AtMMS21 acts in double-strand break amelioration and stem cell niche maintenance during Arabidopsis root development.
Abstract
Plants maintain stem cells in meristems to sustain lifelong growth; these stem cells must have effective DNA damage responses to prevent mutations that can propagate to large parts of the plant. However, the molecular links between stem cell functions and DNA damage responses remain largely unexplored. Here, we report that the small ubiquitin-related modifier E3 ligase AtMMS21 (for methyl methanesulfonate sensitivity gene21) acts to maintain the root stem cell niche by mediating DNA damage responses in Arabidopsis (Arabidopsis thaliana). Mutation of AtMMS21 causes defects in the root stem cell niche during embryogenesis and postembryonic stages. AtMMS21 is essential for the proper expression of stem cell niche-defining transcription factors. Moreover, mms21-1 mutants are hypersensitive to DNA-damaging agents, have a constitutively increased DNA damage response, and have more DNA double-strand breaks (DSBs) in the roots. Also, mms21-1 mutants exhibit spontaneous cell death within the root stem cell niche, and treatment with DSB-inducing agents increases this cell death, suggesting that AtMMS21 is required to prevent DSB-induced stem cell death. We further show that AtMMS21 functions as a subunit of the STRUCTURAL MAINTENANCE OF CHROMOSOMES5/6 complex, an evolutionarily conserved chromosomal ATPase required for DNA repair. These data reveal that AtMMS21 acts in DSB amelioration and stem cell niche maintenance during Arabidopsis root development.
In plants and animals, small pools of stem cells are maintained as a population of undifferentiated cells that can generate differentiated descendants to sustain growth or replace tissues (Sablowski, 2004). Simple systems, such as the root meristem of Arabidopsis (Arabidopsis thaliana), make excellent experimental models for studying the mechanisms controlling stem cell identity (Nawy et al., 2005). In Arabidopsis roots, quiescent center (QC) cells and the surrounding stem cells together form a stem cell niche (Aida et al., 2004). The root stem cell niche is positioned where the activity of the transcription factors SHORT-ROOT (SHR) and SCARECROW (SCR) overlaps with the expression of PLETHORA (PLT), which in turn is controlled by an apical-basal gradient of auxin maintained by PIN transporters (Sabatini et al., 2003; Galinha et al., 2007). Downstream of SCR, the Arabidopsis RETINOBLASTOMA-RELATED protein and the QC-specific transcription factor WUSCHEL-RELATED HOMEOBOX5 (WOX5) also regulate the maintenance of stem cells in plants (Wildwater et al., 2005; Sarkar et al., 2007).
In addition to transcription factors, chromatin factors also govern stem cell self-renewal and differentiation (Orkin and Hochedlinger, 2011). For example, in Arabidopsis, the histone acetyltransferase GCN5 and its cofactor ADA2b are required for root stem cell maintenance through the regulation of PLT expression (Kornet and Scheres, 2009). The chromatin-remodeling factor PICKLE and the PcG protein CURLY LEAF antagonistically determine root meristem activity by controlling the expression of stem cell fate genes (Aichinger et al., 2011). Another chromatin factor implicated in root stem cell maintenance is the CHROMATIN ASSEMBLY FACTOR1 (CAF1) complex. Mutation of the FASCIATA (FAS) genes, which encode subunits of the CAF1 complex, leads to a loss of root stem cells and failure to maintain SCR expression (Kaya et al., 2001). Furthermore, fas mutants show an increased number of DNA double-strand breaks (DSBs; Endo et al., 2006; Kirik et al., 2006), indicating that the Arabidopsis CAF1 complex is required for genome stability.
Recent findings reveal that plant stem cells have specialized mechanisms to maintain genomic stability (Sablowski, 2011). Treatments with DNA-damaging agents preferentially kill stem cells in the shoot and root meristem, a response that requires the transduction of DNA damage signals by ATAXIA-TELANGIECTASIA MUTATED (ATM), ATM/RAD3-RELATED, and SUPPRESSOR OF γ RESPONSE1 (Fulcher and Sablowski, 2009; Furukawa et al., 2010; Sablowski, 2011). In agreement with this, recently characterized mutants involved in DNA repair showed spontaneous death of root stem cells. For example, the accumulation of DNA damage in rad50 and mre11 mutants led to stem cell death and thus to developmental defects in growing plants (Amiard et al., 2010). MERISTEM DISORGANIZATION1 (MDO1) is required for the maintenance of stem cells through a reduction in DNA damage (Hashimura and Ueguchi, 2011). These data indicate that the protection of genomic stability is an important feature of the plant stem cell niche.
SUMO (for small ubiquitin-related modifier) has emerged as a significant regulator of genomic stability (Dou et al., 2011). MMS21 (for methyl methanesulfonate sensitivity gene21) encodes a SUMO E3 ligase that forms a critical component of the STRUCTURAL MAINTENANCE OF CHROMOSOMES5/6 (SMC5/6) complex, which is an evolutionarily conserved chromosomal ATPase required for cell growth and DNA repair (Duan et al., 2009). The SUMO ligase activity of MMS21 contributes to the roles of the SMC5/6 complex in recombinational repair, collapsed replication fork restart, and ribosomal DNA and telomere maintenance, with some relevant substrates having been identified (Potts and Yu, 2005; Zhao and Blobel, 2005). Although protein sumoylation plays important roles in the maintenance of genomic stability from yeast (Saccharomyces cerevisiae) to humans (Dou et al., 2011), it is unknown whether plants possess sumoylation mechanisms that act in the response to DNA damage. In Arabidopsis, AtMMS21/HIGH PLOIDY2 (HPY2) is a functional SUMO ligase and plays an important role in plant root development (Huang et al., 2009; Ishida et al., 2009), but the precise mechanism of AtMMS21 in regulating root meristem function is unclear.
Here, we show that AtMMS21 regulates root stem cell niche maintenance during embryogenesis and postembryonic stages. We provide evidence that AtMMS21 is required for the stable expression of stem cell niche-defining transcription factors. In addition, AtMMS21 maintains the normal cellular organization of the root stem cell niche by preventing cell death. Mutation of AtMMS21 caused increased DSBs and DSB-inducible gene transcription, showing that AtMMS21 is involved in DNA damage responses during root development. We further demonstrate that AtMMS21 acts as a component of the SMC5/6 complex through its interaction with SMC5, thus revealing critical roles of AtMMS21 in maintaining the root stem cell niche and genome stability by reducing DNA damage.
RESULTS
mms21-1 Mutants Show Altered Cell Division and Cell Differentiation in the Root Meristem
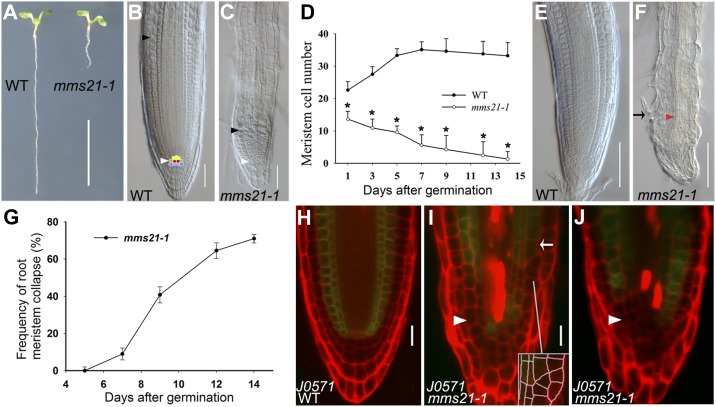
AtMMS21/HYP2 acts in root meristem development (Huang et al., 2009; Ishida et al., 2009). To investigate the mechanisms by which AtMMS21 affects root growth, we examined the pattern of cell division and cell differentiation in wild-type and mms21-1 (transferred DNA insertion mutant) roots at different days after germination (DAG). At 5 DAG, mms21-1 mutants showed shorter roots with smaller meristems (Fig. 1, A–D). Time-course analysis showed that mms21-1 meristems reached their maximum size at 1 DAG (Fig. 1D), but wild-type meristems reached their maximum size at 5 to 7 DAG by a balance of cell division and cell differentiation (Moubayidin et al., 2010). Furthermore, in mms21-1, we observed meristem collapse that occurred as the cells of the root meristem differentiated at 7 to 14 DAG, as shown by the formation of root hairs and xylem strands in the root tips (Fig. 1, F and G). To further establish the role of AtMMS21 in root meristem maintenance, we monitored the expression of markers that express GFP in specific cell types in the root meristems. For example, the J0571 marker specifically expressed GFP in the endodermis and cortex (Fig. 1H). By contrast, the cell files expressing GFP were not continuous in the mms21-1 roots (Fig. 1, I and J, arrowhead), and the expression often occurred in three layers adjacent to each other (Fig. 1I, arrow). Furthermore, abnormal planes of cell division were often observed in the region of GFP expression (Fig. 1I, inset). Collectively, these results indicated that AtMMS21 is required for maintaining the pattern of cell division and cell differentiation in the root meristem.
Figure 1.
The pattern of cell division and cell differentiation is defective in the mms21-1 root meristem. A, Phenotypes of wild-type (WT) and mms21-1 seedlings at 5 DAG. Bar = 1 cm. B and C, Root tips of the wild type and mms21-1 at 5 DAG. The QC is marked in red, and the QC is surrounded by stem cells: endodermal/cortical stem cells (green), vascular stem cells (yellow), and CSCs (blue). White and black arrowheads indicate the QC and the first elongated cortex cell, respectively. Bars = 50 μm. D, Root meristem cell number of the wild type and mms21-1 from 1 to 14 DAG. Data shown are averages ± sd (n = 30), and asterisks indicate significant differences compared with control plants (P < 0 0.005; Student’s t test). E and F, Root tips of the wild type and mms21-1 at 9 DAG. The arrow and arrowhead in F indicate root hair and mature protoxylem cells, respectively. Bars = 100 μm. G, Meristem collapse frequency at different time points for the wild type and mms21-1. Values are means ± sd (n = 20) of three biological replicates. H to J, Expression of ground tissue marker J0571 in the wild type and mms21-1 at 5 DAG. The region with an altered cell division plane is magnified in the inset in I. The arrowheads and arrow indicate discontinuous expression and ectopic expression of J0571, respectively. Bars = 20 μm.
mms21-1 Mutants Show Defective Cellular Organization of the Root Stem Cell Niche
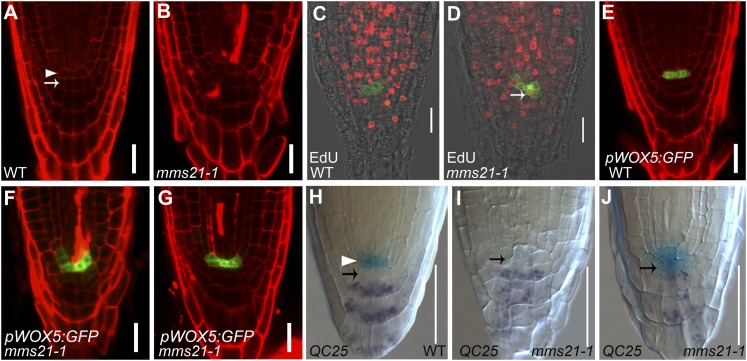
Our finding that AtMMS21 is crucial for maintaining cell fate in the root meristem prompted us to investigate its possible effect on the cellular organization of the QC and its surrounding stem cells. In wild-type roots, the QC cells are mitotically inactive and are easily discernible by confocal microscopy (Fig. 2A). However, in mms21-1, the pattern of cells in the root tips was disrupted, and the QC could not be identified morphologically (Fig. 2B). Because AtMMS21 is an important regulator of cell cycle progression (Huang et al., 2009; Ishida et al., 2009), we checked whether cell cycle activities were altered in the QC of mms21-1 roots. For this purpose, we cultured 2-DAG seedlings for 24 h in the presence of 5-ethynyl-2′-deoxyuridine (EdU). Incorporation of EdU in the nuclei indicates S-phase progression, so EdU can be used to mark cell division in the root meristem (Vanstraelen et al., 2009). After coupling of EdU with the fluorescent substrate Apollo 567, root tips were observed by confocal microscopy. In wild-type root meristems, most meristematic cells and the QC-surrounding stem cells incorporated EdU, but red fluorescent nuclei were observed only occasionally in the QC cells (marked by pWOX5:GFP), indicating that the QC cells had low mitotic activity (Fig. 2C). By contrast, red fluorescent nuclei were frequently observed in the mms21-1 QC cells (11 of 15 in mms21-1 versus two of 15 in the wild type; n = 15; Fig. 2D). These results indicated that the absence of AtMMS21 induces high mitotic activity of the QC cells, and the irregularly dividing QC cells result in a disordered stem cell niche.
Figure 2.
AtMMS21 is essential for QC organization, identity, and function. A and B, Longitudinal view of the stem cell niche of wild-type (WT) and mms21-1 roots at 5 DAG stained with PI showing cells of the QC (arrowhead) and CSCs (arrow). C and D, EdU incorporation assays in wild-type and mms21-1 root meristems. The QC cells are marked with green fluorescence in pWOX5:GFP, and EdU-positive nuclei are marked by red fluorescence. Wild-type QC cells do not have red fluorescent nuclei, indicating that the QC is not mitotically active; by contrast, red fluorescent EdU-positive nuclei in the mms21-1 QC cells indicate a state of active division (arrow). E to F, Expression pattern of pWOX5:GFP in the wild type and mms21-1 at 5 DAG. H to J, Double staining of the QC25:GUS marker and starch granules (purple) in 5-DAG wild type and mms21-1 seedlings. The arrowhead and arrow in the wild type indicate QC and columella initials, respectively. Arrows in mms21-1 indicate cells in the region of the QC and CSCs. Bars = 20 μm (A–G) and 50 μm (H–J).
The irregular QC organization of mms2-1 was also clearly observed by the altered expression pattern of the QC-specific marker pWOX5:GFP (Blilou et al., 2005; Sarkar et al., 2007). pWOX5:GFP showed the expected QC-specific expression pattern in wild-type roots (Fig. 2E). However, the GFP expression domain in the root tips of mms21-1 mutants showed lateral expansion (88%; n = 27) or occasional absence (Fig. 2, F and G; data not shown), which again indicated that QC identity was not stably maintained. Consistent with the expansion of the WOX5 expression pattern, quantitative reverse transcription (qRT)-PCR analysis also revealed that in mms21-1 roots WOX5 transcript levels increased by nearly 2-fold compared with wild-type roots (Supplemental Fig. S1D). To further investigate whether the disorganization of the stem cell niche was associated with misspecification of the QC, we monitored the expression of three independent QC-specific GUS markers (QC25, QC46, and QC184) in mms21-1 mutants. QC25 was expressed in QC cells in wild-type plants (Fig. 2H), but in mms21-1 roots, the QC25 expression was either absent (33.3%; n = 78) or highly reduced (19.2%; n = 78; Fig. 2I) and diffuse (47.4%; n = 78; Fig. 2J). Similarly, two other QC markers, QC46 and QC184, also showed aberrant expression in the mms21-1 roots (Supplemental Fig. S1A; Supplemental Table S1). These results showed that AtMMS21 is essential for proper QC identity.
To test whether QC function was defective in mms21-1 mutants, we performed Lugol staining using the QC marker line. In wild-type roots, a layer of columella stem cells (CSCs) is present between the QC and differentiated columella cells that contain starch granules (Fig. 2H; Supplemental Fig. S1A). However, mms21-1 roots showed starch granules at the stem cell position (Fig. 2, I and J; Supplemental Fig. S1A). The mms21-1 roots also showed irregular expression of the CSC marker J2341 and had defective columella layers (Supplemental Fig. S1, B and C), revealing that the stem cell state of CSCs was disrupted. Furthermore, in the mms21-1 roots, when QC markers were expressed, some GUS-staining cells also accumulated starch granules (Fig. 2J; Supplemental Fig. S1A), indicating that the cells derived from aberrant QC divisions undergo differentiation and then fail to execute the QC function of maintaining CSCs in an undifferentiated state. Taken together, these results indicated that AtMMS21 is required for the maintenance of QC organization, identity, and function and thus is essential for root stem cell niche maintenance.
Mutation of AtMMS21 Affects the Expression of Stem Cell Niche-Defining Transcription Factors
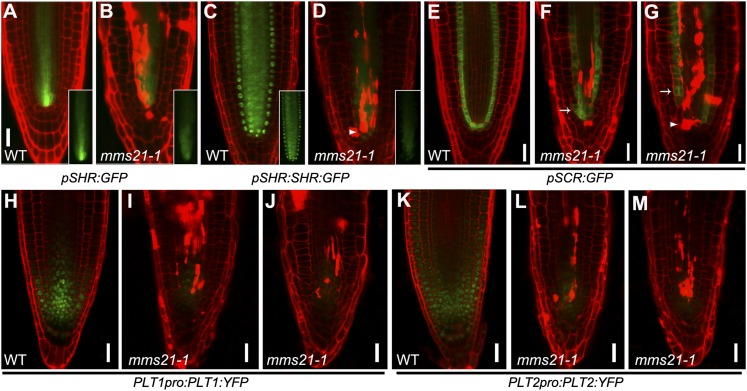
Root stem cell niche maintenance depends on the highly specific expression and subcellular localization of the transcriptional regulators SHR and SCR, which provide the radial position of the stem cell niche, and of PLT1 and PLT2, which provide the longitudinal position (Dinneny and Benfey, 2008). To determine whether the stem cell niche defect in the mms21-1 mutants was caused by misregulation of these stem cell niche-defining transcription factors, we examined the expression pattern of these genes in mms21-1 at 5 DAG. We first monitored the expression of pSHR:GFP and the protein localization of pSHR:SHR:GFP in the mms21-1 mutants. The expression region of the SHR transcript in the stele of mms21-1 roots was slightly reduced compared with the wild type (Fig. 3, A and B; Supplemental Fig. S1D). Also, the expression level of the SHR protein was reduced (Fig. 3, C and D), and SHR localization, which is in endodermal cells, QC cells, and cortical/endodermal stem cells in the wild type, seemed to be missing in some cell files in mms21-1 roots (Fig. 3D, arrowhead).
Figure 3.
Mutation of AtMMS21 caused misexpression of stem cell niche-defining transcription factors. The expression pattern of stem cell marker genes is shown in the roots of the wild type (WT) and mms21-1 at 5 DAG. A and B, pSHR:GFP. C and D, pSHR:SHR:GFP. E to G, pSCR:GFP. H to J, PLT1pro:PLT1:YFP. K to M, PLT2pro:PLT2:YFP. The insets in A to D show the expression of SHR, observed in the GFP channel. The arrowhead in D indicates that SHR localization was missing in some cell files in the mms21-1 roots. The arrowhead and arrows in F and G indicate discontinuous expression and ectopic expression of pSCR:GFP, respectively. Bars = 20 μm.
SHR expressed in the stele moves into the adjacent cell layer, where it controls SCR transcription and endodermis specification (Cui et al., 2007). Therefore, we next analyzed the effect of AtMMS21 on SCR expression. In wild-type roots, the pSCR:GFP-expressing cells were contiguous and appeared to form a single cell layer with the endodermal cells, QC cells, and cortical/endodermal stem cells (Fig. 3E). By contrast, although the expression of SCR was observed in the corresponding cells in most mms21-1 roots, some root stem cell niche or endodermal cells showed ectopic expression of SCR (Fig. 3, F and G, arrow). In extreme cases, SCR expression was abolished in some endodermal cells and the stem cell niche, which displayed a discontinuous pattern (Fig. 3G, arrowhead). In addition, to examine the effect of the SHR/SCR pathway on AtMMS21 expression, we analyzed AtMMS21 expression in the shr1 and scr1 mutants by qRT-PCR. The results showed that the shr1 and scr1 mutations have little effect on the expression of AtMMS21 (Supplemental Fig. S2).
PLT1/PLT2 acts in parallel with the SHR/SCR pathway to pattern the root stem cell niche (Aida et al., 2004). The expression patterns of HPY2/AtMMS21 proteins largely overlap with those of PLT1 and PLT2 (Ishida et al., 2009); together with our finding that AtMMS21 functions in the maintenance of the root stem cell niche, these results imply that AtMMS21 may have a role in defining PLT expression and/or accumulation. We then analyzed the transcript levels of PLT1/PLT2 in wild-type and mms21-1 roots using qRT-PCR. The expression levels of PLT1 and PLT2 were not significantly reduced in mms21-1 roots (Supplemental Fig. S1D). Surprisingly, in mms21-1 root tips, the yellow fluorescent protein (YFP) levels of the translational fusions PLT1pro:PLT1:YFP and PLT2pro:PLT2:YFP were dramatically reduced or weakly expressed compared with the wild type (Fig. 3, H–M), establishing that the loss of AtMMS21 significantly affected the accumulation of PLT protein in the root meristem. Considering that the correct PLT protein dosage is necessary for root stem cell maintenance and root formation (Aida et al., 2004; Galinha et al., 2007), our results revealed that the defective stem cell niche maintenance in the mms21-1 mutants occurs along with the dramatic misregulation of PLT1/PLT2 accumulation.
mms21-1 Mutants Show Defects in the Root Stem Cell Niche from Embryogenesis Onward
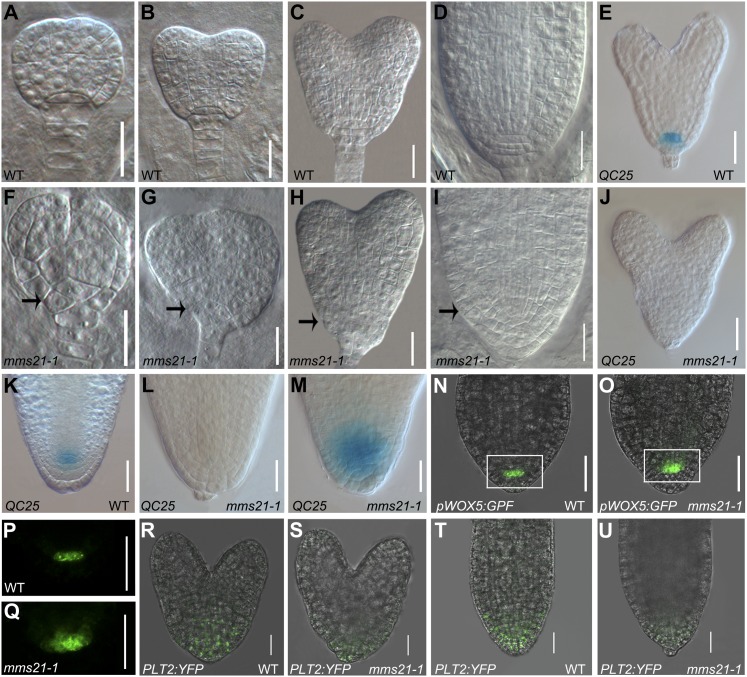
Root stem cell niches are established during embryogenesis and provide cells for postembryonic growth (Weigel and Jurgens, 2002; Aida et al., 2004). In postembryonic mms21-1 roots, the visible defects in stem cell niche organization and marker gene expression were already observed from 1 DAG onward (Supplemental Fig. S3). These findings prompted us to investigate whether AtMMS21 is required for initiation of the stem cell niche during embryogenesis. The cell division pattern is strictly regulated during wild-type embryogenesis, but in the mms21-1 mutants, many types of abnormal embryos with disturbed cellular organization at the basal embryo pole were frequently observed, starting at the early globular stage (Fig. 4; Supplemental Fig. S4, A–G). At the late globular stage of the wild type, the hypophysis had divided asymmetrically, producing a basal cell and a smaller lens-shaped apical cell (Fig. 4A). By contrast, mms21-1 embryos exhibited abnormal cellular organization of the hypophyseal derivatives (Fig. 4F). Before the heart stage, nearly 16% of analyzed embryos (31 of 188) showed aberrant divisions at the basal pole (Supplemental Table S2). At the heart stage of the wild type, the lens-shaped cell undergoes vertical divisions to generate the QC, while the lower cells divide horizontally to form CSCs and the root cap (Fig. 4, B and C). However, 46% of the mms21-1 embryos examined (62 of 134) showed irregular planes of cell division in the hypophysis descendants, leading to disorganization in the basal embryo region where the root stem cell niche initiates (Fig. 4, G and H; Supplemental Table S2). At later stages of embryo development, aberrant divisions were still apparent at the root stem cell niche (Fig. 4I), which translates into seedlings without a primary root in 16% of germinated mms21-1 mutants (35 of 216). Thus, in mms21-1 mutants, the disturbed cellular organization in the root stem cell niche occurs beginning in embryogenesis.
Figure 4.
mms21-1 shows defects in the root stem cell niche during embryogenesis. A to D, Wild-type (WT) embryos in late globular (A), early heart (B), middle heart (C), and cotyledon (D) stages. F to I, mms21-1 mutant embryos at stages equivalent to A to D. Arrows point to aberrant cell divisions in the basal embryo domain of mms21-1 mutants. E to J, QC25 expression in heart-stage embryos of the wild type and mms21-1. K to M, QC25 expression in mature-stage embryos of the wild type and mms21-1. N to Q, pWOX5:GFP expression in mature-stage embryos of the wild type and mms21-1 (P shows the outlined area in N and Q shows the outlined area in O, observed in the GFP channel). R to U, Expression patterns of PLT2pro:PLT2:YFP in wild-type and mms21-1 embryos. Bars = 20 μm.
To further analyze cell identity in the embryonic stem cell region of mms21-1 mutants, we examined the expression of QC-specific markers in the embryo. In wild-type embryos, QC25 was expressed in the QC precursor cell from the heart stage (Fig. 4E) and later in the QC cells (Fig. 4K). However, the majority (67%; n = 42) of the mms21-1 embryos showed no detectable QC25 expression in the corresponding cells at the heart stage (Fig. 4J). In the mature embryo, QC25 expression was absent or diffuse (Fig. 4, L and M), similar to what was observed in postembryonic mms21-1 roots. These data indicated that AtMMS21 is essential for proper specification of the QC.
We also examined the expression of pWOX5:GFP, a QC-specific marker with expression that is restricted to the QC precursor cell of the heart-stage embryo and, subsequently, becomes highly localized to the QC in normal embryogenesis (Fig. 4, N and P; Supplemental Fig. S4H; Haecker et al., 2004; Sarkar et al., 2007). In line with the altered expression of pWOX5:GFP in postembryonic mutant roots, in mms21-1 embryos, WOX5 expression was diffuse and expanded to the adjacent cells (Fig. 4, O and Q; Supplemental Fig. S4H). Maintenance of WOX5 expression depends on the asymmetric nature of the hypophyseal division (Song et al., 2008); therefore, the diffuse expression is consistent with our findings that mms21 mutants showed disorganized cell division in the hypophysis, supporting a role for AtMMS21 in QC organization and correct cell division patterns of neighboring cells.
Finally, we examined whether the mms21-1 embryo phenotype defects were consistent with defects in PLT expression, which is necessary for embryonic specification of the root stem cell (Aida et al., 2004). As observed in postembryonic mms21-1 roots, the expression domains of PLT1 and PLT2 proteins in the basal embryo of mms21-1 were restricted to the heart-stage embryo compared with the wild type (Fig. 4S; Supplemental Fig. S4I). Furthermore, reductions in the expression of PLT1 and PLT2 were more pronounced in mature embryos (Fig. 4U; Supplemental Fig. S4J). Collectively, our results demonstrate that AtMMS21 function is required for the specification and organization of the root stem cell niche during embryogenesis.
AtMMS21 Is Required to Prevent Cell Death in the Root Stem Cell Niche
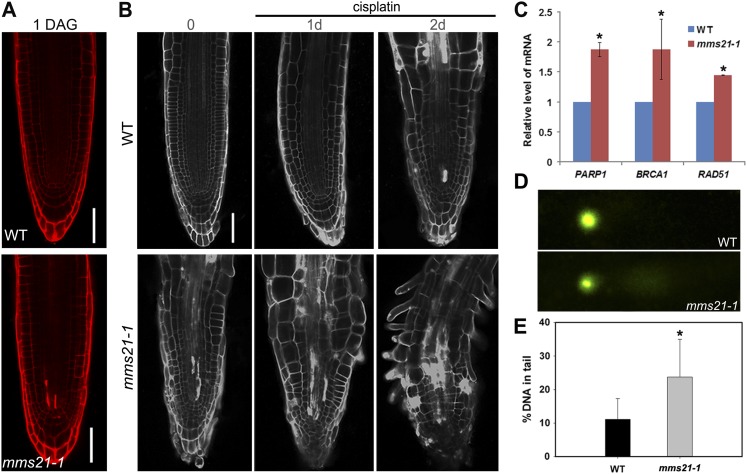
Interestingly, we noticed that in several mms21-1 root meristem cells, the cytoplasm was stained with propidium iodide (PI; Fig. 3). PI stains the walls of living plant cells and is also used as a marker for dead cells that have a loss of membrane integrity (Truernit and Haseloff, 2008). To investigate the spatial occurrence of cell death, plants were stained at different time points with PI. At 1 DAG, no PI-stained cells were observed in wild-type root meristems, but in mms21-1 roots, several stem cells and the early descendants were stained (Fig. 5A). To confirm that cells stained with PI are correctly labeled as dead cells, another cell death marker, Sytox Orange, was used to stain the root tips. As expected, Sytox Orange staining confirmed that the stem cells and their daughter cells showed cell death in the mms21 roots (Supplemental Fig. S5B). At 5 DAG, dead cells in the root meristem of mms21-1 occur in QC cells, surrounding stem cells, and the transiently amplifying region of the vascular tissue, but not in the wild type (Fig. 3; Supplemental Fig. S5D). Detecting the expression of the QC marker pWOX5:GFP in mms21-1 roots confirmed that the dead cells were found in the stem cell niche (Fig. 2, F and G). Interestingly, the expansion of WOX5 expression and the spatial cell death pattern observed in mms21-1 were similar to phenotypes observed in wild-type root meristems after treatment with DNA-damaging agents (Supplemental Fig. S6, B and H; Fulcher and Sablowski, 2009; Furukawa et al., 2010).
Figure 5.
mms21-1 roots show a stem-cell-death phenotype and elevated levels of DSBs. A, Root stem cells and their early descendants stained with PI in mms21-1 roots at 1 DAG, indicating cell death. B, PI staining of the wild type (WT; top) and mms21-1 (bottom) after treatment with 15 μm cisplatin for 0 h (left), 24 h (center), and 48 h (right). Bar = 20 μm. C, qRT-PCR analysis of the expression of DSB-inducible genes in wild-type and mms21-1 roots. Data are means ± sd of three biological repeats. Asterisks indicate significant differences compared with control plants (P < 0 0.005; Student’s t test). D, Representative examples of nuclei seen in the comet assay from wild-type (top) and mms21-1 (bottom) roots. E, DNA damage as measured by the percentage of DNA in the tail of nuclei in comet assays for wild-type and mms21-1 roots. The mean value of more than 200 nuclei is shown with error bars indicating sd. The asterisk indicates a statistically significant difference according to Student’s t test (P < 0 0.01).
Given that mms21-1 mutants showed spontaneous cell death even in the absence of DNA-damaging treatments, we hypothesized that mms21-1 had naturally increased levels of DNA damage that were sufficient to activate cell death and also that external genotoxic treatment that increased the cumulative levels of DNA damage caused more extensive death in the root meristem. Indeed, after growth for 1 d in the presence of cisplatin (which forms DNA cross links and leads to DSBs), the number of dead cells in mms21-1 root tips increased, and many dead cells were observed throughout the mms21-1 meristem, but dead cells were not observed in wild-type plants (Fig. 5B). After treatment for 2 d with cisplatin (15 μm), the root meristems of mms21-1 mutants were not maintained and differentiated; by contrast, only a few dead cells were found in the wild-type stem cell niche (Fig. 5B). Thus, the absence of AtMMS21 triggered cell death in the stem cell niche, and this cell death was associated with DNA damage.
To clarify whether the misregulation of stem cell niche-defining transcription factors is an effect of DNA damage, we examined the expression pattern of these genes after cisplatin treatments. Seedlings were treated with cisplatin (15 μm) for 2 d until external genotoxic stress was sufficient to induce cell death in the stem cell niche. Although the expansion of WOX5 expression was smaller than that observed in mms21-1 mutants, DNA damage had an effect on WOX5 expression (compare Fig. 2, F and G, versus Supplemental Fig. S6, B and H). In addition, the expression of SHR, SCR, PLT1, and PLT2 was slightly reduced after cisplatin treatment, as in the mms21-1 mutants (Supplemental Fig. S6). Thus, these data indicate that the misregulation of stem cell factors in mms21-1 mutants may be a secondary effect due to DNA damage.
mms21-1 Mutant Root Tip Cells Have Increased DSBs
Although experiments in yeast and human cells have demonstrated that MMS21 is required for efficient DSB repair (Potts and Yu, 2005; Zhao and Blobel, 2005), the effect of mms21 mutation on the DNA damage response in plants is unclear. To examine whether the mms21-1 roots had constitutive activation of DNA damage responses, we first measured the expression of the DSB-inducible genes PARP1, BRCA1, and RAD51 by qRT-PCR (Inagaki et al., 2006; Sakamoto et al., 2011). Plants were grown under normal growth conditions, and the transcript levels of all three genes were higher in the mms21-1 roots than in the wild-type plants (Fig. 5C). These data suggested that the mms21-1 root tip cells are exposed to DSBs and showed constitutively activated DNA damage responses, even without additional genotoxic stress.
Next, we directly compared the levels of DSBs in the roots of the wild type and mms21-1 mutants using comet assays (Menke et al., 2001). Levels of DSBs in wild-type and mms21-1 roots were measured by the percentage of DNA in the tail of the comet in the neutral comet assay. As expected, a higher accumulation of DSBs was observed in mms21-1 root tips than in wild-type plants under normal conditions (Fig. 5, D and E). Taken together, our observations that the mms21-1 mutation up-regulated DSB-inducible gene transcription and DSBs indicated that AtMMS21 is involved in reducing the incidence of DSBs in Arabidopsis roots.
mms21-1 Mutants Show Increased Sensitivity to DNA-Damaging Treatments
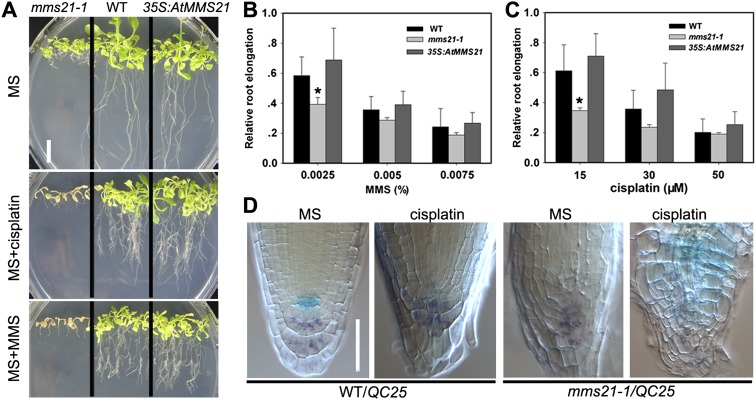
To examine the sensitivity of the mms21-1 mutants to DSB-inducing agents, we evaluated the effect of the DNA cross-linking agents cisplatin and MMS, which cause DSBs, on the growth of mms21-1 and 35S:AtMMS21 plants (Huang et al., 2009). Seedlings at 3 DAG were transferred to medium containing cisplatin (50 μm) or MMS (0.0075%) for 14 d. The mms21-1 plants treated with DNA-damaging agents did not produce additional true leaves, and these plants died 10 to 14 d after treatment, whereas the wild-type plants were less affected (Fig. 6A).
Figure 6.
mms21-1 roots show increased sensitivity to DNA-damaging agents. A, Phenotypes of the wild type (WT) and mms21-1 grown on MS medium (top) and medium containing 50 μm cisplatin (middle) or 0.0075% MMS (bottom). Seedlings at 3 DAG were transplanted onto medium for 14 d. Bar = 1 cm. B and C, Sensitivity of root growth to DNA-damaging agents. Seedlings at 3 DAG were transplanted onto medium containing various concentrations of MMS or cisplatin for 3 d. Relative root elongation is the ratio of root elongation in medium containing the DNA-damaging agents to root elongation in control medium. Each point represents the average of the results from 20 to 30 seedlings, and error bars represent sd. Asterisks indicate significant differences (P < 0 0.01; Student’s t test) relative to the wild type. D, Effects of cisplatin on the root meristem and the expression of QC25:GUS in the wild type (top) and mms21-1 (bottom). Treatments were as described for B and C. Bar = 50 μm.
We next evaluated the effect of short-term treatment with DNA-damaging agents on the growth of mms21-1 roots. Seedlings at 3 DAG grown on vertical plates were transferred to medium containing various concentrations of DSB-inducing agents and incubated for another 3 d. The more severe inhibitory effects on root elongation were observed in the mms21-1 mutants compared with wild-type plants (Fig. 6, B and C). We further found that the mms21-1 root meristems were completely collapsed after exposure to DNA damage, while the application of cisplatin to wild-type roots phenocopied the phenotypes of mms21-1 mutants, including smaller meristems, loss of QC activity, and disorganized stem cell niches and root caps (Fig. 6D; Supplemental Fig. S7). These results indicated that the developmental defects in the mms21 roots were involved in DNA damage responses.
Arabidopsis MMS21 Is a Subunit of the SMC5/6 Complex
Animal and yeast MMS21 form part of the SMC5/6 complex through their interactions with SMC5, and this complex functions in DSB repair (Potts and Yu, 2005; Zhao and Blobel, 2005). Recently, a similar SMC5/6 complex, which also acts in DNA repair, was reported in plant cells (Watanabe et al., 2009). However, non-SMC components of this complex have not been characterized in terms of their interactions with the SMC5/6 complex.
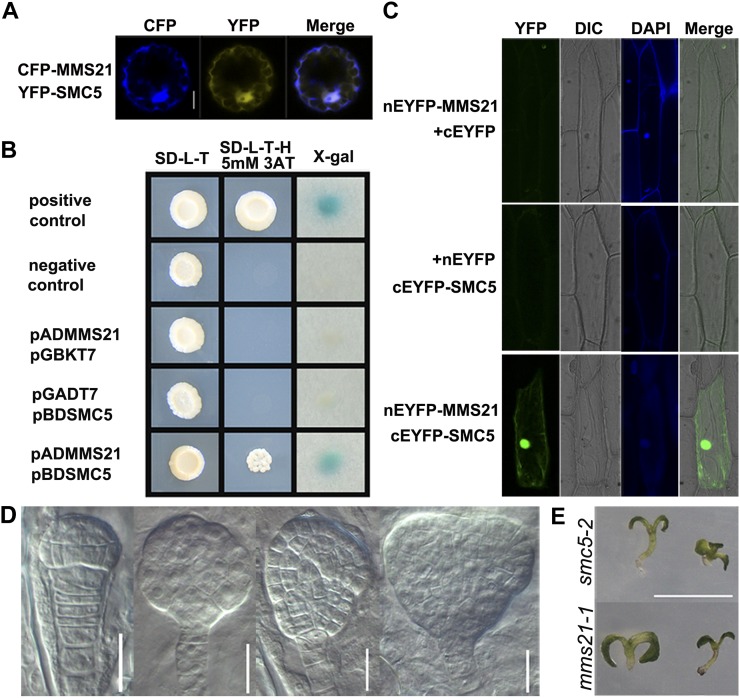
To examine their in vivo protein interaction, we first tested whether AtSMC5 and AtMMS21 colocalize in Arabidopsis protoplasts. When cyan fluorescent protein (CFP)-AtMMS21 was coexpressed with YFP-AtSMC5, predominantly nuclear signals were observed, along with a low signal in the cytosol (Fig. 7A). We next used yeast two-hybrid assays to analyze the direct physical interaction of AtMMS21 with AtSMC5. We found that AtMMS21 interacted with AtSMC5 in yeast (Fig. 7B). This interaction was confirmed using bimolecular fluorescence complementation (BiFC) in plant cells. When the BiFC constructs pSAT6-nEYFP:MMS21 and pSAT6-cEYFP:SMC5 were cotransformed into onion (Allium cepa) epidermal cells, YFP fluorescence was observed in the nucleus, whereas the controls showed only a background signal (Fig. 7C), indicating that MMS21 physically interacts with SMC5 in plant cells. Therefore, Arabidopsis MMS21 is likely the ortholog of the yeast/human MMS21 and is a subunit of the SMC5/6 complex.
Figure 7.
Arabidopsis MMS21 interacts with SMC5. A, AtMMS21 colocalizes with AtSMC5 predominantly in the nucleus. Arabidopsis protoplasts were cotransfected with CFP-AtMMS21 and YFP-AtSMC5. CFP/YFP signals were visualized by confocal microscopy using different channels. Bar = 10 μm. B, AtMMS21 interacts with AtSMC5 in a yeast two-hybrid assay. Yeast cells were cotransformed with pBD-53+pAD-T (positive control), pBD-Lam+pAD-T (negative control), pADMMS2+pGBKT7, pGADT7+pBDSMC5, or pADMMS21+pBDSMC5. Blue colonies in a 5-bromo-4-chloro-3-indolyl-d-galactopyranoside assay show an interaction in vivo between AtMMS21 and AtSMC5. C, BiFC analysis of the interaction between AtMMS21 and AtSMC5 in onion epidermal cells by confocal microscopy. Images were acquired in the YFP and differential interference contrast (DIC) channels, respectively, and then merged together. Cell nuclei were stained with 4′,6-diamino-phenylindole (DAPI), showing a strong interaction between AtMMS21 and AtSMC5 in the nucleus. D and E, Functional similarity between mms21-1 and smc5-2 mutants. As in mms2-1 mutants (for mms21-1 embryos, see Fig. 4; Supplemental Fig. S4), the embryos of SMC5/smc5-2 siliques showed aberrant cell plate formation in the basal embryo region from the early stages of embryogenesis (D) and likely give rise to seedlings germinating without a main root (E). Bars = 20 μm (D) and 1 cm (E).
Additionally, we examined the phenotypic similarities between mms21-1 and smc5-2 mutants. A previous study showed that heterozygotes for a transferred DNA insertion in the Arabidopsis SMC5 gene did not yield homozygous mutants and that siliques of heterozygous plants contained 25% shrunken seeds, indicating that SMC5 is essential for seed development (Watanabe et al., 2009). These findings prompted us to speculate that SMC5 might function in embryogenesis. Similar to mms21-1 mutants, aberrant embryos with abnormal cell division planes were observed in SMC5/smc5-2 siliques (27%, 58 of 218; Fig. 7D). These aberrant divisions were apparent at the basal pole where the root meristem initiates and resulted in seedlings without a main root in 9% (16 of 188) of germinated SMC5/smc5-2 mutants (Fig. 7E). Thus, mms21-1 and smc5-2 mutants exhibited strikingly similar phenotypes, including abnormal embryogenesis and hypersensitivity to DNA damage (Watanabe et al., 2009; this study), suggesting that MMS21 and SMC5 likely function in common or similar pathways to maintain genomic stability and embryo development in plants.
DISCUSSION
AtMMS21 Is Required for Maintenance of the Root Stem Cell Niche
AtMMS21 encodes a SUMO E3 ligase involved in root development (Huang et al., 2009; Ishida et al., 2009); further pursuing the implications of this result, here we demonstrated that AtMMS21 acts in regulating stem cell niche maintenance during root development. The fate of stem cells surrounding the QC can be used as a readout of the QC’s organizing activity (Gonzalez-Garcia et al., 2011). By measuring the expression of QC-specific markers and the differentiation of stem cells, we provide two types of evidence supporting the requirement for AtMMS21 in the maintenance of the root stem cell niche. On the one hand, the irregular QC organization (observed by microscopy and pWOX5:GFP expression), the aberrant expression of QC-specific markers (QC25, QC46, and QC184), and the mitotic activation of QC cells all indicated that AtMMS21 is essential for the proper organization and identity of the QC (Fig. 2; Supplemental Fig. S1). On the other hand, the appearance of starch granules in the region of the QC and CSCs, together with defective columella layers and ground tissue, indicated that QC function and the cell fate of root stem cells are also not maintained properly in the mms21-1 roots, leading to a disorganized meristem pattern and ultimately to short roots (Figs. 1 and 2; Supplemental Fig. S1). Hence, our observations confirmed that AtMMS21 acts in stem cell niches to regulate correct root meristem pattering and function. The stem cell niche is established early in embryogenesis (Weigel and Jurgens, 2002), and the master regulators of root development, such as SHR/SCR and PLT1/PLT2, affect the establishment of the root stem cell niche during embryonic pattern formation (Di Laurenzio et al., 1996; Aida et al., 2004). Similar to shr, scr, and plt1/plt2 mutants, disorganized cell division in the hypophysis region and the defective expression of QC-specific markers were observed in mms21-1 embryos (Fig. 4; Supplemental Fig. S4), indicating that AtMMS21 is required for initiation of the stem cell niche during embryogenesis. Taken together, our data indicate that AtMMS21 is important for the maintenance of root stem cell niches, both during embryogenesis and postembryonic stages.
In mammals, the pluripotency transcription factors Oct4, Nanog, and Sox2 regulate the fate of embryonic stem cells (Orkin and Hochedlinger, 2011). Although the activities of Oct4 and Sox2 are regulated by sumoylation (Hietakangas et al., 2006; Wei et al., 2007; Van Hoof et al., 2009), it is unclear whether this sumoylation is mediated by SUMO E3 ligase. In plants, root stem cell niches are also regulated by pluripotency transcription factors, including PLT1, PLT2, SHR, SCR, and WOX5 (Sablowski, 2011). In the mms21-1 roots, the normal restricted spatial expression pattern of WOX5/SHR/SCR was partially lost, ectopic expression of WOX5/SCR in adjacent cells was observed, and protein levels of PLT1/PLT2 were severely reduced (Fig. 3). In light of the finding that MMS21 is required for chromatin organization during cell division in budding yeast (Rai et al., 2011), AtMMS21 might be targeted to chromatin through its interaction with the SMC5/6 complex and function to ensure the stable epigenetic state of the chromosomes, which is required for the stable expression of pluripotency transcription factors within the stem cell niche (Fig. 3; Supplemental Fig. S3). Intriguingly, PLT1/PLT2 positively regulates the expression and/or accumulation of AtMMS21/HPY2 proteins in the root meristem (Ishida et al., 2009). Therefore, it will be interesting to examine whether a feedback loop exists in which PLT1/PLT2 modulates AtMMS21 accumulation and AtMMS21 influences the accumulation of PLT proteins to maintain pluripotency in the Arabidopsis stem cell niche. Similar to such a scenario, a regulatory loop exists between chromatin factors and stem cell transcription factors in mammals (Loh et al., 2007; Orkin and Hochedlinger, 2011). It is worth noting that MMS21 has yeast, plant, and animal orthologs, suggesting that this mechanism is ancient and conserved. Hence, our results provide an important clue for the further investigation of whether the animal MMS21 also plays a role in the maintenance of stem cell niches.
AtMMS21 Functions in Reducing DNA Damage
The protection of stem cells against DNA damage is crucial for normal development, but little is known about the interplay between DNA damage responses and stem cell niche maintenance (Hashimura and Ueguchi, 2011). Here, we provide three pieces of evidence showing that AtMMS21 plays a crucial role in the maintenance of stem cell niches through a reduction in DNA damage. First, the mms21-1 mutants showed clear cell death in root stem cells and their daughters. Also, external genotoxic treatment caused more extensive death in mms21-1 root meristems (Fig. 5, A and B), suggesting that mutation of AtMMS21 caused increased DNA damage in the cells. In support of this, two recent reports have shown that Arabidopsis root stem cells and their descendants undergo cell death upon DNA damage (Fulcher and Sablowski, 2009; Furukawa et al., 2010). Second, even under normal conditions, a higher accumulation of DSBs was observed in root tips of mms21-1 than in those of wild-type plants. Consistent with this, several genes known to be associated with increased DSB damage were induced in the roots of mms21-1 (Fig. 5, C–E). Third, the mms21-1 mutants exhibited increased sensitivity to the effects of DSB-inducing agents on both root elongation and meristem activity (Fig. 6). Therefore, a possible explanation for the growth defects observed in mms21-1 mutants is that stem cell death increased as a consequence of the accumulation of unrepaired DSBs. This idea is consistent with previous reports that defects in DNA repair genes (e.g. MDO1, MRE11, FAS, BRU1/MGO3/TSK, and TEB) result in similar phenotypes, including elevated levels of DSBs, hypersensitivity to DNA-damaging agents, stem cell death, and misexpression of stem cell regulators, thus causing defective meristem structure with fasciated stems and short roots (Kaya et al., 2001; Bundock and Hooykaas, 2002; Takeda et al., 2004; Inagaki et al., 2006; Amiard et al., 2010; Hashimura and Ueguchi, 2011). All of these developmental defects were also observed in the mms21-1 mutants (Huang et al., 2009; Ishida et al., 2009; this study), suggesting that AtMMS21 is important for root stem cell niche maintenance through the reduction of DSBs.
Although the precise mechanism by which Arabidopsis MMS21 ameliorates DNA damage (Figs. 5 and 6) is unclear, animal and yeast studies provide numerous possible explanations. Experiments in yeast and human cells have demonstrated that MMS21 plays crucial roles in facilitating DSB repair, collapsed replication fork restart, and telomere elongation by homologous recombination (HR; Duan et al., 2009). Although knowledge of the role of the SMC5/6 complex in plants is still limited, a recent study in Arabidopsis indicated that the SMC5/6 complex enhances sister chromatid alignment after DNA damage and thereby facilitates correct DSB repair via HR between sister chromatids (Watanabe et al., 2009). It is likely that AtMMS21 acts in the meristem to efficiently repair DSBs by HR, which is consistent with the finding that DSBs are preferentially repaired by HR between sister chromatids in meristematic cells (Watanabe et al., 2009). Detailed analysis of HR frequency and chromatid behaviors in mms21-1 might help further our understanding of AtMMS21 function in DSB repair and meristem maintenance. In this respect, meristematic cells afflicted by DSBs are faced with the choice of inducing stem cells and their early descendants to die preferentially, or delaying cell division to repair the damage, and endocycles are thought to be an alternative strategy protecting against DSBs (Fulcher and Sablowski, 2009; Adachi et al., 2011). Therefore, in line with the premature onset of endocycles (Ishida et al., 2009) and the early differentiation of root meristematic cells (Fig. 1), we propose that AtMMS21 is important for the prevention of cell death, early differentiation, and premature endocycle onset through a reduction in DSBs (Fig. 8). Additionally, unlike the known SUMO E3, SIZ1, which functions as single proteins, yeast and mammalian MMS21/NSE2 is a conserved subunit of the SMC5/6 complex, which associates with the long coiled-coil region of SMC5 and regulates SMC5/6 complex function (Duan et al., 2009). Here, we demonstrated that Arabidopsis MMS21 physically interacts with SMC5 (Fig. 7), suggesting that the mechanism of MMS21 in DNA repair may represent a conserved function in different organisms. Furthermore, the phenotypic overlap between mutants of AtMMS21 and AtSMC5 suggests a functional overlap between these genes in the maintenance of genomic integrity and the specification of stem cell niches during embryogenesis.
Figure 8.
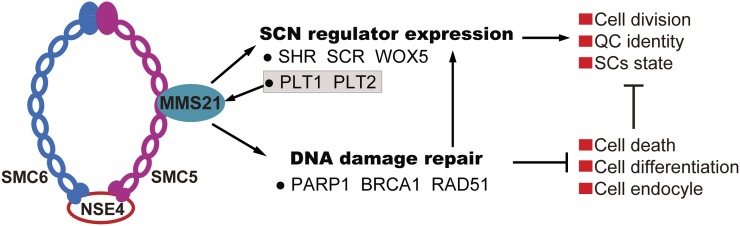
Model for AtMMS21 functions in stem cell niche maintenance and DNA damage responses in the root meristem. The subunit composition of SMC5/6 complexes in Arabidopsis is based on the model of Watanabe et al. (2009), and SMC5-SMC6 heterodimers associate with the δ-kleisin NSE4. Here, we demonstrate that AtMMS21 is an additional subunit of the SMC5/6 complex via direct binding of SMC5. AtMMS21 controls two important processes influencing root stem cell niche maintenance. First, AtMMS21 is essential for the proper identity of QC and stem cells by stable expression of the stem cell niche-defining transcription factors within the niches. Ishida et al. (2009) showed that AtMMS21/HPY2 functions downstream of PLT1/PLT2; therefore, we suggest that a feedback loop exists between AtMMS21 and PLT1/PLT2 in maintaining the root stem cell niche. Second, AtMMS21 is required to prevent cell death, early differentiation, premature endocycle onset, and misregulation of stem cell factors in the root meristem through a reduction in DSBs. Current knowledge about AtMMS21 highlights that AtMMS21-mediated regulation of the balance between cell division, cell endocycle, cell differentiation, and cell death is critical for the normal cellular organization and function of the root meristem. SCs, Stem cells; SCN, stem cell niche. [See online article for color version of this figure.]
CONCLUSION
This study uncovers a cellular mechanism to explain how AtMMS21 defines the stem cell niche and ameliorates DSBs (Fig. 8). AtMMS21 encodes a SUMO E3 ligase expressed in the root meristem (Huang et al., 2009; Ishida et al., 2009), where AtMMS21 mediates the expression levels and patterns of stem cell niche-defining transcription factors; these transcription factors in turn regulate root stem cell niche maintenance. Moreover, AtMMS21 acts as a subunit of the SMC5/6 complex and is essential for the role of the SMC5/6 complex in the reduction of DNA damage. In this regard, AtMMS21 maintains root stem cell survival by ameliorating DSB damage and thus ensures the normal structure and function of the root stem cell niche. Although this explanation has not yet provided definitive evidence for the connection between DSB responses and stem cell niche maintenance, it confirms that AtMMS21 is required for the maintenance of stem cells by reducing DNA damage. In this context, a significant direction for future research will be to identify AtMMS21 target proteins and elucidate the molecular mechanism of AtMMS21 in the maintenance of stem cells and genomic stability, which will help our understanding of how sumoylation controls the developmental process and genomic stability in plants.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
The mms21-1 mutant and 35S:AtMMS21 Arabidopsis (Arabidopsis thaliana; Columbia-0 ecotype) were isolated as described previously (Huang et al., 2009). The following marker lines and mutants were used: QC25, QC46, and QC184 (Sabatini et al., 2003); smc5-2 (Watanabe et al., 2009); pWOX5:GFP (Blilou et al., 2005); pSHR:GFP (Helariutta et al., 2000); pSHR:SHR:GFP (Nakajima et al., 2001); pSCR:GFP (Wysocka-Diller et al., 2000); PLT1pro:PLT1:YFP and PLT2pro:PLT2:YFP (Galinha et al., 2007); shr1 (Benfey et al., 1993); and scr1 (Di Laurenzio et al., 1996). J1092, J0571, and J2341 were from the Haseloff enhancer trap GFP line collection (http://www.plantsci.cam.ac.uk/Haseloff).
Seeds were surface sterilized for 2 min in 75% ethanol followed by 5 min in 1% NaClO solution, rinsed five times with sterile water, plated on Murashige and Skoog (MS) medium with 1% Suc and 0.8% agar, and then stratified at 4°C in the dark for 2 d. Plants were grown under long-day conditions (16 h of light/8 h of dark) at 22°C in a phytotron.
Root Meristem Size Analysis
Seeds were germinated and grown on vertically oriented plates from 1 to 14 d. Roots were examined at different DAG depending on the experiment. Approximately 30 to 50 seedlings were examined in at least three independent experiments, which gave similar results. Roots were mounted in chloral hydrate, and then root meristem size was determined by counting the number of cortex cells in a file extending from the QC to the first elongated cell, which was excluded (Perilli and Sabatini, 2010).
Marker Gene Expression Analysis
The markers QC25, QC46, QC184, J1092, J0571, J2341, pWOX5:GFP, pSHR:GFP, pSHR:SHR:GFP, pSCR:GFP, PLT1pro:PLT1:YFP, and PLT2pro:PLT2:YFP were crossed to the mms21-1 mutants. Homozygous plants for both mms21-1 and marker genes were obtained from F2 populations and analyzed in the next generations.
For qRT-PCR, root tips of 5-DAG seedlings were harvested, and total RNA was extracted using the Plant Easy Spin RNA Miniprep Kit (BioMIGA). RNA was treated with DNaseI and reverse transcribed using a PrimeScript RT reagent kit (Takara). After the RT reaction, the complementary DNA (cDNA) template was subjected to PCR using SYBR Premix ExTaq (Perfect Real Time; Takara). qRT-PCR was performed using three replicates and ACTIN2 (At3g18780) as a reference gene. Real-time measurements of PCR product accumulation were carried out using a 7500 Fast Real-Time PCR system (Applied Biosystems). Data presented are averages from three biological replicates with sd (Inagaki et al., 2006). The statistical significance was evaluated by Student’s t test. The primer sets are listed in Supplemental Table S3.
Histological Analysis
Columella root cap cells specifically accumulate starch granules that are visualized by Lugol staining. Roots were incubated in Lugol solution for 5 min, washed in water once, and then mounted in chloroacetaldehyde:water:glycerol (HCG) solution (8:3:1) for 10 to 20 min before microscopy (Zhou et al., 2010).
Histochemical analysis of GUS activity in enhancer trap lines used as QC markers was performed according to the described method (Stahl et al., 2009) with some modifications. GUS stock solution [0.05 m NaPO4 buffer (pH 7.0), 5 mm K3Fe(CN)6, 5 mm K4Fe(CN)6, and 10 mm X-glucuronide] was made as described previously (Stahl et al., 2009). The seedling roots were stained in a 3:10 dilution of the stock solution at 37°C for 8 to 10 h, rinsed with water, and mounted in HCG solution for microscopy.
For confocal laser imaging of roots, cell walls were labeled with PI as described (Truernit and Haseloff, 2008). Roots were counterstained with 10 μg mL−1 PI (Sigma) for 1 min, washed once in distilled water, and mounted in water for confocal microscopy. Another cell death marker, Sytox Orange (250 nm; Invitrogen), was also used to stain the roots for 5 min before imaging.
The EdU incorporation assay was performed as described previously (Vanstraelen et al., 2009; Zhou et al., 2010) with slight modifications. Seedlings at 2 DAG were grown in liquid MS medium containing 10 μm EdU (Cell-Light EdU Apollo 567 In Vitro Imaging Kit; RiboBio) for 24 h and were fixed in 4% formaldehyde in phosphate-buffered saline (PBS; pH 7.4) for 30 min. Root tips of seedlings were harvested, washed in 3% bovine serum albumin in PBS for 5 min, treated with 0.5% Triton in PBS for 20 min, and washed again. Coupling of EdU to Apollo 567 was performed according to the manufacturer’s instructions, and samples were imaged using confocal microscopy.
Microscopy
Seedling roots and ovules were cleared and mounted in HCG solution and Hoyer’s solution, respectively. Samples were captured using Nomarski optics on a Leica DM2500 with a DFC420 digital camera (Leica) and processed with Leica Application Suite software. Confocal images were taken using a Zeiss LSM 710 laser scanning microscope with the following excitation/emission wavelengths: 561 nm/591 to 635 nm for PI, 543 nm/580 to 610 nm for Sytox Orange, 488 nm/505 to 530 nm for GFP, 514 nm/530 to 600 nm for YFP, 458 nm/475 to 525 nm for CFP, and 550 nm/565 nm for EdU Apollo. At least 15 to 30 seedlings or embryos were examined, and three independent experiments were performed. The seedling roots on MS medium were photographed using the SONY DSC-H50 camera. Images were processed with Photoshop CS4 software (Adobe).
DNA-Damaging Treatments and Comet Assay
For DNA-damaging sensitivity assay, seedlings at 3 DAG were transplanted onto the surface of MS agar plates containing MMS or cisplatin (Sigma) and marked to show the initial positions of their root tips. They were incubated vertically for an additional 14 or 3 d, and the lengths of the newly elongated primary roots were determined using Digimizer 4.2 (http://www.digimizer.com). Root growth was measured and expressed as a percentage of the average length of roots on control plates as described previously (Inagaki et al., 2006). Root meristem size and QC activity were measured as described above. To investigate whether the defective meristem and stem cell niche phenotype of mms21-1 mutants is a DSB stress effect, different concentrations of cisplatin and MMS were tested, and the lowest concentration that gave apparent phenotypes was used. The results presented are averages of 15 to 25 seedlings. Root growth experiments were repeated three times with similar results.
DSBs were measured using comet assays with the N/N protocol as described (Menke et al., 2001). Root tips of 5-DAG seedlings were harvested and prepared as described (Menke et al., 2001). Images of SYBR Green I-stained comets were captured using a fluorescence microscope (DM2500; Leica). DNA damage was calculated by averaging the values for the percentage of DNA in tails from three individual slides, scoring 80 comets per slide (Takahashi et al., 2010). The comet analysis was performed using CASP software (http://www.casp.of.pl/). The statistical significance was evaluated by Student’s t test.
Protein Colocalization and Interaction between AtMMS21 and AtSMC5
For protein colocalization analysis, the coding sequence of AtMMS21 was amplified and fused in a modified pBluescript SK vector containing 35Spro:CFP (Tao et al., 2005), and the coding sequence of AtSMC5 was amplified and fused in the modified pBluescript SK vector containing 35Spro:YFP (Tao et al., 2005). Primers are listed in Supplemental Table S3. This resulted in the CFP-AtMMS21 and YFP-AtSMC5 constructs, which were cotransfected into Arabidopsis protoplasts. Arabidopsis mesophyll protoplasts were prepared and transfected as described previously (Yoo et al., 2007). The fluorescence signals in transfected protoplasts were examined by confocal microscopy.
For yeast (Saccharomyces cerevisiae) two-hybrid analysis, the coding sequence of AtMMS21 was amplified and fused to the GAL4 activation domain in the pGADT7 vector (Clontech), and the coding sequence of AtSMC5 was amplified and fused to the GAL4 DNA-binding domain in the pGBKT7 vector (Clontech). Primers are listed in Supplemental Table S3. The bait and prey constructs were transformed into the yeast strain AH109 (Clontech). The interaction between pAD-T and pBD-53 was used as the positive control, and that between pAD-T and pBD-Lam was used as the negative control. Cotransformed yeast strains were selected on synthetic dextrose/–Leu/–Trp medium. Protein-protein interactions were tested using selective synthetic dextrose/–Leu/–Trp/–His medium and supplied with 5 mm 3-amino-1,2,4-triazole (Clontech). β-Galactosidase activity was measured to assay for the interaction between AtMMS21 and AtSMC5 in a GAL4 two-hybrid system according to the manufacturer’s protocol (Clontech).
For BiFC analysis, the full-length cDNA of AtMMS21 was amplified and fused with the N-terminal part of the YFP coding sequence to give rise to the plasmid pSAT6-nEYFPN1-AtMMS21, and the full-length cDNA of AtSMC5 was amplified and fused with the C-terminal part of YFP to result in the plasmid pSAT6-cEYFPN1-AtSMC5. Primers used for plasmid construction are listed in Supplemental Table S3. Vector combinations (samples and controls) were used, and BiFC assays were performed as described (Walter et al., 2004; Huang et al., 2009). YFP fluorescence after bombardment of onion (Allium cepa) epidermal cells was visualized by confocal microscopy.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AtMMS21 (At3g15150), PLT1 (At3g20840), PLT2 (At1g51190), SCR (At3g54220), SHR (At4g37650), WOX5 (At3g11260), and SMC5 (At5g15920).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Marker gene expression in mms21-1 roots.
Supplemental Figure S2. The shr1 and scr1 mutants show little effect on the expression of AtMMS21.
Supplemental Figure S3. Maintenance of the stem cell niche is defective in mms21-1 roots from 1 DAG.
Supplemental Figure S4. Defects in the stem cell niche in mms21-1 embryos.
Supplemental Figure S5. Sytox Orange staining of mms21-1 mutants shows dead cells in the root meristems.
Supplemental Figure S6. Effects of DNA-damaging treatments on the expression of stem cell niche-defining genes.
Supplemental Figure S7. The root meristem of mms21-1 mutants shows increased sensitivity to DNA-damaging treatments.
Supplemental Table S1. AtMMS21 is required for root stem cell niche maintenance.
Supplemental Table S2. Frequency of embryos exhibiting a defective basal region among the progeny of mms21-1 mutants.
Supplemental Table S3. Primers used in this study.
Acknowledgments
We thank Philip Benfey (Duke University), Ben Scheres (Utrecht University), Lieven De Veylder (Ghent University), A.B. Britt (University of California), Lizhen Tao (South China Agricultural University), and the Arabidopsis Biological Resource Center for kindly providing seeds used in this study.
Glossary
- QC
quiescent center
- DSB
double-strand break
- DAG
d after germination
- EdU
5-ethynyl-2′-deoxyuridine
- qRT
quantitative reverse transcription
- CSC
columella stem cell
- YFP
yellow fluorescent protein
- PI
propidium iodide
- CFP
cyan fluorescent protein
- BiFC
bimolecular fluorescence complementation
- HR
homologous recombination
- MS
Murashige and Skoog
- HCG
chloroacetaldehyde:water:glycerol
- PBS
phosphate-buffered saline
- MMS
methyl-methane sulfonate
- cDNA
complementary DNA
References
- Adachi S, Minamisawa K, Okushima Y, Inagaki S, Yoshiyama K, Kondou Y, Kaminuma E, Kawashima M, Toyoda T, Matsui M, et al. (2011) Programmed induction of endoreduplication by DNA double-strand breaks in Arabidopsis. Proc Natl Acad Sci USA 108: 10004–10009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aichinger E, Villar CB, Di Mambro R, Sabatini S, Kohler C. (2011) The CHD3 chromatin remodeler PICKLE and polycomb group proteins antagonistically regulate meristem activity in the Arabidopsis root. Plant Cell 23: 1047–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aida M, Beis D, Heidstra R, Willemsen V, Blilou I, Galinha C, Nussaume L, Noh YS, Amasino R, Scheres B. (2004) The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119: 109–120 [DOI] [PubMed] [Google Scholar]
- Amiard S, Charbonnel C, Allain E, Depeiges A, White CI, Gallego ME. (2010) Distinct roles of the ATR kinase and the Mre11-Rad50-Nbs1 complex in the maintenance of chromosomal stability in Arabidopsis. Plant Cell 22: 3020–3033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benfey PN, Linstead PJ, Roberts K, Schiefelbein JW, Hauser MT, Aeschbacher RA. (1993) Root development in Arabidopsis: four mutants with dramatically altered root morphogenesis. Development 119: 57–70 [DOI] [PubMed] [Google Scholar]
- Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I, Friml J, Heidstra R, Aida M, Palme K, Scheres B. (2005) The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433: 39–44 [DOI] [PubMed] [Google Scholar]
- Bundock P, Hooykaas P. (2002) Severe developmental defects, hypersensitivity to DNA-damaging agents, and lengthened telomeres in Arabidopsis MRE11 mutants. Plant Cell 14: 2451–2462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H, Levesque MP, Vernoux T, Jung JW, Paquette AJ, Gallagher KL, Wang JY, Blilou I, Scheres B, Benfey PN. (2007) An evolutionarily conserved mechanism delimiting SHR movement defines a single layer of endodermis in plants. Science 316: 421–425 [DOI] [PubMed] [Google Scholar]
- Di Laurenzio L, Wysocka-Diller J, Malamy JE, Pysh L, Helariutta Y, Freshour G, Hahn MG, Feldmann KA, Benfey PN. (1996) The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell 86: 423–433 [DOI] [PubMed] [Google Scholar]
- Dinneny JR, Benfey PN. (2008) Plant stem cell niches: standing the test of time. Cell 132: 553–557 [DOI] [PubMed] [Google Scholar]
- Dou H, Huang C, Van Nguyen T, Lu LS, Yeh ET. (2011) SUMOylation and de-SUMOylation in response to DNA damage. FEBS Lett 585: 2891–2896 [DOI] [PubMed] [Google Scholar]
- Duan X, Sarangi P, Liu X, Rangi GK, Zhao X, Ye H. (2009) Structural and functional insights into the roles of the Mms21 subunit of the Smc5/6 complex. Mol Cell 35: 657–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo M, Ishikawa Y, Osakabe K, Nakayama S, Kaya H, Araki T, Shibahara K, Abe K, Ichikawa H, Valentine L, et al. (2006) Increased frequency of homologous recombination and T-DNA integration in Arabidopsis CAF-1 mutants. EMBO J 25: 5579–5590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulcher N, Sablowski R. (2009) Hypersensitivity to DNA damage in plant stem cell niches. Proc Natl Acad Sci USA 106: 20984–20988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa T, Curtis MJ, Tominey CM, Duong YH, Wilcox BW, Aggoune D, Hays JB, Britt AB. (2010) A shared DNA-damage-response pathway for induction of stem-cell death by UVB and by gamma irradiation. DNA Repair (Amst) 9: 940–948 [DOI] [PubMed] [Google Scholar]
- Galinha C, Hofhuis H, Luijten M, Willemsen V, Blilou I, Heidstra R, Scheres B. (2007) PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature 449: 1053–1057 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Garcia MP, Vilarrasa-Blasi J, Zhiponova M, Divol F, Mora-Garcia S, Russinova E, Cano-Delgado AI. (2011) Brassinosteroids control meristem size by promoting cell cycle progression in Arabidopsis roots. Development 138: 849–859 [DOI] [PubMed] [Google Scholar]
- Haecker A, Gross-Hardt R, Geiges B, Sarkar A, Breuninger H, Herrmann M, Laux T. (2004) Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana. Development 131: 657–668 [DOI] [PubMed] [Google Scholar]
- Hashimura Y, Ueguchi C. (2011) The Arabidopsis MERISTEM DISORGANIZATION 1 gene is required for the maintenance of stem cells through the reduction of DNA damage. Plant J 68: 657–669 [DOI] [PubMed] [Google Scholar]
- Helariutta Y, Fukaki H, Wysocka-Diller J, Nakajima K, Jung J, Sena G, Hauser MT, Benfey PN. (2000) The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell 101: 555–567 [DOI] [PubMed] [Google Scholar]
- Hietakangas V, Anckar J, Blomster HA, Fujimoto M, Palvimo JJ, Nakai A, Sistonen L. (2006) PDSM, a motif for phosphorylation-dependent SUMO modification. Proc Natl Acad Sci USA 103: 45–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Yang S, Zhang S, Liu M, Lai J, Qi Y, Shi S, Wang J, Wang Y, Xie Q, et al. (2009) The Arabidopsis SUMO E3 ligase AtMMS21, a homologue of NSE2/MMS21, regulates cell proliferation in the root. Plant J 60: 666–678 [DOI] [PubMed] [Google Scholar]
- Inagaki S, Suzuki T, Ohto MA, Urawa H, Horiuchi T, Nakamura K, Morikami A. (2006) Arabidopsis TEBICHI, with helicase and DNA polymerase domains, is required for regulated cell division and differentiation in meristems. Plant Cell 18: 879–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida T, Fujiwara S, Miura K, Stacey N, Yoshimura M, Schneider K, Adachi S, Minamisawa K, Umeda M, Sugimoto K. (2009) SUMO E3 ligase HIGH PLOIDY2 regulates endocycle onset and meristem maintenance in Arabidopsis. Plant Cell 21: 2284–2297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaya H, Shibahara KI, Taoka KI, Iwabuchi M, Stillman B, Araki T. (2001) FASCIATA genes for chromatin assembly factor-1 in Arabidopsis maintain the cellular organization of apical meristems. Cell 104: 131–142 [DOI] [PubMed] [Google Scholar]
- Kirik A, Pecinka A, Wendeler E, Reiss B. (2006) The chromatin assembly factor subunit FASCIATA1 is involved in homologous recombination in plants. Plant Cell 18: 2431–2442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornet N, Scheres B. (2009) Members of the GCN5 histone acetyltransferase complex regulate PLETHORA-mediated root stem cell niche maintenance and transit amplifying cell proliferation in Arabidopsis. Plant Cell 21: 1070–1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh YH, Zhang WW, Chen X, George J, Ng HH. (2007) Jmjd1a and Jmjd2c histone H3 Lys 9 demethylases regulate self-renewal in embryonic stem cells. Genes Dev 21: 2545–2557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menke M, Chen I, Angelis KJ, Schubert I. (2001) DNA damage and repair in Arabidopsis thaliana as measured by the comet assay after treatment with different classes of genotoxins. Mutat Res 493: 87–93 [DOI] [PubMed] [Google Scholar]
- Moubayidin L, Perilli S, Dello Ioio R, Di Mambro R, Costantino P, Sabatini S. (2010) The rate of cell differentiation controls the Arabidopsis root meristem growth phase. Curr Biol 20: 1138–1143 [DOI] [PubMed] [Google Scholar]
- Nakajima K, Sena G, Nawy T, Benfey PN. (2001) Intercellular movement of the putative transcription factor SHR in root patterning. Nature 413: 307–311 [DOI] [PubMed] [Google Scholar]
- Nawy T, Lee JY, Colinas J, Wang JY, Thongrod SC, Malamy JE, Birnbaum K, Benfey PN. (2005) Transcriptional profile of the Arabidopsis root quiescent center. Plant Cell 17: 1908–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkin SH, Hochedlinger K. (2011) Chromatin connections to pluripotency and cellular reprogramming. Cell 145: 835–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perilli S, Sabatini S. (2010) Analysis of root meristem size development. Methods Mol Biol 655: 177–187 [DOI] [PubMed] [Google Scholar]
- Potts PR, Yu H. (2005) Human MMS21/NSE2 is a SUMO ligase required for DNA repair. Mol Cell Biol 25: 7021–7032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai R, Varma SP, Shinde N, Ghosh S, Kumaran SP, Skariah G, Laloraya S. (2011) Small ubiquitin-related modifier ligase activity of Mms21 is required for maintenance of chromosome integrity during the unperturbed mitotic cell division cycle in Saccharomyces cerevisiae. J Biol Chem 286: 14516–14530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini S, Heidstra R, Wildwater M, Scheres B. (2003) SCARECROW is involved in positioning the stem cell niche in the Arabidopsis root meristem. Genes Dev 17: 354–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sablowski R. (2004) Plant and animal stem cells: conceptually similar, molecularly distinct? Trends Cell Biol 14: 605–611 [DOI] [PubMed] [Google Scholar]
- Sablowski R. (2011) Plant stem cell niches: from signalling to execution. Curr Opin Plant Biol 14: 4–9 [DOI] [PubMed] [Google Scholar]
- Sakamoto T, Inui YT, Uraguchi S, Yoshizumi T, Matsunaga S, Mastui M, Umeda M, Fukui K, Fujiwara T. (2011) Condensin II alleviates DNA damage and is essential for tolerance of boron overload stress in Arabidopsis. Plant Cell 23: 3533–3546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar AK, Luijten M, Miyashima S, Lenhard M, Hashimoto T, Nakajima K, Scheres B, Heidstra R, Laux T. (2007) Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 446: 811–814 [DOI] [PubMed] [Google Scholar]
- Song SK, Hofhuis H, Lee MM, Clark SE. (2008) Key divisions in the early Arabidopsis embryo require POL and PLL1 phosphatases to establish the root stem cell organizer and vascular axis. Dev Cell 15: 98–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl Y, Wink RH, Ingram GC, Simon R. (2009) A signaling module controlling the stem cell niche in Arabidopsis root meristems. Curr Biol 19: 909–914 [DOI] [PubMed] [Google Scholar]
- Takahashi N, Quimbaya M, Schubert V, Lammens T, Vandepoele K, Schubert I, Matsui M, Inze D, Berx G, De Veylder L. (2010) The MCM-binding protein ETG1 aids sister chromatid cohesion required for postreplicative homologous recombination repair. PLoS Genet 6: e1000817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda S, Tadele Z, Hofmann I, Probst AV, Angelis KJ, Kaya H, Araki T, Mengiste T, Mittelsten Scheid O, Shibahara K, et al. (2004) BRU1, a novel link between responses to DNA damage and epigenetic gene silencing in Arabidopsis. Genes Dev 18: 782–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao LZ, Cheung AY, Nibau C, Wu HM. (2005) RAC GTPases in tobacco and Arabidopsis mediate auxin-induced formation of proteolytically active nuclear protein bodies that contain AUX/IAA proteins. Plant Cell 17: 2369–2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truernit E, Haseloff J. (2008) A simple way to identify non-viable cells within living plant tissue using confocal microscopy. Plant Methods 4: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hoof D, Munoz J, Braam SR, Pinkse MWH, Linding R, Heck AJR, Mummery CL, Krijgsveld J. (2009) Phosphorylation dynamics during early differentiation of human embryonic stem cells. Cell Stem Cell 5: 214–226 [DOI] [PubMed] [Google Scholar]
- Vanstraelen M, Baloban M, Da Ines O, Cultrone A, Lammens T, Boudolf V, Brown SC, De Veylder L, Mergaert P, Kondorosi E. (2009) APC/C-CCS52A complexes control meristem maintenance in the Arabidopsis root. Proc Natl Acad Sci USA 106: 11806–11811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter M, Chaban C, Schutze K, Batistic O, Weckermann K, Nake C, Blazevic D, Grefen C, Schumacher K, Oecking C, et al. (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 40: 428–438 [DOI] [PubMed] [Google Scholar]
- Watanabe K, Pacher M, Dukowic S, Schubert V, Puchta H, Schubert I. (2009) The STRUCTURAL MAINTENANCE OF CHROMOSOMES 5/6 complex promotes sister chromatid alignment and homologous recombination after DNA damage in Arabidopsis thaliana. Plant Cell 21: 2688–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei F, Scholer HR, Atchison ML. (2007) Sumoylation of Oct4 enhances its stability, DNA binding, and transactivation. J Biol Chem 282: 21551–21560 [DOI] [PubMed] [Google Scholar]
- Weigel D, Jurgens G. (2002) Stem cells that make stems. Nature 415: 751–754 [DOI] [PubMed] [Google Scholar]
- Wildwater M, Campilho A, Perez-Perez JM, Heidstra R, Blilou I, Korthout H, Chatterjee J, Mariconti L, Gruissem W, Scheres B. (2005) The RETINOBLASTOMA-RELATED gene regulates stem cell maintenance in Arabidopsis roots. Cell 123: 1337–1349 [DOI] [PubMed] [Google Scholar]
- Wysocka-Diller JW, Helariutta Y, Fukaki H, Malamy JE, Benfey PN. (2000) Molecular analysis of SCARECROW function reveals a radial patterning mechanism common to root and shoot. Development 127: 595–603 [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J. (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Zhao X, Blobel G. (2005) A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization. Proc Natl Acad Sci USA 102: 4777–4782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Wei L, Xu J, Zhai Q, Jiang H, Chen R, Chen Q, Sun J, Chu J, Zhu L, et al. (2010) Arabidopsis tyrosylprotein sulfotransferase acts in the auxin/PLETHORA pathway in regulating postembryonic maintenance of the root stem cell niche. Plant Cell 22: 3692–3709 [DOI] [PMC free article] [PubMed] [Google Scholar]