A simple technique permits fluorescent proteins to be imaged in resin-embedded plant material.
Abstract
Fluorescent proteins (FPs) were developed for live-cell imaging and have revolutionized cell biology. However, not all plant tissues are accessible to live imaging using confocal microscopy, necessitating alternative approaches for protein localization. An example is the phloem, a tissue embedded deep within plant organs and sensitive to damage. To facilitate accurate localization of FPs within recalcitrant tissues, we developed a simple method for retaining FPs after resin embedding. This method is based on low-temperature fixation and dehydration, followed by embedding in London Resin White, and avoids the need for cryosections. We show that a palette of FPs can be localized in plant tissues while retaining good structural cell preservation, and that the polymerized block face can be counterstained with cell wall probes. Using this method we have been able to image green fluorescent protein-labeled plasmodesmata to a depth of more than 40 μm beneath the resin surface. Using correlative light and electron microscopy of the phloem, we were able to locate the same FP-labeled sieve elements in semithin and ultrathin sections. Sections were amenable to antibody labeling, and allowed a combination of confocal and superresolution imaging (three-dimensional-structured illumination microscopy) on the same cells. These correlative imaging methods should find several uses in plant cell biology.
The localization of fluorescent proteins (FPs) in cells and tissues has become one of the major tools in cell biology (Tsien, 1998; Shaner et al., 2005). Advances in confocal microscopy have meant that many proteins can be tagged with appropriate fluorescent markers and tracked as they move within and between cells (Chapman et al., 2005). Additional approaches involving photobleaching and photoactivation of FPs have opened up new avenues for exploring protein dynamics and turnover within cells (Lippincott-Schwartz et al., 2003). However, not all cells are amenable to live-cell imaging, which in plants is usually restricted to surface cells such as the leaf epidermis. An example is the phloem. The delicate nature of sieve elements and companion cells, which are under substantial hydrostatic pressure, has made studies of the fine structure of these cells particularly difficult (Knoblauch and van Bel, 1998). Despite this, significant advances have been made in imaging the phloem through inventive use of imaging protocols that allow living sieve elements to be observed as they translocate assimilates (for review, see Knoblauch and Oparka, 2012). However, determining the precise localization of the plethora of proteins located within the sieve element (SE)-companion cell (CC) complex remains a technical challenge. The phloem is the conduit for long-distance movement of macromolecules in plants, including viral genomes. For several viruses, the entry into the SE-CC complex is a crucial step that determines the capacity for long-distance movement. Identifying the cell types within the phloem that restrict the movement of some viruses is technically challenging due to the small size of phloem cells and their location deep within plant organs (Nelson and van Bel, 1998).
The problems associated with imaging proteins in phloem tissues prompted us to explore methods for retaining the fluorescence of tagged proteins within tissues not normally amenable to confocal imaging. Previously, we used superresolution imaging techniques on fixed phloem tissues sectioned on a Vibroslice, providing information on the association between a viral movement protein (MP) and plasmodesmata (PD) within the SE-CC complex (Fitzgibbon et al., 2010). However, we wished to explore the same cells using correlative light and electron microscopy (CLEM), necessitating the development of methods that would allow sequential imaging of cells using fluorescence microscopy and transmission electron microscopy (TEM). To this end, we developed a protocol that retains fluorescent proteins through aldehyde fixation and resin embedding.
In the last 10 years there has been significant interest in imaging fluorescent proteins in semithin sections (for review, see Cortese et al., 2009). Luby-Phelps and colleagues (2003) first described a method for retaining GFP fluorescence after fixation and resin embedding, but their method has not seen widespread application. The advent of superresolution imaging techniques (for review, see Bell and Oparka, 2011) has stimulated considerable interest in this field as the retention of fluorescence in thin sections means that cells can be imaged using techniques such as photoactivation light microscopy and stochastic optical reconstruction microscopy, allowing a lateral resolution of less than 10 nm to be achieved (Subach et al., 2009; Xu et al., 2012). A number of studies have described CLEM on the same cells (Luby-Phelps et al., 2003; Betzig et al., 2006; Watanabe et al., 2011). Advances in this field were reviewed recently (Jahn et al., 2012; see contributions in Muller-Reichert and Verkade, 2012). For example, Pfeiffer et al. (2003) were able to image SEs and CCs using high-pressure freezing, followed by freeze substitution in acetone and resin embedding. They then used thick optical sections of the tissue to locate cells of interest, and these were subsequently imaged using TEM. However, there have been few attempts to retain FPs in resin-embedded plant tissues. Thompson and Wolniak (2008) described the retention of mCitrine fused to an SE-plasma membrane protein in glycol methacrylate sections. The fluorescent signal was stable using wide-field microscopy but bleached rapidly under the confocal microscope.
To date, cryosections have been the preferred choice for CLEM in mammalian tissues (Watanabe et al., 2011). Recently, Lee et al. (2011) chemically fixed Arabidopsis (Arabidopsis thaliana) seedlings, cut 50-μm sections, and examined these with a confocal microscope. After confocal mapping the sections were embedded in resin and thin sectioned. These authors were able to locate the same PD pit fields using confocal and TEM, providing important information on the localization of a novel PD protein. As general rule, cryosectioning is a time-consuming process, and subcellular details may be obscured in cryosections because of poor tissue contrast (Watanabe et al., 2011). A major problem with imaging FPs in resin sections has been that GFP and its derivatives are quenched by the acidic, oxidizing conditions required for fixation, dehydration, and embedding of delicate specimens (Tsien, 1998; Keene et al., 2008). Recently, however, Watanabe et al. (2011) explored the retention of FPs in Caenorhabditis elegans cells after fixation by different aldehydes and embedding media. These authors tested a range of resins and found that Citrine and tandem dimer Eos (tdEos) could be retained in methacrylate plastic sections. This material was difficult to cut thinly (<70 nm) compared to epoxy-based resins, but the authors obtained valuable correlative images using stimulated emission depletion microscopy and photoactivation light microscopy followed by low-voltage scanning electron microscopy.
Because the retention of fluorescent proteins may differ between plant and animal cells, we explored a number of approaches for retaining fluorescent proteins in resin. Using low-temperature conditions (<8°C) during fixation and dehydration, we could retain strong fluorescence prior to tissue embedding. We also explored different embedding media and found that tissue could be effectively polymerized in London Resin (LR) White while retaining sufficient fluorescence for confocal imaging. Using water-dipping lenses, we were able to detect fluorescent proteins in optical sections up to 40 μm below the surface of the block face. Ultrathin sections from the same blocks showed good structural preservation and allowed CLEM. Subsequently, we cut 1- to 2-μm sections and examined these using confocal microscopy and three-dimensional-structured illumination microscopy (3D-SIM). Sections could be counterstained with a number of conventional fluorophores and antibodies, allowing colocalization studies. These simple methods allow successive imaging of FPs with the light and electron microscope, combining the strengths of both imaging platforms. We believe this approach will have significant utility for tissues that are recalcitrant to conventional confocal imaging.
RESULTS
Fixation and Dehydration
Because a variety of methods have been used to retain FPs in semithin sections, we conducted a series of tests on tobacco (Nicotiana tabacum) and Arabidopsis plants expressing different FPs. For these tests we used a transgenic tobacco line in which HDEL:GFP was expressed under the SEO2 promoter (Knoblauch and Peters, 2010), which is active only in sieve elements. In this line, GFP is targeted to the sieve element reticulum (SER), a specialized form of endoplasmic reticulum that exists as an anastomosing network of tubules and parietal stacked aggregates (Knoblauch and Peters, 2010). This line has discrete GFP fluorescence readily visible in freehand sections (Fig. 1A). This signal was monitored through fixation and embedding and allowed fluorescence levels to be assessed during optimization of the method. Previously we showed that GFP fluorescence, antigenicity, and structural integrity were well preserved in tissue fixed using a combination of 4% paraformaldehyde and 0.25% glutaraldehyde (Fitzgibbon et al., 2010). The aim here was to preserve the tissue sufficiently so that it could withstand the rigors of both light and electron microscopy. As expected, we found that fixation and dehydration at room temperature eliminated fluorescence before the samples were embedded (Keene et al., 2008). However, fixation and dehydration at low temperature (<8°C) successfully retained fluorescence. We were able to increase the glutaraldehyde concentration as high as 2%, while still retaining good fluorescence preservation. However, we noticed a concomitant increase in the autofluorescence of the tissues, particularly the xylem, at this higher glutaraldehyde concentration. However, the fluorescent signal from GFP was easily visible above background (Fig. 1B). To limit background autofluorescence during processing we included dithiothreitol (DTT) during dehydration and infiltration. When used in combination with low temperature processing, DTT reduces background autofluorescence (Brown et al., 1989), preserves antigenicity during chemical fixation (Baskin et al., 1992, 1996), and may prevent quenching of fluorescent proteins (Thompson and Wolniak, 2008).
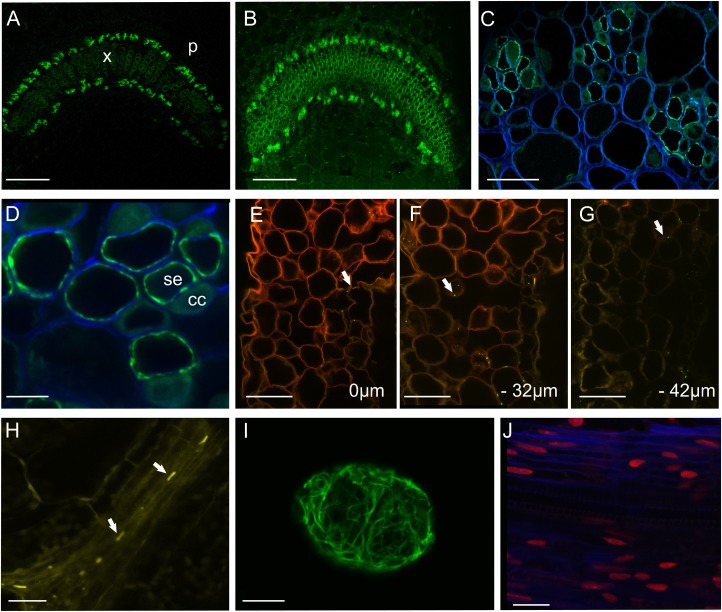
Figure 1.
En bloc imaging of FPs using confocal microscopy. A, Unprocessed, free-hand section of a tobacco petiole expressing pSEO2.HDEL:GFP (shown in green; Knoblauch and Peters, 2010). In this construct, GFP highlights the SER but at this magnification reveals general fluorescence from phloem bundles. p, Phloem, x, xylem. Scale = 600 μm. B, Petiole expressing pSEO2.HDEL:GFP imaged in a polymerized block of LR. Scale = 600 μm. C, An embedded petiole expressing pSEO2.HDEL:GFP imaged with a 63× water-dipping lens. The SER is clearly visible at this magnification. Cell walls (blue) were highlighted with calcofluor white, which was added directly to the block face. Scale = 40 μm. D, A region of the phloem at higher magnification. SEs (se) show conspicuous labeling of the SER, while CCs (cc) show background autofluorescence. Scale = 10 μm. E–G, Imaging of an Arabidopsis line expressing a viral movement protein fused to GFP (MP17:GFP; Vogel et al., 2007). GFP signal is evident from plasmodesmata (arrow) in mesophyll cells of the leaf. Cell walls were counterstained en bloc with propidium iodide (red). The block was optically sectioned and images captured at the block surface (0 μm; E), at −32 µm (F), and at −42 µm (G) below the block surface. Note that GFP fluorescence from PD is apparent to a depth greater than the penetration of the propidium iodide stain. Scale = 50 μm. H, En bloc imaging of SEOR1 protein (arrows) tagged with YFP (see Froelich et al., 2011) in the phloem of the midvein of an Arabidopsis leaf. Scale = 25 μm. I, En bloc reconstruction of a viral X-body produced by a PVX vector modified to express GFP fused to its coat protein (CP:GFP; Santa-Cruz et al.,1996). Scale = 25 μm. J, Nuclei in the hypocotyl of Arabidopsis expressing a histone 2B fused to RFP (H2B:RFP; Federici et al., 2012). Cell walls were counterstained with calcofluor. Scale = 25 μm.
Resin Embedding
We tested a number of resins including LR White, methacrylate, and Durcupan, a water-miscible resin. Following dehydration, we attempted low-temperature, ultraviolet-, and heat-polymerizing protocols. Material embedded in Durcupan did not section well in our hands. Methacrylate retained fluorescence well after low-temperature/ultraviolet polymerization, but significant tissue collapse was evident (data not shown). Our best results were obtained by polymerizing samples at 50°C in LR White following low-temperature fixation and dehydration. We monitored loss of fluorescence from tissue sections using ImageJ software and found a 27.5% (±6, n = 11) loss of fluorescence relative to fresh tissue during fixation and embedding. We deemed this to be an acceptable loss and pursued optimization of subsequent steps. It is likely that plant tissues respond differently to fixatives and embedding media. However, the protocol detailed in the methods section was suitable for most of the tissues we examined.
Imaging FPs en Bloc and in Semithin Sections
We showed previously that plant cell walls can be imaged successfully en bloc following polymerization of tissues in Araldite (Prior et al., 1999). After fixation and embedding in LR White, we imaged the polymerized block face using confocal microscopy. In tobacco phloem tissues expressing HDEL:GFP, we could detect fluorescent phloem bundles en bloc at magnifications as low as 5× (Fig. 1B). We were able to counterstain the cell walls en bloc by adding 10 µg mL−1 calcofluor white (Hahne et al., 1983) or 1 µg mL−1 propidium iodide (Pighin et al., 2004) directly to the block face as droplets. Using a 63× lens, we obtained a strong GFP signal from the SER while the calcofluor staining clearly delineated the cell walls (Fig. 1C). We found that glutaraldehyde fixation caused a faint background autofluorescence from the cytoplasm, allowing CCs to be identified (Fig. 1D). When we used propidium iodide as a wall stain, we found that the cell walls became labeled to a depth of more than 30 μm into the tissue, allowing deep confocal imaging using water-dipping lenses. In a transgenic line expressing a viral MP fused to GFP (MP17-GFP; Vogel et al., 2007), we were able to image fluorescent PD in leaf mesophyll cells to a depth of more than 40 μm into the resin block (Fig. 1E–G). At this depth, the propidium iodide signal had faded significantly, but the MP17-GFP signal remained strong (Fig. 1G).
SEs in Arabidopsis are extremely small (Mullendore et al., 2010), making SE substructures difficult to detect in semithin sections. Using an Arabidopsis line expressing yellow fluorescent protein (YFP) fused to the sieve-element occlusion related (SEOR1) protein (Froelich et al., 2011), we were able to image phloem protein bodies within individual sieve elements en bloc (Fig. 1H). Next we embedded tobacco leaf petioles infected with a Potato Virus X (PVX) vector in which GFP is fused to the viral coat protein (PVX.CP-GFP; Santa Cruz et al., 1996). In this virus, the GFP forms a virion “overcoat,” allowing the virus to be tracked as it moves. As expected, we found large aggregates of virus particles associated with the viral X-bodies, structures that harbor a range of viral and host components (Tilsner et al., 2012). Using en bloc imaging we could reconstruct individual X-bodies using optical sectioning and reconstruction (Fig. 1I). We also embedded an Arabidopsis line expressing a histone 2B-red fluorescent protein (RFP) fusion (Federici et al., 2012). Here we were able to image RFP-labeled nuclei in resin sections of the hypocotyl (Fig. 1J).
CLEM
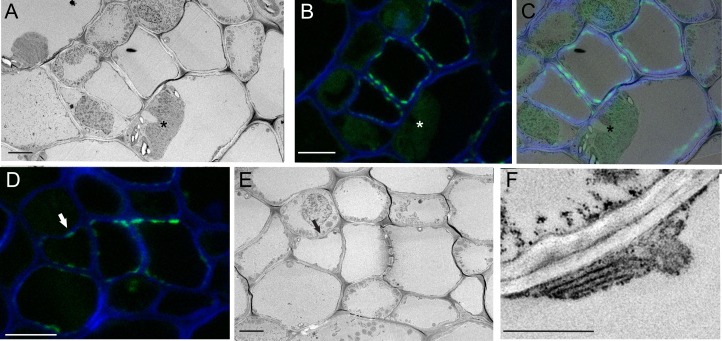
Following observation of the block face in the confocal microscope, we cut ultrathin sections (60 nm) for electron microscopy and stained these with uranyl acetate and lead citrate. We attempted to image these thin sections in the confocal microscope, prior to heavy-metal staining, but were unable to detect a GFP signal (see also Keene et al., 2008). Ultrathin sections of the phloem expressing HDEL:GFP showed good structural preservation, despite the lack of osmication (Fig. 2A). We imaged several phloem bundles in the petiole using TEM and were able to identify the same cells in the block face in the confocal microscope (Fig. 2B). In Figure 2C, note that in addition to the fluorescent SER, small vacuoles in the cytoplasm of parenchyma cells can be seen in both the TEM and confocal images. In a number of sections, we were able to identify fluorescent parietal SER aggregates (Fig. 2D) that could also be detected in ultrathin sections with the TEM (Fig. 2, E and F).
Figure 2.
CLEM of pSEO2.HDEL:GFP. A, TEM image of an ultrathin section of petiole from a plant expressing pSEO2.HDEL:GFP. The section was poststained with uranyl acetate and lead citrate. Scale = 5 μm. B, A semithin section acquired immediately after the TEM section, imaged with the confocal microscope, showing the same field of view. Note that small vacuoles in the cytoplasm can be seen in both the TEM and confocal images (stars in A and B). Scale = 5 μm. C, Overlay image of A and B showing alignment of sieve elements in the confocal and TEM images. D, A Semithin section of the phloem imaged in the confocal microscope shows conspicuous SER stacks (arrow). Scale = 10 μm. E, The TEM image of the same field of view. The same SER stack arrowed in D is apparent in E (arrow). Scale = 5 μm. F, An enlarged image of the SER stack arrowed in E. Scale = 1 μm.
Correlative 3D-SIM, Confocal Microscopy, and TEM
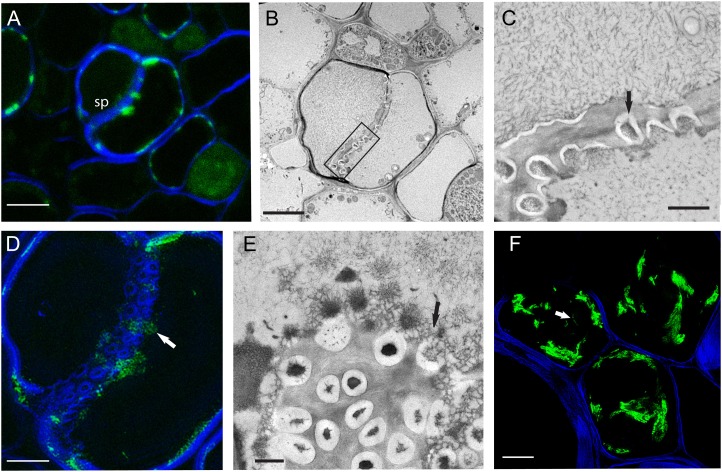
Most superresolution imaging approaches require that the cells of interest lie close to the coverslip to maximize spatial resolution (Huang et al., 2009; Bell and Oparka, 2011). The retention of GFP in semithin sections meets these requirements and allows superresolution imaging. After confocal imaging of the block face, we cut ultrathin sections and a semithin section from the same region of the block. Using 3D-SIM, we obtained images of sieve plate pores that revealed spatial information not present using confocal microscopy or TEM. Figure 3, A–D, shows sieve plates imaged sequentially by confocal laser scanning microscopy (CLSM), TEM, and 3D-SIM. Note that each method reveals different information on the structure of the sieve plate. Using CLSM we could detect, but not resolve, sieve plate pores and the SER associated with them (Fig. 3A). In the thin-section TEM image, we could resolve sieve plate pores and their callose collars, but only partial pore transects were encountered due to the section thickness (Fig. 3, B and C). In the 3D-SIM image, in which we were able to take sequential Z-sections at 125-nm spacing, we were able to reconstruct portions of the sieve plate within the thickness of the section (Fig. 3D). 3D-SIM resolved the sieve plate pores and revealed distinct cellulose collars that were not apparent in either the TEM or CLSM images (Fig. 3D). Our fixation protocol preserved the fine structure of the SER, which in glancing at sections of sieve plates appeared as a fine mesh of interconnected tubules (Fig. 3E). Using 3D-SIM imaging, we could detect similar fine tubules of fluorescent SER associated with sieve plates (Fig. 3D). These correlative imaging approaches reveal that the three forms of microscopy adopted here (CLSM, 3D-SIM, and TEM) are complementary, each revealing important information on subcellular structure.
Figure 3.
Correlative 3D-SIM, confocal microscopy, and TEM of the phloem. A, A semithin section of the phloem from a tobacco petiole expressing pSEO2.HDEL:GFP counterstained with calcofluor white to highlight cell walls and a sieve plate (SP). The sieve plate and SER are visible but not resolved. Scale = 5 μm. B, The TEM image of the same field reveals details of the sieve plate and resolves sieve plate pores. Scale = 5 μm. C, Enlargement of the sieve plate region boxed in B, revealing callose collars (arrow) around the pores. Scale = 1 μm. D, A 3D-SIM image of the same sieve plate was taken using the section shown in A. The 3D-SIM image was reconstructed from 20 serial Z-sections and, unlike the confocal image, resolves distinct cellulose collars around the sieve plate pores. The SER is visible at the sieve plate (arrow). Scale = 5 μm. E, The fine structure of the tubular SER (arrow) is apparent in a glancing transverse section of a sieve plate imaged using TEM. Scale = 1 μm. F. 3D-SIM image of PVX X-body (see also en bloc image in Fig. 1). 3D-SIM resolves fine viral filaments at around 100 nm in diameter (arrow). Scale = 5 μm.
We also examined sections of tissue infected by PVX.CP-GFP using 3D-SIM. We were able to image fine bundles of virus filaments that could be resolved to about 100 nm in diameter (Fig. 3F). Significantly, we saw very little bleaching of the GFP signal during the multiple acquisitions required to generate 3D-SIM images.
Immunofluorescence
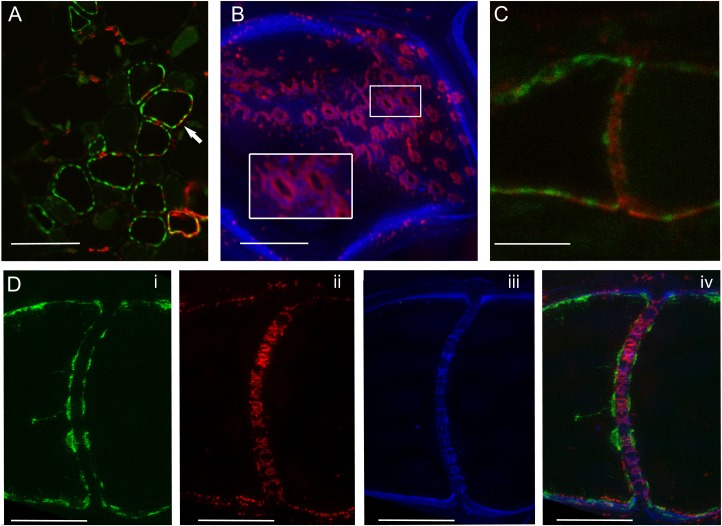
LR is compatible with fluorescent antibody labeling, so we were able to achieve triple labeling of sieve elements by cutting 1- to 2-μm sections from the blocks and labeling these with an antibody against callose. Callose is a cell wall constituent found at the neck of PD (Simpson et al., 2009) and sieve plates (Fitzgibbon et al., 2010). Using the confocal microscope, we could detect callose at the lateral sieve plates (Alexa 594 secondary antibody) along with the SER (HDEL:GFP; Fig. 4A). Figure 4B is a 3D-SIM image of a sieve plate in transverse orientation. The sieve plate callose was immunolabeled and the cellulose highlighted using calcofluor white. The inset is an enlarged view of two of the pores and shows that the cellulose collars (see Fig. 3D) form outside the callose pore linings. Figure 4C shows a confocal image of an immunolabeled sieve tube in longitudinal orientation. We used 3D-SIM to image the same sieve tube following staining with calcofluor white (Fig. 4D). This revealed the arrangements of the SER, callose, and cellulose, respectively, on the sieve plate (Fig. 4D, i–iv).
Figure 4.
Immunodetection of callose in semithin sections of resin-embedded material. A, SER stacks (pSEO2.HDEL:GFP) are seen in tranverse sections of sieve elements. Alexa 594-conjugated secondary antibody reveals callose at the sites of lateral sieve areas (red; arrow). Scale = 25 μm. B, 3D-SIM image of a sieve plate in transverse orientation. The sieve plate callose was labeled with anticallose antibody and visualized using an Alexa 594 secondary antibody (red). The inset is an enlarged view of two of the pores and shows that the cellulose collars form outside the callose pore linings. C, Confocal image of a sieve element in longitudinal orientation. Callose labeling appears at the sieve plate as well as the lateral areas. Scale = 10 μm. D, 3D-SIM image of the same sieve plate shown in C. The arrangements of the SER, callose, and cellulose are revealed. SER is shown in green (pSEO2.HDEL:GFP; i), callose in red (anticallose antibody and Alexa 594 secondary; ii), cellulose in blue (calcofluor; iii). The merge of all three channels is shown (iv). Scale = 5 μm.
DISCUSSION
The field of correlative microscopy has undergone considerable expansion in recent years (Muller-Reichert and Verkade, 2012). Correlative microscopy is the application of two or more microscopy techniques to the same region of a sample, generating complementary structural and chemical information that would not be possible using a single technique (Jahn et al., 2012). In this field, new superresolution imaging instruments have bridged the gap between light and electron microscopy (Watanabe et al., 2011). While confocal microscopy has become the mainstay of modern cell biology, there is a growing need to image the localization of proteins with increasing subcellular accuracy (Betzig et al., 2006). Two distinct types of correlative microscopy approaches have been identified; “combinatorial labeling,” in which two or more labels are identified using different forms of microscopy (e.g. confocal and TEM) and “noncombinatorial labeling,” in which the label appears in only one type of imaging method but allows identification of the same cells using a second method (Jahn et al., 2012). Generally, noncombinatorial labeling involves faster and simpler sample preparation. The technique we have described here, in which FPs are retained in fixed and embedded plant cells, is an example of a noncombinatorial approach that allows the same FP-containing cells to be identified using TEM. However, our method could be adapted to a combinatorial one if the proteins of interest were first labeled with a probe (e.g. fluoronanogold or quantum-dot complexes) that would produce fluorescent and electron-dense signals in both confocal microscopy and TEM, respectively.
A major goal has been to determine precisely the structures within which fluorescent proteins reside. The technique we have described here retains fluorescent proteins in resin blocks and semithin sections, and allows imaging of those plant tissues that are problematic with conventional live-cell fluorescence microscopy. To achieve CLEM we first examined the block face, ahead of cutting a semithin section, followed by ultrathin sections for TEM. These sections showed good structural preservation when fixed with a combination of glutaraldehyde and paraformaldehyde. As in previous studies (Keene et al., 2008), we were unable to retain sufficient fluorescence in the ultrathin sections to achieve correlative imaging on the same ultrathin section. However, using sequential sectioning we were able to locate the same cells and fluorescently tagged structures using confocal imaging and TEM. The retention of fluorescent proteins in semithin sections also allowed us to use superresolution imaging on the same sections as those used for confocal imaging, extending the range of imaging protocols that can be brought to bear on a single sample. Interestingly, sequential imaging did not simply extend the resolution range but also provided new information on subcellular structure. For example, using 3D-SIM we were able to resolve distinct cellulose collars surrounding sieve plate pores. These collars were situated outside the central callose collar but were not visible in either the confocal or TEM images. They may have been generated by the formation of sieve plate callose that, as it expanded, compressed the cellulose microfibrils around the pore.
Autofluorescence
In the material we used here, the FP signal was strong and detected easily above background autofluorescence. With increasing glutaraldehyde concentrations we found that the autofluorescence of cells increased. This was particularly true of the xylem, but other cells showed a degree of cytoplasmic fluorescence. In confocal images this was useful in identifying different cell types in the phloem, such as companion cells (e.g. Fig. 1D). It is unlikely that our method will work on cells that show a low and/or diffuse FP signal. Ultimately, the method requires a trade off between fixation and fluorescence that will depend on the questions being addressed.
Variability in LR White pH
Watanabe et al. (2011) noted that the pH of batches of LR White varied considerably. Generally, FPs are quenched at low pH (<6; Tsien, 1998). We noted also that the pH of LR White batches was variable but obtained good FP retention in plant tissues embedded in a LR White pH range extending from 4.6 to 6.5 (data not shown). Higher-pH resin batches are to be preferred because of potential quenching of fluorophores. LR White may be buffered to higher pH using ethanolamine (Watanabe et al., 2011). However, this may cause the blocks to become brittle and difficult to section (data not shown). Checking the pH of the resin before attempting FP localization is advisable.
CONCLUSION
The greatest utility of our method is likely to be in the imaging of FP-labeled structures that are difficult to image using conventional fluorescence microscopy. Such imaging is usually conducted on surface cells, or to a depth that can be accommodated by serial optical imaging. While multiphoton microscopy may extend the depth to which such sectioning is possible (Zipfel et al., 2003), resolution becomes limited. We have shown that by imaging plant tissues en bloc using appropriate counterstains, small structures such as PD (<50 nm) can be viewed to a depth of more than 40 μm in leaf cells. Such deep imaging is helpful when trying to locate structures in the block for subsequent electron microscope imaging (Prior et al., 1999) and also permits optical reconstruction of cells without painstaking serial sectioning. When required, semithin sections of the tissue can be cut, and these can be stained with conventional fluorophores and antibodies. Despite multiple imaging steps on both confocal and 3D-SIM microscopes, the fluorescent proteins we studied retained strong fluorescence and showed little photobleaching. We have been able to return to the same blocks over a period of several months, and so the method is likely to allow long-term preservation of fluorescent proteins in resin when sectioning is required at a later date.
MATERIALS AND METHODS
Plant Material
Tobacco (Nicotiana tabacum) plants expressing pSEO2.HDEL:GFP (Knoblauch and Peters, 2010) and Nicotiana benthamiana were grown from seed in a heated glasshouse and used in experiments between 30 and 55 d old. Arabidopsis (Arabidopsis thaliana) seedlings expressing SEOR1:YFP (Froelich et al., 2011), MP17:GFP (Vogel et al., 2007), and H2B:RFP (Federici et al., 2012) were germinated and grown on Murashige and Skoog media. Arabidopsis plants were used between 3 and 5 d post germination.
Fixation and Embedding
For tobacco, the petiole was cut and immediately submerged in 4% (w/v) formaldehyde (Agar Scientific), 2% (w/v) glutaraldehyde (TAAB), 50 mm PIPES, and 1 mm CaCl2 and then trimmed further under fixative to eliminate any potential airlocks. The petioles were allowed to transpire the fixative solution via the xylem for 60 min at room temperature in an illuminated fume hood (see Fitzgibbon et al., 2010). The petiole was then sectioned transversely into about 2-mm transverse slices using a double-edged razor blade. The sections were then returned to the fixative and incubated for 16 h on a rolling-bed platform in the dark at 8°C. Further tissue processing was done at 8°C in the dark unless stated otherwise. The sections were then washed in buffer (50 mm PIPES, 1 mm CaCl2) three times for 10 min before dehydration in a graded ethanol series (50% [v/v], 70% [v/v], and 90% [v/v] twice, each for 15 min). The ethanol solutions also contained 1 mm DTT to reduce tissue autofluorescence (Brown et al., 1989). The tissue sections were then infiltrated in medium grade LR (London Resin Company) at 1:1, 1:2, and 1:3 ratios of 90% ethanol (supplemented by 1 mm DTT) to resin for 45 min each before two 60-min changes in 100% LR. The final embedding step was done at ambient temperature. The samples were then polymerized in gelatin capsules (TAAB) at 50°C for 24 h.
In the case of Arabidopsis, the seedlings were processed intact and embedded as described above. The only deviation was that the H2B:RFP seedlings were fixed with 1% glutaraldehyde to maximize RFP fluorescence.
Loss of Tissue Fluorescence
To measure losses in FP fluorescence during the above steps, tissue slices were removed at each stage of the fixation and embedding process and their fluorescence compared with fresh hand sections of the petiole. For each section of the line expressing pSEO2.HDEL:GFP, a region of the phloem was photographed at 5× under identical settings on the confocal microscope. For each stage of fixation and embedding the fluorescence was measured in 11-phloem bundles, using ImageJ (Schneider et al., 2012), and the mean fluorescence value calculated as a percentage of the initial fluorescence.
En Bloc Staining
En bloc staining of cell walls was done with 1 µg mL−1 propidium iodide (Invitrogen) and 10 µg mL−1 calcofluor white (Sigma) by adding a drop of dyes to trimmed block faces and allowing them to penetrate into the tissue for 20 min. The remaining dye was then rinsed from the block face by immersing it in distilled water for 2 min.
Confocal Microscopy
Semithin sections (1 to 2 µm) were cut using a glass knife on a Leica Ultracut UCT ultramicrotome (Leica Microsystems). These, and also the intact block faces, were imaged using a Leica SP2 confocal laser scanning microscope (Leica Microsystems) with either a 5× (HC PL FLUOTAR; Leica Microsystems) or a 63× water-immersion lens (HCX PLAPO CS; Leica Microsystems). Calcofluor was excited at 405 nm, GFP at 488 nm, YFP at 514 nm, RFP at 561 nm, and Alexa 594 at 591 nm.
Transmission Electron Microscopy
Ultrathin sections (60 nm) were cut using a Diatome diamond knife, stained in 1% (w/v) aqueous uranyl acetate and Reynolds lead citrate (Reynolds, 1963) and viewed in a Philips CM120 transmission electron microscope (FEI). Representative images were taken on a Gatan Orius CCD camera (Gatan).
Immunolocalization of Callose
Semithin sections were cut as above and then affixed to a slide using a coating of poly-l-Lys (Invitrogen) and heated briefly (1 to 2 min) on a slide warmer. Sections were then incubated for 10 min in blocking solution (3% [w/v] bovine serum albumin; 50 mm Gly in 1% [w/v] phosphate buffered saline [PBS]) before being rinsed with 1% PBS three times for 1 min each. They were then incubated for 90 min in mouse anticallose antibody (Biosupplies) at a 1:400 dilution. Sections were rinsed three times for 1 min in the blocking solution before incubation with secondary antibody, anti-mouse Alexa 594 (Invitrogen) diluted 1:500, for 1 h. All antibodies were diluted in 1% bovine serum albumin, 0.02% (v/v) Tween in 1% (w/v) PBS and incubated at 37°C. The sections were rinsed in 1% PBS three times for 1 min each before staining with 10 mm calcofluor (Invitrogen). After 2 min, excess stain was rinsed off using 1% PBS. The sections were then allowed to air dry before mounting with Citifluor AF1 antifade agent (Agar Scientific) under a number 1 coverslip and then sealed with nail varnish. Tissue sections were then imaged with a Leica SP2 confocal scanning microscope, using 59-nm excitation for Alexa 594 and 405-nm excitation for Calcofluor, as described above.
PVX Infections
Infections were initiated in N. benthamiana using a 35S promoter-driven PVX.GFP-CP bombardment construct (C. Lacomme, unpublished data). Microprojectile bombardments were carried out with a custom-built gene gun similar to the one described in Gaba and Gal-On (2005).
3D-SIM
3D-SIM was conducted as described by Fitzgibbon et al. (2010), using the protocol initially described by Schermelleh et al. (2008). Images were acquired on an OMX microscope (Applied Precision) equipped with 405-, 488-, and 593-nm solid-state lasers and a UPlanSApochromat 100× 1.4 numerical aperture, oil immersion objective lens (Olympus). Samples were illuminated by a coherent scrambled laser light source that had passed through a diffraction grating to generate the structured illumination by interference of light orders in the image plane to create a three-dimensional sinusoidal pattern, with lateral stripes approximately 0.2 µm apart. The pattern was shifted laterally through five phases and through three angular rotations of 60° for each Z-section, separated by 0.125 µm. Exposure times were typically between 100 and 200 ms, and the power of each laser was adjusted to achieve optimal intensities of between 2,000 and 4,000 counts in a raw image of 16-bit dynamic range, at the lowest possible laser power to minimize photo bleaching. Raw images were processed and reconstructed to reveal structures with greater resolution (Gustafsson et al., 2008) implemented on SoftWorx, version 6.0 (Applied Precision). The channels were then aligned in x, y, and rotationally using predetermined shifts as measured using 100-nm TetraSpeck (Invitrogen) beads with the SoftWorx alignment tool (Applied Precision).
Acknowledgments
We thank Dr. Christophe Lacomme for the use of the 35S promoter-driven PVX.GFP-CP bombardment construct. We thank Dr. Michael Knoblauch for providing the SEO lines used in this study and Dr. Jim Haseloff for providing the histone 2B:RFP line.
Glossary
- SER
sieve element reticulum
- 3D-SIM
3D-structured illumination microscopy
- TEM
transmission electron microscopy
- CLEM
Correlative light and electron microscopy
- SE
sieve element
- CC
companion cell
- PD
plasmodesmata
- LR
London Resin
- PVX
Potato Virus X
- CLSM
confocal laser scanning microscopy
- DTT
dithiothreitol
- PBS
phosphate buffered saline
- FP
fluorescent protein
- MP
movement protein
- YFP
yellow fluorescent protein
- RFP
red fluorescent protein
References
- Baskin TI, Busby CH, Fowke LC, Sammut M, Gubler F. (1992) Improvements in immunostaining samples embedded in methacrylate: localisation of microtubules and other antigens throughout developing organs in plants of diverse taxa. Planta 187: 405–413 [DOI] [PubMed] [Google Scholar]
- Baskin TI, Miller DD, Vos JW, Wilson JE, Hepler PK. (1996) Cryofixing single cells and multicellular specimens enhances structure and immunocytochemistry for light microscopy. J Microsc 182: 149–161 [DOI] [PubMed] [Google Scholar]
- Bell K, Oparka KJ. (2011) Imaging plasmodesmata. Protoplasma 248: 9–25 [DOI] [PubMed] [Google Scholar]
- Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF. (2006) Imaging intracellular fluorescent proteins at nanometer resolution. Science 313: 1642–1645 [DOI] [PubMed] [Google Scholar]
- Brown RC, Lemmon BE, Mullinax JB. (1989) Immunofluorescent staining of microtubules in plant tissues: improved embedding and sectioning techniques using polyethylene glycol (PEG) and Steedman’s wax. Bot Acta 102: 54–61 [Google Scholar]
- Chapman S, Oparka KJ, Roberts AG. (2005) New tools for in vivo fluorescence tagging. Curr Opin Plant Biol 8: 565–573 [DOI] [PubMed] [Google Scholar]
- Cortese K, Diaspro A, Tacchetti C. (2009) Advanced correlative light/electron microscopy: current methods and new developments using Tokuyasu cryosections. J Histochem Cytochem 57: 1103–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Federici F, Dupuy L, Laplaze L, Heisler M, Haseloff J. (2012) Integrated genetic and computation methods for in planta cytometry. Nat Methods 9: 483–485 [DOI] [PubMed] [Google Scholar]
- Fitzgibbon J, Bell K, King E, Oparka K. (2010) Super-resolution imaging of plasmodesmata using three-dimensional structured illumination microscopy. Plant Physiol 153: 1453–1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froelich DR, Mullendore DL, Jensen KH, Ross-Elliott TJ, Anstead JA, Thompson GA, Pélissier HC, Knoblauch M. (2011) Phloem ultrastructure and pressure flow: sieve-element-occlusion-related agglomerations do not affect translocation. Plant Cell 23: 4428–4445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaba V, Gal-On A. (2005). Inoculation of plants using bombardment. Curr Protoc Microbiol 16: B3.1–B3.14 [DOI] [PubMed] [Google Scholar]
- Gustafsson MGL, Shao L, Carlton PM, Wang RCJ, Golubovskaya IN, Cande ZW, Agard DA, Sedat JW. (2008) Three-dimensional resolution doubling in wide-field fluorescence microscopy by structured illumination. Biophys J 94: 4957–4970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahne G, Herth W, Hoffman F. (1983) Wall formation and cell-division in fluorescence-labelled plant protoplasts. Protoplasma 115: 217–221 [Google Scholar]
- Huang B, Bates M, Zhuang XW. (2009) Super-resolution fluorescence microscopy. Annu Rev Biochem 78: 993–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn KA, Barton DA, Kobayashi K, Ratinac KR, Overall RL, Braet F. (2012) Correlative microscopy: providing new understanding in the biomedical and plant sciences. Micron 43: 565–582 [DOI] [PubMed] [Google Scholar]
- Keene DR, Tufa SF, Lunstrum GP, Holden P, Horton WA. (2008) Confocal/TEM overlay microscopy: a simple method for correlating confocal and electron microscopy of cells expressing GFP/YFP fusion proteins. Microsc Microanal 14: 342–348 [DOI] [PubMed] [Google Scholar]
- Knoblauch M, Oparka K. (2012) The structure of the phloem: still more questions than answers. Plant J 70: 147–156 [DOI] [PubMed] [Google Scholar]
- Knoblauch M, Peters WS. (2010) Münch, morphology, microfluidics: our structural problem with the phloem. Plant Cell Environ 33: 1439–1452 [DOI] [PubMed] [Google Scholar]
- Knoblauch M, van Bel AJE. (1998) Sieve tubes in action. Plant Cell 10: 35–50 [Google Scholar]
- Lee JY, Wang X, Cui W, Sager R, Modla S, Czymmek K, Zybaliov B, van Wijk K, Zhang C, Lu H, et al. (2011) A plasmodesmata-localized protein mediates crosstalk between cell-to-cell communication and innate immunity in Arabidopsis. Plant Cell 23: 3353–3373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott-Schwartz J, Altan-Bonnet N, Patterson GH. (2003) Photobleaching and photoactivation: following protein dynamics in living cells. Nat Cell Biol 5(Suppl): S7–S14 [PubMed] [Google Scholar]
- Luby-Phelps K, Ning G, Fogerty J, Besharse JC. (2003) Visualization of identified GFP-expressing cells by light and electron microscopy. J Histochem Cytochem 51: 271–274 [DOI] [PubMed] [Google Scholar]
- Mullendore DL, Windt CW, Van As H, Knoblauch M. (2010) Sieve tube geometry in relation to phloem flow. Plant Cell 22: 579–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller-Reichert T, Verkade P, editors (2012) Correlative Light and Electron Microscopy. Methods in Cell Biology, Vol 111. Academic Press, Oxford [DOI] [PubMed] [Google Scholar]
- Nelson RS, van Bel AJE. (1998) The mystery of virus trafficking into, through and out of vascular tissue. Prog Bot 59: 476–533 [Google Scholar]
- Pfeiffer S, Beese M, Boettcher M, Kawaschinski K, Krupinska K. (2003) Combined use of confocal laser scanning microscopy and transmission electron microscopy for visualisation of identical cells processed by cryotechniques. Protoplasma 222: 129–137 [DOI] [PubMed] [Google Scholar]
- Pighin JA, Zheng HQ, Balakshin LJ, Goodman IP, Western TL, Jetter R, Kunst L, Samuels AL. (2004) Plant cuticular lipid export requires an ABC transporter. Science 306: 702–704 [DOI] [PubMed] [Google Scholar]
- Prior DAM, Oparka KJ, Roberts IM. (1999) En bloc optical sectioning of resin-embedded specimens using a confocal laser scanning microscope. J Microsc 193: 20–27 [DOI] [PubMed] [Google Scholar]
- Reynolds ES. (1963) The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J Cell Biol 17: 208–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santa Cruz S, Chapman S, Roberts AG, Roberts IM, Prior DAM, Oparka KJ. (1996) Assembly and movement of a plant virus carrying a green fluorescent protein overcoat. Proc Natl Acad Sci USA 93: 6286–6290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schermelleh L, Carlton PM, Haase S, Shao L, Winoto L, Kner P, Burke B, Cardoso MC, Agard DA, Gustafsson MGL, et al. (2008) Subdiffraction multicolor imaging of the nuclear periphery with 3D structured illumination microscopy. Science 320: 1332–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9: 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner NC, Steinbach PA, Tsien RY. (2005) A guide to choosing fluorescent proteins. Nat Methods 2: 905–909 [DOI] [PubMed] [Google Scholar]
- Simpson C, Thomas C, Findlay K, Bayer E, Maule AJ. (2009) An Arabidopsis GPI-anchor plasmodesmal neck protein with callose binding activity and potential to regulate cell-to-cell trafficking. Plant Cell 21: 581–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subach FV, Patterson GH, Manley S, Gillette JM, Lippincott-Schwartz J, Verkhusha VV. (2009) Photoactivatable mCherry for high-resolution two-color fluorescence microscopy. Nat Methods 6: 153–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson MV, Wolniak SM. (2008) A plasma membrane-anchored fluorescent protein fusion illuminates sieve element plasma membranes in Arabidopsis and tobacco. Plant Physiol 146: 1599–1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilsner J, Linnik O, Wright KM, Bell K, Roberts AG, Lacomme C, Santa Cruz S, Oparka KJ. (2012) The TGB1 movement protein of potato virus X reorganizes actin and endomembranes into the X-body, a viral replication factory. Plant Physiol 158: 1359–1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsien RY. (1998) The green fluorescent protein. Annu Rev Biochem 67: 509–544 [DOI] [PubMed] [Google Scholar]
- Vogel FD, Hofius D, Sonnewald U. (2007) Intracellular trafficking of potato leafroll virus movement protein in transgenic Arabidopsis. Traffic 8: 1205–1214 [DOI] [PubMed] [Google Scholar]
- Watanabe S, Punge A, Hollopeter G, Willig KI, Hobson RJ, Davi MW, Hell SW, Jorgensen EM. (2011) Protein localization in electron micrographs using fluorescence microscopy. Nat Methods 8: 80–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Babcock HP, Zhuang X. (2012) Dual-objective STORM reveals three-dimensional filament organization in the actin cytoskeleton. Nat Methods 9: 185–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel WR, Williams RM, Webb WW. (2003) Nonlinear magic: multiphoton microscopy in the biosciences. Nat Biotechnol 21: 1369–1377 [DOI] [PubMed] [Google Scholar]