Autophagic recycling of proteins supplies nighttime energy sources such as amino acids under conditions of limited sugar availability.
Abstract
Autophagy is an intracellular process leading to the vacuolar degradation of cytoplasmic components. Autophagic degradation of chloroplasts is particularly activated in leaves under conditions of low sugar availability. Here, we investigated the importance of autophagy in the energy availability and growth of Arabidopsis (Arabidopsis thaliana). autophagy-deficient (atg) mutants showed reduced growth under short-day conditions. This growth inhibition was largely relieved under continuous light or under short-day conditions combined with feeding of exogenous sucrose, suggesting that autophagy is involved in energy production at night for growth. Arabidopsis accumulates starch during the day and degrades it for respiration at night. Nighttime energy availability is perturbed in starchless mutants, in which a lack of starch accumulation causes a transient sugar deficit at night. We generated starchless and atg double mutants and grew them under different photoperiods. The double mutants showed more severe phenotypes than did atg or starchless single mutants: reduced growth and early cell death in leaves were observed when plants were grown under 10-h photoperiods. Transcript analysis of dark-inducible genes revealed that the sugar starvation symptoms observed in starchless mutants became more severe in starchless atg double mutants. The contents of free amino acids (AAs) increased, and transcript levels of several genes involved in AA catabolism were elevated in starchless mutant leaves. The increases in branched-chain AA and aromatic AA contents were partially compromised in starchless atg double mutants. We conclude that autophagy can contribute to energy availability at night by providing a supply of alternative energy sources such as AAs.
Plants get energy for growth and survival via photosynthetic carbon assimilation during the day. Energy availability needs to be maintained to allow continuous plant growth throughout the day/night cycle (Smith and Stitt, 2007). In Arabidopsis (Arabidopsis thaliana), a portion of the photoassimilate is accumulated as starch and degraded during respiration at night. When diel energy availability is perturbed, a transient sugar deficit can occur, leading to temporal interruption of plant growth (Gibon et al., 2004; Graf et al., 2010). Thus, plants have to adapt to fluctuations of sugar availability due to changes in day/night periods or growth environments. Energy availability can be strongly perturbed by several suboptimal conditions, such as shading, flooding, drought, and extreme temperatures, that interfere with carbon assimilation and/or respiration (Baena-González et al., 2007; Baena-González and Sheen, 2008). Such low-energy stress is mimicked in starchless mutants, in which the lack of starch accumulation causes a sugar deficit at night (Gibon et al., 2004; Bläsing et al., 2005). Since plants cannot simply move out of suboptimal environments, they must metabolically produce alternative respiratory substrates as an energy source for survival and growth under these energy-limited conditions (Araújo et al., 2011).
Protein is an important respiratory substrate in heterotrophic mammalian systems. Because plant respiration is primarily dependent on sugar oxidation, the importance of protein and amino acids (AAs) in plant respiration remains unclear (Plaxton and Podesta, 2006). However, enzymatic pathways for the catabolism of alternative respiratory substrates seem to be induced in vegetative organs of plants when sugar availability strongly declines (Buchanan-Wollaston et al., 2005). It has been reported that an electron-transfer flavoprotein (ETF)/ETF:ubiquinone oxidoreductase (ETFQO) complex functions to transfer electrons to the ubiquinone pools in order to support respiration during continuous darkness (Ishizaki et al., 2005, 2006). Isovaleryl-CoA dehydrogenase (IVDH) and 2-hydroxyglutarate dehydrogenase (D2HGDH) have been identified as enzymes for AA breakdown and electron donors to the ETF/ETFQO complex (Araújo et al., 2010). Glu dehydrogenase (GDH) has also been suggested to play a key role in AA catabolism (Miyashita and Good, 2008). These reports demonstrate that AAs can be catabolized and become a respiratory substrate in plant systems.
Autophagy is a ubiquitous recycling system in eukaryotic cells by which proteins and organelles are transported for degradation in the vacuoles of yeast and plants or the lysosomes of animals (for review, see Nakatogawa et al., 2009; Li and Vierstra, 2012; Liu and Bassham, 2012; Yoshimoto, 2012). During autophagy, a portion of the cell’s cytoplasmic contents is sequestered by a double-membrane vesicle called an autophagosome and delivered to the vacuole/lysosome. The outer membrane of the autophagosome then fuses with the vacuolar/lysosomal membrane, and the inner membrane structure (called the autophagic body) is degraded by resident hydrolases. Autophagy-related genes (ATGs) are well conserved across yeast, plants, and animals (Meijer et al., 2007). Numerous analyses using RNA interference knockdown and transfer DNA-insertional knockout mutants of ATGs in Arabidopsis have revealed that the core machinery for autophagosomal membrane elongation is conserved in plants (Yoshimoto et al., 2004; Thompson et al., 2005; Xiong et al., 2005; Phillips et al., 2008; Chung et al., 2010; Suttangkakul et al., 2011). Autophagy is considered to play an important role in nutrient recycling under starvation conditions similar to its role in yeast and animals (Li and Vierstra, 2012; Liu and Bassham, 2012; Yoshimoto, 2012). Recently, Guiboileau et al. (2012) demonstrated the importance of autophagy in nitrogen remobilization of Arabidopsis.
We have demonstrated that chloroplastic proteins are degraded by autophagy via Rubisco-containing bodies (RCBs), a type of autophagic body containing chloroplast stroma (Chiba et al., 2003; Ishida et al., 2008). The chloroplast is an organelle specific to photoautotrophs and has a central role not only in photosynthesis but also in the assimilation of several mineral nutrients. Chloroplasts are the major source of material for autophagic recycling in plants, because the majority of plant nutrients are distributed to chloroplasts, such that chloroplastic proteins account for 75% to 80% of total leaf nitrogen in C3 plants (Makino et al., 2003). The RCB/autophagy system contributes to Rubisco degradation during leaf senescence (Ono et al., 2013), and RCB production is particularly activated in leaves under conditions of low sugar availability, such as in individually darkened leaves, leaves at the end of the night in a diurnal cycle, or leaves of starchless mutants (Wada et al., 2009; Izumi et al., 2010), suggesting the involvement of the RCB/autophagy system in energy production. Although autophagy is an important catabolic pathway regulating energy homeostasis in mammalian systems (Singh and Cuervo, 2011), a substantial role for autophagy in energy availability has not been reported in plant systems.
In this study, we investigated the importance of autophagy in diel energy availability and growth of Arabidopsis. autophagy-deficient (atg) mutants showed more severe phenotypes when sugar availability during the night was reduced due to shorter photoperiods and/or starchless mutations. Metabolic and transcript analyses indicated that autophagy provides a supply of energy sources such as AAs as an alternative to sugars. Our data demonstrate that autophagic recycling can contribute to plant energy availability.
RESULTS
Growth Retardation of Autophagy-Deficient Mutants Occurs under Short-Day Conditions But Not under Continuous Light
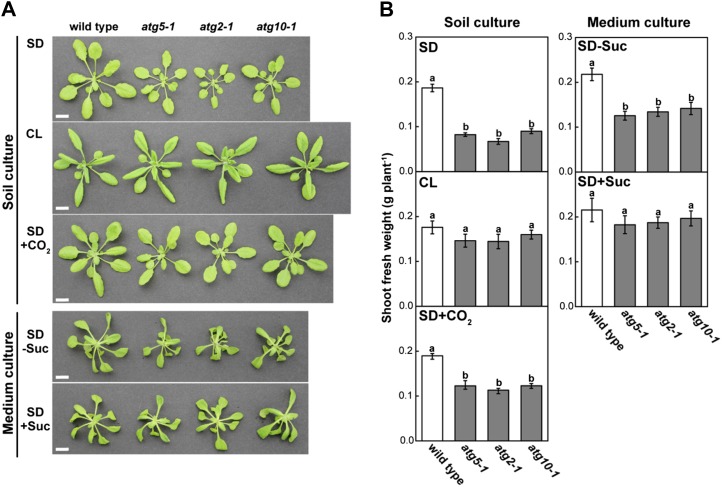
Photoperiod is a major factor controlling plant energy availability under nonstressed conditions: sugar availability from photosynthetic carbon assimilation is reduced under short days (SD; Gibon et al., 2004, 2009; Smith and Stitt, 2007). Although atg mutants of Arabidopsis can complete their life cycle irrespective of photoperiod conditions (Doelling et al., 2002; Hanaoka et al., 2002), their growth rates are clearly lower than those of wild-type plants under SD conditions (Guiboileau et al., 2012). First, we examined the vegetative growth rates of several atg mutants under SD (10 h of light/14 h of dark) and continuous light (CL) conditions (Fig. 1). All atg mutants grown in soil culture under SD displayed growth retardation: shoot fresh weight in atg mutants was 36% to 48% of the wild-type level (Fig. 1B). This reduced growth was not observed under CL. Furthermore, we grew wild-type and atg mutants under SD conditions with elevated CO2 concentrations (1,000–1,200 μL L−1), which increase the CO2 assimilation rate during the day and sugar availability (Cheng et al., 1998). Shoot fresh weight of atg mutants was 60% to 65% of the wild-type level; therefore, the SD-dependent reduced growth of the atg mutants was partially but not fully compensated for by elevated CO2 concentration. These results suggest that SD-dependent growth retardation of atg mutants is especially related to nighttime carbon utilization.
Figure 1.
SD-dependent growth retardation of atg single mutant plants. Wild-type, atg5-1, atg2-1, and atg10-1 plants were grown in soil culture for 30 d under SD (SD), for 23 d under CL (CL), or for 24 d with CO2 supply under SD (SD+CO2). The same lines were also grown in mineral-rich medium culture under SD for 28 d without Suc (SD−Suc) or for 23 d with 1% (w/v) Suc (SD+Suc). Images (A) and shoot fresh weight (B) were obtained when the shoot fresh weight of the wild type was approximately 0.2 g under the indicated conditions. Bars = 10 mm. The data represent means ± se (n = 5–8). Statistical analysis was performed using Tukey’s test; values with the same letter were not significantly different among individual genotypes (P ≤ 0.05).
We grew wild-type and atg mutant plants on mineral-rich medium with or without Suc to create sugar-excess conditions throughout the day/night cycle of SD (Fig. 1). Reduced growth of atg mutants was observed in Suc-free medium, similar to that observed in soil; however, atg growth was not significantly different from the wild type in Suc-rich medium (Fig. 1B). Shoot fresh weight for the wild type and atg mutants under all conditions corresponded to the observed plant size (Fig. 1). These findings suggest that autophagy contributes to plant growth under SD conditions via carbon metabolism at night.
Autophagy Is Required for Adaptation and Growth during Fluctuations in Energy Availability Due to Starchless Carbohydrate Metabolism
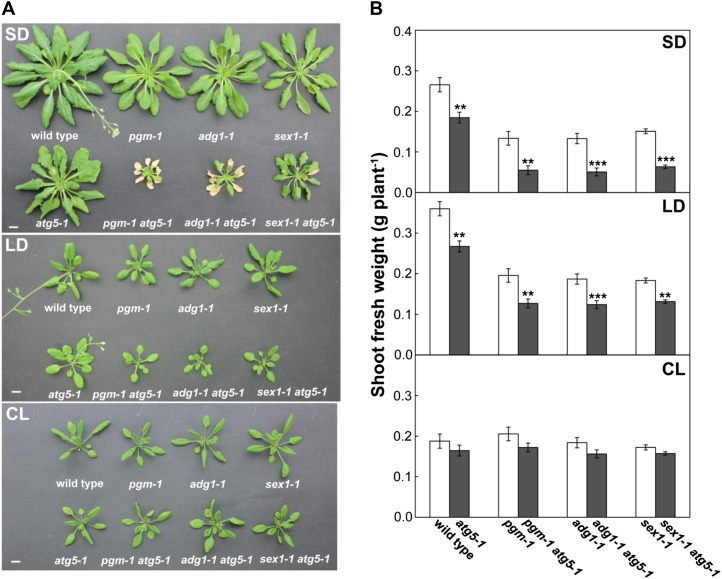
Sugar availability at night is severely perturbed in starchless mutants due to the lack of starch accumulation and partially perturbed in starch-excess mutants due to a reduced capacity for starch degradation, resulting in reduced energy availability at night (Caspar et al., 1985, 1991; Lin et al., 1988). To characterize further the role of autophagy in carbon metabolism, we generated double mutants containing a starchless mutation, phosphoglucomutase (pgm; Caspar et al., 1985) or ADP-glucose pyrophosphorylase1 (adg1; Lin et al., 1988), with an atg5 mutation (pgm atg5 and adg1 atg5). The starch-excess1 (sex1; Caspar et al., 1991) and atg5 double mutant was also generated (sex1 atg5). The wild-type and mutant plants were grown under different photoperiod conditions (SD, long days [LD; 14 h of light/10 h of dark], or CL) in soil culture. Under SD, pgm atg5 and adg1 atg5 exhibited early cell death in leaves before the transition to the reproductive stage (Fig. 2A). This phenotype was not observed under LD or CL conditions or in sex1 atg5 even under SD (Fig. 2A). Although a similar cell death phenomenon of early senescence is observed in atg single mutants (Yoshimoto et al., 2009), almost all the leaves of pgm atg5 and adg1 atg5 died visibly earlier than those of atg5, despite slower growth (Fig. 2A). Early cell death in leaves under SD was also observed in double mutants of adg1 with another atg mutation, atg2 (adg1 atg2; Supplemental Fig. S1).
Figure 2.
Starchless and autophagy-deficient double mutants display an early-cell-death phenotype in leaves and reduced growth under SD. A, The wild type, single mutant lines (atg5-1, pgm-1, adg1-1, sex1-1), and double mutant lines (pgm-1 atg5-1, adg1-1 atg5-1, sex1-1 atg5-1) were grown in nutrient-rich soil for 60 d under SD, for 30 d under LD, or for 23 d under CL. Bars = 10 mm. B, The same lines were grown for 40 d under SD, for 30 d under LD, or for 23 d under CL, and plant shoot fresh weight was measured. The data represent means ± se (n = 4–5). Statistical analysis was performed by Student’s t test; asterisks indicate significant differences between the lines with and without the atg5 mutation at the 5% (*), 1% (**), and 0.1% (***) levels, respectively.
The starchless atg double mutant plants were much smaller than wild-type plants and the respective single mutants under SD but were not different under CL conditions (Fig. 2A). To compare plant growth, we measured shoot fresh weight of the mutant lines (Fig. 2B). Shoot fresh weight in pgm, adg1, and sex1 was lower than that in the wild type under SD and LD. Thus, the growth retardation observed in starchless and starch-excess single mutants under shortened photoperiods was confirmed, and the atg5 mutation led to further reductions of shoot fresh weight in all backgrounds under SD and LD conditions. Under CL, there were no differences in shoot fresh weight among the different genotypes. The shoot fresh weight of the respective single and double mutants corresponded to the observed plant size (Fig. 2A). The growth retardation observed under shortened photoperiods was similar between atg mutants and starchless mutants, and the phenotypes were additive in the starchless atg double mutants under SD (Fig. 2). These findings also suggest that autophagy is involved in carbon utilization at night, particularly when sugar availability is perturbed.
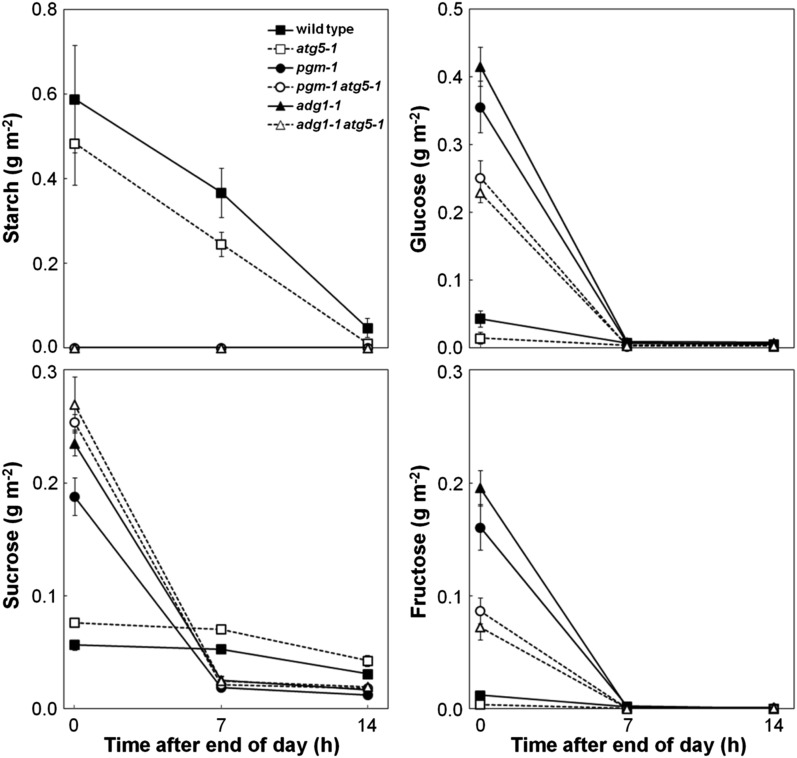
We measured leaf starch and sugar contents during the 14-h night under SD in the wild type, atg5, starchless mutants, and starchless atg double mutants to check the nighttime sugar availability (Fig. 3). In the wild type and atg5, starch accumulated at the end of the day gradually decreased during the night and was almost entirely consumed at the end of the night. Suc levels were maintained throughout the night in both lines. These diurnal changes represent normal carbohydrate metabolism in Arabidopsis (Smith and Stitt, 2007). In pgm and adg1, a lack of accumulation of starch led to an obvious accumulation of soluble sugars at the end of the day; however, these sugars rapidly decreased at the beginning of the night, and sugars were almost exhausted before the middle of the night (Fig. 3). In pgm atg5 and adg1 atg5, carbohydrate metabolism during the night was the same as that observed in starchless single mutants, with the exception that Suc contents tended to be higher and Glc and Fru contents were lower than in starchless single mutants at the end of the day (Fig. 3). Thus, the occurrence of sugar exhaustion during the night was obvious in starchless atg double mutants.
Figure 3.
Sugar exhaustion in starchless mutant leaves at night. Starch, Suc, Glc, and Fru contents were measured in the leaves of the wild type, single mutant lines (atg5-1, pgm-1, adg1-1), and double mutant lines (pgm-1 atg5-1, adg1-1 atg5-1) at 0, 7, and 14 h after the end of the day during the 14-h night 40 d after sowing. The data represent means ± se (n = 4).
Autophagy Deficiency Makes Starvation Symptoms More Severe under a Transient Sugar Deficit
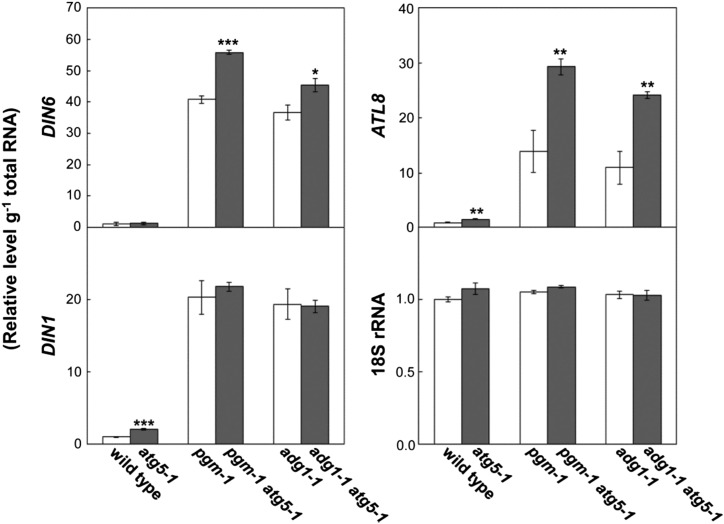
Next, to assess the extent of the starvation symptoms during the night, we measured transcript levels of DIN6 (Fujiki et al., 2001b), ATL8 (Graf et al., 2010), and DIN1 (Oh et al., 1996), as dark-inducible genes (DINs), in leaves at the end of the night of SD (Fig. 4). DIN transcripts are acutely induced upon the initiation of sugar starvation, and the DINs analyzed here have been established as appropriate reporters of sugar starvation symptoms (Baena-González et al., 2007; Graf et al., 2010). Transcript levels of ATL8 and DIN1 were significantly elevated in atg5 compared with the wild type. However, transcript levels of analyzed DINs were much higher in pgm and adg1 than in the wild type and atg5, indicating that sugar starvation was indeed occurring in leaves of starchless mutants at the end of the night. In addition, transcript levels of DIN6 and ATL8 were further elevated in pgm atg5 and adg1 atg5, implying that the extent of the starvation symptoms was more severe in starchless atg double mutants than in starchless single mutants.
Figure 4.
Transcript levels of DINs as a marker of sugar starvation symptoms during the night. Total RNA was isolated from leaves of the wild type, single mutant lines (atg5-1, pgm-1, adg1-1), and double mutant lines (pgm-1 atg5-1, adg1-1 atg5-1) at the end of the night 40 d after sowing and subjected to qRT-PCR analysis to measure transcript levels of DIN6, DIN1, and ATL8. 18S rRNA was used as an internal control. mRNA levels are shown relative to the wild-type level, which is represented as 1. The data represent means ± se (n = 4). Statistical analysis was performed by Student’s t test; asterisks indicate significant differences between lines with or without the atg5 mutation at the 5% (*), 1% (**), and 0.1% (***) levels, respectively.
To provide additional support for this conclusion, we analyzed transcript levels of DINs during extended night in the atg single mutant (Supplemental Fig. S2). Wild-type and atg5 plants were grown for 30 d under SD and then subjected to a 6-h extended night beyond the normal 14-h night. The extended night treatment has been established as an experimental system for temporal sugar starvation symptoms in Arabidopsis, because it causes starch exhaustion and acute responses to sugar deficits (Smith and Stitt, 2007; Graf et al., 2010). While DIN6, ATL8, and DIN1 transcript levels were strongly elevated in wild-type leaves during the extended night, this elevation occurred earlier in atg5 (Supplemental Fig. S2). Two hours after the start of the extended night, DIN6, ATL8, and DIN1 transcript levels in atg5 leaves were 2.1-, 2.2-, and 1.5-fold greater than in wild-type leaves, respectively. These results support the notion that autophagy deficiency causes more severe starvation symptoms during a sugar deficit.
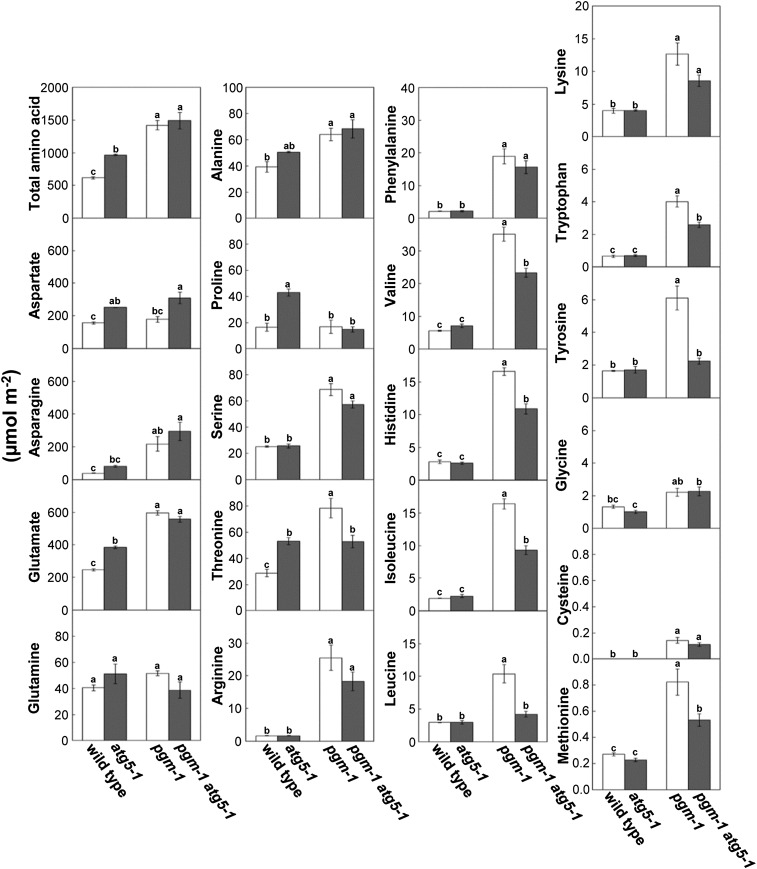
Increases in AA Contents of Starchless Mutants Were Partially Compromised by Deficiency in Autophagy
We hypothesized that autophagic protein degradation supplies free AAs as an alternative respiratory substrate. To examine the effects of starchless and/or atg mutations on the free AA pool, we performed capillary electrophoresis time-of-flight mass spectrometry (CE-TOFMS) analysis and measured AA contents in the leaves of wild-type, atg5, pgm, and pgm atg5 plants (Fig. 5). The contents of the individual AAs largely increased in pgm compared with the wild type, with the exception of Asp, Gln, and Pro. Therefore, the total AA content in pgm leaves increased to 2.3-fold of the wild-type amount. The increases in branched-chain amino acids (BCAAs; Leu, Ile, and Val), aromatic AAs (Phe, Trp, and Tyr) except Phe, and Thr, His, and Met observed in pgm were partially compromised in pgm atg5. Because the content of Asp, a quantitatively major AA, was higher in pgm atg5 than in pgm, the total AA content did not differ between the two genotypes. In atg5 single mutants, individual AA contents in leaves did not decrease compared with the wild type.
Figure 5.
Increases in leaf AA contents in starchless mutants were partially compromised in starchless atg double mutants. AA contents were measured in leaves of the wild type, atg5-1, pgm-1, and pgm-1 atg5-1 at the end of the night under SD by CE-TOFMS analysis. The data represent means ± se (n = 3). Statistical analysis was performed by Tukey’s test; values with the same letter were not significantly different among the individual genotypes (P ≤ 0.05).
Free AAs can be catabolized, and the resulting carbon skeletons are possibly supplied to the tricarboxylic acid (TCA) cycle for respiration (Araújo et al., 2011). Among TCA cycle-related organic acids, the contents of fumarate, succinate, 2-oxoglutarate, pyruvate, and isocitrate increased in pgm and pgm atg5 leaves (Supplemental Fig. S3), suggesting that AA catabolism was activated in starchless leaves.
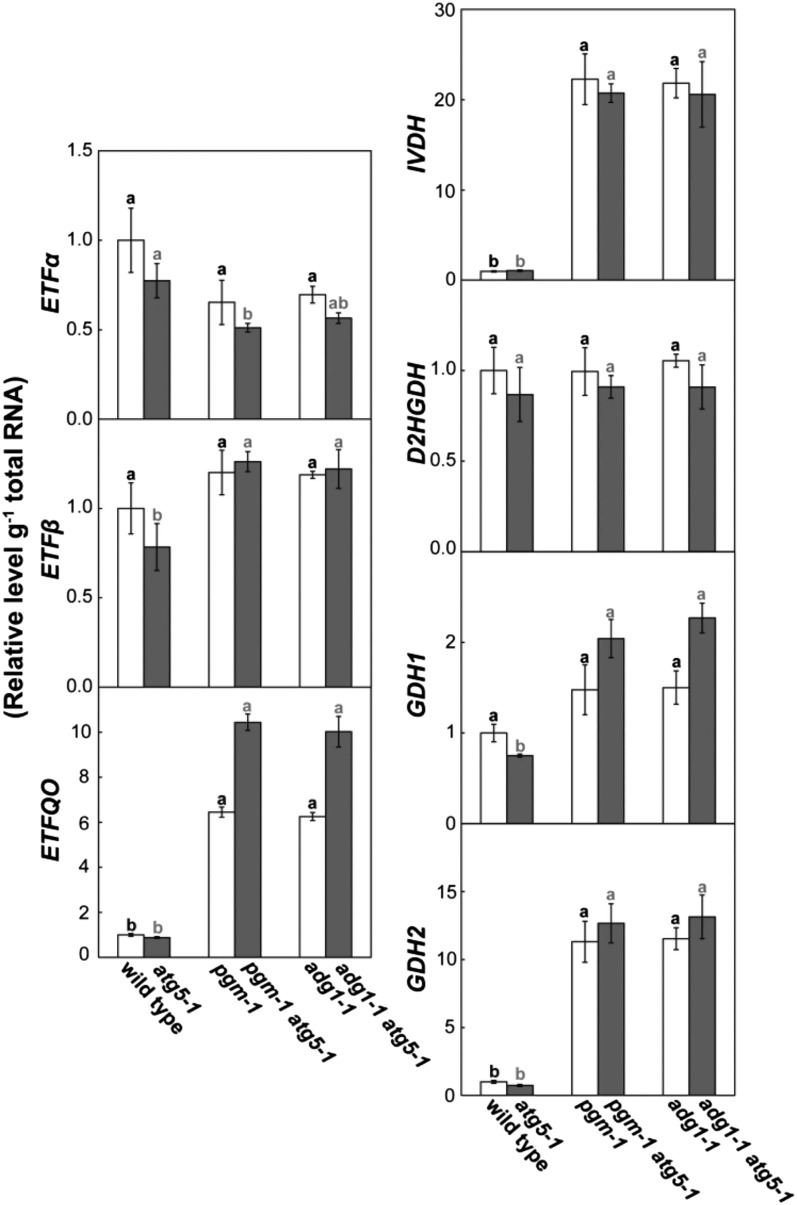
Recently, the importance of BCAAs, aromatic AAs, and Lys for respiration during sugar starvation has been demonstrated through the identifications of IVDH, D2HGDH, ETF, and ETFQO as enzymes and proteins related to the catabolism of these AAs (Ishizaki et al., 2005, 2006; Araújo et al., 2010). In addition, GDH has been suggested to play an important role in AA catabolism by supplying 2-oxoglutarate for deamination reactions (Miyashita and Good, 2008). Thus, we measured transcript levels of ETFα, ETFβ, ETFQO, IVDH, D2HGDH, GDH1, and GDH2 in leaves of the wild type, atg5, starchless mutants, and starchless atg5 double mutants at the end of the night (Fig. 6). The transcript levels of IVDH, ETFQO, and GDH2 were much higher in starchless lines than in the wild type and atg5. The transcript pattern of ETFQO was similar to that of DIN6 and ATL8 (Fig. 4).
Figure 6.
Transcripts of several genes involved in AA catabolism were elevated in starchless leaves during the night. Total RNA was isolated from leaves of the wild type, single mutant lines (atg5-1, pgm-1, adg1-1), and double mutant lines (pgm-1 atg5-1, adg1-1 atg5-1) at the end of the night 40 d after sowing and subjected to qRT-PCR analysis to measure transcript levels of ETFα, ETFβ, ETFQO, D2HGDH, IVDH, GDH1, and GDH2. mRNA levels are shown relative to the wild-type level, which is represented as 1. The data represent means ± se (n = 4). Statistical analysis was performed by Tukey’s test; columns with the same letter were not significantly different among the wild type, pgm-1, and adg1-1 (black letters; P ≤ 0.05) or among atg5-1, pgm-1 atg5-1, and adg1-1 atg5-1 (gray letters; P ≤ 0.05).
To examine the changes of autophagic activity in starchless mutants, we measured transcript levels of ATG8 gene family members (ATG8a to ATG8i) as ATGs (Supplemental Fig. S4). The ATG8 protein is a component of the autophagosome membrane and is turned over with autophagic bodies via vacuolar degradation (Yoshimoto et al., 2004; Suttangkakul et al., 2011). ATG8e, ATG8f, and ATG8h mRNA levels in starchless lines were approximately twice those in the wild type and atg5 (Supplemental Fig. S4). We also examined RCB production in the mutant leaves under SD (Supplemental Fig. S5), using concanamycin A treatment to inhibit the vacuolar degradation of RCBs (Izumi et al., 2010). Leaves were excised at the end of the day and incubated with concanamycin A in darkness for 14 h, the same period as the night under SD. Small vesicles exhibiting GFP fluorescence without chlorophyll fluorescence, which represent RCBs, accumulated in the central vacuole of wild-type, pgm, and adg1 leaves (Supplemental Fig. S5A). The number of accumulated RCBs in pgm and adg1 was 3.2- and 2.5-fold higher than in the wild type, respectively (Supplemental Fig. S5B). We also confirmed that RCBs were absent in atg5, pgm atg5, and adg1 atg5 leaves (Supplemental Fig. S5). These results support the notion that autophagy and AA catabolism are activated for the supply of alternative respiratory substrates during a sugar deficit in starchless mutant leaves.
Other Effects of Autophagy Deficiency in the Catabolism of Possible Energy Sources
CE-TOFMS analysis also revealed large metabolic changes other than in AAs due to autophagy deficiency (Supplemental Data S1). We focused on the changes in intermediates of the catabolism of possible energy sources (Table I). The contents of several intermediates in AA catabolism significantly decreased in pgm atg5 compared with pgm (Table I). These included 4-methyl-2-oxovaleric acid and 3-methyl-2-oxovaleric acid, which are initial intermediates of Leu and Ile catabolic processes (Binder et al., 2007). Saccharopine and 2-aminoadipic acid, intermediates of Lys catabolism (Galili et al., 2001), were also decreased in pgm atg5 relative to pgm, as was Lys itself (Fig. 5). These findings support our conclusion that autophagy provides a supply of AAs as alternative energy sources.
Table I. Selected metabolites involved in the catabolism of possible energy sources that were significantly increased or decreased in leaves of the Arabidopsis pgm atg5 mutant compared with pgm.
| Metabolite Description | Fold Change (pgm-1 atg5-1/pgm-1) | Involved Catabolic Pathway |
|---|---|---|
| Glycerophosphocholine | 2.65 | Phospholipid degradation |
| Phosphorylcholine | 1.67 | Phospholipid degradation |
| Choline | 1.40 | Phospholipid degradation |
| 4-Methyl-2-oxovaleric acid | 0.69 | Leu and Ile catabolism |
| 3-Methyl-2-oxovaleric acid | ||
| 2-Aminoadipic acid | 0.55 | Lys catabolism |
| Saccharopine | 0.50 | Lys catabolism |
| Allantoin | 0.33 | Purine catabolism |
| Urate | 0.19 | Purine catabolism |
Decreases in allantoin and urate, which are intermediates of purine catabolism (Zrenner et al., 2006), were also observed (Table I). Nucleotides are also possible energy sources (Zrenner et al., 2006) and could be targets of autophagic degradation, so that ribosomes containing ribosomal RNA (rRNA) would be degraded by an autophagic process similar to yeast ribophagy (Hillwig et al., 2011). Although fatty acids and lipids can be alternative respiratory substrates during sugar starvation (Kunz et al., 2009), the contents of choline, glycerophosphocholine, and phosphorylcholine, which are phospholipid-related metabolites, increased with atg5 mutation (Table I). These metabolites are derived from phospholipase-mediated degradation of phosphatidylcholine, a major phospholipid in plants (Bargmann and Munnik, 2006; Inoue and Moriyasu, 2006). Phospholipid degradation may thus be compensatorily accelerated by deficiency in autophagy.
DISCUSSION
Recent work has greatly developed our understanding of the role of autophagy in several biological processes of plants (Liu et al., 2005, 2012; Fujiki et al., 2007; Xiong et al., 2007; Ishida et al., 2008; Hofius et al., 2009; Wada et al., 2009; Yoshimoto et al., 2009; Kwon et al., 2010; Derrien et al., 2012; Guiboileau et al., 2012; Ono et al., 2013). Although the importance of autophagy in carbon utilization has been suggested based on the observed activation of autophagy during sugar starvation and the sensitivity of atg mutants to continuous darkness (Thompson et al., 2005; Phillips et al., 2008; Chung et al., 2010; Izumi et al., 2010; Suttangkakul et al., 2011), a substantial role for autophagy in the energy availability of plant systems has not been established experimentally. In this study, we found that the growth retardation of atg mutants under SD is closely related to nighttime energy availability. Thus, we generated starchless atg double mutants because sugar availability at night is reduced in starchless mutants. The double mutants showed further severe starvation symptoms and phenotypes: reduced growth and early cell death in leaves were observed under SD. CE-TOFMS analysis indicated that autophagy provides a supply of energy sources such as AAs instead of sugars. Our data thus support a model in which autophagy contributes to energy availability at night for growth by supplying energy sources under conditions of limited sugar availability.
The Importance of Autophagy during Fluctuations in Energy Availability
We established the importance of autophagy under unstable energy availability using starchless mutant plants. Fluctuations or disruptions in energy availability can be caused during several biotic or abiotic stresses (Baena-González et al., 2007; Baena-González and Sheen, 2008). In starchless mutants, photosynthesis is not limited, but a transient sugar deficit of several hours occurs at night (Fig. 3). Starchless mutation, therefore, seems to mimic energy limitation caused by environmental stresses more closely than artificial sugar starvation in which plants are grown in darkness for several days.
Starchless atg double mutants exhibited a more severe phenotype under shortened photoperiods (Fig. 2). The early-cell-death phenotype of atg single mutants has been shown to be dependent on the excessive accumulation of salicylic acid (SA; Yoshimoto et al., 2009). SA was only detected in pgm atg5 leaves by CE-TOFMS (Supplemental Data S1), suggesting that SA accumulation was directly responsible for the cell death phenomenon in starchless atg double mutants. However, the trigger for the excessive accumulation of SA remains unknown. Arabidopsis plants seem to decrease respiration activity in order to maintain an energy supply and survive when they are exposed to extended darkness (Keech et al., 2007). The proper balance of several catabolic pathways for controlled energy supply is likely to be disrupted by autophagy deficiency, such as an acceleration of phospholipid degradation (Table I). This imbalanced energy supply may stimulate cell damage and SA accumulation and ultimately may cause early cell death. Increased oxidative damage in pgm atg5 and atg5 was implied by the increases in the oxidized form of glutathione compared with the wild type and pgm (Supplemental Data S1; Meyer and Hell, 2005).
When energy availability is perturbed due to interference with photosynthesis, photosynthetic proteins in chloroplasts cannot fully function and are likely an optimal energy source via autophagic degradation. Partial degradation via RCBs without dismantling of the whole chloroplast would appear to be an efficient mechanism for maintaining some basal functions of the chloroplast until growth conditions improve. Whereas autophagy has generally been considered to be a bulk degradation system for nutrient recycling, several lines of evidence for selectivity in plant autophagy have recently been provided (Floyd et al., 2012; Li and Vierstra, 2012). In terms of organelle selectivity, autophagic degradation of the endoplasmic reticulum during endoplasmic reticulum stress was recently reported (Liu et al., 2012). In addition, chloroplastic proteins may be selectively degraded via the RCB/autophagy system for proper carbon recycling, although the mechanism underpinning such selectivity is not known.
The Link between Autophagic Protein Degradation and AA Catabolism
Recently, etfβ, etfqo, and ivdh, which are mutants of genes involved in BCAA or aromatic AA catabolism, were identified and found to show increased accumulation of BCAA and aromatic AAs compared with the wild type during extended darkness of several days (Ishizaki et al., 2005, 2006; Araújo et al., 2010). Here, increases of these AA contents in starchless mutants were partially compromised in starchless atg double mutants (Fig. 5). These findings imply that autophagy contributes to AA catabolism via free AA supply. BCAAs and aromatic AAs are quantitatively minor in AA pools; however, the contents of these AAs and transcript levels of several genes for their catabolism drastically increase during sugar starvation (Fujiki et al., 2001a, 2002; Buchanan-Wollaston et al., 2005; Araújo et al., 2010). Furthermore, etfβ, etfqo, and ivdh mutants show reduced tolerance to sugar starvation: these mutants die earlier than wild-type plants during extended darkness (Ishizaki et al., 2005, 2006; Araújo et al., 2010). Although GDH catalyzes the conversion between Glu and 2-oxoglutarate, the gdh1 gdh2 double mutants also display high accumulation of BCAA and early cell death during extended darkness, which is suggested to be a result of decreased 2-oxoglutarate production for the initial step of BCAA catabolism (Miyashita and Good, 2008). These phenomena clearly indicate a key role of BCAAs and aromatic AAs in the metabolic adaptation to sugar starvation conditions. Autophagy deficiency appears to affect BCAA and aromatic AA contents in starchless atg double mutants, because these AAs were actively catabolized in starchless mutants (Fig. 5).
Although high sensitivity to extended darkness of several days is also observed in several atg mutants (Thompson et al., 2005; Phillips et al., 2008; Chung et al., 2010), metabolic changes have not been investigated during such conditions. Further metabolic analysis using mutants of autophagy and AA catabolism are needed to confirm the metabolic link between autophagic protein degradation and AA catabolism.
Contribution of Autophagy to Energy Availability at Night for Normal Growth
The growth retardation with shortened photoperiods in atg single mutants is similar to that observed in starchless and starch-excess mutants in which nighttime sugar availability is reduced (Figs. 1 and 2; Caspar et al., 1985, 1991; Lin et al., 1988). This suggests that autophagy supports energy availability at night for normal growth. However, a major regulator of diel energy availability in response to photoperiods is the rate of starch synthesis and starch degradation (Smith and Stitt, 2007; Graf et al., 2010). In fact, the extent of growth retardation with shortened photoperiod was more severe in starchless mutants than in atg mutants (Fig. 2). In our CE-TOFMS analysis, clear evidence that autophagy provides AAs as energy sources at night during SD was not observed in atg5 single mutants (Fig. 4). It is conceivable that autophagic protein degradation constitutively supports carbon utilization and plant growth each night under SD conditions at lower levels than during a sugar deficit.
Plant productivity was recently shown to be closely associated not only with carbon assimilation capacity during the day but carbon utilization at night (Sulpice et al., 2009; Graf et al., 2010). When plant growth is environmentally restricted by carbon utilization, such as under SD conditions, it may be more efficient to have a portion of proteins degraded and consumed as an energy source. In fact, a metabolic shift that is dependent on light intensity for growth conditions was shown to result in increases in BCAA, aromatic AA, and Lys contents under low light compared with normal and high light in Arabidopsis leaves (Jänkänpää et al., 2012). In addition, some reports have suggested the involvement of protein degradation in diel carbon metabolism. GDH activity increases with photoperiod shortening (Gibon et al., 2009). Global gene expression analysis of diurnal changes showed that ATG8e transcripts elevate toward the end of the night, along with some genes involved in ubiquitin-dependent protein degradation (Bläsing et al., 2005). The carbon concentration in seeds is lower in atg5 than in the wild type (Guiboileau et al., 2012), suggesting the possible involvement of autophagy in whole-plant carbon utilization. The link between protein degradation and carbon metabolism during photoautotrophic growth has been uncharacterized in plant systems. Our findings suggest a metabolic strategy in which autophagy can contribute to carbon utilization and plant growth. However, protein is a less efficient respiratory substrate than carbohydrate in plants (Araújo et al., 2011). The role of autophagy in energy availability should be further characterized not only during severe stress conditions but also during nonstressed conditions that moderately alter sugar availability.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) ecotype Columbia was used for all experiments. Transgenic plants expressing stroma-targeted GFP (CT-GFP) and pgm-1, adg1-1, and sex1-1 background lines with CT-GFP were described previously (Izumi et al., 2010). The autophagy-deficient mutants atg5-1 (SAIL_129_B07), atg2-1 (SALK_076727), and atg10-1 (SALK_084434) were described previously (Thompson et al., 2005; Phillips et al., 2008; Yoshimoto et al., 2009). Double mutant lines were generated by sexual crossing. Plants were grown in soil (Metro-Mix 350; SunGro Horticulture) or on mineral-rich medium in chambers at 23°C to 24°C with fluorescent lamps (100 µmol quanta m−2 s−1) under different photoperiods (10 h for SD, 14 h for LD, and 24 h for CL). A 1:1,000 dilution of Hyponex (Hyponex Japan) was added to the soil once per week as a source of mineral nutrients. Mineral-rich medium was produced using a previously described hydroponic solution (Izumi et al., 2012) with or without 1% (w/v) Suc. Growth analysis was performed after different numbers of days of growth depending on photoperiod or culture conditions. For the atg single mutants (Fig. 1), shoot fresh weight and images were obtained at the point when the shoot fresh weight of wild-type plants was approximately 0.2 g in the various conditions. In medium culture, individual plants were germinated and grown in Suc-rich medium for 1 week and transplanted to the same Suc-rich medium or Suc-free medium. For the double mutant lines (Fig. 2), images and shoot fresh weight were obtained approximately 5 d after bolting of wild-type plants under CL and LD conditions. Under SD, images of plants were obtained 60 d after sowing and shoot fresh weight was obtained 40 d after sowing because the start of bolting was later under SD compared with the other conditions. Plants grown for 40 d were used in the other experiments using starchless atg double mutants under SD.
Measurement of Carbohydrates
Third to sixth rosette leaves were harvested at 0, 7, and 14 h after the end of the day during the 14-h night of SD. Starch, Suc, Glc, and Fru contents were measured using an F-kit for α-Glc/Suc/Fru (Roche Diagnostics) and an F-kit for starch (Roche Diagnostics) as described previously (Izumi et al., 2010).
Quantitative Reverse Transcription-PCR Analysis
Total RNA was isolated from third to sixth rosette leaves using the RNeasy kit (Qiagen). Isolated RNA was treated with DNase (DNA-free; Ambion), and complementary DNA was synthesized using random hexamer and oligo(dT) primers with the PrimeScript reverse transcription reagent kit (Takara). Quantitative reverse transcription (qRT)-PCR analysis was performed as described previously (Izumi et al., 2012) using an aliquot of the synthesized complementary DNA derived from 1.8 ng of total RNA. The sequences of the primers used are shown in Supplemental Table S1. Several primers were used previously (Baena-González et al., 2007; Miyashita and Good, 2008; Carbonell-Bejerano et al., 2010; Graf et al., 2010; Kwon et al., 2010). Amplicon sequences of original primers were confirmed by sequencing after TA cloning. The level of 18S rRNA was measured as an internal standard (Fig. 4; Supplemental Fig. S2), and rRNA levels did not differ between wild-type and mutant plants, indicating the validity of the method employed.
Measurement of Ionic Metabolites by CE-TOFMS
Third to sixth rosette leaves were homogenized with methanol (internal control). After additions of water and chloroform, the upper aqueous layer was filtered through a Millipore 5-kD cutoff filter to remove proteins. The filtrate was lyophilized, dissolved in water, and analyzed by CE-TOFMS using the Agilent CE Capillary Electrophoresis System (Agilent Technologies). Cationic metabolites were analyzed with a fused silica capillary (50 μm i.d. × 80 cm total length), with Cation Buffer Solution (Human Metabolome Technologies) as the electrolyte (Soga and Heiger, 2000). Anionic metabolites were analyzed with a fused silica capillary (50 μm i.d. × 80 cm total length), with Anion Buffer Solution (Human Metabolome Technologies) as the electrolyte (Soga et al., 2007). Raw data obtained by CE-TOFMS were processed with MasterHands (Sugimoto et al., 2010). Signal peaks corresponding to isotopomers of 108 compounds including intermediates of the glycolytic system, intermediates of the TCA cycle, and AAs were extracted. Their migration times were normalized using those of the internal standards, and the resultant relative area values were further normalized by sample leaf area.
Analysis of RCB Production Activity
RCB production was analyzed using the previously described method with a slight modification (Supplemental Fig. S5; Izumi et al., 2010). Fourth rosette leaves were excised at the end of the day from three to four independent CT-GFP plants. The leaves were incubated in 10 mm MES-NaOH (pH 5.5) with 1 µm concanamycin A for 14 h, the same period as the usual night period of SD, at 23°C in darkness. After incubation, each leaf was divided into four parts, and two square regions (188 µm × 188 µm each) in each section were monitored by laser scanning confocal microscopy and images were obtained. The number of accumulated RCBs in the images was counted. Each image was considered an independent data point and subjected to statistical analysis. Laser scanning confocal microscopy was performed with a Nikon C1si system equipped with a CFI Plan Apo VC 60× water-immersion objective (numerical aperture = 1.20; Nikon) as described previously in detail (Ishida et al., 2008).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Early-cell-death phenotype in leaves of atg2 and adg1 double mutants under SD.
Supplemental Figure S2. Early induction of DIN transcripts by autophagy deficiency during an extended night.
Supplemental Figure S3. Contents of organic acids of the TCA cycle were increased in starchless mutant leaves.
Supplemental Figure S4. Transcript levels of several members of the ATG8 gene family were elevated in starchless mutant leaves.
Supplemental Figure S5. Higher RCB production activity in starchless mutant leaves.
Supplemental Table S1. List of primer sequences for qRT-PCR analysis.
Supplemental Data S1. Changes in metabolite contents caused by atg5 mutation according to CE-TOFMS analysis.
Acknowledgments
We thank Dr. Kohki Yoshimoto and Dr. Yoshinori Ohsumi for providing the Arabidopsis atg mutants and Dr. Maureen R. Hanson for the use of CT-GFP Arabidopsis. We appreciate the Arabidopsis Biological Resource Center and original donors for the use of Arabidopsis transfer DNA insertional knockout mutants and starch-related mutants.
Glossary
- AA
amino acid
- RCB
Rubisco-containing body
- SD
short day
- CL
continuous light
- LD
long day
- CE-TOFMS
capillary electrophoresis time-of-flight mass spectrometry
- BCAA
branched-chain amino acid
- TCA
tricarboxylic acid
- SA
salicylic acid
- CT-GFP
stroma-targeted GFP
- qRT
quantitative reverse transcription
- rRNA
ribosomal RNA
References
- Araújo WL, Ishizaki K, Nunes-Nesi A, Larson TR, Tohge T, Krahnert I, Witt S, Obata T, Schauer N, Graham IA, et al. (2010) Identification of the 2-hydroxyglutarate and isovaleryl-CoA dehydrogenases as alternative electron donors linking lysine catabolism to the electron transport chain of Arabidopsis mitochondria. Plant Cell 22: 1549–1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araújo WL, Tohge T, Ishizaki K, Leaver CJ, Fernie AR. (2011) Protein degradation: an alternative respiratory substrate for stressed plants. Trends Plant Sci 16: 489–498 [DOI] [PubMed] [Google Scholar]
- Baena-González E, Rolland F, Thevelein JM, Sheen J. (2007) A central integrator of transcription networks in plant stress and energy signalling. Nature 448: 938–942 [DOI] [PubMed] [Google Scholar]
- Baena-González E, Sheen J. (2008) Convergent energy and stress signaling. Trends Plant Sci 13: 474–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargmann BO, Munnik T. (2006) The role of phospholipase D in plant stress responses. Curr Opin Plant Biol 9: 515–522 [DOI] [PubMed] [Google Scholar]
- Binder S, Knill T, Schuster J. (2007) Branched-chain amino acid metabolism in higher plants. Physiol Plant 129: 68–78 [Google Scholar]
- Bläsing OE, Gibon Y, Günther M, Höhne M, Morcuende R, Osuna D, Thimm O, Usadel B, Scheible WR, Stitt M. (2005) Sugars and circadian regulation make major contributions to the global regulation of diurnal gene expression in Arabidopsis. Plant Cell 17: 3257–3281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan-Wollaston V, Page T, Harrison E, Breeze E, Lim PO, Nam HG, Lin JF, Wu SH, Swidzinski J, Ishizaki K, et al. (2005) Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. Plant J 42: 567–585 [DOI] [PubMed] [Google Scholar]
- Carbonell-Bejerano P, Urbez C, Carbonell J, Granell A, Perez-Amador MA. (2010) A fertilization-independent developmental program triggers partial fruit development and senescence processes in pistils of Arabidopsis. Plant Physiol 154: 163–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspar T, Huber SC, Somerville C. (1985) Alterations in growth, photosynthesis, and respiration in a starchless mutant of Arabidopsis thaliana (L.) deficient in chloroplast phosphoglucomutase activity. Plant Physiol 79: 11–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspar T, Lin TP, Kakefuda G, Benbow L, Preiss J, Somerville C. (1991) Mutants of Arabidopsis with altered regulation of starch degradation. Plant Physiol 95: 1181–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng SH, Moore BD, Seemann JR. (1998) Effects of short- and long-term elevated CO2 on the expression of ribulose-1,5-bisphosphate carboxylase/oxygenase genes and carbohydrate accumulation in leaves of Arabidopsis thaliana (L.) Heynh. Plant Physiol 116: 715–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba A, Ishida H, Nishizawa NK, Makino A, Mae T. (2003) Exclusion of ribulose-1,5-bisphosphate carboxylase/oxygenase from chloroplasts by specific bodies in naturally senescing leaves of wheat. Plant Cell Physiol 44: 914–921 [DOI] [PubMed] [Google Scholar]
- Chung T, Phillips AR, Vierstra RD. (2010) ATG8 lipidation and ATG8-mediated autophagy in Arabidopsis require ATG12 expressed from the differentially controlled ATG12A and ATG12B loci. Plant J 62: 483–493 [DOI] [PubMed] [Google Scholar]
- Derrien B, Baumberger N, Schepetilnikov M, Viotti C, De Cillia J, Ziegler-Graff V, Isono E, Schumacher K, Genschik P. (2012) Degradation of the antiviral component ARGONAUTE1 by the autophagy pathway. Proc Natl Acad Sci USA 109: 15942–15946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doelling JH, Walker JM, Friedman EM, Thompson AR, Vierstra RD. (2002) The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J Biol Chem 277: 33105–33114 [DOI] [PubMed] [Google Scholar]
- Floyd BE, Morriss SC, Macintosh GC, Bassham DC. (2012) What to eat: evidence for selective autophagy in plants. J Integr Plant Biol 54: 907–920 [DOI] [PubMed] [Google Scholar]
- Fujiki Y, Ito M, Itoh T, Nishida I, Watanabe A. (2002) Activation of the promoters of Arabidopsis genes for the branched-chain alpha-keto acid dehydrogenase complex in transgenic tobacco BY-2 cells under sugar starvation. Plant Cell Physiol 43: 275–280 [DOI] [PubMed] [Google Scholar]
- Fujiki Y, Ito M, Nishida I, Watanabe A. (2001a) Leucine and its keto acid enhance the coordinated expression of genes for branched-chain amino acid catabolism in Arabidopsis under sugar starvation. FEBS Lett 499: 161–165 [DOI] [PubMed] [Google Scholar]
- Fujiki Y, Yoshikawa Y, Sato T, Inada N, Ito M, Nishida I, Watanabe A. (2001b) Dark-inducible genes from Arabidopsis thaliana are associated with leaf senescence and repressed by sugars. Physiol Plant 111: 345–352 [DOI] [PubMed] [Google Scholar]
- Fujiki Y, Yoshimoto K, Ohsumi Y. (2007) An Arabidopsis homolog of yeast ATG6/VPS30 is essential for pollen germination. Plant Physiol 143: 1132–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galili G, Tang GL, Zhu XH, Gakiere B. (2001) Lysine catabolism: a stress and development super-regulated metabolic pathway. Curr Opin Plant Biol 4: 261–266 [DOI] [PubMed] [Google Scholar]
- Gibon Y, Bläsing OE, Palacios-Rojas N, Pankovic D, Hendriks JHM, Fisahn J, Höhne M, Günther M, Stitt M. (2004) Adjustment of diurnal starch turnover to short days: depletion of sugar during the night leads to a temporary inhibition of carbohydrate utilization, accumulation of sugars and post-translational activation of ADP-glucose pyrophosphorylase in the following light period. Plant J 39: 847–862 [DOI] [PubMed] [Google Scholar]
- Gibon Y, Pyl ET, Sulpice R, Lunn JE, Höhne M, Günther M, Stitt M. (2009) Adjustment of growth, starch turnover, protein content and central metabolism to a decrease of the carbon supply when Arabidopsis is grown in very short photoperiods. Plant Cell Environ 32: 859–874 [DOI] [PubMed] [Google Scholar]
- Graf A, Schlereth A, Stitt M, Smith AM. (2010) Circadian control of carbohydrate availability for growth in Arabidopsis plants at night. Proc Natl Acad Sci USA 107: 9458–9463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiboileau A, Yoshimoto K, Soulay F, Bataillé MP, Avice JC, Masclaux-Daubresse C. (2012) Autophagy machinery controls nitrogen remobilization at the whole-plant level under both limiting and ample nitrate conditions in Arabidopsis. New Phytol 194: 732–740 [DOI] [PubMed] [Google Scholar]
- Hanaoka H, Noda T, Shirano Y, Kato T, Hayashi H, Shibata D, Tabata S, Ohsumi Y. (2002) Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol 129: 1181–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillwig MS, Contento AL, Meyer A, Ebany D, Bassham DC, Macintosh GC. (2011) RNS2, a conserved member of the RNase T2 family, is necessary for ribosomal RNA decay in plants. Proc Natl Acad Sci USA 108: 1093–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofius D, Schultz-Larsen T, Joensen J, Tsitsigiannis DI, Petersen NHT, Mattsson O, Jørgensen LB, Jones JDG, Mundy J, Petersen M. (2009) Autophagic components contribute to hypersensitive cell death in Arabidopsis. Cell 137: 773–783 [DOI] [PubMed] [Google Scholar]
- Inoue Y, Moriyasu Y. (2006) Autophagy is not a main contributor to the degradation of phospholipids in tobacco cells cultured under sucrose starvation conditions. Plant Cell Physiol 47: 471–480 [DOI] [PubMed] [Google Scholar]
- Ishida H, Yoshimoto K, Izumi M, Reisen D, Yano Y, Makino A, Ohsumi Y, Hanson MR, Mae T. (2008) Mobilization of Rubisco and stroma-localized fluorescent proteins of chloroplasts to the vacuole by an ATG gene-dependent autophagic process. Plant Physiol 148: 142–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizaki K, Larson TR, Schauer N, Fernie AR, Graham IA, Leaver CJ. (2005) The critical role of Arabidopsis electron-transfer flavoprotein:ubiquinone oxidoreductase during dark-induced starvation. Plant Cell 17: 2587–2600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizaki K, Schauer N, Larson TR, Graham IA, Fernie AR, Leaver CJ. (2006) The mitochondrial electron transfer flavoprotein complex is essential for survival of Arabidopsis in extended darkness. Plant J 47: 751–760 [DOI] [PubMed] [Google Scholar]
- Izumi M, Tsunoda H, Suzuki Y, Makino A, Ishida H. (2012) RBCS1A and RBCS3B, two major members within the Arabidopsis RBCS multigene family, function to yield sufficient Rubisco content for leaf photosynthetic capacity. J Exp Bot 63: 2159–2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi M, Wada S, Makino A, Ishida H. (2010) The autophagic degradation of chloroplasts via Rubisco-containing bodies is specifically linked to leaf carbon status but not nitrogen status in Arabidopsis. Plant Physiol 154: 1196–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jänkänpää HJ, Mishra Y, Schröder WP, Jansson S. (2012) Metabolic profiling reveals metabolic shifts in Arabidopsis plants grown under different light conditions. Plant Cell Environ 35: 1824–1836 [DOI] [PubMed] [Google Scholar]
- Keech O, Pesquet E, Ahad A, Askne A, Nordvall D, Vodnala SM, Tuominen H, Hurry V, Dizengremel P, Gardeström P. (2007) The different fates of mitochondria and chloroplasts during dark-induced senescence in Arabidopsis leaves. Plant Cell Environ 30: 1523–1534 [DOI] [PubMed] [Google Scholar]
- Kunz HH, Scharnewski M, Feussner K, Feussner I, Flügge UI, Fulda M, Gierth M. (2009) The ABC transporter PXA1 and peroxisomal β-oxidation are vital for metabolism in mature leaves of Arabidopsis during extended darkness. Plant Cell 21: 2733–2749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon SI, Cho HJ, Jung JH, Yoshimoto K, Shirasu K, Park OK. (2010) The Rab GTPase RabG3b functions in autophagy and contributes to tracheary element differentiation in Arabidopsis. Plant J 64: 151–164 [DOI] [PubMed] [Google Scholar]
- Li FQ, Vierstra RD. (2012) Autophagy: a multifaceted intracellular system for bulk and selective recycling. Trends Plant Sci 17: 526–537 [DOI] [PubMed] [Google Scholar]
- Lin TP, Caspar T, Somerville C, Preiss J. (1988) Isolation and characterization of a starchless mutant of Arabidopsis thaliana (L.) Heynh lacking ADPglucose pyrophosphorylase activity. Plant Physiol 86: 1131–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Burgos JS, Deng Y, Srivastava R, Howell SH, Bassham DC. (2012) Degradation of the endoplasmic reticulum by autophagy during endoplasmic reticulum stress in Arabidopsis. Plant Cell 24: 4635–4651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Czymmek K, Tallóczy Z, Levine B, Dinesh-Kumar SP. (2005) Autophagy regulates programmed cell death during the plant innate immune response. Cell 121: 567–577 [DOI] [PubMed] [Google Scholar]
- Liu YM, Bassham DC. (2012) Autophagy: pathways for self-eating in plant cells. Annu Rev Plant Biol 63: 215–237 [DOI] [PubMed] [Google Scholar]
- Makino A, Sakuma H, Sudo E, Mae T. (2003) Differences between maize and rice in N-use efficiency for photosynthesis and protein allocation. Plant Cell Physiol 44: 952–956 [DOI] [PubMed] [Google Scholar]
- Meijer WH, van der Klei IJ, Veenhuis M, Kiel JAKW. (2007) ATG genes involved in non-selective autophagy are conserved from yeast to man, but the selective Cvt and pexophagy pathways also require organism-specific genes. Autophagy 3: 106–116 [DOI] [PubMed] [Google Scholar]
- Meyer AJ, Hell R. (2005) Glutathione homeostasis and redox-regulation by sulfhydryl groups. Photosynth Res 86: 435–457 [DOI] [PubMed] [Google Scholar]
- Miyashita Y, Good AG. (2008) NAD(H)-dependent glutamate dehydrogenase is essential for the survival of Arabidopsis thaliana during dark-induced carbon starvation. J Exp Bot 59: 667–680 [DOI] [PubMed] [Google Scholar]
- Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. (2009) Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol 10: 458–467 [DOI] [PubMed] [Google Scholar]
- Oh SA, Lee SY, Chung IK, Lee CH, Nam HG. (1996) A senescence-associated gene of Arabidopsis thaliana is distinctively regulated during natural and artificially induced leaf senescence. Plant Mol Biol 30: 739–754 [DOI] [PubMed] [Google Scholar]
- Ono Y, Wada S, Izumi M, Makino A, Ishida H. (2013) Evidence for contribution of autophagy to Rubisco degradation during leaf senescence in Arabidopsis thaliana Plant Cell Environ (in press) [DOI] [PubMed] [Google Scholar]
- Phillips AR, Suttangkakul A, Vierstra RD. (2008) The ATG12-conjugating enzyme ATG10 is essential for autophagic vesicle formation in Arabidopsis thaliana. Genetics 178: 1339–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaxton WC, Podesta FE. (2006) The functional organization and control of plant respiration. Crit Rev Plant Sci 25: 159–198 [Google Scholar]
- Singh R, Cuervo AM. (2011) Autophagy in the cellular energetic balance. Cell Metab 13: 495–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AM, Stitt M. (2007) Coordination of carbon supply and plant growth. Plant Cell Environ 30: 1126–1149 [DOI] [PubMed] [Google Scholar]
- Soga T, Heiger DN. (2000) Amino acid analysis by capillary electrophoresis electrospray ionization mass spectrometry. Anal Chem 72: 1236–1241 [DOI] [PubMed] [Google Scholar]
- Soga T, Ishikawa T, Igarashi S, Sugawara K, Kakazu Y, Tomita M. (2007) Analysis of nucleotides by pressure-assisted capillary electrophoresis-mass spectrometry using silanol mask technique. J Chromatogr A 1159: 125–133 [DOI] [PubMed] [Google Scholar]
- Sugimoto M, Hirayama A, Ishikawa T, Robert M, Baran R, Uehara K, Kawai K, Soga T, Tomita M. (2010) Differential metabolomics software for capillary electrophoresis-mass spectrometry data analysis. Metabolomics 6: 27–41 [Google Scholar]
- Sulpice R, Pyl ET, Ishihara H, Trenkamp S, Steinfath M, Witucka-Wall H, Gibon Y, Usadel B, Poree F, Piques MC, et al. (2009) Starch as a major integrator in the regulation of plant growth. Proc Natl Acad Sci USA 106: 10348–10353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suttangkakul A, Li FQ, Chung T, Vierstra RD. (2011) The ATG1/ATG13 protein kinase complex is both a regulator and a target of autophagic recycling in Arabidopsis. Plant Cell 23: 3761–3779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson AR, Doelling JH, Suttangkakul A, Vierstra RD. (2005) Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol 138: 2097–2110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada S, Ishida H, Izumi M, Yoshimoto K, Ohsumi Y, Mae T, Makino A. (2009) Autophagy plays a role in chloroplast degradation during senescence in individually darkened leaves. Plant Physiol 149: 885–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Contento AL, Bassham DC. (2005) AtATG18a is required for the formation of autophagosomes during nutrient stress and senescence in Arabidopsis thaliana. Plant J 42: 535–546 [DOI] [PubMed] [Google Scholar]
- Xiong Y, Contento AL, Nguyen PQ, Bassham DC. (2007) Degradation of oxidized proteins by autophagy during oxidative stress in Arabidopsis. Plant Physiol 143: 291–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto K. (2012) Beginning to understand autophagy, an intracellular self-degradation system in plants. Plant Cell Physiol 53: 1355–1365 [DOI] [PubMed] [Google Scholar]
- Yoshimoto K, Hanaoka H, Sato S, Kato T, Tabata S, Noda T, Ohsumi Y. (2004) Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell 16: 2967–2983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto K, Jikumaru Y, Kamiya Y, Kusano M, Consonni C, Panstruga R, Ohsumi Y, Shirasu K. (2009) Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. Plant Cell 21: 2914–2927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zrenner R, Stitt M, Sonnewald U, Boldt R. (2006) Pyrimidine and purine biosynthesis and degradation in plants. Annu Rev Plant Biol 57: 805–836 [DOI] [PubMed] [Google Scholar]