading
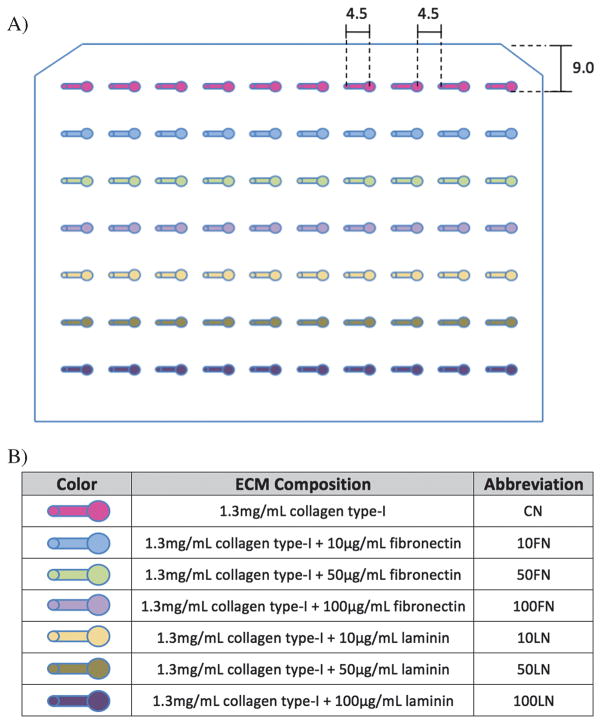
Polydimethylsiloxane (PDMS) tubeless microfluidic devices were fabricated as described previously18. The dimensions of the PDMS microchannels array (MCA) with straight microchannels (0.75 mm wide, 0.25 mm high, and 4.5 mm long) are shown in Figure 2.
Figure 2.
A) A microchannel array was used to culture T47D cells in monocultures (first 5 microchannels per row) and in co-cultures with human mammary fibroblasts (HMF, last 5 microchannels per row) with different ECM compositions. Microchannels dimensions are shown in millimeters. B) Seven different ECM compositions were used to culture T47D and HMF cells in the microchannel array.
The automated liquid handler used to load microchannels was optimized and described previously18. In this work, the same platform was used to culture T47D cells in different ECM compositions in the presence and absence of HMF cells. The ECM/cell mixtures were manually pipetted to seven wells of a 96-well plate and the automated platform was used to load microchannels as described previously18. Seven combinations of three different ECM proteins (CN, FN and LN) were used to culture T47D cells in monocultures (first 5 microchannels per row, Figure 2A) and in co-cultures with HMF cells (last 5 microchannels per row, Figure 2A). Between each loading with different ECM compositions, the probe was rinsed in a 50% DMSO and water solution, and washed with deionized water. The MCA included 5 replicates for each ECM/cell combination that resulted in a total of 70 microchannels. After the loading was done, the MCA was kept inside a 37°C incubator for seven days. Media changes were done every other day. The data discussed in this work comes from at least 2 separate MCA experiments.
Immunofluorescent staining
For the quantification of T47D cluster size, cells in the MCA were stained using the automated platform as described previously18. T47D and HMF cells were stained with primary antibodies against pan-cytokeratin (CK, 1:75 dilution ratio, mouse monoclonal antihuman pan-cytokeratin; LabVision, Fremont, CA), and vimentin (VM, 1:150 dilution ratio, rabbit polyclonal antihuman vimentin; LabVision, Fremont, CA). Secondary antibodies were added in a 1:150 dilution ratio (Alexa Fluor 594 goat antimouse; Alexa Fluor 488 goat antirabbit; Invitrogen, Carlsbad, CA). For counterstaining the nuclei, Hoechst 33342 was used at 20μg/mL (H3570; Invitrogen, Carlsbad, CA).
Image acquisition and analysis
Fluorescence imaging of T47D and HMF cells was performed on an inverted microscope (Eclipse Ti, Nikon Instruments, Melville, NY) using the NIS-Element imaging system (Diagnostic Instruments, Sterling Heights, MI). The high-throughput data analysis platform, JeXperiment (http://jexperiment.wikidot.com), was used to perform the microscopy image processing and data mining. The JeXperiment platform allowed importing data collected from each microchannel into a database, and managed the data processing for each microchannel with custom user algorithms or functions chosen from a library. Custom analysis algorithms were made to plug into the JeXperiment workflow and enabled the quantification of circularity, aspect ratio, cluster size, and total staining area of CK-positive clusters. Circularity was measured with the formula of 4π × area/perimeter2. Aspect ratio was defined as the ratio of major axis over minor axis. A rolling-ball background-subtraction algorithm was applied to determine a threshold value to obtain binary masks (Figure 3). The ImageJ (Rasband, W.S., ImageJ; U.S. National Institutes of Health, Bethesda, MD, http://rsb.info.nih.gov/ij/, 1997–2009) particle analyzer was applied to obtain the circularity, aspect ratio and size of T47D clusters. The bin sizes for the cluster size histograms were determined by dividing the range of cluster sizes identified using the automated image analysis software into 7 bins. This number provided a relevant number of clusters per bin, allowing good separation between conditions while having a sufficient number of clusters in each bin. Further, the minimum (369 μm2) and maximum (2583μm2) area values occurred while converting images (0.440 pixel/μm) from pixel areas (71 pixels-500 pixels) into micrometers squared. All data was analyzed using the pair-wise Wilcoxon rank sum test, and conditions significantly different (p<0.05) were used in the results and discussion.
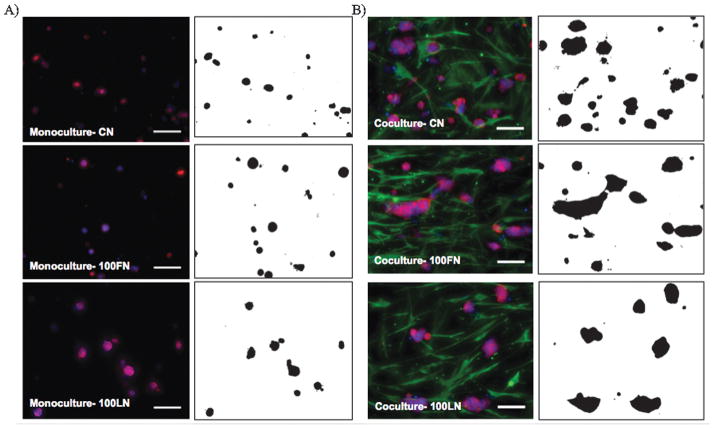
Figure 3.
Immunofluorescence images (left panels) of T47D cells in A) monocultures, and B) co-cultures of with HMF cells inside microchannels with 1.3mg/mL of collagen (CN), 1.3 mg/mL of collagen and 100 μg/mL of fibronectin (100FN), or 1.3 mg/mL of collagen and 100 μg/mL of laminin (100LN). Cells were cultured for 7 days, fixed, and T47D cells were stained against cytokeratin (red), HMF against vimentin (green), and nuclei with Hoechst dye (blue). Binary images (right panels) were produced with JeXperiment to analyze the morphology of T47D clusters. Scale bars represent 100 μm.
RESULTS AND DISCUSSION
The complexity of the 3D microenvironment and its constant remodeling during breast carcinoma progression represent a challenge for identifying ECM components and mechanisms involved in breast cancer. However, screening with different ECM compositions can help elucidate candidate microenvironmental components that support malignancy. An automated microfluidic platform previously developed for 3D cell culture18 was expanded in this work to include 3D microenvironments of different ECM composition, monocultures of T47D cells, and co-cultures of T47D and HMF cells. The platform is used to treat monocultures and co-cultures separately, and to screen for the effect of the 3D ECM composition on the phenotype, behavior, and proliferation of T47D cell clusters.
Figure 2A shows a representation of a 10 by 7 MCA used to culture T47D cells in monoculture (first 5 channels in a row) and in co-culture with HMF cells (last 5 channels in a row). ECM compositions contained CN as the major ECM protein, and FN and LN were mixed with CN to obtain seven different ECM compositions (see Figure 2B). Previously, the morphology19–20 and proliferation17, 21 of breast cancer cells showed to be useful readouts to investigate breast cancer cell behavior. The total cytokeratin (CK) and nuclei staining area of T47D clusters showed a correlation with T47D cell number and have been used as readouts for T47D cell growth18, 21. Moreover, traditional fluorescence microscopy can be used to examine T47D growth in 3D since T47D cells showed to grow evenly distributed along the horizontal and vertical dimensions of microchannels22. Therefore, the total staining area of T47D clusters and traditional 2D imaging are used here to examine the morphology and proliferation of T47D cell populations growing in the 3D gels.
Morphology of T47D clusters in different ECM compositions
Examining the morphology of breast cancer cells can provide important information. For example, 3D microenvironments have shown to affect the morphology and gene expression patterns of different breast cancer cell lines20. Also, the rigidity of the microenvironment affected the morphology of T47D cells, resulting in the down-regulation of Rho and FAK function23. More recently, changes in the circularity (Circ.) and aspect ratio (AR.) of MCF10DCIS.com cells were used as primary readouts for studying the transition from ductal carcinoma in situ (DCIS) to the invasive ductal carcinoma (IDC)19. In this work, we hypothesized that changes in the ECM composition would affect the morphology of T47D clusters. In order to test this hypothesis, the morphology of T47D clusters cultured in 3D microenvironments of different ECM compositions was examined in the presence and absence of HMF cells.
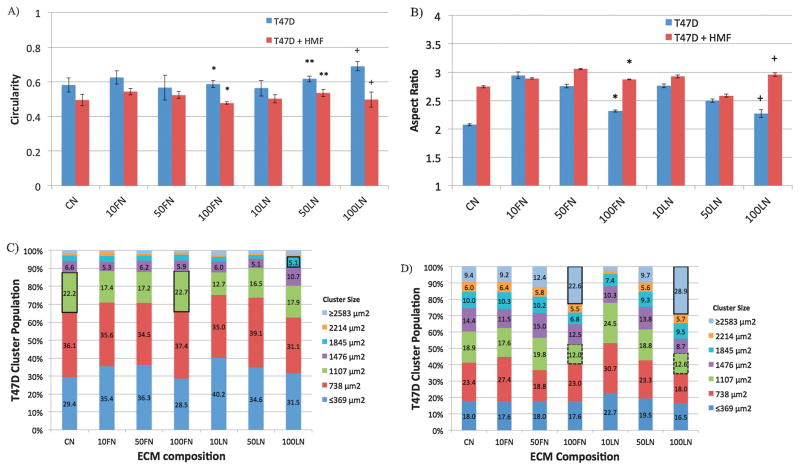
The morphology of T47D clusters was examined for different ECM compositions via immunofluorescence microscopy and binary mask image generation. Figure 3 shows immunofluorescence images of T47D cells in monocultures (left panel, Figure 3A), and in co-cultures with HMF cells (left panel, Figure 3B) inside microchannels. Binary mask images (right panels, Figure 3A and 3B) facilitated the automated analysis of T47D cluster morphology shown in Figure 3. Figures 4A and 4B show the circularity and aspect ratio of T47D clusters in monocultures (blue bars) and in co-cultures (red bars) with different ECM compositions. Circular clusters were defined as clusters with circularity (Circ.) or aspect ratio (AR.) values close to 1. As noticed in Figures 3A and 3B, in 1.3 mg/mL collagen type-I gels (CN), T47D cells formed bigger clusters in co-cultures compared to monocultures, which agreed with previously reported data21. However, in collagen gels containing 100 μg/mL of fibronectin (100FN), T47D clusters were bigger, but less circular (Circ. 0.48±0.01, *p<0.02, Figure 4A), and more elongated (AR. 2.87±0.01, *p<0.02, Figure 4B) in co-cultures than in monocultures (Circ. 0.58± 0.02, AR 2.31 ± 0.02, Figure 4A and 4B). Increasing laminin concentration in collagen type-I gels (e.g. 100LN) also increased the circularity of clusters in monocultures (Circ. 0.69±0.05, +p<0.05; AR 2.27±0.07, +p<0.02, Figures 4A and 4B) compared to co-cultures (Circ.0.50±0.04, +p<0.05, AR 2.95±0.03, +p<0.02, Figures 4A and 4B). T47D clusters in CN (Figure 3), 10FN, 50FN, 10LN or 50LN had similar morphologies between monocultures (Figure S1) and co-cultures (Figure S2), and no significant differences were found in the circularity or aspect ratio of T47D clusters (Figures 4A and 4B, p>0.05). Therefore, these results show that specific ECM compositions affected the morphology of T47D clusters, and suggest that T47D clusters became more elongated when co-cultured with HMF cells at high FN or LN concentrations (i.e. 100FN, 50LN or 100LN).
Figure 4.
The circularity (A), and aspect ratio (B) of T47D clusters were examined in monocultures (blue bars) and in co-cultures with HMF cells (red bars) for different ECM compositions after 7 days of culture inside microchannels, p<0.02, **p<0.05, +p<0.05, n=4. T47D cluster size population was examined in monocultures (C), and in co-cultures with HMF cells (D) under the effect of different ECM compositions. Black outlined boxes represent significant increases in clusters of the same size between different ECM compositions, p<0.05, n=4. Black dashed lines represent significant decreases in T47D cluster size between ECM compositions of the same cluster size, p<0.05, n=4.
Additionally, the size of T47D clusters was examined in monocultures and co-cultures under the influence of different ECM compositions. Figures 4C and 4D show the cluster size distribution of T47D cells cultured in different ECM compositions as monocultures (Figure 4C) or co-cultures with HMF cells (Figure 4D). The total population of T47D clusters was divided into 7 groups (represented by different colors) that included clusters of 369 μm2 in size to 2583 μm2. Clusters smaller than 369 μm2 or bigger than 2583 μm2 were also included, and statistical information about differences in cluster sizes between the different ECM compositions was analyzed (Figures S3 and S4). In monocultures (Figure 4C), no significant differences were found among different ECM compositions with small (i.e. ≤369 to 738 μm2) or big (2214 to ≥2583 μm2) clusters. However, mid-sized (1107 μm2) T47D clusters cultured with CN and 100FN displayed a modest increase in size (22.2%±0.2 CN, 22.7%±2.2 100FN, green outlined boxes, Figure 4C) compared to other ECM compositions (p<0.05 with 17.4%±1.0 10FN, 17.2%±1.0 50FN, 12.7%±1.8 10LN, and 16.5%±1.2 50LN). Similarly, 100LN also produced more clusters of size 1845 μm2 (5.1%±0.8, blue outlined box, Figure 4C) than 50LN (3.0%±0.6, Figure 3C, p<0.05). In co-cultures (Figure 4D), an increase in cluster size was observed across the board, and much stronger differences between ECM compositions were found, mostly in large cluster sizes (i.e. 1845 μm2 to ≥2583 μm2, Figure 4D). ECM compositions 100FN and 100LN displayed a depletion of mid size clusters around 1107 μm2 (12.0%±1.4 100FN, 12.6%±3.3 100LN, p<0.05, green outlined boxes, Figure 4D), largely counterbalanced by a significant increase in the number of large clusters. For example, by increasing the concentration of FN from 10FN to 100FN, the percentage of clusters sized ≥2583 μm2 was increased from 9.2%±2.1 to 22.6%±1.6 (blue outlined box, p<0.05, Figure 4D). The increase in the percentage of clusters larger than 2583 μm2 was even more significant with increments of LN concentration. 10LN had only 2.5%±0.9 of clusters with a size of 2583 μm2, whereas 100LN had 28.9%±4.3 (p<0.05, blue outlined box, Figure 4D). Moreover, the majority of clusters had a size of 738 μm2 at low LN concentrations (i.e. 10LN, 30.7%±2.9, Figure 4D), but as LN concentration was increased to 100LN, the highest population of T47D clusters was shifted to 2583 μm2 (28.9%±4.3, Figure 4D).
In summary, ECM compositions containing CN, 100FN or 100LN induced an increase from small to medium size T47D clusters in monocultures. These effects were compounded in co-culture conditions, as the ECM compositions containing 100FN and 100LN induced an increase of large T47D clusters. These results show that the ECM composition affects the 3D morphology and size of T47D clusters and suggest that ECM compositions (particularly 100FN and 100LN), could impact the proliferation of T47D cells differently in monocultures and in co-cultures with HMF cells.
Proliferation of T47D cells in different ECM compositions
Measuring the proliferation of breast cancer cells has shown to be useful for, predicting clinical response, providing a prognosis indicator, and studying stroma-to-carcinoma cell signaling. For example, a decrease in the proliferation index of tumor cells previously showed to be predictive of good clinical response24, and the proliferation of tumor cells in conjunction with tumor size, grade, nodal status, and steroid receptor status were used as useful prognostic indicators25. Also, a study of T47D breast carcinoma proliferation revealed that the overexpression of syndecan-1 (Sdc-1) in stromal fibroblasts stimulated T47D proliferation in vivo26 and in vitro27. More recently, a co-culture system of T47D and HMF cells in 3D co-culture systems was used to decipher specific mechanisms involved in T47D growth stimulation using traditional well-plates21 and microchannels28.
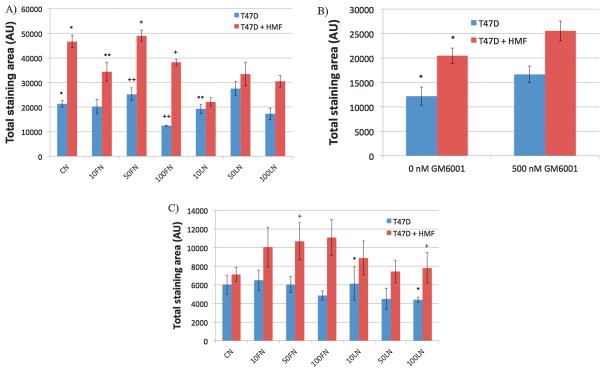
In this work, the culture of T47D cells was expanded with 3D microenvironments of different ECM compositions to screen for specific ECM compositions that affect T47D growth in monoculture and co-culture conditions. Figure 5A shows the screening results for monocultures of T47D cells (blue bars) and co-cultures of T47D and HMF cells (red bars) in 3D microenvironments of different ECM compositions. As expected, co-cultures in CN supported a 2.2-fold increase in T47D cell number compared to monocultures in CN (*p<0.02, Figure 5A), which agreed with previously reported data18, 21. In contrast, 2D cultures (cells on ECM-coated microchannels) only showed a 1.4-fold increase in T47D growth and were not able to support T47D growth as much as the 3D microenvironments (*p<0.05, Figure S5). Moreover, most differences in T47D growth in 2D cultures were found between monocultures and co-cultures, independently of the ECM composition (Figure S5). On the other hand, T47D cells were more sensitive to 3D microenvironments containing FN or low concentrations of LN (i.e. 10LN), but not to 3D microenvironments containing 50LN or 100LN. For example, a significant decrease in CK-positive area was observed between co-cultures containing collagen only (CN) and samples containing 10 μg/mL of FN (10FN) or 10 μg/mL of LN (10LN) (**p<0.05, Figure 5A). Interestingly, no effect was observed between monocultures in CN, and monocultures in 10FN or 10LN. Moreover, at higher FN concentrations (i.e. 50FN versus 100FN), the growth of T47D cells was reduced in both co-cultures (+p<0.02, Figure 5A), and monocultures (++p<0.02, Figure 5A). Therefore, monocultures needed higher doses of FN (i.e. 100FN) in order to decrease T47D cell growth significantly, whereas in co-cultures 10FN was sufficient to decrease the growth of T47D cells. Surprisingly, an inverse effect in T47D growth was noted in 2D cultures. In 2D cultures, increasing FN concentration in monocultures also increased T47D growth (10FN versus 50FN, *p<0.04, Figure S5). However, no differences were noticed in all other ECM compositions. In conclusion, 3D microenvironments influenced T47D proliferation more significantly than 2D cultures, and were able to inverse the effect of FN in 2D monocultures. In 3D microenvironments, FN affected T47D growth more than CN or LN microenvironments. These results highlight the fact that cells respond differently in 2D and 3D microenvironments, and suggest that FN could be an important ECM protein that interferes with paracrine signals between T47D and HMF cells that are necessary for supporting T47D growth.
Figure 5.
The proliferation of T47D cells was analyzed after 7 days of culture in monocultures (blue bars) and co-cultures with HMF cells (red bars) in different ECM compositions. Total CK-staining area represents the total cell number of T47D cells inside microchannels. Averages were calculated from at least 4 replicates, and 2 separate experiments. A) Screening for the influence of ECM composition in T47D proliferation. n=4, *p<0.02, **p<0.05 compared to CN co-cultures, +p<0.02, ++p<0.02. B) Screening for the effect of the GM6001 MMP inhibitor in HMF-induced T47D growth in CN-only gels. n=5, *p<0.05. C) Screening for the effect of the GM6001 MMP inhibitor in HMF-induced T47D growth in different ECM compositions, n=5, *p<0.02, +p<0.01.
Matrix metalloproteinase (MMP) activity inhibition in different ECM compositions
An MMP inhibitor was used to demonstrate the capability of the automated microfluidic platform to perform small molecule screenings. The broad-spectrum MMP inhibitor GM6001 was previously used to abolish HMF-induced T47D growth in CN-only gels28. In this study, the inhibition of HMF-induced T47D growth was screened for different ECM compositions in monocultures and in co-cultures with HMF cells. GM6001 (500nM) was added to the gels and to the cell culture medium, and GM6001-containing samples were loaded in the MCA. Figure 5B shows the total CK staining area of T47D cells cultured inside microchannels as monocultures (blue bars) or co-cultures with HMF cells (red bars) in CN-only gels. A significant increase in T47D growth was observed in co-cultures of T47D and HMF cells compared to T47D monocultures (*p<0.05, Figure 5B) when the inhibitor GM6001 was not added. However, no significant differences were found between monocultures and co-cultures in the presence of GM6001, which agreed with data reported previously28.
Figure 5C shows the effect of the MMP enzyme activity inhibitor in different ECM compositions. As expected, the stimulation of T47D cell growth by HMF cells was blocked in CN samples, and no significant differences were observed between monocultures and co-cultures (Figure 5C). Moreover, the increase of T47D cell growth by HMF cells was blocked by GM6001 in all the ECM compositions tested. This indicated that MMP activity was dominant over ECM composition to support breast cancer cell growth. However, in monocultures, 10LN supported T47D cell growth better than 100LN (*p<0.05, Figure 5C), and in co-cultures, FN increased T47D cell growth more significantly than compositions containing LN (e.g. 50FN versus 100LN, +p<0.02, Figure 5C). Therefore, although the ECM composition did not affect the growth of T47D cells between monocultures and co-cultures, significant differences in the growth of T47D cells within monocultures or co-cultures show that the ECM composition influences T47D growth even in the presence of a MMP inhibitor.
CONCLUSION
An automated microfludic platform for 3D cell culture18 was expanded in this work to culture cells in 3D microenvironments of different ECM compositions, and to screen for ECM compositions that influenced the morphology of T47D clusters, the proliferation of T47D cells, and the effect of a broad-spectrum MMP inhibitor in T47D growth. The morphology quantification revealed ECM-specific differences (particularly in 100FN and 100LN) in the circularity of T47D clusters between monocultures and co-cultures. Also, differences in the size-distribution of T47D clusters were found within co-cultures in CN and co-cultures containing 100FN or 100LN. These results suggested a compounded effect of the ECM composition and culture conditions on the proliferation of T47D cells. In fact, a proliferation screening showed that T47D cell growth decreased only in co-cultures containing 10μgmL of FN (10FN condition) and not in monocultures. Moreover, 3D microenvironments influenced T47D growth more significantly than 2D cultures, thus highlighting the importance of the 3D microenvironment. Finally, an MMP inhibition screening showed that although blocking the MMP activity reduced the growth of T47D cells in co-cultures in all the ECM compositions tested, FN still supported T47D growth better than LN.
The microfluidic platform presented in this work provides an information-rich in vitro assay that presents many advantages over current ECM screening platforms. First, culturing cells embedded in 3D microenvironments rich in both stromal cells and ECM molecules increased the biological relevance of the screening. Second, the small volumes of ECM proteins and cells required for loading each microchannel (approximately 2 μL per microchannel) allows for a cost-effective screening of ECM compositions when compared to the volumes required in traditional 3D cell culture assays (approximately 50 μL per well). Third, the enclosed compartments provided by microchannels allowed the individualized treatment of monocultures and co-cultures within a single MCA, and the analysis of paracrine interactions between T47D and HMF cells. Finally, an MMP inhibitor screening showed the capability of the platform to perform small molecule inhibitor screenings.
This new platform promises to be useful for advancing the development of more in vivo-like screening platforms. For example, screening in 3D cultures can provide relevant information about the performance of cancer drugs by using more biologically relevant 3D cultures. Also, the reduced amount of reagents required in this platform, and its ability to culture and treat cells in separate compartments can be expanded to incorporate different cancer cell lines (normal and malignant), primary cells, ECM molecules, and soluble formulations. Defined microenvironmental compositions within the MCA will also expedite the identification of important ECM molecules and mechanisms involved in cancer. Finally, increasing the number of microchannels in the MCA will provide a higher throughput analysis to further study the role of ECM and stromal components, and identify new drug targets in breast cancer.
Supplementary Material
Acknowledgments
This work was supported by the NIH grants R33CA137673, DOD/BRCP W81XWH-11-1-0208, DOD BCRP W81XWH-10-BCRP-CA, KRF-2008-220-D00133 and NLM5T15LM007359.
References
- 1.Lochter A, Bissell MJ. INVOLVEMENT OF EXTRACELLULAR-MATRIX CONSTITUENTS IN BREAST-CANCER. Semin Cancer Biol. 1995;6(3):165–173. doi: 10.1006/scbi.1995.0017. [DOI] [PubMed] [Google Scholar]
- 2.Barcelloshoff MH, Aggeler J, Ram TG, Bissell MJ. Functional-Differentiation and Alveolar Morphogenesis of Primary Mammary Cultures on Reconstituted Basement-Membrane. Development. 1989;105(2):223. doi: 10.1242/dev.105.2.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mueller H, Kassack MU, Wiese M. Comparison of the usefulness of the MTT, ATP, and calcein assays to predict the potency of cytotoxic agents in various human cancer cell lines. J Biomol Screen. 2004;9(6):506–515. doi: 10.1177/1087057104265386. [DOI] [PubMed] [Google Scholar]
- 4.Kasibhatla S, Gourdeau H, Meerovitch K, Drewe J, Reddy S, Oiu L, Zhang H, Bergeron F, Bouffard D, Yang Q, Herich J, Lamothe S, Cai SX, Tseng B. Discovery and mechanism of action of a novel series of apoptosis inducers with potential vascular targeting activity. Mol Cancer Ther. 2004;3(11):1365–1373. [PubMed] [Google Scholar]
- 5.Debnath J, Brugge JS. Modelling glandular epithelial cancers in three-dimensional cultures. Nature reviews. Cancer. 2005;5(9):675–88. doi: 10.1038/nrc1695. [DOI] [PubMed] [Google Scholar]
- 6.Lee GY, Kenny PA, Lee EH, Bissell MJ. Three-dimensional culture models of normal and malignant breast epithelial cells. Nat Methods. 2007;4(4):359–65. doi: 10.1038/nmeth1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flaim CJ, Chien S, Bhatia SN. An extracellular matrix microarray for probing cellular differentiation. Nature Methods. 2005;2(2):119–125. doi: 10.1038/nmeth736. [DOI] [PubMed] [Google Scholar]
- 8.LaBarge MA, Nelson CM, Villadsen R, Fridriksdottir A, Ruth JR, Stampfer MR, Petersen OW, Bissell MJ. Human mammary progenitor cell fate decisions are products of interactions with combinatorial microenvironments. Integrative Biology. 2009;1(1):70–79. doi: 10.1039/b816472j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flaim CJ, Teng D, Chien S, Bhatia SN. Combinatorial signaling microenvironments for studying stem cell fate. Stem Cells and Development. 2008;17(1):29–39. doi: 10.1089/scd.2007.0085. [DOI] [PubMed] [Google Scholar]
- 10.Kunz-Schughart LA, Freyer JP, Hofstaedter F, Ebner R. The use of 3-D cultures for high-throughput screening: The multicellular spheroid model. J Biomol Screen. 2004;9(4):273–285. doi: 10.1177/1087057104265040. [DOI] [PubMed] [Google Scholar]
- 11.Hirschhaeuser F, Menne H, Dittfeld C, West J, Mueller-Klieser W, Kunz-Schughart LA. Multicellular tumor spheroids: An underestimated tool is catching up again. J Biotechnol. 2010;148(1):3–15. doi: 10.1016/j.jbiotec.2010.01.012. [DOI] [PubMed] [Google Scholar]
- 12.Tung YC, Hsiao AY, Allen SG, Torisawa YS, Ho M, Takayama S. High-throughput 3D spheroid culture and drug testing using a 384 hanging drop array. Analyst. 2011;136(3):473–478. doi: 10.1039/c0an00609b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kunz-Schughart LA, Kreutz M, Knuechel R. Multicellular spheroids: a three-dimensional in vitro culture system to study tumour biology. Int J Exp Pathol. 1998;79(1):1–23. doi: 10.1046/j.1365-2613.1998.00051.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoffmann TK, Schirlau K, Sonkoly E, Brandau S, Lang S, Pivarcsi A, Balz V, Muller A, Homey B, Boelke E, Reichert T, Friebe-Hoffmann U, Greve J, Schuler P, Scheckenbach K, Schipper J, Bas M, Whiteside TL, Bier H. A novel mechanism for anti-EGFR antibody action involves chemokine-mediated leukocyte infiltration. Int J Cancer. 2009;124(11):2589–2596. doi: 10.1002/ijc.24269. [DOI] [PubMed] [Google Scholar]
- 15.Truong HH, de Sonneville J, Ghotra VPS, Xiong JL, Price L, Hogendoorn PCW, Spaink HH, van de Water B, Danen EHJ. Automated microinjection of cell-polymer suspensions in 3D ECM scaffolds for high-throughput quantitative cancer invasion screens. Biomaterials. 2012;33(1):181–188. doi: 10.1016/j.biomaterials.2011.09.049. [DOI] [PubMed] [Google Scholar]
- 16.Broutyboye D, Raux H. Differential Influence of Stromal Fibroblasts from Different Breast Tissues on Human Breast-Tumor Cell-Growth in Nude-Mice. Anticancer Res. 1993;13(4):1087–1090. [PubMed] [Google Scholar]
- 17.Sadlonova A, Novak Z, Johnson MR, Bowe DB, Gault SR, Page GP, Thottassery JV, Welch DR, Frost AR. Breast fibroblasts modulate epithelial cell proliferation in three-dimensional in vitro co-culture. Breast Cancer Res. 2005;7(1):R46–R59. doi: 10.1186/bcr949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Montanez-Sauri SI, Sung KE, Puccinelli JP, Pehlke C, Beebe DJ. Automation of three-dimensional cell culture in arrayed microfluidic devices. J Lab Autom. 2011;16(3):171–85. doi: 10.1016/j.jala.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sung KE, Yang N, Pehlke C, Keely PJ, Eliceiri KW, Friedl A, Beebe DJ. Transition to invasion in breast cancer: a microfluidic in vitro model enables examination of spatial and temporal effects. Integr Biol (Camb) doi: 10.1039/c0ib00063a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kenny PA, Lee GY, Myers CA, Neve RM, Semeiks JR, Spellman PT, Lorenz K, Lee EH, Barcellos-Hoff MH, Petersen OW, Gray JW, Bissell MJ. The morphologies of breast cancer cell lines in three-dimensional assays correlate with their profiles of gene expression. Molecular Oncology. 2007;1(1):84–96. doi: 10.1016/j.molonc.2007.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Su G, Blaine SA, Qiao DH, Friedl A. Shedding of syndecan-1 by stromal fibroblasts stimulates human breast cancer cell proliferation via FGF2 activation. Journal of Biological Chemistry. 2007;282(20):14906–14915. doi: 10.1074/jbc.M611739200. [DOI] [PubMed] [Google Scholar]
- 22.Su G, Sung KE, Beebe DJ, Friedl A. Functional Screen of Paracrine Signals in Breast Carcinoma Fibroblasts. Plos One. 2012;7(10) doi: 10.1371/journal.pone.0046685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wozniak MA, Desai R, Solski PA, Der CJ, Keely PJ. ROCK-generated contractility regulates breast epithelial cell differentiation in response to the physical properties of a three-dimensional collagen matrix. Journal of Cell Biology. 2003;163(3):583–595. doi: 10.1083/jcb.200305010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang J, Powles TJ, Allred DC, Ashey SE, Clark GM, Makris A, Assersohn L, Gregory RK, Osborne CK, Dowsett M. Biologic markers as predictors of clinical outcome from systemic therapy for primary operable breast cancer. J Clin Oncol. 1999;17(10):3058–3063. doi: 10.1200/JCO.1999.17.10.3058. [DOI] [PubMed] [Google Scholar]
- 25.Beresford MJ, Wilson GD, Makris A. Measuring proliferation in breast cancer: practicalities and applications. Breast Cancer Res. 2006;8(6) doi: 10.1186/bcr1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maeda T, Desouky J, Friedl A. Syndecan-1 expression by stromal fibroblasts promotes breast carcinoma growth in vivo and stimulates tumor angiogenesis. Oncogene. 2006;25(9):1408–12. doi: 10.1038/sj.onc.1209168. [DOI] [PubMed] [Google Scholar]
- 27.Maeda T, Alexander CM, Friedl A. Induction of syndecan-1 expression in stromal fibroblasts promotes proliferation of human breast cancer cells. Cancer Research. 2004;64(2):612–621. doi: 10.1158/0008-5472.can-03-2439. [DOI] [PubMed] [Google Scholar]
- 28.Bauer M, Su G, Beebe DJ, Friedl A. 3D microchannel co-culture: method and biological validation. Integrative Biology. 2(7–8):371–378. doi: 10.1039/c0ib00001a. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.