A novel pattern of in vivo posttranslational modifications activates phosphoenolpyruvate carboxylase in maturing proteoid roots of harsh hakea.
Abstract
Accumulating evidence indicates important functions for phosphoenolpyruvate (PEP) carboxylase (PEPC) in inorganic phosphate (Pi)-starved plants. This includes controlling the production of organic acid anions (malate, citrate) that are excreted in copious amounts by proteoid roots of nonmycorrhizal species such as harsh hakea (Hakea prostrata). This, in turn, enhances the bioavailability of mineral-bound Pi by solubilizing Al3+, Fe3+, and Ca2+ phosphates in the rhizosphere. Harsh hakea thrives in the nutrient-impoverished, ancient soils of southwestern Australia. Proteoid roots from Pi-starved harsh hakea were analyzed over 20 d of development to correlate changes in malate and citrate exudation with PEPC activity, posttranslational modifications (inhibitory monoubiquitination versus activatory phosphorylation), and kinetic/allosteric properties. Immature proteoid roots contained an equivalent ratio of monoubiquitinated 110-kD and phosphorylated 107-kD PEPC polypeptides (p110 and p107, respectively). PEPC purification, immunoblotting, and mass spectrometry indicated that p110 and p107 are subunits of a 430-kD heterotetramer and that they both originate from the same plant-type PEPC gene. Incubation with a deubiquitinating enzyme converted the p110:p107 PEPC heterotetramer of immature proteoid roots into a p107 homotetramer while significantly increasing the enzyme’s activity under suboptimal but physiologically relevant assay conditions. Proteoid root maturation was paralleled by PEPC activation (e.g. reduced Km [PEP] coupled with elevated I50 [malate and Asp] values) via in vivo deubiquitination of p110 to p107, and subsequent phosphorylation of the deubiquitinated subunits. This novel mechanism of posttranslational control is hypothesized to contribute to the massive synthesis and excretion of organic acid anions that dominates the carbon metabolism of the mature proteoid roots.
Phosphoenolpyruvate (PEP) carboxylase (PEPC; EC 4.1.1.31) is a ubiquitous and tightly regulated cytosolic enzyme of vascular plants that is also widely distributed in green algae and bacteria. PEPC catalyzes the irreversible β-carboxylation of PEP to form oxaloacetate (OAA) and inorganic phosphate (Pi). Vascular plant PEPCs belong to a small multigene family encoding several closely related plant-type PEPCs (PTPCs), along with a distantly related bacterial-type PEPC (BTPC; O’Leary et al., 2011a). PTPC genes encode 105- to 110-kD polypeptides that typically assemble as approximate 400-kD Class-1 PEPC homotetramers. In contrast, BTPC genes encode larger 116- to 118-kD polypeptides owing to a unique intrinsically disordered region that mediates BTPC’s tight interaction with coexpressed PTPC subunits. This association results in the formation of unusual Class-2 PEPC heterooctameric complexes that are largely desensitized to allosteric effectors and that dynamically associate with the surface of mitochondria in vivo (O’Leary et al., 2009, 2011a; Igawa et al., 2010; Park et al., 2012).
The critical role of Class-1 PEPC in assimilating atmospheric CO2 during C4 and Crassulacean acid metabolism photosynthesis has been studied extensively. Class-1 PEPCs also fulfill a wide range of crucial nonphotosynthetic functions, particularly the anaplerotic replenishment of tricarboxylic acid cycle intermediates consumed during biosynthesis (O’Leary et al., 2011a). Class-1 PEPCs are subject to a complex set of posttranslational controls including allosteric effectors, covalent modification via phosphorylation or monoubiquitination, and protein-protein interactions (Uhrig et al., 2008; O’Leary et al., 2009, 2011a, 2011b). Allosteric activation by Glc-6-P and inhibition by l-malate are routinely observed, whereas phosphorylation and dephosphorylation are catalyzed by a Ca2+-independent PEPC protein kinase (PPCK) and a protein phosphatase type-2A (PP2A), respectively (O’Leary et al., 2011a). Phosphorylation at a conserved N-terminal seryl residue activates Class-1 PEPCs by decreasing inhibition by malate while increasing activation by Glc-6-P. By contrast, Class-1 PEPC is subject to inhibitory monoubiquitination during castor oil (Ricinus communis) seed (COS) germination, or following depodding of developing COS (Uhrig et al., 2008; O’Leary et al., 2011b). Immunoblots of germinating COS extracts revealed a 1:1 ratio of immunoreactive 110- and 107-kD PTPC polypeptides (p110 and p107, respectively). PEPC purification and mass spectrometry (MS) demonstrated that (1) p110 and p107 are subunits of a 440-kD Class-1 PEPC heterotetramer, (2) both subunits arise from the same PTPC gene (RcPpc3) that also encodes the phosphorylated 410-kD Class-1 PEPC homotetramer of intact developing COS, and (3) p110 is a monoubiquitinated form of p107 (Uhrig et al., 2008). The monoubiquitination site (Lys-628) of COS p110 is conserved in vascular plant PEPCs and is proximal to a PEP-binding/catalytic domain. Incubation with a deubiquitinating enzyme converted the Class-1 PEPC p110:p107 heterotetramer into a p107 homotetramer while exerting significant effects on the enzyme’s kinetic properties (Uhrig et al., 2008). PTPC monoubiquitination rather than phosphorylation is widespread throughout the astor plant and appears to be the predominant posttranslational modification (PTM) of Class-1 PEPC that occurs in unstressed plants (O’Leary et al., 2011b). The distinctive developmental patterns of Class-1 PEPC phosphoactivation versus monoubiquitination-inhibition indicated that these PTMs might be mutually exclusive in the castor plant (O’Leary et al., 2011a, 2011b).
Substantial evidence indicates that PEPC plays a pivotal role in plant acclimation to nutritional Pi deficiency (Duff et al., 1989; Vance et al., 2003; O’Leary et al., 2011a; Plaxton and Tran, 2011; Supplemental Fig. S1), a common abiotic stress that frequently limits plant growth in natural ecosystems. The marked induction of Class-1 PEPCs during Pi stress has been linked to the synthesis and excretion of large amounts of organic acid anions by roots of Pi-starved (–Pi) plants (O’Leary et al., 2011a; Uhde-Stone et al., 2003; Vance et al., 2003; Shane et al., 2004a). The excreted organic acids chelate metal cations such as Al3+ and Ca2+ that immobilize Pi in the soil, thus increasing soluble Pi concentrations by up to 1,000-fold (Vance et al., 2003). Harsh hakea (Hakea prostrata) is a perennial nonmycotroph that has evolved a host of traits that allow it to thrive in the nutrient-impoverished, ancient soils of western Australia. A crucial adaptation of harsh hakea is its proteoid roots, which excrete copious quantities of citrate and malate to mediate Pi solubilization and acquisition from the soil’s mineral-bound Pi (Supplemental Figs. S1 and S2; Shane et al., 2003, 2004a, 2004b; Shane and Lambers, 2005). Shane and coworkers (2004a) correlated proteoid root development in –Pi harsh hakea with marked increases in respiration, internal carboxylate concentrations, and rates of carboxylate exudation. Immunoblotting indicated that PEPC abundance remained relatively constant during proteoid root development, except in senescing 3-week-old roots, where it showed a marked decline. The PEPC immunoblots also revealed approximately 110- and 100-kD immunoreactive polypeptides that were of equal intensity in young proteoid roots, whereas mature proteoid roots showed a marked reduction in the p110 (Shane et al., 2004a). The possible contribution of PTMs such as phosphorylation to the in vivo activation of proteoid root PEPCs is currently unclear (e.g. see Uhde-Stone et al., 2003). However, this is feasible since the pronounced induction of PPCK genes coupled with the reversible phosphorylation-activation of a Class-1 PEPC isozyme (AtPPC1) has been conclusively demonstrated in –Pi Arabidopsis (Arabidopsis thaliana) suspension cells and seedlings (Gregory et al., 2009).
The goal of the current study was to test the hypothesis that PEPC PTMs contribute to the metabolic adaptations of harsh hakea proteoid roots. We report a novel metabolic control paradigm that involves the in vivo deubiquitination and consequent kinetic activation of a phosphorylated form of a C3 plant Class-1 PEPC.
RESULTS AND DISCUSSION
Influence of Pi-Supply on Growth, Leaf Phosphate Concentration, and Root Development
The influence of optimal Pi (+Pi) versus suboptimal Pi supply (+Pi [25 μm] and –Pi [1 μm], respectively) on harsh hakea growth and leaf free Pi versus organic P (Po) levels was assessed. Consistent with previous studies (Shane et al., 2004b), the –Pi plants showed a 43% reduction in total fresh weight, a 2-fold greater root mass ratio (root mass:total plant mass), and prolific growth of proteoid roots (approximately 50% of total root biomass; Supplemental Figs. S2 and S3A). The total leaf phosphate concentration ([P]) of –Pi plants was 23-fold lower than that of +Pi plants, and 86% of total leaf [P] was Po. Conversely, 64% of the total leaf [P] of +Pi plants was free Pi (Supplemental Fig. S3B).
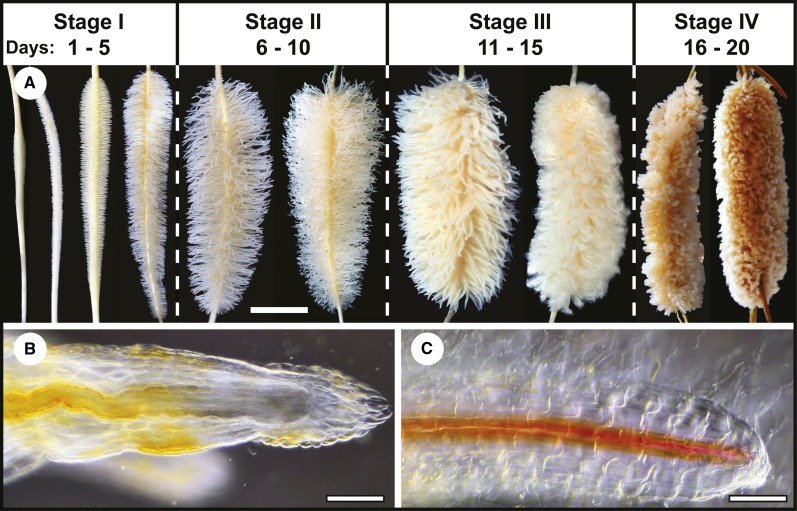
As reported by Shane and coworkers (2004a), (1) proteoid roots only formed under –Pi conditions (Supplemental Fig. S2) and (2) proteoid root life span was approximately 20 d as judged from rootlet initiation to senescence (Fig. 1A). In stages I and II, thousands of white rootlets with meristems and root caps emerged from each proteoid root (Fig. 1, A and B). As rootlets matured during stage III, growth and elongation ceased and meristems were lost, vascular tissues became fully differentiated to tips, and rootlets, which were now bright white, formed numerous root hairs (Fig. 1, A and C). During stage IV, rootlets, but not parent axes, senesced, changing color from white to gray and eventually turning dark brown (Fig. 1A).
Figure 1.
Time course of proteoid root development for –Pi plants supplemented with 1 µm Pi. A, Each developmental stage is labeled according to the number of days following rootlet emergence observed at day 1 until senescence at day 20. Scale bar = 1 cm. B and C, Rootlets from immature stage I (B) and mature stage III (C) proteoid roots were stained for lignified tissues, cleared, and imaged as described in “Materials and Methods.” Scale bar = 0.35 mm.
Influence of Proteoid Root Development on Key Enzymes of Organic Acid Metabolism
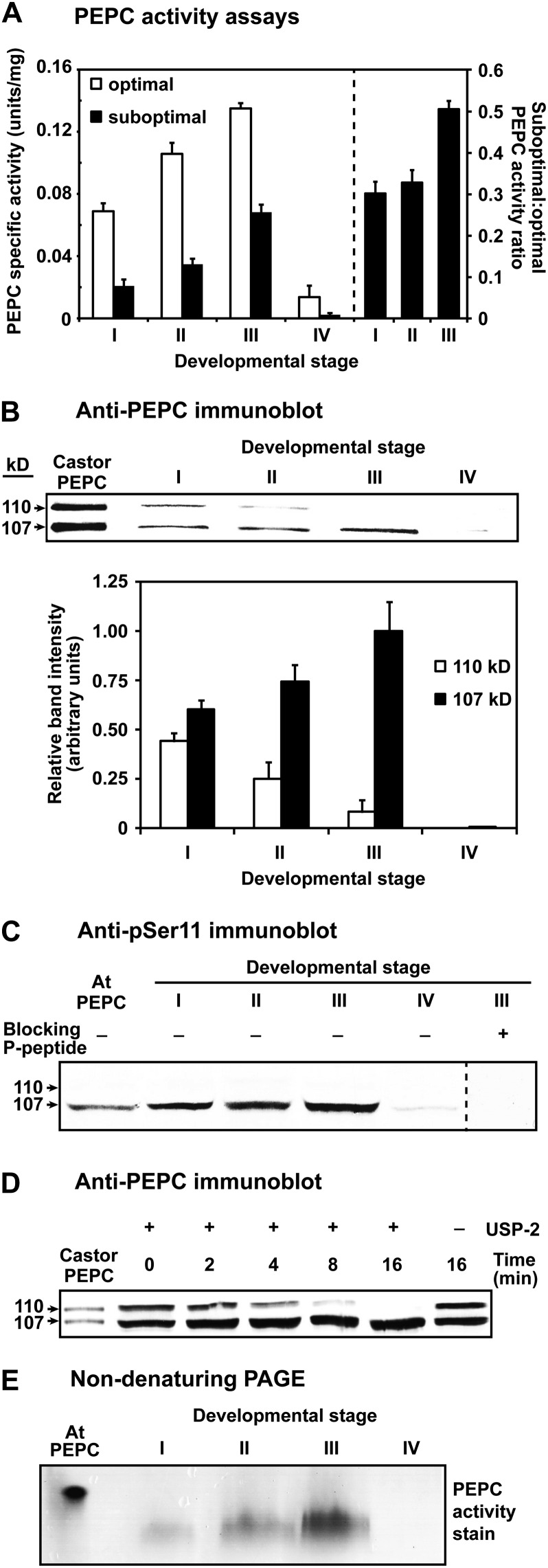
A significant increase in PEPC specific activity (2- and 3.5-fold under optimal and suboptimal assay conditions, respectively) occurred during proteoid root development, becoming maximal at stage III (Fig. 2A). Interestingly, the ratio of PEPC specific activity under suboptimal (physiological) versus optimal conditions significantly increased as stage I roots matured into stage III roots (Fig. 2A). This suggests that PEPC might have been activated by some type of PTM. In the well-studied model crop white lupin (Lupinus albus), optimal PEPC activity also increases as its proteoid roots attain maturity (Johnson et al., 1996; Massonneau et al., 2001; Uhde-Stone et al., 2003).
Figure 2.
Influence of proteoid root development on PEPC activity, subunit composition, and PTMs. A, All values represent the mean (±se) PEPC-specific activity of duplicate determinations of desalted extracts from n = four biological replicates determined under optimal (pH 8.2, 2.5 mm PEP) or suboptimal (pH 7.2, 0.2 mm PEP, 0.125 mm malate) conditions. All differences in specific activity and suboptimal:optimal activity ratio between stage I and III roots were statistically significant (P < 0.05). B, Hakea extracts were subjected to SDS-PAGE (10% gels; 0.5 μg protein lane−1), electroblotted onto a poly(vinylidene difluoride) membrane, and probed with anti-PEPC. Relative band intensity (±se) was quantified on n = five biological replicates from scanned immunoblots using ImageJ. C, Immunoblots were probed with anti-pSer11 in the presence of 10 μg mL−1 of the corresponding dephosphopeptide or phosphopeptide (20 μg of protein lane−1 for hakea extracts). D, Desalted extracts of stage I roots were incubated with and without 15 μΜ USP-2 and then immunoblotted with anti-PEPC (2 μg protein lane−1). E, In-gel PEPC activity staining followed nondenaturing PAGE of proteoid root extracts (7.5 μg protein lane−1) on 7% gels. The lanes labeled “Castor PEPC” (B and D) contained 50 and 25 ng, respectively, of purified monoubiquitinated Class-1 PEPC (RcPPC3) from germinating COS (Uhrig et al., 2008). The lanes labeled “At PEPC” (C and E) contained 250 and 500 ng, respectively, of purified phosphorylated Class-1 PEPC (AtPPC1) from –Pi Arabidopsis suspension cells (Gregory et al., 2009).
Suc synthase (SUS)- but not alkaline invertase (INV)-specific activity paralleled the PEPC activity profile during proteoid root maturation (Supplemental Fig. S4A). SUS and INV activities were comparable at stage I, but SUS activity was significantly greater at subsequent developmental stages (Supplemental Fig. S4A). SUS activity at stage III was almost 4-fold that at stage I. Similarly, a 2-fold increase in SUS activity occurred during maturation of white lupin proteoid roots (Massonneau et al., 2001). The peak in SUS and PEPC activities at stage III of hakea proteoid root development correlates with their maximal rates of citrate and malate exudation (Shane et al., 2004a). By contrast, citrate synthase (CS) and malate dehydrogenase (MDH) activities remained relatively constant throughout development (except in senescing stage IV roots, where they showed a pronounced decline; Supplemental Fig. S4, A–C).
Influence of Proteoid Root Development on PEPC Subunit Composition and PTMs
The availability of anti-COS Class-1 PEPC (anti-PEPC) and anti-pSer11 phospho-site specific (anti-pSer11)-IgGs allowed us to employ immunoblotting to assess PEPC’s subunit composition and phosphorylation status in root extracts. Consistent with earlier results (Shane et al., 2004a), (1) anti-PEPC immunoblots of stage I root extracts revealed comparable amounts of immunoreactive p110 and p107 that comigrated with the purified, partially monoubiquitinated PEPC heterotetramer from germinating COS, whereas (2) a progressive loss of immunoreactive p110 in stages II to III was paralleled by increased levels of p107 such that the total amount of immunoreactive PEPC polypeptides remained relatively constant (Fig. 2B). Anti-pSer11 immunoblotting indicated that p107 was in vivo phosphorylated at a conserved N-terminal seryl residue in stage I to III roots (Fig. 2C). An approximate 2-fold increase in p107’s relative anti-pSer11 immunoreactive intensity at stage III relative to stage I (1.0 ± 0.18 and 0.6 ± 0.14, respectively, means ± se of n = three biological replicates) indicated that the apparent increase in p107 phosphorylation paralleled its increased abundance at stage III (Fig. 2, B and C). However, immunoblotting with anti-COS BTPC-IgG (O’Leary et al., 2009) failed to reveal any immunoreactive BTPC polypeptides in stage I to IV root extracts.
Incubation with ubiquitin-specific protease-2 (USP-2) core, a recombinant human deubiquitinating enzyme, resulted in p110’s apparent in vitro deubiquitination; e.g. over the 16-min time course, p110’s disappearance was paralleled by an increase in the amount of immunoreactive p107 (Fig. 2D). Because the USP-2 core lacks an N-terminal regulatory domain it can deubiquitinate a large number of substrates in vitro (Lin et al., 2001), including the monoubiquitinated p110 subunit of the Class-1 PEPC heterotetramer from germinating COS (Uhrig et al., 2008). Results shown in Figure 2D indicating that the p110 of stage I roots is a monoubiquitinated form of p107 were confirmed in subsequent experiments (see below). Nondenaturing PAGE of root extracts followed by in-gel activity staining revealed a single PEPC activity band suggesting that the p110 and p107 of the stage I roots may be subunits of a single Class-1 PEPC heteromer (Fig. 2E). The differential migration of proteoid root PEPC activity-staining bands with that of purified PEPC from –Pi Arabidopsis suspension cells (Fig. 2E) may arise from differences in their respective mass-to-charge ratio and/or formation of different PEPC oligomers during nondenaturing PAGE.
Kinetic Properties of Proteoid Root PEPC
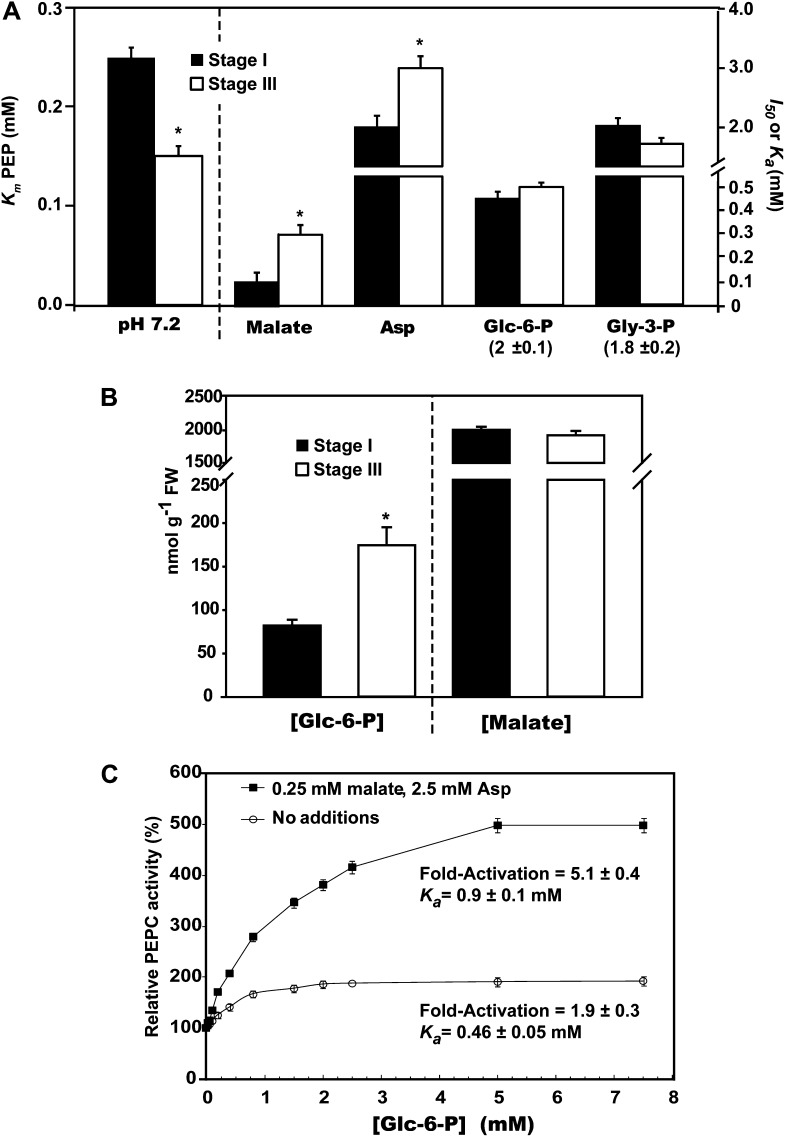
We next compared kinetic properties of PEPC in desalted extracts prepared from stage I versus stage III roots. Hyperbolic PEP saturation kinetics was routinely observed. However, PEPC from stage III roots displayed a significantly (60%) lower Km (PEP) value at a physiologically relevant pH (7.2) relative to PEPC from stage I roots (Fig. 3A).Various compounds were tested as potential PEPC effectors at optimal versus suboptimal pH (8.2 and 7.2, respectively) and subsaturating PEP (0.2 mm). The following metabolites exerted little or no influence (±20% of the control rate) on PEPC’s activity from stage I or III roots: citrate, isocitrate, Glu, and ATP (2.5 mm each). Similar to other plant PEPCs (O’Leary et al., 2011a), the enzyme from stage I and III roots displayed pH-dependent modulation by several metabolites such that they were effective at pH 7.2, but not 8.2. At pH 7.2, Glc-6-P, Glc-1-P, Fru-6-P, and glycerol-3-P (Gly-3-P) functioned as activators, whereas malate and Asp were inhibitory (Supplemental Table S1; Fig. 3A). PEPC from stage I roots was significantly more sensitive to inhibition by malate and Asp, relative to PEPC from stage III roots (Fig. 3A). Stage I and III roots exhibited equivalent malate concentrations of approximately 2 μmol g−1 fresh weight, whereas the concentration of Glc-6-P was significantly elevated (by about 2.5-fold) at stage III, relative to stage I (Fig. 3B).
Figure 3.
Kinetic constants of PEPC in desalted extracts prepared from stage I versus stage III proteoid roots. A and C, All assays were performed at pH 7.2. I50 (malate and Asp) and Ka (Glc-6-P and Gly-3-P) values were determined using subsaturating PEP (0.2 mm). A, Values in parentheses represent mean (±se) fold activation of PEPC by saturating Glc-6-P or Gly-3-P. B, Cellular concentration of malate and Glc-6-P in stage I versus stage III roots. C, Influence of combined presence of 0.25 mm malate and 2.5 mm Asp on Glc-6-P activation of PEPC from stage III roots. All values represent the mean (±se) of duplicate determinations on n = four biological replicates. The asterisks denote statistically significant differences (P < 0.05).
PEPC monoubiquitination during COS germination interferes with PEP binding while enhancing its sensitivity to inhibition by malate and Asp (Uhrig et al., 2008). Conversely, our results indicate that PEPC’s apparent in vivo deubiquitination during proteoid root maturation (Fig. 2, B and D) was accompanied by a significant increase in its affinity for PEP coupled with reduced sensitivity to malate and Asp inhibition (Fig. 3A). Interestingly, Glc-6-P activation of PEPC from stage III roots was markedly enhanced by the combined presence of low concentrations of malate and Asp (0.25 and 2.5 mm, respectively), such that an approximately 5-fold activation was observed at saturating Glc-6-P (Fig. 3C). This finding is consistent with the magnified effects of allosteric activators on anaplerotic PEPC from Escherichia coli in the presence of inhibitors (Xu et al., 2012) and may further contribute to PEPC’s in vivo activation during proteoid root maturation.
PEPC Purification and MS
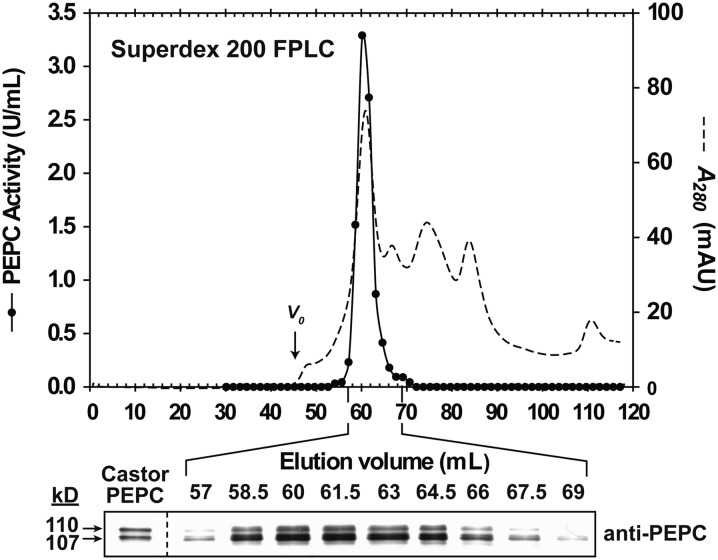
PEPC from stage I and II proteoid roots was purified 24-fold, resulting in an overall yield of 26% and a final specific activity of 3.6 units mg−1 protein (Supplemental Table S2). Assuming our final PEPC preparation was approximately 50% pure (Supplemental Fig. S5A, lane 4), then the specific activity of the fully pure enzyme would be approximately 7.2 units mg−1, comparable to the specific activity of 9.6 units mg−1 reported for the purified, monoubiquitinated PEPC from germinating castor seeds (Uhrig et al., 2008). SDS-PAGE and immunoblotting indicated that the final PEPC preparation consisted of equivalent amounts of protein-staining or immunoreactive p110 and p107 (Supplemental Fig. S5, A and B). PEPC’s native Mr as estimated by FPLC on a calibrated Superdex 200 column was 430 ± 5-kD (mean ± se, n = 3). Fractions eluting with the single peak of PEPC activity resolved during butyl-Sepharose hydrophobic interaction or Superdex 200 gel filtration FPLC contained a constant proportion of immunoreactive p110 and p107 (Supplemental Fig. S6; Fig. 4). Thus, purified PEPC from the stage I and II roots appears to exist as a heterotetramer composed of an approximately 1:1 ratio of p110 and p107 subunits, similar to the monoubiquitinated Class-1 PEPC heterotetramer purified from germinated COS (Uhrig et al., 2008). Interestingly, immunoblotting with anti-soybean (Glycine max) root nodule SUS-IgG indicated that the copurifying 93-kD protein-staining polypeptide (p93) is SUS (Supplemental Fig. S5). This was corroborated by SUS assays of the final preparation (SUS-specific activity = 2.2 units mg−1) and MS.
Figure 4.
Coelution of PEPC activity with p110 and p107 during Superdex 200 FPLC of PEPC from stage I and II proteoid roots. Aliquots (1 μL) from various fractions along with purified PEPC (50 ng) from germinating COS (“Castor PEPC”) were subjected to immunoblotting with anti-PEPC. Vo denotes the void volume.
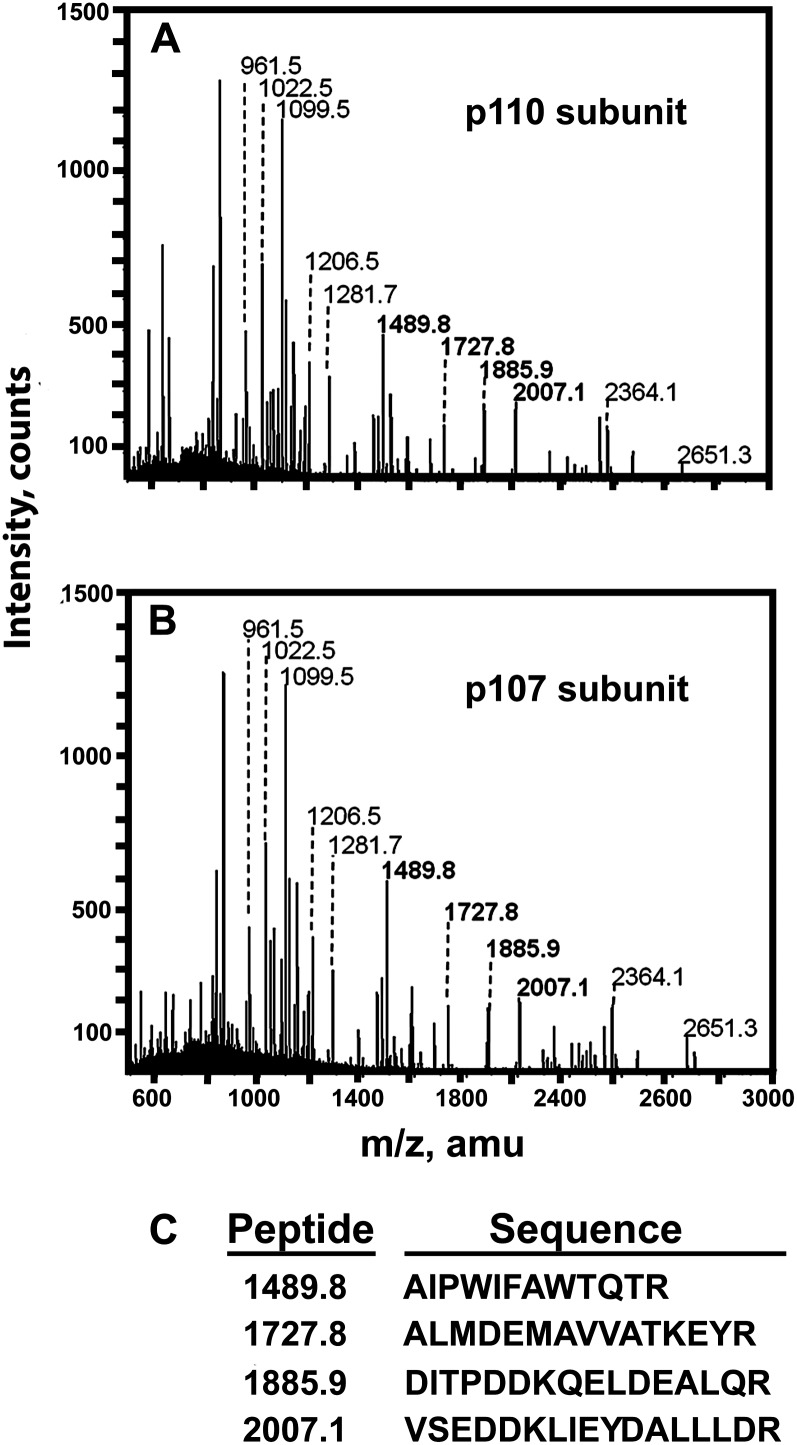
Protein-stained p110, p107, and p93 were excised from SDS-PAGE gels of the final preparation and subjected to tryptic digestion, matrix-assisted laser-desorption ionization (MALDI) quadrupole time-of-flight (Q-TOF) MS peptide mass fingerprinting, and tandem mass spectrometry (MS/MS) sequencing. The best match for p110 and p107 was with a cotton (Gossypium hirsutum) PTPC (Fig. 5; Supplemental Table S3). Apart from displaying virtually identical peptide mass fingerprints, p110 and p107 also shared identical amino acid sequences in four common tryptic peptides (Fig. 5), indicating that they originated from the same PTPC gene. Peptide mass fingerprinting also confirmed that the copurifying p93 is SUS (Supplemental Table S3). Extensive copurification of developing COS Class-1 PEPC with SUS also occurs (Gennidakis et al., 2007). Future research is needed to assess the intriguing possibility that these two key enzymes of cytosolic carbohydrate metabolism might physically interact in vivo.
Figure 5.
MALDI Q-TOF MS spectra of p110 (A) and p107 (B) tryptic peptides derived from purified PEPC of stage I and II proteoid roots. C, MALDI Q-TOF MS/MS analysis of four peptides common to both subunits (boldface in A and B) yielded identical amino acid sequences.
A PTPC amino acid sequence containing 964 residues was recently deduced from the transcriptome of nonproteoid roots of +Pi harsh hakea plants by RNA sequencing using the Illumina HiSEquation 2000 platform (R. Jost, personal communication). The deduced sequence contains: (1) the C-terminal QNTG tetrapeptide characteristic of PTPCs, (2) exact matches to MS/MS derived sequences determined for all four tryptic peptides obtained from the p110 and p107 subunits of the PEPC purified from stage I and II proteoid roots (Fig. 5C), and (3) Ser and Lys residues that align with the positions of the phosphorylated-Ser-11 and monoubiquitinated-Lys-628 of developing and germinating COS PTPC (RcPPC3), respectively, and that appear to be absolutely conserved in all PTPCs (Tripodi et al., 2005; Gennidakis et al., 2007; Uhrig et al., 2008; O’Leary et al., 2011a). Research is in progress to isolate a full-length PTPC complementary DNA from proteoid roots of –Pi harsh hakea to determine if its deduced amino acid sequence matches that derived by sequencing the transcriptome of nonproteoid roots.
Influence of in Vitro Dephosphorylation and Deubiquitination on PEPC Activity
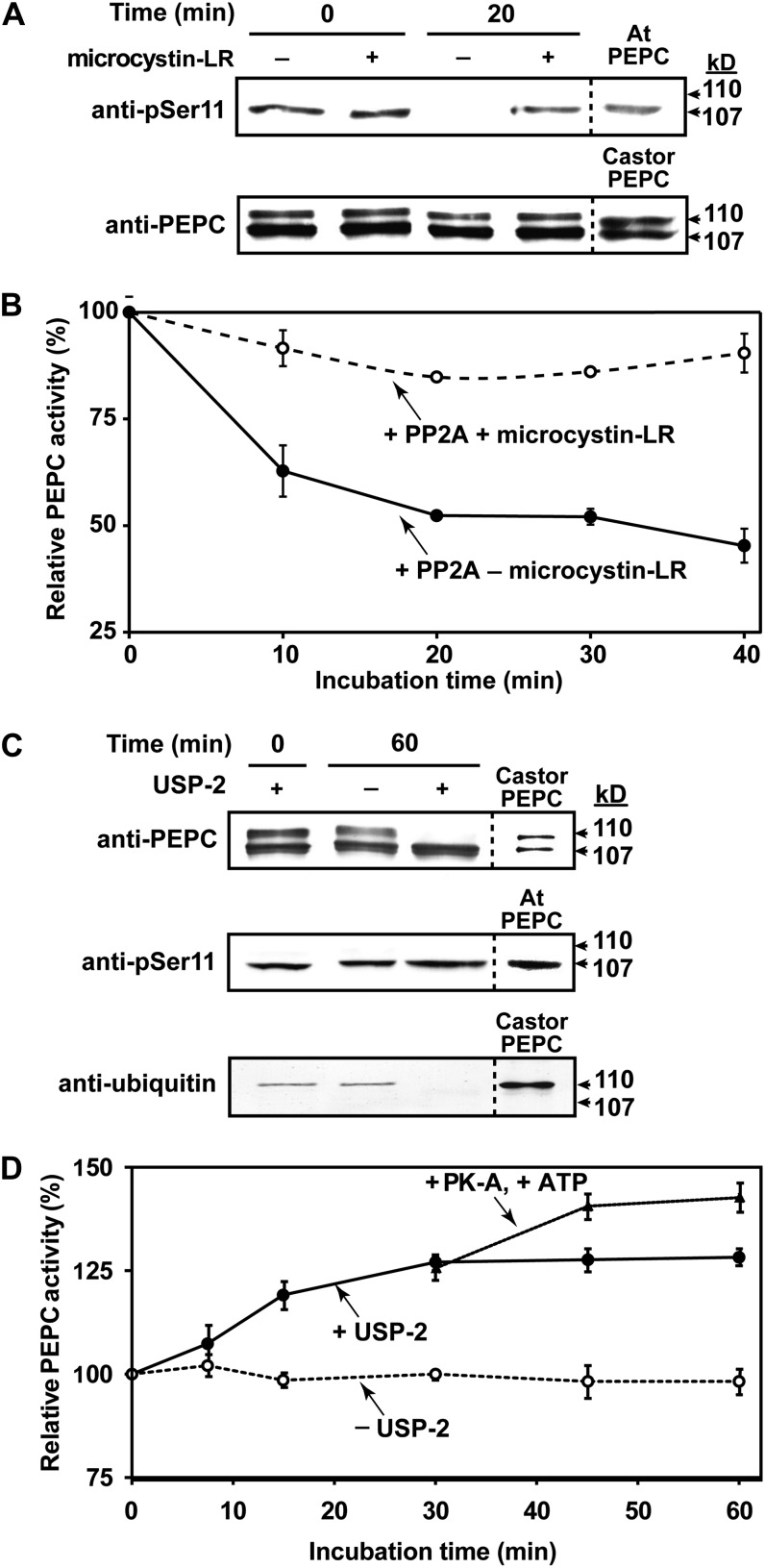
Purified PEPC isolated from stage I and II roots exhibited a broad pH-activity profile with optimal activity occurring in the range of pH 8.0 to 8.5, typical of other Class-1 PEPCs (O’Leary et al., 2011a). The phosphorylated p107 subunit of the purified PEPC was in vitro dephosphorylated when incubated with the catalytic subunit of bovine heart PP2A for 20 min (Fig. 6A). No change in p107 phosphorylation occurred when PEPC was incubated in the presence of PP2A and 50 nm microcystin-LR (a potent PP2A and PP1 inhibitor; Fig. 6A). Immunoblotting with anti-PEPC confirmed that the proportion of p110:p107 was unaffected during PP2A treatment (Fig. 6A). Dephosphorylation of p107 caused an approximate 60% decrease in PEPC activity when assayed under suboptimal conditions (pH 7.2, 0.2 mm PEP, 0.125 mm malate; Fig. 6B). These results support earlier findings that PTPC phosphorylation at a conserved seryl residue near its N terminus serves to activate the enzyme (Tripodi et al., 2005; Gregory et al., 2009; O’Leary et al., 2011a).
Figure 6.
Influence of in vitro dephosphorylation by PP2A (A and B) or deubiquitination by USP-2 (C and D) on activity of PEPC purified from stage I and II proteoid roots. A, Immunoblot analysis was performed using anti-PEPC or anti-pSer11 (75 ng or 1 μg of hakea PEPC lane−1, respectively. “Castor PEPC” denotes 50 ng of monoubiquitinated PEPC (RcPPC3) purified from germinating COS (Uhrig et al., 2008), whereas “At PEPC” denotes 500 ng of phosphorylated-PEPC (AtPPC1) purified from –Pi Arabidopsis (Gregory et al., 2009). B, Time course for PP2A-dependent inhibition of hakea PEPC activity (±50 nm microcystin-LR). C, Immunoblot analysis was performed using anti-PEPC, anti-pSer11, or antiubiquitin (75 ng, 1 μg, or 4 μg of hakea PEPC lane−1, respectively; reference lanes contained 50 ng of germinating COS PEPC, 0.5 μg of –Pi Arabidopsis PEPC, or 1 μg of germinating COS PEPC, respectively). D, Influence of USP-2 mediated deubiquitination, followed by bovine heart protein kinase-A mediated phosphorylation after 30 min, on activity of hakea PEPC. PEPC was assayed using suboptimal conditions (pH 7.2, 0.2 mm PEP, 0.125 mm malate). All values represent the mean (±se) of n = three independent experiments.
Probing immunoblots of the final PEPC preparation with anti-ubiquitin-IgG resulted in monospecific detection of p110 (Fig. 6C). The Mr difference between p110 and p107 is indicative of covalent attachment of a single ubiquitin molecule to p107, thus giving rise to p110. No additional higher Mr immunoreactive PEPC or ubiquitin bands indicative of polyubiquitination were observed. USP-2 catalyzed p110’s deubiquitination without influencing p107’s phosphorylation status, while eliciting a progressive 25% increase in PEPC activity when assayed under suboptimal conditions (Fig. 6, C and D). A further 15% increase in PEPC activity followed the apparent phosphorylation of deubiquitinated p110 subunits (i.e. now p107) by the catalytic subunit of protein kinase-A (Fig. 6D), which effectively in vitro phosphorylates the N-terminal seryl phosphorylation site of Class-1 PEPCs (Tripodi et al., 2005; O’Leary et al., 2011a).
Monoubiquitination was recently discovered to be a novel PTM of vascular plant PTPCs, e.g. Arabidopsis seedlings and most tissues of the castor plant (Uhrig et al., 2008; O’Leary et al., 2011b), maize (Zea mays) roots and leaves (Prinsi et al., 2009), and lily (Lilium spp.) pollen (Igawa et al., 2010). Prinsi and coworkers (2009) showed that nitrate availability may influence monoubiquitination and phosphorylation of PTPC in maize roots and leaves, respectively. Our results indicate that monoubiquitination may play a role in attenuating the in vivo PEPC activity of immature proteoid roots of –Pi harsh hakea.
Influence of Pi Deprivation on PEPC Abundance, Activity, and PTMs in Nonproteoid Roots
Nonproteoid roots of –Pi plants (Supplemental Fig. S2) exhibited significantly greater total PEPC protein abundance, phosphorylation, and specific activity compared to roots of +Pi plants (Supplemental Fig. S7). This is not surprising, since levels of PEPC protein and phosphorylation status and/or activity are known to be markedly up-regulated in response to Pi deprivation in a wide range of plant species (Duff et al., 1989; Johnson et al., 1996; Uhde-Stone et al., 2003; Vance et al., 2003; Gregory et al., 2009; O’Leary et al., 2011a; Plaxton and Tran, 2011). Immunoblots of extracts prepared from meristematic root tips of +Pi and –Pi plants had equivalent ratios of immunoreactive p110:p107 subunits. In older regions of these roots (15 cm proximal to the tips) there was a substantial reduction in the amount of p110 (Supplemental Fig.S7A). Anti-pSer11 immunoblotting indicated that, similar to the PEPC from proteoid roots (Fig. 2C), the p107 subunits from nonproteoid roots of –Pi plants was phosphorylated, but not the p110 subunits Supplemental Fig. S7B).
CONCLUDING REMARKS
PEPC is an important regulatory enzyme situated at a key branch point of primary plant metabolism that is believed to play a pivotal role in the metabolic adaptations of –Pi plants (Supplemental Fig. S1). This study documented a novel developmental pattern of PEPC PTMs as related to carbon metabolism in maturing proteoid roots of –Pi harsh hakea. As summarized in the model presented in Supplemental Figure S8, we discovered that (1) monoubiquitination and phosphorylation can simultaneously occur on different, but otherwise identical, subunits of an oligomeric native enzyme, and (2) deubiquitination of the monoubiquitinated subunits, followed by phosphorylation of the deubiquitinated subunits, appears to function as an in vivo enzyme activation mechanism (Fig. 2, B and C). To the best of our knowledge, both discoveries are unprecedented for any metabolic enzyme in nature, including a C3 PEPC. Monoubiquitination has been more generally implicated in mediating protein-protein interactions and protein localization to control diverse processes including membrane trafficking, gene expression, and DNA repair and replication (Sadowski et al., 2012). Thus, future studies are needed to assess potential ubiquitin-binding domain proteins that might interact with monoubiquitinated plant PEPCs, as well as the possible influence of this PTM on the enzyme’s subcellular localization. It will also be of interest to determine (1) the signaling pathway and E3-ubiquitin ligase that mediates PEPC monoubiquitination, and (2) whether an endogenous deubiquitinating enzyme catalyzes in vivo deubiquitination of Class-1 PEPC’s p110 subunits during proteoid root maturation in –Pi harsh hakea.
Elimination of photosynthate supply to developing COS caused by excision of intact fruit clusters resulted in the endosperm’s phosphorylated Class-1 PEPC homotetramer to be dephosphorylated, concomitant with disappearance of PPCK activity (Tripodi et al., 2005; Murmu and Plaxton, 2007). This was followed by monoubiquitination of 50% of the COS Class-1 PEPC’s p107 subunits to form the p110:p107 heterotetrameric Class-1 PEPC characteristic of germinating COS (Uhrig et al., 2008; O’Leary et al., 2011b). By contrast, the strong carbohydrate sink that develops during maturation of –Pi harsh hakea proteoid roots (Shane and Lambers, 2005) appears to have provoked a completely opposite set of Class-1 PEPC PTMs to those documented following COS depodding, e.g. in vivo deubiquitination of p110 followed by increased p107-phosphorylation (Fig. 2, B and C). Studies of –Pi white lupin have revealed that (1) up to 25% of photosynthetically fixed CO2 is translocated to its proteoid roots to support organic acid synthesis and exudation into the rhizosphere, whereas (2) anaplerotic CO2 fixation in the proteoid roots contributes about 25% and 34% of the carbon excreted as citrate and malate, respectively (Neumann and Martinoia, 2002; Vance et al., 2003). Similarly, the majority of photosynthate produced by leaves of –Pi harsh hakea is exported to support growth, respiration, and organic acid anion synthesis and exudation by the proteoid roots (Shane and Lambers, 2005). Our results support the hypothesis that the combined effects of in vivo deubiquitination and increased phosphorylation, coupled with enhanced levels of the allosteric activator Glc-6-P (Fig. 3B), provide a synergistic metabolic control mechanism that facilitates PEPC’s rapid anaplerotic carboxylation of PEP in support of the massive organic acid synthesis and exudation that dominates the carbon metabolism of mature proteoid roots of –Pi harsh hakea (Shane et al., 2004a).
This study also emphasizes the potential for significant discovery arising from investigations of physiological and biochemical adaptations of native nonmycotrophic plants that have evolved for millennia in habitats extremely deficient in bioavailable Pi. It is of growing concern internationally to capitalize on the enormous diversity of plant form and function to unearth novel adaptive traits that might be eventually genetically engineered into modern crop varieties so they can effectively exploit the significant amounts of insoluble Pi that remain mineral bound in agricultural soils. Clearly, the integration of developmental biology and physiology with genomics, metabolomics, proteomics, and native enzyme biochemistry is needed to achieve a thorough understanding of the intricate mechanisms by which plants acclimate to nutritional Pi deficiency. Our results suggest that while gene expression is important, future research must also focus on protein PTMs and the signaling pathways that control them.
MATERIALS AND METHODS
Plant Material
Roots were obtained from hydroponically grown harsh hakea (Hakea prostrata) plants propagated in glasshouses at the University of Western Australia during summer as described by Shane et al. (2004a) with the following modifications. Initially, 16-week-old plants were transferred from a +Pi soil to –Pi hydroponic media, followed by a 4-week adjustment period. Experimental plants were cultivated for 12 weeks in 2-L pots containing basal nutrient solution supplemented with 1 or 25 μm Pi (supplied as K2HPO4), with the entire nutrient solution replaced daily. Proteoid roots of –Pi plants at stages I, II, III, and IV of development (Fig. 1A) were harvested. Noncluster roots were also harvested from +Pi and –Pi plants at two stages of development (1 cm lengths, including root tips [0–1 cm], and 14–15 cm proximal to root tips). All roots were harvested between 11 am and 2 pm by rapid excision. The roots were blotted with paper towels, weighed, and immediately frozen in liquid nitrogen. Roots were transported on dry ice to Queen’s University (Kingston, Canada) and stored at −80°C.
Whole Root Material for Light Microscopy
Proteoid roots at various stages of development were stained with basic fuchsin and cleared according to Runzin (1999). Excised rootlets were treated for 12 h at 60°C in 1% (v/v) basic fuchsin and 1% (w/v) NaOH, then washed in several changes of water, and cleared for several weeks in a saturated solution of chloral hydrate. Individual rootlets were observed by bright-field optics using a Zeiss Axiophot microscope. Photomicrographs were recorded digitally using a Nikon D700 camera (Martin Microscope) and processed using Photoshop CS5 software.
Enzyme and Protein Concentration Assays and PEPC Kinetic Studies
Enzyme assays were conducted at 25°C using a kinetics microplate spectrophotometer (Spectramax Plus, Molecular Devices) to follow (1) NADH oxidation (for PEPC and MDH) or NAD+ reduction (for SUS and INV) at 340 nm, or (2) the oxidation of acetyl-CoA to CoA by CS at 412 nm. All assays were optimized with respect to pH, substrate, and cofactor concentration, and were linear with respect to time and concentration of enzyme assayed. One unit of activity is defined as the amount of enzyme resulting in the production of 1 µmol product min−1.
PEPC was assayed under optimal or suboptimal conditions. PEPC’s optimal assay reaction mix contained 50 mm HEPES-KOH (pH 8.2), 15% (v/v) glycerol, 2.5 mm PEP, 2 mm KHCO3, 5 mm MgCl2, 1 mm dithiothreitol (DTT), 0.2 mm NADH, and 5 units mL−1 of desalted porcine muscle MDH; suboptimal conditions were the same, except that assays were conducted at pH 7.2 with subsaturating PEP (0.2 mm) and 0.125 mm l-malate. The MDH reaction mix contained 25 mm HEPES-KOH (pH 7.5), 1 mm OAA, and 0.2 mm NADH. The CS reaction mix contained 50 mm HEPES-KOH (pH 7.5), 1 mm 5,5′-dithio-bis-nitrobenzoic acid, 0.5 mm acetyl-CoA, and 2 mm OAA. OAA stocks were stored at −80°C as a 1 m solution in 0.5 m HCl. The SUS reaction mix contained 50 mm HEPES-KOH (pH 7.0), 100 mm Suc, 1 mm UDP, 0.5 mm ATP, 0.5 mm NAD+, 2 mm MgCl2, 2 units mL−1 hexokinase, 1.5 units mL−1 phospho-Glc isomerase, and 1 unit mL−1 of Leuconostoc mesenteroides Glc-6-P dehydrogenase (Roche Canada). SUS activities were corrected for contaminating INV activity by omitting UDP from the reaction mix. Protein concentrations were determined using the Coomassie Blue G-250 dye binding method with bovine γ-globulin as the protein standard.
PEPC’s apparent Km, I50, and Ka values (concentrations of inhibitors and activators producing 50% inhibition and activation, respectively) were calculated using the kinetics program of Brooks (1992). All kinetic parameters are the means of triplicate determinations of at least n = three biological replicates. Stock solutions of all metabolites were made equimolar with MgCl2 and adjusted to pH 7.2.
Preparation of Clarified Extracts
Quick-frozen tissues were ground to a powder under liquid nitrogen using a mortar and pestle and homogenized (1:1.5, w/v) using a PT-3100 Polytron with ice-cold buffer A containing 50 mm imidazole-HCl (pH 7.0), 0.1% (v/v) Triton X-100, 10% (v/v) glycerol, 10 mm thiourea, 2 mm MgCl2, 2% (w/v) polyethylene glycol 8,000, 25 mm NaF, 1 mm NaMoO4, 1 mm NaVO3, 1% (w/v) polyvinyl(polypyrrolidone), 1% (w/v) polyvinylpyrrolidone, and 2.5 μL mL−1 ProteCEASE-100 (G-Biosciences). Homogenates were centrifuged at 17,500g for 15 min at 4°C. Supernatants (0.5 mL) were desalted through 3 mL Sephadex G-25 spin columns equilibrated in extraction buffer [lacking polyvinyl(polypyrrolidone) and polyvinylpyrrolidone].
Buffers Used during PEPC Purification
All buffers were adjusted to their respective pH at 25°C and contained 2 mm MgCl2 and a phosphatase inhibitor cocktail (25 mm NaF, 1 mm EDTA, 1 mm NaMoO4, and 1 mm NaVO3), whereas 50 nm microcystin-LR and 2.5 μl mL−1 ProteCEASE-100 were added to all pooled column fractions to further inhibit any copurifying phosphatase or protease activity. Buffer B contained 50 mm imidazole-HCl (pH 7.1), 1 mm DTT, and 25% (saturation) (NH4)2SO4. Buffer C consisted of buffer B lacking (NH4)2SO4, but containing 10% (v/v) ethylene glycol. Buffer D contained 50 mm imidazole-HCl (pH 7.8), 5 mm KCl, 1 mm DTT, and 15% (v/v) glycerol.
PEPC Purification from Stage I and II Proteoid Roots
Both chromatography steps were carried out at room temperature (25°C) using an ÄKTA FPLC system (GE Healthcare Bio-Sciences Inc.). Quick-frozen stage I and II proteoid roots (8.5 g each) were combined and ground under liquid nitrogen using a mortar and pestle, homogenized in ice-cold buffer A (1:1.5; w/v) using the Polytron, and centrifuged at 4°C and 18,000g for 20 min. The supernatant was brought to 25% (saturation) (NH4)2SO4, stirred for 20 min at 4°C, and centrifuged. The supernatant was adjusted to 60% (saturation) (NH4)2SO4, then stirred and centrifuged. The resulting pellet was resuspended in 5 mL of buffer B, clarified by centrifugation for 15 min, and loaded at 2 mL min−1 on to a column (1 × 2.5 cm) of butyl-Sepharose 4 Fast Flow (GE Healthcare Bio-Sciences Inc.) preequilibrated with buffer B. The column was washed until the A280 decreased to baseline and PEPC eluted with a linear gradient (50 mL) of decreasing concentrations of buffer B (100%–0%) and simultaneously increasing concentrations of buffer C (0%–100%; Supplemental Fig. S6). Pooled peak activity fractions were concentrated to 0.65 mL using AMICON Ultra-15 ultrafiltration devices (30-kD cutoff) and applied at 0.3 mL min−1 onto a Superdex-200 HR 16/60 gel filtration column (GE Healthcare Bio-Sciences Inc.) that had been preequilibrated with buffer D. Peak activity fractions (Fig. 4) were pooled and concentrated as above to 0.3 mL. Aliquots (25 μl) were frozen in liquid nitrogen, and stored at –80°C. PEPC activity remained stable for at least 3 months when stored frozen.
Determination of Native Molecular Mass via Superdex-200 Gel Filtration
PEPC’s native Mr was estimated during FPLC on the Superdex-200 column as described above. Native Mr was calculated from a plot of Kav (partition coefficient) against log Mr for the following protein standards: thyroglobulin (669 kD), ferritin (440 kD), catalase (232 kD), aldolase (158 kD), bovine serum albumin (67 kD), and chymotrypsinogen (25 kD).
Electrophoresis and Immunoblotting
Nondenaturing and SDS-PAGE using a Bio-Rad mini-gel apparatus (7% and 10% separating gels, respectively), in-gel PEPC activity staining, and immunoblotting were conducted as previously described (Tripodi et al., 2005; Gregory et al., 2009; O’Leary et al., 2011b). Antigenic polypeptides were visualized using an alkaline phosphatases-conjugated secondary antibody and chromogenic detection. All gel and immunoblot results were replicated a minimum of three times with representative results shown in the various figures. Preparation of rabbit anti-COS PEPC and pSer11 has been described elsewhere (Tripodi et al., 2005; Gennidakis et al., 2007). For anti-pSer11 immunoblots the corresponding dephosphopeptide (10 µg mL−1) was used to block any nonspecific cross reaction with nonphosphorylated PEPC (Tripodi et al., 2005). Antiubiquitin-IgG (catalog no. 05-944) was purchased from Millipore Ltd.. Purified PEPCs from germinating COS and –Pi Arabidopsis (Arabidopsis thaliana) suspension cells were obtained as previously described (Uhrig et al., 2008; Gregory et al., 2009). The relative amount of PEPC polypeptides was estimated by densitometry of immunoreactive p110 and p107 using ImageJ (http://rsbweb.nih.gov/ij/). Derived values were linear with respect to the amount of the immunoblotted extract.
MS
Excised gel bands were destained with 100 mm NH4HCO3/acetonitrile (ACN; 1:1, v/v), dehydrated in 100% ACN, and dried using a SpeedVac. Following reduction with 10 mm DTT (in 100 mm NH4HCO3) at 56°C for 1 h, proteins were alkylated with an equal volume of 55 mm iodoacetamide at 23°C for another 45 min. Subsequent digestion was performed using 6 ng μL−1 of sequencing grade trypsin (EMD Millipore, Canada) in 25 mm NH4HCO3 (pH 7.6) at 37°C overnight. The resulting peptides were extracted by successive sonication with 0.1% (v/v) trifluoroacetic acid, 0.1% trifluoroacetic acid in 50% (v/v) ACN, and 100% ACN followed by a C18 ZipTip (Millipore) cleanup step. The peptides were deposited on a MALDI target by mixing with an equal volume (0.5 μL) of 2,5-dihydroxybenzoic acid matrix (100 mg mL−1 in 50% ACN). MALDI data were acquired using an Applied Biosystems/MDS Sciex QStar XL Q-TOF mass spectrometer equipped with an oMALDI II source and a nitrogen laser operating at 337 nm. Peptide sequencing of the selected ions was carried out by MALDI Q-TOF MS/MS measurements using argon as the collision gas. All peptide fingerprinting masses and MS/MS ion searches were performed through Mascot search engine (MatrixScience, http://www.matrixscience.com) using the National Center for Biotechnology Information nonredundant database (NCBIrn released 9/12/2008; containing 7,031,513 protein sequences). The mass tolerance between calculated and observed masses used for database search was considered at the range of ±50 ppm for MS peaks and ±0.1 kD for MS/MS fragment ions.
In Vitro Dephosphorylation, Deubiquitination, or Phosphorylation of PEPC
Aliquots (50 μL) of purified PEPC were desalted into a standard dephosphorylation buffer (50 mm Tris-HCl, pH 7.2, containing 5 mm MgCl2, 1 mm DTT, and 20% [v/v] glycerol) using Micro Spin-OUT GT-1200 desalting columns (G-Biosciences) according to the manufacturer’s instructions. Desalted PEPC (35 μg) was incubated at 30°C with 5 milliunits mL−1 of the catalytic subunit of bovine heart PP2A (Gregory et al., 2009) in 50 μL of dephosphorylation buffer in the presence and absence of 50 nm microcystin-LR. Aliquots were withdrawn at specified times and assayed for PEPC activity or analyzed by SDS-PAGE and immunoblotting as described above.
Deubiquitination was performed by incubating aliquots (100 μL) of desalted extracts (400 μg protein) of stage I roots, or purified PEPC (100 μg protein) from stage I to II roots, at 37°C for up to 1 h in the presence and absence of 15 μm USP-2 (Progenra Inc.) in 50 mm Tris-HCl (pH 7.5), 50 mm KCl, 5 mm MgCl2, 20% (v/v) glycerol, 1 mm DTT, phosphatase inhibitors (50 nm microcystin-LR, 0.5 mm NaVO3, and 0.5 mm NaMoO4), and 2.5 μL mL−1 ProteCEASE-100 (G-Biosciences). Aliquots of USP-2-treated PEPC were subsequently incubated at 30°C with 20 units mL−1 of the catalytic subunit of bovine heart protein kinase-A and 1 mm MgATP. Aliquots were withdrawn at specified times and assayed for PEPC activity or analyzed by immunoblotting as described above.
Leaf Free Phosphate and Total Phosphorus Determinations
Mature leaf tissue (200 mg) was either extracted in 5 m H2SO4 for free Pi assays (Delhaize and Randall, 1995), or digested in concentrated HNO3:HClO4 (3:1) at 175°C for total phosphorus assays (Motomizu et al., 1983). Po was estimated by subtracting free Pi from total phosphorus.
Malate and Glc-6-P Determinations
Proteoid roots (100 mg) were extracted (1:5, w/v) with 0.6 m perchloric acid at 4°C and centrifuged for 5 min at 15,000g as described in Delhaize et al. (1993). Extracts were neutralized with 1 m K2CO3 and centrifuged for 5 min. Aliquots of the supernatant were assayed for malate or Glc-6-P using assay kits (catalog no. MAK067 and MAK014; Sigma-Aldrich) following procedures provided by the manufacturer.
Statistics
All values are presented as means ± se. Data were analyzed using GraphPad Prism 5 comprising ANOVA followed by Student’s t test, and deemed significant if P < 0.05.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Model highlighting PEPC’s metabolic functions during plant acclimation to nutritional Pi deprivation.
Supplemental Figure S2. Representative root system of hydroponically grown –Pi harsh hakea after supplementation with 1 µm Pi for 12 weeks.
Supplemental Figure S3. Influence of Pi supply on shoot and root biomass, and free Pi and Po concentrations of mature leaves of harsh hakea.
Supplemental Figure S4. Developmental profiles for activities of SUS, INV, CS, and MDH in proteoid root extracts of –Pi harsh hakea.
Supplemental Figure S5. SDS-PAGE and immunoblot analysis of various fractions obtained during PEPC purification from stage I and II proteoid roots.
Supplemental Figure S6. Coelution of PEPC activity with p110 and p107 during butyl-Sepharose FPLC of PEPC from stage I and II proteoid roots.
Supplemental Figure S7. PEPC activities and immunoblotting of nonproteoid root extracts of –Pi and +Pi harsh hakea plants.
Supplemental Figure S8. Model summarizing harsh hakea proteoid root development in relation to variation in PEPC activity, relative amount, phosphorylation, monoubiquitination, oligomeric subunit composition, and organic acid accumulation/exudation.
Supplemental Table S1. Influence of various metabolites on the activity of PEPC from desalted extracts of stage I and III proteoid roots.
Supplemental Table S2. Purification of PEPC from 17 g of stage I and II proteoid roots.
Supplemental Table S3. MALDI Q-TOF MS analysis of tryptic peptides derived from p110, p107, and p93 of the final preparation of PEPC isolated from stage I and II proteoid roots.
Acknowledgments
We thank Dr. Yamin Ma for assisting with growing and harvesting the hakea plants, Dr. Craig Leach (Progenra, Inc.) for the gift of USP-2, and Dr. Raymond Chollet (University of Nebraska) for the gift of anti-soybean root nodule SUS-IgG. We also thank Drs. Brendan O’Leary (University of Oxford), Uli Mathesius (Australian National University), Harvey Millar (University of Western Australia), and John Raven (University of Dundee) for their helpful comments on earlier versions of the manuscript. We gratefully acknowledge the two anonymous referees for their constructive criticisms and suggestions. MS and MS/MS analyses were performed at the Mass Spectrometry and Proteomics Unit, Department of Chemistry, Queen’s University.
Glossary
- PEPC
phosphoenolpyruvate carboxylase
- Pi
inorganic phosphate
- OAA
oxaloacetate
- PEP
phosphoenolpyruvate
- PTPC
plant-type PEPC
- BTPC
bacterial-type PEPC
- PP2A
protein phosphatase type-2A
- COS
castor oil seed
- MS
mass spectrometry
- MS/MS
tandem mass spectrometry
- PTM
posttranslational modification
- –Pi
Pi-starved
- +Pi
optimal Pi
- SUS
Suc synthase
- INV
invertase
- CS
citrate synthase
- MDH
malate dehydrogenase
- USP-2
ubiquitin-specific protease-2 core
- MALDI
matrix-assisted laser-desorption ionization
- Q-TOF
quadrupole time-of-flight
- anti-PEPC
anti-castor oil seed Class-1 phosphoenolpyruvate carboxylase-IgGs
- anti-pSer11
anti-pSer11 phospho-site specific
- DTT
dithiothreitol
- Gly-3-P
glycerol-3-P
- [P]
phosphate concentration
- ACN
acetonitrile
- Po
organic phosphate
References
- Brooks S. (1992) A simple computer program with statistical tests for the analysis of enzyme kinetics. Biotechniques 13: 906–911 [PubMed] [Google Scholar]
- Delhaize E, Ryan PR, Randall PJ. (1993) Aluminum tolerance in wheat (Triticum aestivum L.). II. Aluminum-stimulated excretion of malic acid from root apices. Plant Physiol 103: 695–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhaize E, Randall PJ. (1995) Characterization of a phosphate-accumulator mutant of Arabidopsis thaliana. Plant Physiol 107: 207–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duff SMG, Moorhead GBG, Lefebvre DD, Plaxton WC. (1989) Phosphate starvation inducible “bypasses” of adenylate and phosphate dependent glycolytic enzymes in Brassica nigra suspension cells. Plant Physiol 90: 1275–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gennidakis S, Rao S, Greenham K, Uhrig RG, O’Leary B, Snedden WA, Lu C, Plaxton WC. (2007) Bacterial- and plant-type phosphoenolpyruvate carboxylase polypeptides interact in the hetero-oligomeric Class-2 PEPC complex of developing castor oil seeds. Plant J 52: 839–849 [DOI] [PubMed] [Google Scholar]
- Gregory AL, Hurley BA, Tran HT, Valentine AJ, She YM, Knowles VL, Plaxton WC. (2009) In vivo regulatory phosphorylation of the phosphoenolpyruvate carboxylase AtPPC1 in phosphate-starved Arabidopsis thaliana. Biochem J 420: 57–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igawa T, Fujiwara M, Tanaka I, Fukao Y, Yanagawa Y. (2010) Characterization of bacterial-type phosphoenolpyruvate carboxylase expressed in male gametophyte of higher plants. BMC Plant Biol 10: 200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JF, Vance CP, Allan DL. (1996) Phosphorus deficiency in Lupinus albus. Altered lateral root development and enhanced expression of phosphoenolpyruvate carboxylase. Plant Physiol 112: 31–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Yin L, Reid J, Wilkinson KD, Wing SS. (2001) Divergent N-terminal sequences of a deubiquitinating enzyme modulate substrate specificity. J Biol Chem 276: 20357–20363 [DOI] [PubMed] [Google Scholar]
- Massonneau A, Langlade N, Léon S, Smutny J, Vogt E, Neumann G, Martinoia E. (2001) Metabolic changes associated with cluster root development in white lupin (Lupinus albus L.): relationship between organic acid excretion, sucrose metabolism and energy status. Planta 213: 534–542 [DOI] [PubMed] [Google Scholar]
- Motomizu S, Wakimoto T, Toei F. (1983) Spectrophotometric determination of phosphate in river waters with molybdate and malachite green. Analyst 108: 361–367 [DOI] [PubMed] [Google Scholar]
- Murmu J, Plaxton WC. (2007) Phosphoenolpyruvate carboxylase protein kinase from developing castor oil seeds: partial purification, characterization, and reversible control by photosynthate supply. Planta 226: 1299–1310 [DOI] [PubMed] [Google Scholar]
- Neumann G, Martinoia E. (2002) Cluster roots—an underground adaptation for survival in extreme environments. Trends Plant Sci 7: 162–167 [DOI] [PubMed] [Google Scholar]
- O’Leary B, Rao SK, Kim J, Plaxton WC. (2009) Bacterial-type phosphoenolpyruvate carboxylase (PEPC) functions as a catalytic and regulatory subunit of the novel class-2 PEPC complex of vascular plants. J Biol Chem 284: 24797–24805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Leary B, Park J, Plaxton WC. (2011a) The remarkable diversity of plant PEPC (phosphoenolpyruvate carboxylase): recent insights into the physiological functions and post-translational controls of non-photosynthetic PEPCs. Biochem J 436: 15–34 [DOI] [PubMed] [Google Scholar]
- O’Leary B, Fedosejevs ET, Hill AT, Bettridge J, Park J, Rao SK, Leach CA, Plaxton WC. (2011b) Tissue-specific expression and post-translational modifications of plant- and bacterial-type phosphoenolpyruvate carboxylase isozymes of the castor oil plant, Ricinus communis L. J Exp Bot 62: 5485–5495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Khuu N, Howard AS, Mullen RT, Plaxton WC. (2012) Bacterial- and plant-type phosphoenolpyruvate carboxylase isozymes from developing castor oil seeds interact in vivo and associate with the surface of mitochondria. Plant J 71: 251–262 [DOI] [PubMed] [Google Scholar]
- Prinsi B, Negri AS, Pesaresi P, Cocucci M, Espen L. (2009) Evaluation of protein pattern changes in roots and leaves of Zea mays plants in response to nitrate availability by two-dimensional gel electrophoresis analysis. BMC Plant Biol 9: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaxton WC, Tran HT. (2011) Metabolic adaptations of phosphate-starved plants. Plant Physiol 156: 1006–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runzin SE (1999) Plant Microtechnique and Microscopy. Oxford University Press, New York. [Google Scholar]
- Sadowski M, Suryadinata R, Tan AR, Roesley SN, Sarcevic B. (2012) Protein monoubiquitination and polyubiquitination generate structural diversity to control distinct biological processes. IUBMB Life 64: 136–142 [DOI] [PubMed] [Google Scholar]
- Shane MW, Cramer MD, Funayama-Noguchi S, Cawthray GR, Millar AH, Day DA, Lambers H. (2004a) Developmental physiology of cluster-root carboxylate synthesis and exudation in harsh hakea. Expression of phosphoenolpyruvate carboxylase and the alternative oxidase. Plant Physiol 135: 549–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shane MW, De Vos M, De Roock S, Cawthray GR, Lambers H. (2003) Effects of external phosphorus supply on internal phosphorus concentration and the initiation, growth and exudation of cluster roots in Hakea prostrata R.Br. Plant Soil 248: 209–219 [Google Scholar]
- Shane MW, McCully ME, Lambers H. (2004b) Tissue and cellular phosphorus storage during development of phosphorus toxicity in Hakea prostrata (Proteaceae). J Exp Bot 55: 1033–1044 [DOI] [PubMed] [Google Scholar]
- Shane MW, Lambers H. (2005) Cluster roots: a curiosity in context. Plant Soil 274: 99–123 [Google Scholar]
- Tripodi KE, Turner WL, Gennidakis S, Plaxton WC. (2005) In vivo regulatory phosphorylation of novel phosphoenolpyruvate carboxylase isoforms in endosperm of developing castor oil seeds. Plant Physiol 139: 969–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhde-Stone C, Gilbert G, Johnson JMF, Litjens R, Zinn KE, Temple SJ, Vance CP, Allan DL. (2003) Acclimation of white lupin to phosphorus deficiency involves enhanced expression of genes related to organic acid metabolism. Plant Soil 248: 99–116 [Google Scholar]
- Uhrig RG, She YM, Leach CA, Plaxton WC. (2008) Regulatory monoubiquitination of phosphoenolpyruvate carboxylase in germinating castor oil seeds. J Biol Chem 283: 29650–29657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance CP, Uhde-Stone C, Allan DL. (2003) Phosphorus acquisition and use: critical adaptations by plants for securing a nonrenewable resource. New Phytol 157: 423–447 [DOI] [PubMed] [Google Scholar]
- Xu YF, Amador-Noguez D, Reaves ML, Feng XJ, Rabinowitz JD. (2012) Ultrasensitive regulation of anapleurosis via allosteric activation of PEP carboxylase. Nat Chem Biol 8: 562–568 [DOI] [PMC free article] [PubMed] [Google Scholar]