Gene functions should be studied not only under stable laboratory conditions, but also in the environment abounding in multiple stresses.
Abstract
There is growing evidence that for a comprehensive insight into the function of plant genes, it is crucial to assess their functionalities under a wide range of conditions. In this study, we examined the role of LESION SIMULATING DISEASE1 (LSD1), ENHANCED DISEASE SUSCEPTIBILITY1 (EDS1), and PHYTOALEXIN DEFICIENT4 (PAD4) in the regulation of photosynthesis, water use efficiency, reactive oxygen species/hormonal homeostasis, and seed yield in Arabidopsis (Arabidopsis thaliana) grown in the laboratory and in the field. We demonstrate that the LSD1 null mutant (lsd1), which is known to exhibit a runaway cell death in nonpermissive conditions, proves to be more tolerant to combined drought and high-light stress than the wild type. Moreover, depending on growing conditions, it shows variations in water use efficiency, salicylic acid and hydrogen peroxide concentrations, photosystem II maximum efficiency, and transcription profiles. However, despite these changes, lsd1 demonstrates similar seed yield under all tested conditions. All of these traits depend on EDS1 and PAD4. The differences in the pathways prevailing in the lsd1 in various growing environments are manifested by the significantly smaller number of transcripts deregulated in the field compared with the laboratory, with only 43 commonly regulated genes. Our data indicate that LSD1, EDS1, and PAD4 participate in the regulation of various molecular and physiological processes that influence Arabidopsis fitness. On the basis of these results, we emphasize that the function of such important regulators as LSD1, EDS1, and PAD4 should be studied not only under stable laboratory conditions, but also in the environment abounding in multiple stresses.
Despite possessing large amounts of knowledge concerning a particular gene’s role obtained from laboratory studies using loss-of-function mutants, it is generally accepted that specific phenotypic effects are influenced by environmental conditions (Tonsor et al., 2005; Ungerer et al., 2008). The main reason for performing experiments in growth chambers is to reduce variations in the measurements of specific plant traits. However, such highly controlled conditions frequently lack many aspects of the natural environment, such as variability in light intensity, temperature, and water availability. Only a few of the laboratory-performed studies take into account the fact that plant responses may be different in variable conditions. Thus, in order to investigate plant responses and acclimation mechanisms, there has been an emerging need to perform experiments in multiple environments.
The context-dependent gene function has been demonstrated in the study of the photosynthetic feedback deexcitation pathway (energy-dependent quenching type of nonphotochemical quenching) and its influence on plant fitness in different light environments. This work has proven that feedback deexcitation is important for plant performance in the field and in fluctuating light, but it does not affect plant fitness under constant light conditions (Külheim et al., 2002). It has also been shown that quantitative trait loci for flowering time differ in the field compared with highly controlled growth chamber conditions. While some quantitative trait loci have been detected in all environments, others have been present only in conditions that shared a similar photoperiod (Weinig et al., 2002). These findings illustrate the environmental dependency of plant metabolic and developmental pathways. Therefore, studies in multivariable field conditions appear to be crucial to fully describe the nature of gene function and to understand the integration of metabolism and development with the external environment.
LESION SIMULATING DISEASE1 (LSD1) has been described to integrate signaling pathways in response to diverse stresses, both biotic (Rustérucci et al., 2001; Wiermer et al., 2005) and abiotic (Mateo et al., 2004; Mühlenbock et al., 2007, 2008). The LSD1 mutant (lsd1) belongs to one of the best characterized Arabidopsis (Arabidopsis thaliana) mutants in the context of deregulated cell death (Jabs et al., 1996; Dietrich et al., 1997; Rustérucci et al., 2001; Epple et al., 2003; Mateo et al., 2004; Torres et al., 2005; Mühlenbock et al., 2007; Mühlenbock et al., 2008). lsd1 was initially characterized for its reactive oxygen species (ROS)- and salicylic acid (SA)-dependent uncontrolled spread of cell death that develops under nonpermissive conditions, such as long (longer than 16 h) or continuous photoperiods, supply of a superoxide ion, or infection with avirulent pathogens (Dietrich et al., 1994; Jabs et al., 1996; Hunt et al., 1997). The runaway cell death (RCD) phenotype of lsd1 is indicative for the failure to stop both the initiation and propagation of cell death. LSD1 was proposed as a negative regulator of RCD, acting as a ROS rheostat and preventing the prodeath pathway below certain ROS levels (Jabs et al., 1996; Dietrich et al., 1997; Kliebenstein et al., 1999). However, our previous results report that LSD1 is also required for acclimation to conditions that promote excess excitation energy (EEE; Mateo et al., 2004; Mühlenbock et al., 2008) and root hypoxia stress (Mühlenbock et al., 2007). The lsd1 mutant shows reduced stomatal conductance and catalase activity in short-day permissive conditions (Mateo et al., 2004) and increased ethylene (ET) and hydrogen peroxide (H2O2) accumulation followed by RCD in nonpermissive conditions (Mühlenbock et al., 2008). The lsd1/chloroplastic signal recognition particle cpSRP43 (cao) double mutant that has reduced PSII antenna size due to the cao mutation displays reduced RCD and higher nonphotochemical quenching (Mateo et al., 2004). Therefore, we linked RCD in lsd1 to several parameters: the amount of light energy absorbed in excess by the PSII light-harvesting complex, the redox changes in PSII and emerging changes in nonphotochemical quenching, stomatal conductance, and, ultimately, the photorespiration-associated production of H2O2.
It is important to note that lsd1 traits depend on ENHANCED DISEASE SUSCEPTIBILITY1 (EDS1) and its interacting partner PHYTOALEXIN DEFICIENT4 (PAD4). The Arabidopsis EDS1 and PAD4 proteins constitute a regulatory hub for gene-mediated and basal resistance and are required for accumulation of SA (Parker et al., 1996; Glazebrook et al., 1997; Falk et al., 1999; Feys et al., 2001; Wiermer et al., 2005). Null mutations in EDS1 and PAD4 revert the lsd1-conditioned limitation of foliar gas exchange, ET and H2O2 accumulation, and RCD (Rustérucci et al., 2001; Mateo et al., 2004; Mühlenbock et al., 2007, 2008).
In this study, we used the cell death-deregulated mutants lsd1, eds1, and pad4 and the respective double mutants eds1/lsd1 and pad4/lsd1 to measure different physiological parameters important for the plants’ fitness. Our results demonstrate that LSD1, EDS1, and PAD4 regulate PSII maximum efficiency (Fv′/Fm′), water use efficiency (WUE), H2O2 and SA foliar concentrations, and seed yield (YS). We prove here that apart from playing an important role in abiotic and biotic stress responses, LSD1, EDS1, and PAD4 also participate in the regulation of photosynthesis, transpiration, cellular signaling, and YS. We also show that the phenotype of the lsd1 mutant strongly depends on the growing conditions, which is exhibited by significant differences in physiological parameters and transcription profiles in laboratory- and field-grown plants. Our results emphasize the importance of examining gene functions not only under stable laboratory conditions, but also in the natural environment abounding in multiple stresses.
RESULTS
LSD1 Negatively Regulates Drought and High-Light Stress Tolerance
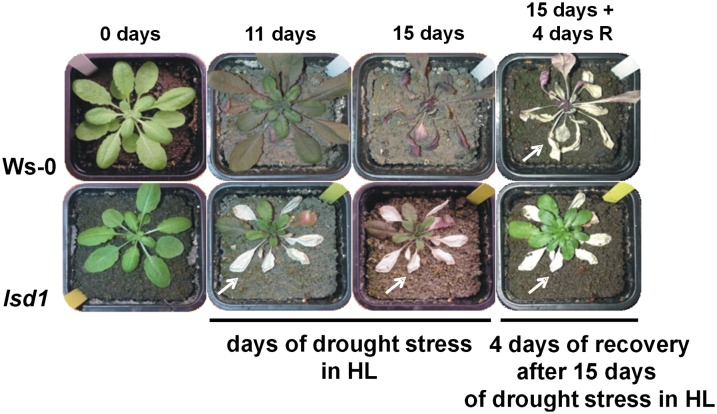
We previously observed that the artificial restriction of stomatal conductance promoting photorespiration in a single leaf was sufficient to induce RCD in other older leaves of the lsd1 mutant rosette cultivated under a permissive short photoperiod and low-light conditions (Mühlenbock et al., 2008). Both drought and high-light stress are factors favoring photorespiration on their own or in combination (Wingler et al., 1999; Noctor et al., 2002; Zhou et al., 2007). Here, we naturally induced photorespiration by exposing the plants simultaneously to drought and high-light stress. After 11 d of water deficiency, RCD was apparent in mature lsd1 leaves, while the younger leaves in the central part of the rosette remained vital (Fig. 1). In the wild-type (Wassilewskija [Ws-0]) plants, no cell death was observed. After 15 d of water deficiency, the wild-type plants were completely wilted, while the younger lsd1 leaves remained vital (Fig. 1). When allowing the plants to recover by restoring the normal watering regime, the lsd1 plants continued growth and formed inflorescences. The survival rate of 5-week-old plants was significantly higher in lsd1 compared with the wild type, and this difference was also clearly reflected in the seed yield of 9-week-old plants (Table I). In the same experiment, both the survival rate and seed production for eds1, pad4, eds1/lsd1, and pad4/lsd1 mutants were determined. After 15 d of growing in water-deficient conditions, all of the tested genotypes except for lsd1 and eds1 were irreversibly wilted (Fig. 1; Supplemental Fig. S1). Although to a lesser extent than lsd1, eds1 could also recover after rewatering. The double mutants eds1/lsd1 and pad4/lsd1 showed a similar sensitivity to drought and high-light stress as the wild-type plants (Table I; Supplemental Fig. S1), which is consistent with previous reports that indicated LSD1 as a negative regulator of both EDS1- and PAD4-dependent cell death (Rustérucci et al., 2001; Mateo et al., 2004; Mühlenbock et al., 2007, 2008). These results show that although lsd1 easily initiates RCD in older leaves in response to nonpermissive conditions, it demonstrates significantly better tolerance to combined drought and high-light stress and produces a higher YS compared with the wild-type plants and other genotypes (Fig. 1; Table I).
Figure 1.
LSD1 negatively regulates simultaneous drought and high-light stress tolerance in the laboratory. Effects of simultaneously acting water deficiency (drought stress) and high-light stress on wild-type (Ws-0) and lsd1 mutant plants. Five-week-old plants were grown for 11 and 15 d without watering, and then they were rewatered and subjected to seed production (see Table I). Arrows indicate RCD. HL, High-light; R, rewatered.
Table I. Regulation of drought/high-light stress tolerance and YS by LSD1, EDS1, and PAD4.
After stress, all plants were rewatered and cultivated in ambient laboratory conditions for an additional 3 weeks until the seeds were harvested. The results are representative of two independent experiments (n = 10–15 ± se). Asterisks indicate a significant difference compared with Ws-0 at the level *P < 0.05 by Tukey’s multiple comparison test.
| 11 d of Water Deficiency and High-Light Stress |
15 d of Water Deficiency and High-Light Stress |
|||
|---|---|---|---|---|
| Genotype | Survival Ratea | YS (No. of Seeds per Plant)b | Survival Ratec | YS (No. of Seeds per Plant)d |
| % | % | |||
| Ws-0 | 100 | 5,996 ± 897 | 0 | 0 |
| lsd1 | 100 | 5,629 ± 708 | 45 | 1,612 ± 762* |
| eds1 | 93 | 6,488 ± 809 | 26 | 1,288 ± 697 |
| pad4 | 93 | 5,002 ± 729 | 0 | 0 |
| eds1 lsd1 | 79 | 3,783 ± 625 | 0 | 0 |
| pad4 lsd1 | 93 | 5,864 ± 616 | 0 | 0 |
Survival rate of the wild type (Ws-0) and mutants after 11 d of drought and high-light (PPFD of 500 ± 50 µmol m–2 s–1) stress. bYS of Ws-0 and mutants after 9 weeks of growth (with 11 d of drought and high-light [PPFD of 500 ± 50 µmol m–2 s–1] stress). cSurvival rate of Ws-0 and mutants after 15 d of drought and high-light (PPFD of 500 ± 50 µmol m–2 s–1) stress. dYS of Ws-0 and mutants after 9 weeks of growth (with 15 d of drought and high-light [PPFD of 500 ± 50 µmol m–2 s–1] stress).
lsd1 Displays Different Maximum Efficiency of PSII and WUE But Similar Seed Yield in Laboratory and Field Conditions
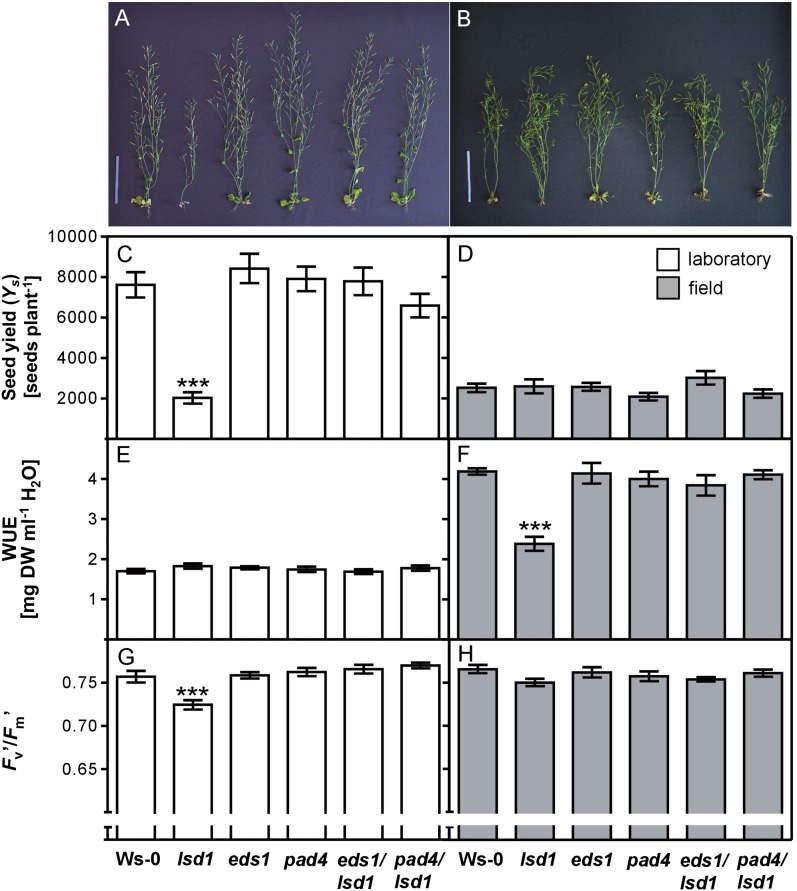
The observation that lsd1 manifests a higher YS compared with wild-type plants and the other mutants under combined drought and high-light stress inspired us to test all of the genotypes in a natural environment, abounding in multiple abiotic and biotic stresses, during several seasons and at various geographical locations (Supplemental Fig. S2). We observed that the lsd1 plants displayed strongly reduced YS in laboratory conditions (Fig. 2, A and C), which was reversed in the double eds1/lsd1 and pad4/lsd1 mutants. In the natural environment, YS in all genotypes was approximately 3 times lower than in the growth chamber, but no significant difference in YS was apparent in lsd1 compared with the wild type (Fig. 2, B and D). In fact, the lsd1 mutant exhibited similar YS in stable laboratory conditions, in laboratory drought and the high-light stress experiment, and in the field (Fig. 2, C and D; Table I). The germination rates of seeds collected from both the laboratory and field experiments were similar in the wild type and mutant lines (Supplemental Fig. S3). We also observed that during the generative stage, the old leaves of the lsd1 plants died (Fig. 2, A and B), which is consistent with the laboratory drought and high-light stress experiment (Fig. 1).
Figure 2.
Condition-dependent regulation of YS, WUE, and photosynthesis by LSD1, EDS1, and PAD4. Morphology of plants grown in controlled laboratory (A) and variable field (B) conditions, and seed production per plant in the laboratory (C) and field (D) of 8-week-old plants. Mean values (± se) are averages of four independent field experiments performed during the 2004 to 2007 and 2010 summer seasons and tested in two different geographical locations (Warsaw and Krakow, Poland) and six independent laboratory experiments (Krakow and Warsaw, Poland and Stockholm, Sweden); at least 13 plants for each experiment per genotype were analyzed (n = 45–89). WUE (mg dry weight × mL–1 water used) of plants grown in the laboratory (E) and field (F). Fv′/Fm′ in the laboratory (G) and field (H). Mean values (± se) are derived from four to 14 separate plants per genotype (n = 4–14). Asterisks above the bars indicate the significant difference from the wild type (Ws-0) at the level ***P < 0.001 as indicated by Tukey’s multiple comparison test. DW, Dry weight. Bars = 10 cm.
In the growth chamber, WUE was similar in all of the tested genotypes (Fig. 2E). On the other hand, field-grown lsd1 displayed significantly lower WUE (Fig. 2F). The lsd1 plants produced approximately 40% less dry mass per 1 mL water utilized compared with the other genotypes, but apparently such low WUE did not affect lsd1 seed production. Moreover, reduced WUE in lsd1 depended on EDS1 and PAD4, since in the eds1/lsd1 and pad4/lsd1 mutants, the WUE was similar to that observed in the wild-type plants (Fig. 2F). The Fv′/Fm′ parameter provides an estimate of the maximum efficiency of PSII at a given light intensity and spectral quality (Baker, 2008). Fv′/Fm′ was significantly reduced in lsd1 in the laboratory, but not in the field (Fig. 2, G and H), and also depended on EDS1 and PAD4. These results imply that LSD1 regulation of WUE and Fv′/Fm′ depends on the conditions (laboratory or field), whereas YS in the lsd1 mutant does not show such dependence. The above results (Figs. 1 and 2; Table I) indicate that drought stress tolerance, WUE, Fv′/Fm′, and YS in Arabidopsis plants are jointly regulated by the LSD1/EDS1/PAD4 molecular node.
Regulation of ROS and Hormonal Homeostasis by LSD1, EDS1, and PAD4
Taking into consideration the fact that LSD1, EDS1, and PAD4 are involved in the regulation of biotic and abiotic stress responses, we determined the levels of foliar SA and H2O2 for wild-type and mutant plants in growth chambers and in the natural environment. Increased foliar concentrations of H2O2 were observed in all genotypes grown in the laboratory compared with the field (Table II). The changes in H2O2 ranged from 2-fold for lsd1, 3-fold for eds1/lsd1 and pad4/lsd1, and 5-fold for Ws-0, eds1, and pad4. Despite these considerable differences in H2O2 content between plants grown in different environments, only lsd1 displayed a significant change in the H2O2 level compared with the other genotypes within a particular condition. The concentration of SA was 5-fold and 3-fold higher in lsd1 compared with the wild-type plants in the laboratory or field conditions, respectively. These lsd1 traits depended on EDS1 and PAD4, since the eds1/lsd1 and pad4/lsd1 mutants showed similar foliar H2O2 and SA levels as the wild type under the same conditions (Table II). Our results indicate that LSD1, together with EDS1 and PAD4, is responsible for the control of H2O2 and SA concentrations in the cell and that this regulation is condition dependent.
Table II. Foliar SA and H2O2 content in plants grown under laboratory or field conditions.
Foliar SA and H2O2 were measured in leaves of 4-week-old Arabidopsis wild-type (Ws-0) and mutant plants grown under laboratory and field conditions. All data are means ± se from at least six to 15 independent plants from two independent experiments, and the statistical significances between genotypes are indicated with different letters (e.g. “a” and “a” are not significantly different, and “a” and “b” are significantly different, according to Duncan’s multiple range test [P < 0.05]).
| Genotype | SA Total | H2O2 |
|---|---|---|
| nmol g–1 fresh wt | ||
| Laboratory conditions | ||
| Ws-0 | 24.89 ± 4.58 a | 38.22 ± 1.28 b |
| lsd1 | 125.72 ± 7.60 d | 75.23 ± 3.25 d |
| eds1 | 36.12 ± 4.88 b | 37.28 ± 2.40 b |
| pad4 | 41.45 ± 4.88 b | 36.74 ± 0.39 b |
| eds1/lsd1 | 23.21 ± 4.97 a | 33.73 ± 1.38 b |
| pad4/lsd1 | 23.74 ± 4.28 a | 34.26 ± 0.94 b |
| Field conditions | ||
| Ws-0 | 22.73 ± 2.64 a | 8.21 ± 0.44 a |
| lsd1 | 69.14 ± 4.96 c | 44.85 ± 1.14 c |
| eds1 | 25.10 ± 0.99 a | 7.21 ± 0.76 a |
| pad4 | 26.72 ± 4.21 a | 6.45 ± 0.46 a |
| eds1/lsd1 | 23.97 ± 3.99 a | 11.12 ± 0.41 a |
| pad4/lsd1 | 20.99 ± 2.89 a | 9.55 ± 1.65 a |
Essential Differences in Laboratory and Field Transcriptome Profiles
For a better understanding of the molecular mechanisms underlying differences in the phenotypes, particularly the condition-dependent phenotype of the lsd1 mutant, a genome-wide transcriptome analysis was performed for wild-type (Ws-0), lsd1, eds1, pad4, eds1/lsd1, and pad4/lsd1 plants grown in laboratory or field environments. RNA obtained from 5-week-old plants harvested from two independent experiments was hybridized to 24 Agilent Arabidopsis Oligo Arrays. After data processing, a two-factor ANOVA was performed to assess the significance of the three major sources of variability affecting mRNA levels: the genotype (mutant versus the wild type), the growth conditions (laboratory versus field), and the interaction between them (combined effect of genotype and growth conditions). Multiple comparisons were corrected by controlling the false discovery rate (Storey and Tibshirani, 2003). Moreover, an independent (different experimental time and place) validation of the microarray results using quantitative PCR confirmed the expression patterns for 10 selected genes (Supplemental Table S3).
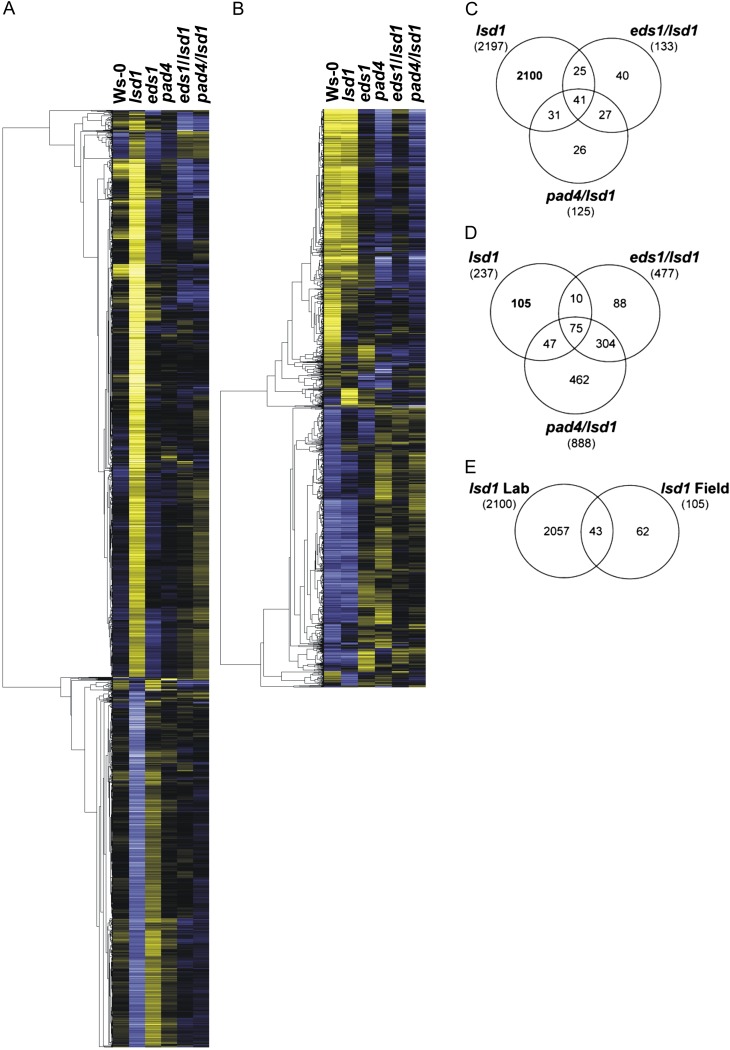
In order to visualize the transcription profiles for each genotype in laboratory and field conditions, we performed a hierarchical average linkage clustering (Fig. 3). The most noticeable was the considerable difference between the lsd1 and wild-type plants grown in the laboratory and only slight changes in the expression profiles of these two genotypes in the field. The number of deregulated transcripts in eds1, pad4, eds1/lsd1, and pad4/lsd1 was larger in the field compared with the laboratory conditions (Fig. 3), which may indicate that the role of LSD1 in the negative regulation of EDS1- and PAD4-dependent pathways is reduced in a multivariable environment compared with stable laboratory conditions. Interestingly, we observed similar transcription profiles between field-grown eds1 and eds1/lsd1, as well as pad4 and pad4/lsd1, which may also suggest the superior role of EDS1 and PAD4 over LSD1 in multivariable conditions.
Figure 3.
Hierarchical average linkage clustering and distribution of transcripts regulated by LSD1, EDS1, and PAD4. Expression profile for laboratory (A) and field (B) conditions. Transcripts significantly regulated in at least one genotype and with at least a 2-fold difference in expression compared with the wild type were selected. Clusters present expression patterns of 2,346 and 1,489 transcripts differentially regulated in laboratory (A) and field (B) conditions, respectively. Each gene is represented by a single row of colored boxes; each genotype is represented by a single column. The color scale ranges from saturated yellow (gene expression induction) to saturated blue (gene expression suppression). Note the different expression profiles between the Ws-0 and lsd1 plants grown in the laboratory and the much smaller difference in field conditions. Validation of the data were made by using quantitative PCR and is presented in Supplemental Table S3. Within the interaction-significant genes, pairwise comparisons between three genotypes, lsd1, eds1/lsd1, and pad4/lsd1, and the wild type were made separately for laboratory (C) and field conditions (D). After exclusion of transcripts that were commonly regulated in lsd1 and double mutants, we obtained two lists with 2,100 and 105 lsd1-specific genes with at least a 2-fold altered expression compared with the wild type for laboratory and field conditions, respectively. A comparison of these lsd1-specific genes in laboratory and field conditions allowed us to identify 43 commonly regulated genes (E) that are presented in Supplemental Table S4.
To distinguish the processes underlying the altered physiological parameters of lsd1 plants in both the growth chamber and in the natural environment, we performed a detailed functional analysis of genes specific for lsd1. After exclusion of transcripts that were commonly regulated in lsd1 and the double mutants eds1/lsd1 and pad4/lsd1, we obtained a list of 2,100 genes for the laboratory conditions and 105 genes for the field conditions specific to lsd1 with at least a 2-fold altered expression level compared with the wild type (Fig. 3). This considerable dissimilarity between the numbers of genes deregulated in the lsd1 mutant (Fig. 3) shows how differential pathways function in Arabidopsis plants grown under stable laboratory conditions compared with natural multivariable field conditions. Interestingly, only 43 transcripts were common for both growth conditions (Fig. 3E; Supplemental Table S4). The functional analysis that was concentrated on the signaling pathways and metabolic processes enabled us to identify the different regulatory strategies being fulfilled by plants grown in stable laboratory or variable natural conditions.
The lsd1 plants from the growth chamber demonstrated altered transcript levels of 67 receptor kinases, among them numerous Leu-rich repeat kinases, a domain of unknown function (DUF26), and wall-associated kinases. Furthermore, a large number of light- and calcium-signaling pathway components (especially calmodulin-binding proteins) were up-regulated in lsd1, but only in the laboratory. Many transcription factor (TF) families also appeared to be deregulated in the lsd1 mutant, most notably the APETALA2/ethylene-responsive element binding proteins (AP2/EREBP), phospho-accepting response regulator (ARR), G2-like, WRKY, JUMONJI, and TCP TFs (Supplemental Table S1). The AP2/EREBP TFs have been shown to be implicated in hormone, sugar, and redox signaling in the context of stress signal integration and retrograde signaling from chloroplasts to the nucleus (Dietz et al., 2010).
The expression level of two genes encoding C-repeat element-binding factor TFs that induce cold-regulated genes and increase plant freezing tolerance was down-regulated in lsd1 both in laboratory and field conditions (Supplemental Tables S1 and S2). This may be one of the reasons why the lsd1 mutant demonstrated a reduced cold tolerance (Huang et al., 2010). Two genes belonging to the G2-like family of TFs, which are postulated to regulate chloroplast development (Fitter et al., 2002), were repressed in lsd1 in the growth chamber.
The transcript levels of numerous genes encoding light reaction chain components, such as light-harvesting complex II and PSII proteins, were decreased in lsd1 (Supplemental Table S1). This might explain, at least partially, the impaired photosynthesis in laboratory-grown lsd1 (Fig. 2) and its higher sensitivity to EEE (Mateo et al., 2004). Excess light energy results in the accumulation of reducing equivalents (NADPH), which are generated by photochemical reactions. In the mitochondria, NADPH is oxidized by the respiratory electron transport chain in two different pathways: the cytochrome oxidase pathway and the alternative oxidase (AOX) pathway (Yoshida et al., 2006). The nonphosphorylating AOX pathway functions as a sink for the excess reducing equivalents generated by photosynthesis (Maxwell et al., 1999; Yoshida et al., 2007). In laboratory-grown lsd1, the induced expression of AOX (Supplemental Table S1) suggests the necessity of eliminating excess reducing equivalents via the AOX pathway.
Moreover, in the lsd1 plants grown in the laboratory, numerous genes (67) associated with biotic stress responses, such as genes encoding Toll/Interleukin-1 Receptor-domain-containing nucleotide-binding site (NBS) leucine-rich repeat (LRR) proteins and coiled-coil-NBS-LRR resistance proteins, pathogenesis-related proteins, proteinase inhibitors, and plant defensins, were altered compared with the wild type, most being positively regulated (Supplemental Table S1). The next overrepresented group (32 genes) in the lsd1 mutant included genes induced by abiotic stresses, such as heat shock proteins, heat shock factors, and germin-like proteins. A great number of genes engaged in redox homeostasis maintenance, such as thioredoxins and glutaredoxins, were also overrepresented in the lsd1. Furthermore, 12 peroxidases and eight glutathione S-transferases, which detoxify ROS derivatives, were up-regulated in the growth chamber (Supplemental Table S1). By comparison, only two and five genes involved in biotic and abiotic responses, respectively, were deregulated in the lsd1 mutant grown in the field (Supplemental Table S2).
In the laboratory-grown lsd1 plants, hormonal homeostasis was disturbed, which is indicated by the fact that many hormone biosynthesis and metabolism genes were deregulated. The data showed changes in the transcript abundance of three genes involved in abscisic acid (ABA) synthesis, the gene encoding 1-aminocyclopropane-1-carboxylic acid synthase (a key enzyme in the ethylene synthesis pathway), and seven genes involved in jasmonic acid (JA) biosynthesis. The SA metabolism was also modified in the lsd1 mutant, which is manifested in the overexpression of enzymes participating in SA conjugation (Supplemental Table S1). The expression levels of the above-mentioned genes in the field-grown lsd1 mutant were not different from those in the wild type (Supplemental Table S2). The remarkable perturbation of hormonal homeostasis in laboratory-grown lsd1 is also clearly manifested by the abundant presence of H2O2 and hormone-responsive transcripts (Supplemental Fig. S4), which indicates that the majority of the lsd1-specific genes is also deregulated by ABA, JA, SA, and H2O2 treatments. The general trend is that most of the transcripts up-regulated in the laboratory-grown lsd1 were also up-regulated by H2O2 or SA treatment and vice versa (Supplemental Fig. S4). It is a well-known fact that ABA together with other hormones, such as indole-3-acetic acid and JA, is responsible for controlling the expression of a broad spectrum of genes. We therefore postulate that in lsd1, the transcription of many genes is altered due to a secondary effect of ROS and hormonal perturbation.
Interestingly, the transcript levels of 14 genes engaged in cell division or cell cycle regulation and 70 genes directly involved in variable development processes were altered in the lsd1 compared with the wild type (Supplemental Table S1), which might account for its impeded growth under laboratory conditions. By comparison, no cell cycle/division regulation genes and only four genes engaged in plant development were differentially regulated in the field-grown lsd1 plants (Supplemental Table S2).
DISCUSSION
It has been previously demonstrated that LSD1 integrates signaling pathways in response to diverse stresses, such as avirulent pathogen infection (Rustérucci et al., 2001; Wiermer et al., 2005), EEE (Mateo et al., 2004; Mühlenbock et al., 2008), and root hypoxia (Mühlenbock et al., 2007), and that it prevents EDS1- and PAD4-dependent execution of the programmed cell death program above a certain threshold of these stresses. Here, we show that LSD1, EDS1, and PAD4 are also involved in the regulation of a combined drought and high-light stress tolerance.
The mutants of immune defense genes are often associated with the deregulation of growth and development (Dietrich et al., 1994; Glazebrook, 2001; Züst et al., 2011). Indeed, in permissive laboratory conditions, the inflorescence of lsd1 is much reduced, and it produces significantly fewer seeds compared with the wild type (Fig. 2, A and C). However, under a simultaneously acting water deficiency and high-light stress, lsd1 plants demonstrate better fitness, manifested in an increased survival rate (Fig. 1; Table I) and seed production (Table I) compared with the wild-type plants. One of the reasons for lsd1 higher water deficiency tolerance may be its ability to dispose of older leaves (Fig. 1). Leaf abscission is one of the adaptations helping to reduce shoot water loss during drought stress (Sinclair, 2000). It has also been proven to be connected with a higher level of H2O2 (Sakamoto et al., 2008) and ET (Gomez-Cadenas et al., 1996), which is in agreement with the increased concentrations of both H2O2 and ET (Mühlenbock et al., 2007) observed in the lsd1 mutant. After 15 d of such unfavorable conditions, all of the tested genotypes, except lsd1 and eds1, induced irreversible withering (Fig. 1; Supplemental Fig. S1), and only these two genotypes were able to continue growth and form inflorescence after rewatering. The ability of lsd1 and eds1 Arabidopsis mutants to survive longer under drought stress may be an argument for testing the effects of these mutations in crop plants under water deficiency conditions.
The above observations inspired us to perform a field experiment to confront the physiological parameters of all the tested genotypes in the laboratory and the natural environment. Under laboratory conditions, our results showed reduced YS and Fv′/Fm′ in the lsd1 mutant compared with the wild type (Fig. 2, C and G). However, we observed that the lsd1 plants grown in the field produced a similar number of seeds per plant as the other tested genotypes (Fig. 2, B and D). In the natural environment, Fv′/Fm′, but not WUE, was similar in the lsd1 and wild-type plants (Fig. 2, F and H). A decreased WUE in the field indicates greater water consumption per produced biomass and may denote a higher transpiration rate and better gas exchange, thus a higher CO2 uptake. Therefore, we suggest that the intensified photorespiration that had previously been proven to be responsible for the RCD phenotype in the laboratory-grown lsd1 (Mateo et al., 2004) may be suppressed in the field. However, further analysis should be performed to confirm this.
We also observed that the levels of SA and H2O2 were significantly elevated in the lsd1 mutant compared with the other genotypes and were higher in the laboratory than in the field (Table II). However, mutations in eds1 and pad4 reverted this pattern in the double mutants eds1/lsd1 and pad4/lsd1. Interestingly, we observed a higher concentration of H2O2 in all of the genotypes grown in growth chambers compared with their outdoor-grown counterparts. These results may suggest that plants acclimated to multivariable field conditions from an early stage of development evolved physiological improvements to meet the requirements of a continuously changing environment. The oxidative burst is one of the earliest and most common plant responses to abiotic and biotic stimuli. To a large extent, both H2O2 and SA in plant cells originate from pathways localized in the chloroplasts (Asada, 1999; Foyer and Noctor, 2005; Vlot et al., 2009). Taking into consideration the fact that RCD in the lsd1 mutant has been linked to the amount of light energy absorbed in excess by both the PSII light-harvesting complex (Mateo et al., 2004) and the elevated SA and H2O2 levels in lsd1 (Table II), LSD1 seems to be indispensable for restraining EEE-triggered ROS and SA production. The perturbation of ROS/hormonal homeostasis in lsd1 was also confirmed by the microarray-based transcriptomic results. They proved that numerous genes engaged in redox homeostasis maintenance, such as thioredoxins, glutaredoxins, peroxidases, and glutathione S-transferases, were induced in the lsd1 mutant grown in laboratory conditions. Furthermore, genes involved in ABA, SA, ET, and JA synthesis and metabolism were also up-regulated (Supplemental Table S1). Meanwhile, the lsd1 plants grown under field conditions did not show much difference in terms of stress-related genes (Supplemental Table S2). This dissimilarity indicates that the lsd1 plants in a growth chamber seem to rearrange their metabolism to overcome stress rather than to reproduce. Such considerable differences between the numbers of genes deregulated in lsd1 (Fig. 3; Supplemental Tables S1 and S2) show how differential signaling pathways function in Arabidopsis plants grown in stable and optimal laboratory conditions compared with a natural multivariable environment. It is important to note, however, that many lsd1 laboratory-specific genes may be deregulated as a secondary effect of the ROS/hormonal lack of adjustment.
Taken together, these results demonstrate that LSD1, EDS1, and PAD4 play important roles in plant fitness regulation, as determined by parameters such as the maximal efficiency of PSII, WUE, ROS/hormonal homeostasis, and YS. Our results also suggest that the LSD1/EDS1/PAD4 hub is important in the integration and regulation of acclimatory and defense responses that underpin plant fitness during growth and development.
LSD1 contains three zinc finger domains that have been demonstrated to be responsible for interacting with many proteins, including metacaspase1 and TF basic leucine zipper10 (Kaminaka et al., 2006; Coll et al., 2010, 2011). LSD1 has also been postulated as being a TF by itself (Dietrich et al., 1997). In this way, it is a probable regulator of a broad spectrum of genes involved in different signaling pathways and metabolic rearrangements during acclimatory and defense responses (Fig. 3; Supplemental Tables S1 and S2). LSD1 has been proven to inhibit the EDS1- and PAD4-mediated hypersensitive response and RCD in laboratory experiments (Rustérucci et al., 2001; Mateo et al., 2004; Mühlenbock et al., 2007, 2008). However, a remarkably lower number of genes deregulated in field-grown lsd1, together with no significant differences in YS compared with the wild-type plants (Figs. 2 and 3), may imply that the suppressive role of LSD1 on EDS1/PAD4 depends on the growing conditions. EDS1 and PAD4 appear to be less responsive to LSD1-dependent regulation in plants grown in the natural environment compared with those grown under laboratory conditions. This would suggest an overriding influence of other pathways that modulate EDS1- and PAD4-dependent responses or the presence of other factors that repress LSD1 function.
Moreover, our results indicate that the function of such important regulatory genes as LSD1, EDS1, and PAD4 should not be tested only under one set of conditions. Since the environment greatly influences the overall plant metabolism and signaling, we need to be cautious when interpreting the effects from growth chamber experiments. Although much has been learned from laboratory experiments on mutants, a holistic understanding of gene function also requires studies in the natural environment. Bearing in mind that plant genomes have been evolving for millions of years and carry a genetic record of their adaptations, measuring the effect in the natural environment ensures a real estimate of the gene function and its consequence on plant fitness. Therefore, we strongly postulate examining the regulatory gene’s impact on survival and reproduction in the face of challenges posed by various ambient conditions.
MATERIALS AND METHODS
Growth Conditions, Seed Yield, and Germination Rate Determination
Wild-type Arabidopsis (Arabidopsis thaliana; Ws-0) and five different mutants of the same accession (lsd1-1, eds1-1, pad4-5, eds1-1/lsd1-1, and pad4-5/lsd1-1) were grown in a growth chamber under standard laboratory conditions (9- or 16-h photoperiod, photosynthetic photon flux density (PPFD) of 100 ± 25 µmol m–2 s–1, 50% relative air humidity, and day/night temperature of 22°C /18°C). Arabidopsis Ws-0 and mutant plants were also grown in the field at two different locations (Krakow, Poland, 50°03'41″ N, 19°56'18″ E and Warsaw, 52°09'38″ N, 21°02'52″ E) during several seasons (June–September, 2004–2007 and 2010). Examples of the average meteorological data are presented in Supplemental Figure S2. The smallest experimental field unit was approximately 100 cm2, in which all six representative genotypes were grown together. For measurements of Fv′/Fm′, H2O2, and SA, 4-week-old plants were used, whereas for the determination of YS, 8-week-old plants were harvested. The mass of 1,000 seeds of Ws-0 and each mutant was determined for four and six independent experiments conducted in field and laboratory conditions, respectively. On this basis, the total number of seeds per plant was calculated. The germination rate was determined as the percentage of germinated seeds after 5 and 7 d. Seeds were stratified for 3 to 4 d at 4°C, placed on wet tissue paper in petri dishes, and transferred to the growth chamber at PPFD of 200 ± 25 µmol m–2s–1, a 12-h photoperiod, and day/night temperature of 18°C /16°C.
Determination of WUE
WUE was determined as dry weight per unit of water used (mg dry weight × mL–1 water) for 4-week-old wild-type and mutant plants grown in a 9-h photoperiod in a growth chamber or in the field. Plants were grown in 50-mL tubes filled with perlite and soil in a 1:1 proportion and 35 mL of water. Seeds were placed in a hole (approximately 1.5 mm wide) made in the cap. After germination, the system was weighed. Plants were decapitated after 4 weeks and dried for 3 h at 105°C, and then each tube was weighed to determine the water loss.
Drought and High-Light Stress Experiment
Five-week-old wild-type and mutant plants grown in a 9-h photoperiod were transferred to high-light conditions (PPFD of 500 ± 50 µmol m–2s–1), and watering was stopped. The survival rate, defined as the percentage of plants that survived, was measured after 11 and 15 d of water deficiency stress and 4 d of rewatering. Moreover, seed production, defined as the number of seeds per plant, was determined in 9-week-old plants.
Biochemical and Physiological Parameter Measurements
Fv′/Fm′, H2O2, and SA concentrations were determined for 4-week-old wild-type and mutant plants grown in a 9-h photoperiod in a growth chamber or in the field. The total H2O2 content was measured by fluorometric assay with homovanillic acid (Ishikawa et al., 1993). To determine SA, 2-methoxybenzoic acid and 3-hydroxybenzoic acid were used as internal standards, respectively (Meuwly and Métraux, 1993). SA was determined by HPLC using an isocratic elution with KH2PO4 buffer (pH 2; adjusted with HCl) and acetonitrile (752:45, v/v) in a Shimadzu HPLC System and a Luna-C 18(2) column (Phenomenex; 250, 4.6, 0.005 mm). Chlorophyll a fluorescence parameter Fv′/Fm′ was measured on FluorImager and associated software (Photon System Instruments). Chlorophyll fluorescence terminology has previously been described in detail (Baker, 2008).
Microarray Analysis, Meta-Analysis, and Quantitative Reverse Transcription-PCR
RNA was obtained from 5-week-old plants harvested during the day/light period from two independent experiments in the laboratory and in the field using TRIzol reagent (Invitrogen) and further purified using the RNeasy MinElute Cleanup Kit (Qiagen). The RNA concentration and quality were determined as described previously (Vanderauwera et al., 2007). Microarray hybridizations were performed in two loop designs (one for the laboratory and one for the field samples), and dye assignments (cyanine dyes Cy5 and Cy3) were balanced across the 24 Arabidopsis 4 Oligo Microarrays (Agilent Technologies). Reverse transcription, labeling, hybridization, and scanning were performed according to the manufacturer’s instructions (Agilent) by the VIB Microarray Facility (http://www.nucleomics.be). Scanning, feature extraction, dye normalization (Linear Lowess), and normalization for array and repeat effects were performed as described previously (Vanderauwera et al., 2011). The residuals from these data normalizations were subjected to a two-factor ANOVA using the TIGR MultiExperiment Viewer of the TM4 software suite (Saeed et al., 2003), and a multiple testing correction was performed on the P values of the F statistics to assess the false discovery rate using the publicly available software QVALUE (http://genomine.org/qvalue; Storey and Tibshirani, 2003). Genes with P values of less than 0.001 and Q values of less than 0.05 were retained for further analysis. Within the interaction-significant profiles, the wild-type and mutant profiles were compared pairwise as well by selecting genes with a P value of less than 0.001 and a Q value of less than 0.05. Significant profiles were further processed. Expression values were obtained by averaging the inverse log2-transformed normalized values of the Cy5 and Cy3 signal intensities of the two replicate samples. Fold changes were obtained using the averaged expression values of the mutants relative to the wild-type samples, and only probes (genes) showing at least a 2-fold difference in expression were retained. For clustering analysis, the data sets were log transformed, median centered across each gene, and subjected to hierarchical average linkage clustering (Euclidian distance) using Cluster and TreeView software (Eisen et al., 1998). Gene annotations were compiled by the Arabidopsis Information Resource (http://www.arabidopsis.org; Rhee et al., 2003), and the agilent_array_elements-2008-9-17 version was used for Agilent array element information. Full access to the microarray data are available at the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=ltihrsqeoymmons&acc=GSE24766).
Transcriptomic data sets were functionally classified using MapMan 3.5.0 Beta (Thimm et al., 2004) in terms of involvement in various cellular pathways relevant in stress responses, signal transduction, and development.
For the microarray meta-analysis, data sets of lsd1-specific genes from laboratory and field conditions were compared with relevant publicly available microarray experiments by means of the Perturbations tool in the Condition Search toolset of Genevestigator (Hruz et al., 2008). To explore SA responsiveness, wild-type (Columbia-0) samples in response to exogenously applied SA were selected (van Leeuwen et al., 2007), whereas for the other hormone responses, the 3-h time point of the AtGenExpress hormone treatment data set were used (Goda et al., 2008). For oxidative stress responses, we selected the H2O2-sprayed wild-type (Columbia-0) samples (Davletova et al., 2005) and the 8-h time point of high-light-treated catalase-deficient plants (CAT2HP1; Vanderauwera et al., 2005).
To describe the variability among samples, the Pearson correlation coefficient was calculated between two biological replicates for laboratory and field samples separately and was plotted using GPLOTs and RColorBrewer in R. The graphical representation of sample variability is presented as Supplemental Figure S5. To verify the microarray results, the expression levels of 10 genes were validated using samples of two additional independent repeats (Supplemental Table S3). Quantitative reverse transcription-PCR analyses were done as described (Vanderauwera et al., 2007). Actin-related protein7 (ARP7, AT3G60830) and CAP-binding protein20 (CBP20, AT5G44200) were used as the housekeeping genes. Quantitative PCR analysis was performed to check the responsiveness of the following genes: catalase1 (CAT1, AT1G20630), Uridine diphosphate glycosyltransferase74E2 (UGT74E2, AT1G05680), glutathione S-transferase TAU24 (GSTU24, AT1G17170), tolB-related protein (TolB, AT4G01870), flavin-containing monooxygenase1 (FMO1, AT1G19250), pathogenesis-related protein1 (PR1, AT2G14610), auxin-responsive protein (IAA5, AT1G15580), lesion simulating disease1 (LSD1, AT4G20380), phytoalexin deficient4 (PAD4, AT3G52430), and dehydration-responsive element-binding protein1F (DDF1, AT1G12610). Primers used for quantitative PCR experiments were designed using ProbeFinder 2.45 (Roche Diagnostics; Supplemental Table S5).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Simultaneous drought and high-light stress tolerance in the laboratory.
Supplemental Figure S2. Seasonal precipitation, temperature, and insolation.
Supplemental Figure S3. Germination rate.
Supplemental Figure S4. Expression analysis of lsd1-specific genes in response to various hormone treatments and oxidative stress.
Supplemental Figure S5. The Pearson correlation coefficient between two biological repeats for laboratory and field samples.
Supplemental Table S1. Functional categorization of differentially expressed genes in lsd1 in laboratory conditions.
Supplemental Table S2. Functional categorization of differentially expressed genes in lsd1 in field conditions.
Supplemental Table S3. Validation of microarray results.
Supplemental Table S4. Expression characteristics of 43 lsd1-specific genes commonly regulated in laboratory and field conditions.
Supplemental Table S5. Sequence of primers used for microarray results validation by quantitative PCR.
Acknowledgments
We thank Wiesława Przewoźniczuk from the Department of Meteorology and Climatology (Warsaw University of Life Sciences) for providing the meteorological data for Warsaw for the year 2010.
Glossary
- ROS
reactive oxygen species
- SA
salicylic acid
- RCD
runaway cell death
- ET
ethylene
- EEE
excess excitation energy
- H2O2
hydrogen peroxide
- Fv′/Fm′
PSII maximum efficiency
- WUE
water use efficiency
- YS
seed yield
- Ws-0
Wassilewskija
- TF
transcription factor
- AOX
alternative oxidase
- ABA
abscisic acid
- JA
jasmonic acid
- PPFD
photosynthetic photo flux density
- Ws-0
Wassilewskija
References
- Asada K. (1999) THE WATER-WATER CYCLE IN CHLOROPLASTS: scavenging of active oxygens and dissipation of excess photons. Annu Rev Plant Physiol Plant Mol Biol 50: 601–639 [DOI] [PubMed] [Google Scholar]
- Baker NR. (2008) Chlorophyll fluorescence: a probe of photosynthesis in vivo. Annu Rev Plant Biol 59: 89–113 [DOI] [PubMed] [Google Scholar]
- Coll NS, Epple P, Dangl JL. (2011) Programmed cell death in the plant immune system. Cell Death Differ 18: 1247–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll NS, Vercammen D, Smidler A, Clover C, Van Breusegem F, Dangl JL, Epple P. (2010) Arabidopsis type I metacaspases control cell death. Science 330: 1393–1397 [DOI] [PubMed] [Google Scholar]
- Davletova S, Schlauch K, Coutu J, Mittler R. (2005) The zinc-finger protein Zat12 plays a central role in reactive oxygen and abiotic stress signaling in Arabidopsis. Plant Physiol 139: 847–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich RA, Delaney TP, Uknes SJ, Ward ER, Ryals JA, Dangl JL. (1994) Arabidopsis mutants simulating disease resistance response. Cell 77: 565–577 [DOI] [PubMed] [Google Scholar]
- Dietrich RA, Richberg MH, Schmidt R, Dean C, Dangl JL. (1997) A novel zinc finger protein is encoded by the Arabidopsis LSD1 gene and functions as a negative regulator of plant cell death. Cell 88: 685–694 [DOI] [PubMed] [Google Scholar]
- Dietz K-J, Vogel MO, Viehhauser A. (2010) AP2/EREBP transcription factors are part of gene regulatory networks and integrate metabolic, hormonal and environmental signals in stress acclimation and retrograde signalling. Protoplasma 245: 3–14 [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 95: 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epple P, Mack AA, Morris VRF, Dangl JL. (2003) Antagonistic control of oxidative stress-induced cell death in Arabidopsis by two related, plant-specific zinc finger proteins. Proc Natl Acad Sci USA 100: 6831–6836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk A, Feys BJ, Frost LN, Jones JD, Daniels MJ, Parker JE. (1999) EDS1, an essential component of R gene-mediated disease resistance in Arabidopsis has homology to eukaryotic lipases. Proc Natl Acad Sci USA 96: 3292–3297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys BJ, Moisan LJ, Newman MA, Parker JE. (2001) Direct interaction between the Arabidopsis disease resistance signaling proteins, EDS1 and PAD4. EMBO J 20: 5400–5411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitter DW, Martin DJ, Copley MJ, Scotland RW, Langdale JA. (2002) GLK gene pairs regulate chloroplast development in diverse plant species. Plant J 31: 713–727 [DOI] [PubMed] [Google Scholar]
- Foyer CH, Noctor G. (2005) Redox homeostasis and antioxidant signaling: a metabolic interface between stress perception and physiological responses. Plant Cell 17: 1866–1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook J. (2001) Genes controlling expression of defense responses in Arabidopsis—2001 status. Curr Opin Plant Biol 4: 301–308 [DOI] [PubMed] [Google Scholar]
- Glazebrook J, Zook M, Mert F, Kagan I, Rogers EE, Crute IR, Holub EB, Hammerschmidt R, Ausubel FM. (1997) Phytoalexin-deficient mutants of Arabidopsis reveal that PAD4 encodes a regulatory factor and that four PAD genes contribute to downy mildew resistance. Genetics 146: 381–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goda H, Sasaki E, Akiyama K, Maruyama-Nakashita A, Nakabayashi K, Li W, Ogawa M, Yamauchi Y, Preston J, Aoki K, et al. (2008) The AtGenExpress hormone and chemical treatment data set: experimental design, data evaluation, model data analysis and data access. Plant J 55: 526–542 [DOI] [PubMed] [Google Scholar]
- Gomez-Cadenas A, Tadeo FR, Talon M, Primo-Millo E. (1996) Leaf abscission induced by ethylene in water-stressed intact seedlings of Cleopatra mandarin requires previous abscisic acid accumulation in roots. Plant Physiol 112: 401–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P. (2008) Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinforma 2008: 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Li Y, Zhang X, Zuo J, Yang S. (2010) The Arabidopsis LSD1 gene plays an important role in the regulation of low temperature-dependent cell death. New Phytol 187: 301–312 [DOI] [PubMed] [Google Scholar]
- Hunt MD, Delaney TP, Dietrich RA, Weymann KB, Dangl JL, Ryals JA. (1997) Salicylate-independent lesion formation in Arabidopsis lsd mutants. Mol Plant Microbe Interact 10: 531–536 [DOI] [PubMed] [Google Scholar]
- Ishikawa T, Takeda T, Shigeoka S, Hirayama O, Mitsunaga T. (1993) Hydrogen peroxide generation in organelles of Euglena gracilis. Phytochemistry 33: 1297–1299 [Google Scholar]
- Jabs T, Dietrich RA, Dangl JL. (1996) Initiation of runaway cell death in an Arabidopsis mutant by extracellular superoxide. Science 273: 1853–1856 [DOI] [PubMed] [Google Scholar]
- Kaminaka H, Näke C, Epple P, Dittgen J, Schütze K, Chaban C, Holt BF, III, Merkle T, Schäfer E, Harter K, et al. (2006) bZIP10-LSD1 antagonism modulates basal defense and cell death in Arabidopsis following infection. EMBO J 25: 4400–4411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein DJ, Dietrich RA, Martin AC, Last RL, Dangl JL. (1999) LSD1 regulates salicylic acid induction of copper zinc superoxide dismutase in Arabidopsis thaliana. Mol Plant Microbe Interact 12: 1022–1026 [DOI] [PubMed] [Google Scholar]
- Külheim C, Agren J, Jansson S. (2002) Rapid regulation of light harvesting and plant fitness in the field. Science 297: 91–93 [DOI] [PubMed] [Google Scholar]
- Mateo A, Mühlenbock P, Rustérucci C, Chang CC-C, Miszalski Z, Karpinska B, Parker JE, Mullineaux PM, Karpinski S. (2004) LESION SIMULATING DISEASE 1 is required for acclimation to conditions that promote excess excitation energy. Plant Physiol 136: 2818–2830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell DP, Wang Y, McIntosh L. (1999) The alternative oxidase lowers mitochondrial reactive oxygen production in plant cells. Proc Natl Acad Sci USA 96: 8271–8276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuwly P, Métraux JP. (1993) Ortho-anisic acid as internal standard for the simultaneous quantitation of salicylic acid and its putative biosynthetic precursors in cucumber leaves. Anal Biochem 214: 500–505 [DOI] [PubMed] [Google Scholar]
- Mühlenbock P, Plaszczyca M, Plaszczyca M, Mellerowicz E, Karpinski S. (2007) Lysigenous aerenchyma formation in Arabidopsis is controlled by LESION SIMULATING DISEASE1. Plant Cell 19: 3819–3830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühlenbock P, Szechynska-Hebda M, Plaszczyca M, Baudo M, Mateo A, Mullineaux PM, Parker JE, Karpinska B, Karpinski S. (2008) Chloroplast signaling and LESION SIMULATING DISEASE1 regulate crosstalk between light acclimation and immunity in Arabidopsis. Plant Cell 20: 2339–2356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor G, Veljovic-Jovanovic S, Driscoll S, Novitskaya L, Foyer CH. (2002) Drought and oxidative load in the leaves of C3 plants: a predominant role for photorespiration? Ann Bot (Lond) 89: 841–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker JE, Holub EB, Frost LN, Falk A, Gunn ND, Daniels MJ. (1996) Characterization of eds1, a mutation in Arabidopsis suppressing resistance to Peronospora parasitica specified by several different RPP genes. Plant Cell 8: 2033–2046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee SY, Beavis W, Berardini TZ, Chen G, Dixon D, Doyle A, Garcia-Hernandez M, Huala E, Lander G, Montoya M, et al. (2003) The Arabidopsis Information Resource (TAIR): a model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Res 31: 224–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustérucci C, Aviv DH, Holt BF, III, Dangl JL, Parker JE. (2001) The disease resistance signaling components EDS1 and PAD4 are essential regulators of the cell death pathway controlled by LSD1 in Arabidopsis. Plant Cell 13: 2211–2224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, et al. (2003) TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34: 374–378 [DOI] [PubMed] [Google Scholar]
- Sakamoto M, Munemura I, Tomita R, Kobayashi K. (2008) Involvement of hydrogen peroxide in leaf abscission signaling, revealed by analysis with an in vitro abscission system in Capsicum plants. Plant J 56: 13–27 [DOI] [PubMed] [Google Scholar]
- Sinclair TR. (2000) Model analysis of plant traits leading to prolonged crop survival during severe drought. Field Crop Res 68: 211–217 [Google Scholar]
- Storey JD, Tibshirani R. (2003) Statistical significance for genomewide studies. Proc Natl Acad Sci USA 100: 9440–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, Selbig J, Müller LA, Rhee SY, Stitt M. (2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37: 914–939 [DOI] [PubMed] [Google Scholar]
- Tonsor SJ, Alonso-Blanco C, Koornneef M. (2005) Gene function beyond the single trait: natural variation, gene effects, and evolutionary ecology in Arabidopsis thaliana. Plant Cell Environ 28: 2–20 [Google Scholar]
- Torres MA, Jones JDG, Dangl JL. (2005) Pathogen-induced, NADPH oxidase-derived reactive oxygen intermediates suppress spread of cell death in Arabidopsis thaliana. Nat Genet 37: 1130–1134 [DOI] [PubMed] [Google Scholar]
- Ungerer MC, Johnson LC, Herman MA. (2008) Ecological genomics: understanding gene and genome function in the natural environment. Heredity (Edinb) 100: 178–183 [DOI] [PubMed] [Google Scholar]
- van Leeuwen H, Kliebenstein DJ, West MAL, Kim K, van Poecke R, Katagiri F, Michelmore RW, Doerge RW, St Clair DA. (2007) Natural variation among Arabidopsis thaliana accessions for transcriptome response to exogenous salicylic acid. Plant Cell 19: 2099–2110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderauwera S, De Block M, Van de Steene N, van de Cotte B, Metzlaff M, Van Breusegem F. (2007) Silencing of poly(ADP-ribose) polymerase in plants alters abiotic stress signal transduction. Proc Natl Acad Sci USA 104: 15150–15155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderauwera S, Suzuki N, Miller G, van de Cotte B, Morsa S, Ravanat J-L, Hegie A, Triantaphylidès C, Shulaev V, Van Montagu MCE, et al. (2011) Extranuclear protection of chromosomal DNA from oxidative stress. Proc Natl Acad Sci USA 108: 1711–1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderauwera S, Zimmermann P, Rombauts S, Vandenabeele S, Langebartels C, Gruissem W, Inzé D, Van Breusegem F. (2005) Genome-wide analysis of hydrogen peroxide-regulated gene expression in Arabidopsis reveals a high light-induced transcriptional cluster involved in anthocyanin biosynthesis. Plant Physiol 139: 806–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlot AC, Dempsey DA, Klessig DF. (2009) Salicylic acid, a multifaceted hormone to combat disease. Annu Rev Phytopathol 47: 177–206 [DOI] [PubMed] [Google Scholar]
- Weinig C, Ungerer MC, Dorn LA, Kane NC, Toyonaga Y, Halldorsdottir SS, Mackay TFC, Purugganan MD, Schmitt J. (2002) Novel loci control variation in reproductive timing in Arabidopsis thaliana in natural environments. Genetics 162: 1875–1884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiermer M, Feys BJ, Parker JE. (2005) Plant immunity: the EDS1 regulatory node. Curr Opin Plant Biol 8: 383–389 [DOI] [PubMed] [Google Scholar]
- Wingler A, Quick WP, Bungard RA, Bailey KJ, Lea PJ, Leegood RC. (1999) The role of photorespiration during drought stress: an analysis utilizing barley mutants with reduced activities of photorespiratory enzymes. Plant Cell Environ 22: 361–373 [Google Scholar]
- Yoshida K, Terashima I, Noguchi K. (2006) Distinct roles of the cytochrome pathway and alternative oxidase in leaf photosynthesis. Plant Cell Physiol 47: 22–31 [DOI] [PubMed] [Google Scholar]
- Yoshida K, Terashima I, Noguchi K. (2007) Up-regulation of mitochondrial alternative oxidase concomitant with chloroplast over-reduction by excess light. Plant Cell Physiol 48: 606–614 [DOI] [PubMed] [Google Scholar]
- Zhou Y, Lam HM, Zhang J. (2007) Inhibition of photosynthesis and energy dissipation induced by water and high light stresses in rice. J Exp Bot 58: 1207–1217 [DOI] [PubMed] [Google Scholar]
- Züst T, Joseph B, Shimizu KK, Kliebenstein DJ, Turnbull LA. (2011) Using knockout mutants to reveal the growth costs of defensive traits. Proc Biol Sci 278: 2598–2603 [DOI] [PMC free article] [PubMed] [Google Scholar]