Expression of a set of related proteins in Arabidopsis and its relative Lepidium papillosum (Brassicaceae) precede the onset of seed dormancy.
Abstract
Seed dormancy is a block to the completion of germination of an intact viable seed under favorable conditions and is an adaptive and agronomically important trait. Thus, elucidating conserved features of dormancy mechanisms is of great interest. The worldwide-distributed genus Lepidium (Brassicaceae) is well suited for cross-species comparisons investigating the origin of common or specific early-life-history traits. We show here that homologs of the seed dormancy-specific gene DELAY OF GERMINATION1 (DOG1) from Arabidopsis (Arabidopsis thaliana) are widespread in the genus Lepidium. The highly dormant Lepidium papillosum is a polyploid species and possesses multiple structurally diversified DOG1 genes (LepaDOG1), some being expressed in seeds. We used the largely elongated and well-structured infructescence of L. papillosum for studying primary dormancy induction during seed development and maturation with high temporal resolution. Using simultaneous germination assays and marker protein expression detection, we show that LepaDOG1 proteins are expressed in seeds during maturation prior to dormancy induction. Accumulation of LepaDOG1 takes place in seeds that gain premature germinability before and during the seed-filling stage and declines during the late maturation and desiccation phase when dormancy is induced. These analyses of the Lepidium DOG1 genes and their protein expression patterns highlight similarities and species-specific differences of primary dormancy induction mechanism(s) in the Brassicaceae.
Seed dormancy mechanisms are intrinsic blocks to the completion of germination during (temporary) favorable environmental conditions (Finch-Savage and Leubner-Metzger, 2006; Alonso-Blanco et al., 2009; Donohue et al., 2010). These blocks to germination have evolved differently across species through adaptation to the prevailing environment, so that germination occurs when conditions for establishing a new plant generation are likely to be suitable. Therefore, dormancy is important for the adaptation of a plant’s earliest developmental stages to local environments and is, together with flowering time, a major key trait for plant fitness. Germination timing depends largely on seed dormancy mechanisms and is a target for intense natural selection early in the colonization process. In general, genetic variation at individual gene loci, together with single-gene and whole-genome duplication events, are the origin of evolutionary novelties important for angiosperm diversification and adaptation to environmental cues and ecological niches (Tonsor et al., 2005; Franzke et al., 2011; Gossmann and Schmid, 2011; Wang et al., 2011). This has been thoroughly investigated in the case of flowering time but rarely regarding seed dormancy.
Quantitative trait locus (QTL) analyses of the Brassicaceae model species Arabidopsis (Arabidopsis thaliana) have been used to investigate the existing natural genetic variation for seed dormancy and flowering time, and major quantitative trait genes underlying these QTLs include DELAY OF GERMINATION1 (DOG1) and FLOWERING LOCUS C (FLC), respectively (Koornneef et al., 1998; Alonso-Blanco et al., 2009). For flowering time, natural genetic variation at the FLC locus has also been shown for the Brassicaceae species Capsella rubella and Brassica rapa (Zhao et al., 2010; Guo et al., 2012). In addition, duplications of the FLC gene in polyploid relatives of the diploid species Arabidopsis and B. rapa have been reported to underlay the observed natural variation (Schranz and Osborn, 2004; Nah and Chen, 2010).
For seed dormancy, analysis of Arabidopsis natural genetic variation has led to the cloning of AtDOG1 as the first specific seed dormancy gene (Bentsink et al., 2006) and has been shown to be important for local adaptation to different environments (Huang et al., 2010; Chiang et al., 2011; Footitt et al., 2011; Kendall et al., 2011; Kronholm et al., 2012). Homologs of the AtDOG1 gene are also known for the Brassicaceae species B. rapa (BrDOG1) and Lepidium sativum (LesaDOG1; Graeber et al., 2010). However, in contrast to the situation for flowering time and the FLC gene, neither the natural genetic variation at the DOG1 loci of these Arabidopsis relatives nor the distribution of the DOG1 gene in diploid and polyploid Brassicaceae relatives have been investigated. The work of Graeber et al. (2010) indicated that LesaDOG1 has functions beyond dormancy (i.e. during the germination of nondormant seeds). This leaves the prevalence and diversity of DOG1-mediated mechanisms in seed-related processes between species an open question.
The DOG1 gene encodes a protein of unknown function, and the Arabidopsis dog1 loss-of-function mutant is nondormant with no obvious pleiotropic phenotypes (Bentsink et al., 2006; Graeber et al., 2012). In Arabidopsis, the AtDOG1 gene is a member of a small gene family together with the four DOG1-Like genes (DOG1-Like1 to DOG1-Like4), but in contrast to AtDOG1, none of the DOG1-Like genes provides a seed phenotype (Bentsink et al., 2006). Seed dormancy is induced during seed maturation, and it has been shown in Arabidopsis that seed-specific DOG1 transcript expression starts during seed development 9 d after pollination (DAP) and reaches its highest level during seed maturation. Furthermore, AtDOG1 as well as LesaDOG1 transcripts are present in dry seeds of Arabidopsis and L. sativum, respectively, and transcript levels decline rapidly upon seed imbibition in both species (Bentsink et al., 2006; Graeber et al., 2010). The temporal, spatial, and hormonal LesaDOG1 transcript expression patterns suggest a role of this gene in the control of germination timing of nondormant L. sativum seeds (Graeber et al., 2010). Recent work shows that the AtDOG1 protein accumulates during seed maturation and, unlike the transcript, remains stable throughout imbibition of Arabidopsis seeds (Nakabayashi et al., 2012). The mother plant environment, especially the ambient temperature during seed development, controls AtDOG1 gene expression during seed maturation as well as the seed dormancy status (Kendall et al., 2011; Nakabayashi et al., 2012).
Arabidopsis seed development occurs in siliques (fruit longer than three times the width), each containing 40 to 60 seeds, while the typical fruit of Lepidium spp. is a silicle (fruit shorter than 3 times the width), each containing only two seeds (Ferrándiz et al., 1999; Mummenhoff et al., 2009). Characteristic Brassicaceae inflorescences are racemes, and upon fruit ripening they develop into infructescences in which the fruits are arranged in a spatial order that more or less reflects the temporal fruit-ripening pattern. The spatiotemporal seed maturation pattern is a combination of between-fruit (i.e. each fruit is in a slightly different maturation stage) and within-fruit (i.e. different seeds within the same fruit are in slightly different maturation stages) variation. In terms of resolving the temporal maturation process, spatially “well-ordered” infructescences with just a few seeds per fruit but many fruits in total provide technical advantages for studying different seed maturation stages with high resolution.
The genus Lepidium (cress) is highly suited for cross-species comparative research of early-life-history traits that are either present or absent in Arabidopsis. Lepidium spp. sensu lato, as it has three major lineages with approximately 250 species, is a monophyletic genus and represents one of the largest Brassicaceae genera (Mummenhoff et al., 2009). The three main phylogenetic lineages of Lepidium spp. sensu lato (Fig. 1) are supported by both internal transcribed spacers (ITS) and chloroplast DNA data, as described and discussed in detail by Mummenhoff et al. (2009, and refs. therein). Intercontinental dispersal of mucilaginous seeds adhering to birds led to a relatively rapid worldwide distribution, forming the basis for adaptations to very different habitats (Mummenhoff et al., 2004; Dierschke et al., 2009). Comparative work with L. sativum and Arabidopsis seeds (Linkies et al., 2009; Graeber et al., 2010, 2011; Voegele et al., 2011) and fruits (Mummenhoff et al., 2009; Mühlhausen et al., 2010) highlights the suitability of the genus Lepidium for cross-species research on diverse developmental aspects. Here, we exploit the experimental advantage of the highly structured Lepidium papillosum infructescence (i.e. a definable seed maturation gradient from the bottom [most mature] to the top [most immature] fruits within the largely elongated infructescence) to study the developmental and molecular mechanisms of seed dormancy induction and DOG1 protein expression during seed maturation.
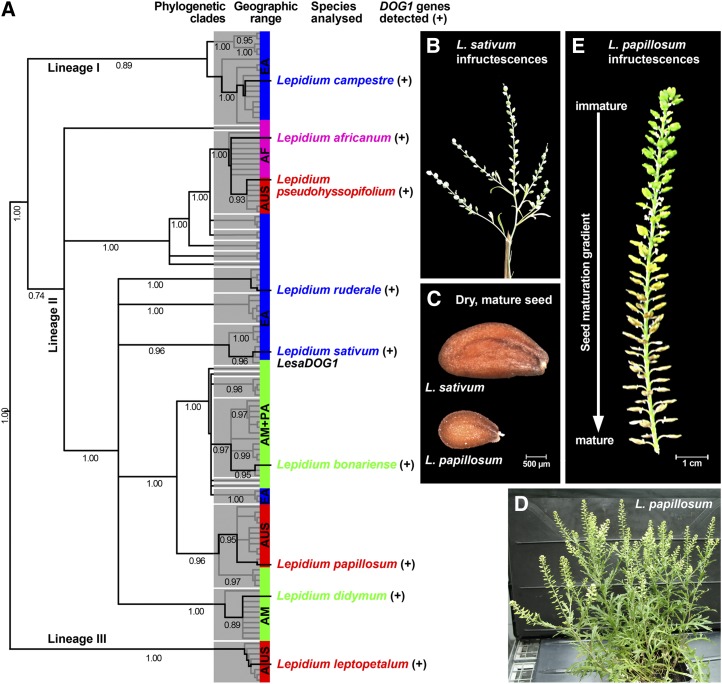
Figure 1.
Phylogeny of the genus Lepidium, the distribution of the DOG1 gene, and infructescence morphology of L. sativum and L. papillosum. A, Distribution of DOG1 homologs in representatives of all major lineages, subclades, and geographic ranges of Lepidium spp. Shown is the Bayesian 50% majority rule consensus tree based on ITS sequence analysis, with mean posterior probability values (clade credibilities) below branches. Phylogeny was redrawn from Mummenhoff et al. (2009). For DOG1 gene sequences, see Supplemental Table S1. AF, Africa; AM, America; AUS, Australia and New Zealand; EA, Eurasia; PA, Pacific Islands (Hawaii). B to E, Comparison of infructescences and seeds of L. sativum and L. papillosum. Within the well-ordered infructescences of L. papillosum, the seed maturation gradient is accompanied by primary dormancy induction.
RESULTS
Lepidium spp. as a Monophyletic Clade Suitable for Comparative Studies of Biogeographical, Developmental, and Molecular Mechanisms of Seed Dormancy
Despite evidence for the presence of putative orthologs of the AtDOG1 gene in the Brassicaceae species L. sativum and B. rapa, it is still unclear how this gene evolved, how broadly it is phylogenetically distributed among Brassicaceae, and whether it is a key factor of a conserved seed dormancy mechanism shared between species (Graeber et al., 2010, 2012). Several species of the monophyletic genus Lepidium (cress) have been shown to vary considerably in their degree of physiological dormancy (Graeber et al., 2010). Within the genus Lepidium, we found DOG1 homologs in species from all major lineages and distinct geographic ranges (Fig. 1A). The similarities of the obtained sequences to the known L. sativum DOG1 gene LesaDOG1 varied and were between 66% and 84% for the more than 1-kb Lepidium spp. genomic fragments obtained, each largely covering exons 1 and 2 including the whole intron 1 when aligned to LesaDOG1; some Lepidium species seem to have more than one DOG1 gene (Supplemental Table S1). Thus, DOG1 homologs are present in species with completely nondormant seeds, such as the domesticated L. sativum, as well as in dormant wild species such as L. papillosum, which raises questions about the role(s) of these genes and their involvement in primary dormancy induction.
The typical Lepidium spp. inflorescence is an elongating raceme with pedicellate flowers (Fig. 1B; Thellung, 1906; Hewson, 1981). In some Lepidium species, such as L. papillosum, this type of inflorescence develops into a remarkably elongated infructescence in which the individual fruits are well ordered and separated along the fruit axis (Fig. 1E). This arrangement of fruits within the L. papillosum infructescence provides a seed maturation gradient from the bottom (most mature) to the top (most immature). The gradient spatially reflects the temporal process of seed development and maturation on the mother plant. We exploited the highly structured infructescence with its numerous fruits (as compared with L. sativum; Fig. 1, B and D) to study the physiological and molecular mechanisms of primary dormancy induction during L. papillosum seed maturation.
The Architecture of the L. papillosum Infructescence Provides a Well-Ordered Spatial Arrangement of Fruits to Study the Temporal Pattern of Seed Maturation and Dormancy Induction
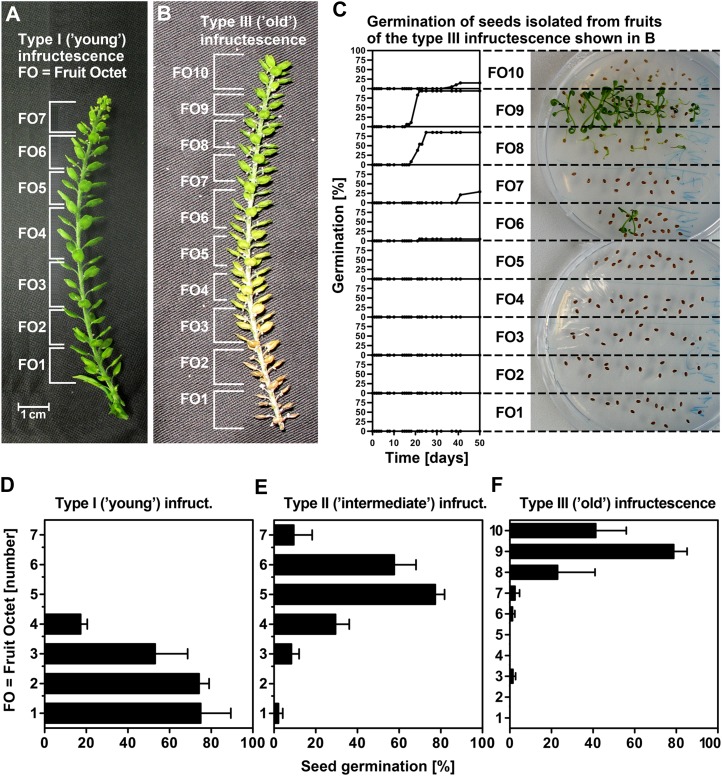
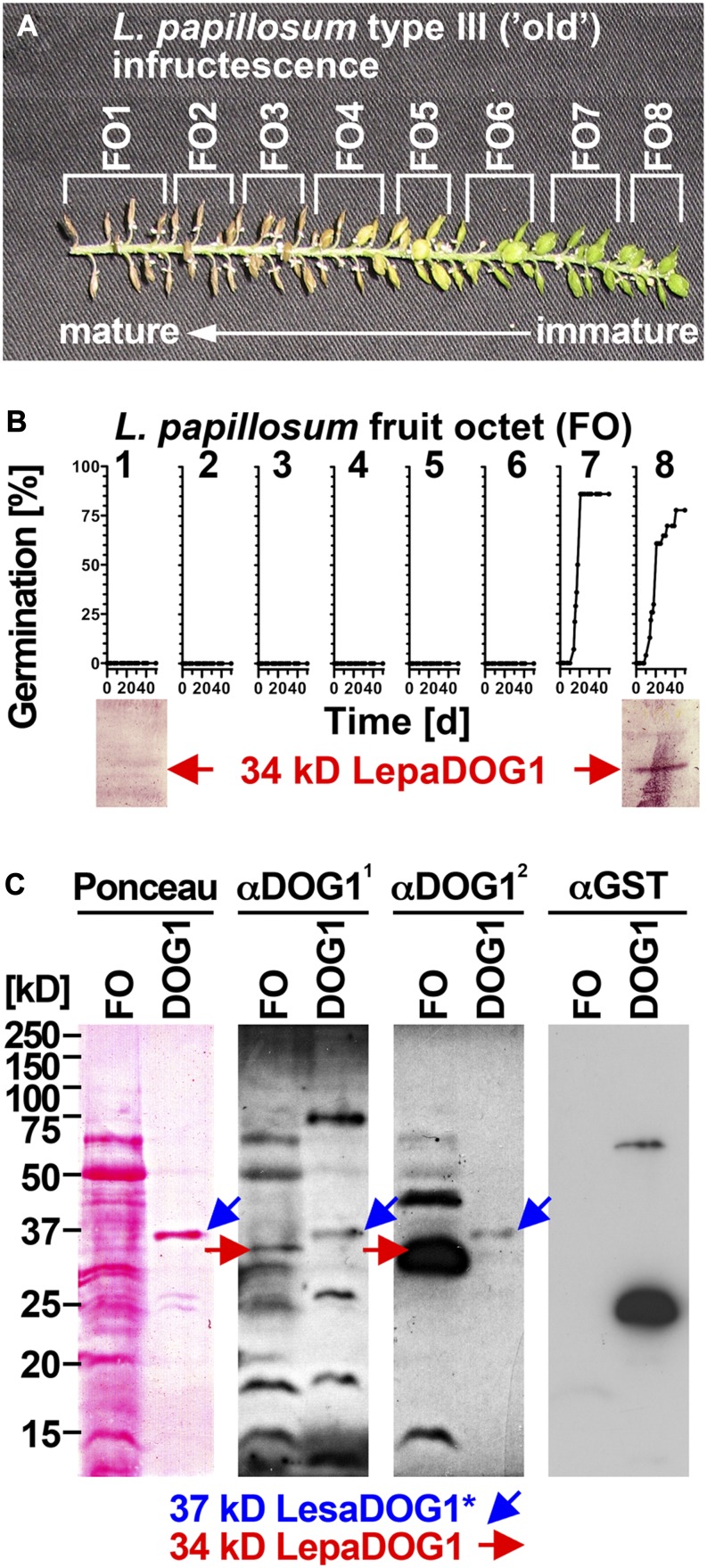
To study primary dormancy induction, we utilized three developmental stages of L. papillosum infructescences (Figs. 1E and 2, A and B) classified as follows: type I (“young”: all bottom fruits still green and several flower buds at the top), type II (“intermediate”: first bottom fruit yellow), and type III (“old”: bottom fruits brown and dried, no flower buds at the top). For our experiments, the well-ordered infructescence was divided into fruit octets (FOs): each FO contained eight fruits (each fruit contains two seeds), which were consecutively numbered from the bottom (most mature FOs) to the top (most immature FOs). Figure 2C shows the time course for the germination of seeds from individual FOs of one type III (old) infructescence. Within this type III infructescence, all seeds of FO1 to FO6 are dormant, whereas all seeds of FO8 and FO9 germinate; therefore, FO7 provides the transition stage when primary dormancy is induced. Germination analyses of multiple type I to III infructescences revealed that, in contrast to type III, for which the bottom FOs harbor seeds after dormancy induction (Fig. 2F), type I bottom FOs harbor seeds prior to dormancy induction (Fig. 2D) and type II exhibits an intermediate pattern (Fig. 2E). Furthermore, the top FO seeds from type I infructescences are too immature for germination, whereas seeds from type III infructescences germinate, indicating that they are sufficiently developed and are just prior to dormancy induction (Fig. 2, D–F).
Figure 2.
Germination and dormancy as related to the seed maturation gradient within L. papillosum infructescences. A to C, Experimental separation of type I (young) and type III (old) infructescences into FOs and FO-specific analysis of their germination time courses. Note that lack of germination in bottom-part FOs (low numbers) is due to primary dormancy, whereas lack of germination in top-part FOs (high numbers) is due to immaturity of seeds. Exemplary seeds from type III infructescence incubated on germination plates (at day 39) and germination kinetics are shown in C. D and E, Final germination frequency gradients indicative for primary dormancy induction of seeds from FOs of type I (young), type II (intermediate), and type III (old) infructescences. Mean values ± se of four type I, six type II, and four type III infructescences from different plants are shown. Final germination percentages were taken from FO (each containing eight fruits and thereby 16 seeds) time-course analyses at 50 d.
L. papillosum develops within 5 months from a seedling to a mature plant with completely dried infructescence, in which seeds from all FOs represent the dormant state of the bottom-part FOs from type III infructescences. To further characterize the dormancy of L. papillosum, we used seeds from completely dried infructescences and show in Figure 3 that L. papillosum primary dormancy can be categorized as physiological coat dormancy (for categories, see Finch-Savage and Leubner-Metzger, 2006). This physiological dormancy is released by seed after-ripening combined with cold stratification, and scarification (mechanical slitting/removal of the “coats” covering the embryo) can replace after-ripening and cold stratification (Fig. 3). Our medium contains 4 mm nitrate to synchronize the germination process (Finch-Savage and Leubner-Metzger 2006; Müller et al., 2006), but as for the coat dormancy of tobacco (Nicotiana tabacum) seeds (Leubner-Metzger, 2002, and refs. therein), this concentration did not release the coat dormancy established during seed development of L. papillosum (Figs. 2 and 3). To investigate a potential effect of this low concentration of nitrate, we verified that scarified seeds germinate to 100% in less then 100 h of imbibition on water or 4 or 25 mm nitrate, whereas nonscarified seeds of the dormant population do not, which indicates the absence of embryo dormancy in these seeds.
Figure 3.
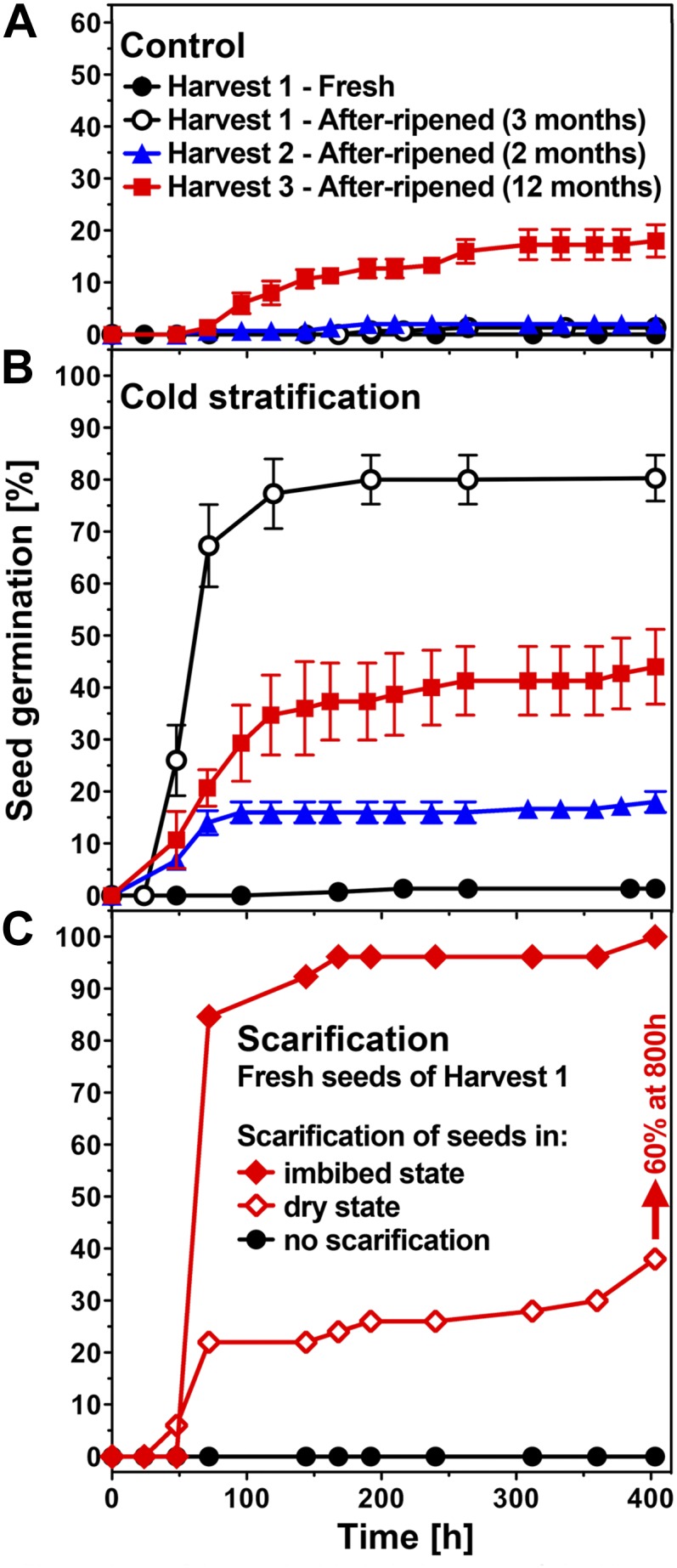
Primary physiological dormancy of L. papillosum seeds and its release by after-ripening, cold stratification, and scarification. A, Germination kinetics of imbibed L. papillosum seed populations. The primary dormancy of freshly harvested mature (fresh) seeds was not appreciably released by seed after-ripening. Different harvests and dry storage periods/conditions were compared. Harvest 1 (phytochamber-grown) seeds were after-ripened following removal from the mother plant by dry storage at room temperature for 3 months. Harvest 2 (phytochamber-grown) seeds were after-ripened on the dried mother plant for 2 months. Harvest 3 (greenhouse-grown, “natural” conditions) seeds were after-ripened on the dried mother plant for 12 months. B, The effect of cold stratification (7 d at 4°C preincubation) on seed dormancy of fresh and after-ripened seeds. Note that cold stratification did not release the dormancy of fresh seeds and that differences in the effectiveness of cold stratification depend on after-ripening storage conditions. C, Effect of scarification on imbibed (coat removal; i.e. embryo rescue) and dry (coat slitting) fresh seeds. L. papillosum seeds are coat dormant, as scarification releases the dormancy of fresh seeds; note further that in contrast, the release of dormancy by cold stratification was only possible for after-ripened but not for fresh seeds. Mean values ± se of 3 × 50 seeds are shown. A part of the harvest 1 results is shown for comparison and has been shown earlier by Graeber et al. (2010). [See online article for color version of this figure.]
Molecular Analyses of L. papillosum Genome Constitution and Cloning of LepaDOG1 Genes
The systematics of Australian Lepidium spp. is complex (Mummenhoff et al., 2001, 2004, and refs. therein). The abundant presence of polyploid Lepidium spp. in Australia implied a reticulate history of the genus on this continent characterized by hybridization/polyploidization events (Lee et al., 2002; Mummenhoff et al., 2004; Dierschke et al., 2009). Lepidium oxytrichum and L. papillosum differ only in trichome type. Thus, L. oxytrichum was not recognized (and segregated from L. papillosum) until 1915 (Hewson, 1981). In molecular phylogenetic analysis, L. oxytrichum and L. papillosum were recognized as separate species, and they do form a monophyletic lineage (Mummenhoff et al., 2004, 2009). Therefore, we verified by trichome morphology and ITS sequence analysis (Supplemental Figs. S1 and S2) that our accession Mummenhoff 2415 (Supplemental Table S1) is undoubtedly L. papillosum and not L. oxytrichum, as reported previously (Graeber et al., 2010).
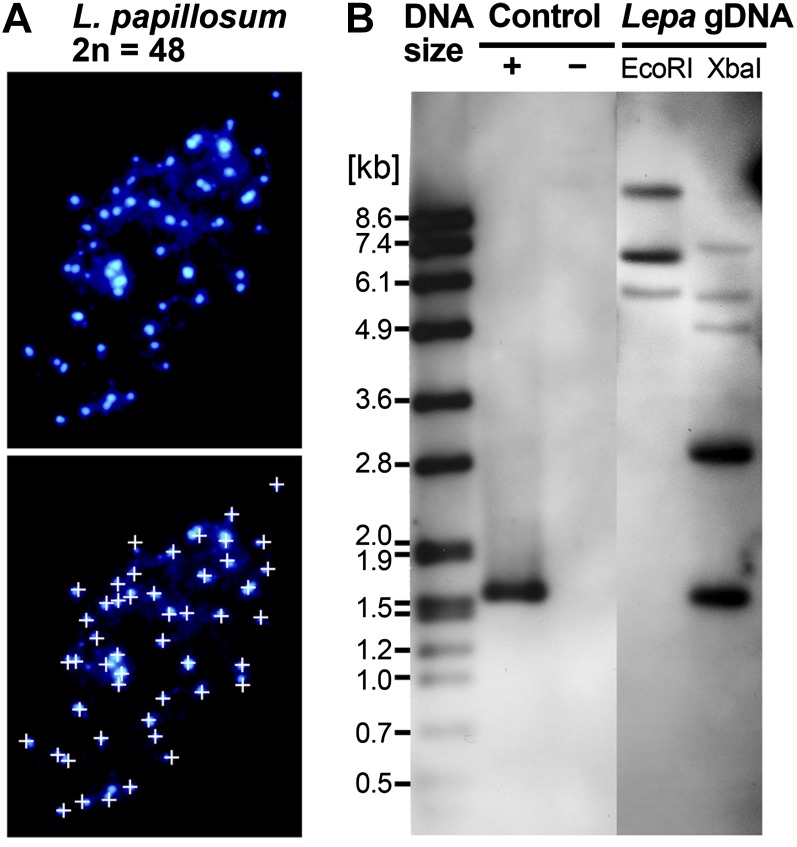
We found by analyzing mitotic spreads that L. papillosum has 2n = 48 chromosomes (Fig. 4A), whereas its sister species L. oxytrichum is characterized by 2n = 24 chromosomes (Hurka et al., 1992). Based on n = 8 (Al-Shehbaz, 1986) or n = 12 (Murín, 1986) as chromosome base numbers for Lepidium spp., it seems that L. papillosum (2n = 48) is hexaploid or tetraploid, dependent on a chromosome base number of 8 or 12, respectively. As a result of all these data, we suggest that L. papillosum evolved by polyploidization from the very closely related L. oxytrichum (or its progenitor), most probably by autoploidization.
Figure 4.
Chromosome number and DOG1 gene analysis of the L. papillosum genome. A, 4′,6-Diamino-2-phenylindole-stained mitotic chromosome spreads from flower bud tissue exhibiting 2n = 48 chromosomes. For better counting, chromosomes are labeled by small white crosses in the bottom image. B, Southern analysis of L. papillosum genomic DNA (Lepa gDNA) digested with EcoRI or XbaI and hybridized with a 0.35-kb LesaDOG1 exon 1 probe shows the presence of three to five hybridizing bands indicative of LepaDOG1 genes. Note that neither LesaDOG1 nor the cloned genomic LepaDOG1 fragments (Supplemental Table S1) contain EcoRI or XbaI restriction sites within the region covered by the Southern probe. Hybridization controls are plasmids with (+) or without (−) LesaDOG1 full-length genomic DNA inserts; the left lane (DNA size) shows a DNA molecular mass ladder. [See online article for color version of this figure.]
With regard to its polyploid origin, we expected that L. papillosum possesses several DOG1 genes in contrast to the one DOG1 gene present in the genome of Arabidopsis (AtDOG1). To investigate this, we used a 0.35-kb DNA fragment covering exon 1 of the DOG1 homolog LesaDOG1 from L. sativum as a probe to perform Southern analysis with L. papillosum genomic DNA. In agreement with the polyploid nature of this species, the Southern analysis (Fig. 4B) indicates that L. papillosum has three to five genomic sequences homologous to the LesaDOG1 exon 1 probe. These are either L. papillosum DOG1 homologs or other genes with sufficient sequence similarity to provide a hybridization signal, such as homologs of the DOG1-Like genes. We cloned four genomic fragments from L. papillosum representing full-length or nearly full-length homologs of LesaDOG1 (Fig. 5; Supplemental Table S1). We did not obtain any DOG1-Like gene sequences for L. papillosum (Fig. 5; Supplemental Table S1) or any other Lepidium spp. (Fig. 1; Supplemental Table S1), demonstrating that our PCR strategy was indeed specific for the Lepidium DOG1 genes. Based on further analysis summarized below, these L. papillosum sequences were named LepaDOG1a to LepaDOG1d, and their alignment is shown in Figure 5A in comparison with the LesaDOG1 gene.
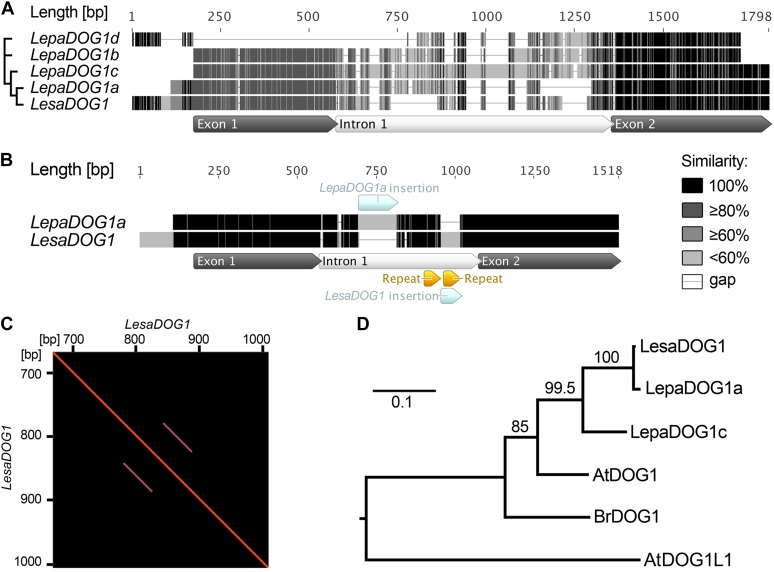
Figure 5.
Comparison of L. papillosum and L. sativum DOG1 genomic sequences and phylogenetic analysis of predicted Brassicaceae DOG1 protein sequences. A, Multiple alignment of the genomic LepaDOG1a (JX512183), LepaDOG1b (JX512184), LepaDOG1c (JX512185), and LepaDOG1d (JX512186) DNA fragments (for details, see Supplemental Table S1) as compared with the LesaDOG1 gene (Graeber et al., 2010). We applied a grayscale coloring scheme according to the similarity of the residues in the alignment. A certain residue in a sequence is colored black if it is 100% identical to the other residues in the same alignment column. A residue is colored dark gray if it occurs identically in 80% of the positions in a column (e.g. four out of five position in a column). A residue is colored moderate gray if it occurs identically in 60% of the positions in a column (e.g. three out of five positions) or light gray if it occurs identically in less than 60% of the positions in a column (e.g. one or two out of five positions in a column). Internal gaps are treated as unknown characters, and end gaps are not considered in the similarity calculation. Gaps are shown as horizontal lines. The annotation of exon-intron structure below the alignment is based on LesaDOG1. Note that the position of the intron splice site is conserved, as verified by comparison with LepaDOG1a/LepaDOG1c cDNAs. The relationship of the sequences to each other is indicated on the left by a maximum-likelihood cladogram. B, Pairwise alignment of the genomic sequences of LepaDOG1a with LesaDOG1. Only minor nucleotide changes are present in the two exon regions, and the positions of the introns with InDels are indicated. Note that in this pairwise alignment, only identical residues are colored black; otherwise, they are light gray. C, Dot-blot analysis demonstrating the presence of repeat elements in the LesaDOG1 intron sequence indicated as parallel lines. The LesaDOG1 sequence used for this self-dot-blot analysis corresponds to that shown in B, and the detected repeat locations are marked there. Note that the first repeat sequence is present in LesaDOG1 and LepaDOG1a, while the second repeat sequence is missing in LepaDOG1a. D, Maximum-likelihood phylogenetic tree of predicted protein sequences for Brassicaceae DOG1 genes for which transcript expression in seeds has been demonstrated by Bentsink et al. (2006; AtDOG1, α-splice variant), Graeber et al. (2010; LesaDOG1, BrDOG1), and in this work (LepaDOG1a, LepaDOG1c) in relation to the Arabidopsis DOG1-Like1 gene (AtDOG1L1). The scale bar indicates substitutions per site, and values on branches show bootstrap support. Phylogenetic analysis was performed using PhyML (for details, see “Materials and Methods”). [See online article for color version of this figure.]
LepaDOG1a, LepaDOG1b, and LepaDOG1c show the same exon-intron structure as LesaDOG1, with high sequence similarity in the exon regions and diversity in the intron regions. The introns of LepaDOG1a, LepaDOG1b, and LepaDOG1c strongly vary in length (438, 558, and 737 bp, respectively) compared with LesaDOG1 (383 bp), but the putative intron splice sites of these genes are conserved. In contrast to LepaDOG1a, LepaDOG1b, and LepaDOG1c, the LepaDOG1d sequence that we obtained (Fig. 5A) displays high sequence similarity only to LesaDOG1 exon 2 and to the LesaDOG1 200-bp 5′ upstream region from the ATG (i.e. the proximal gene promoter region), but interestingly, the exon 1 region is completely missing.
Of the four LepaDOG1 genomic sequences, LepaDOG1a exhibits over its entire length of 1.3 kb the highest sequence similarity to LesaDOG1 (Fig. 5A). The pairwise alignment of the LesaDOG1 and LepaDOG1a genomic sequences shows only minor nucleotide changes in exon 1 and 2 regions, while there are species-specific differences in the intron region, including two insertions/deletions (InDels; Fig. 5B). The larger 121-bp InDel is present as an insertion within the LepaDOG1a intron, while the smaller 62-bp InDel is present as an insertion in the LesaDOG1 intron. Interestingly, the 62-bp InDel seems to result from an insertion of a repeat region within the LesaDOG1 intron, as indicated by the dot-blot analysis shown in Figure 5C. This supports the close relationship between LepaDOG1a and LesaDOG1, as only a few single-nucleotide changes accumulated, and the differences between the two genes seem to result largely from insertions of repeat regions within the intron (Fig. 5B). Therefore, we propose that LepaDOG1a and LesaDOG1 are putative orthologs. Taken together, the four diversified LepaDOG1 sequences that we cloned (Fig. 5) further support the findings from chromosome and Southern analysis (Fig. 4) that L. papillosum is polyploid and that, especially, the LepaDOG1 introns accumulate polymorphisms regarding length and sequence.
To test whether the LepaDOG1 genes are expressed in seeds, we performed reverse transcription-PCR experiments with L. papillosum seed RNA and obtained complementary DNA (cDNA) sequences for LepaDOG1a and LepaDOG1c (Supplemental Table S1). These transcripts exhibit the same splice sites for intron 1 that are known to be conserved for the seed-expressed transcripts of AtDOG1, LesaDOG1, and BrDOG1 in Arabidopsis, L. sativum, and B. rapa, respectively (Bentsink et al., 2006; Graeber et al., 2010).
Molecular phylogenetic analysis of the deduced full-length Brassicaceae DOG1 protein sequences for which seed-specific transcript expression is known (Bentsink et al., 2006; Graeber et al., 2010; this work) showed that the LepaDOG1a and LepaDOG1c proteins are more closely related to LesaDOG1 than to other Brassicaceae spp. DOG1 proteins (Fig. 5D). This relationship suggests that polyclonal antibodies that detect the AtDOG1 and LesaDOG1 proteins will also detect LepaDOG1 proteins. In the next section, we show that polyclonal antibodies raised against AtDOG1 detect purified LesaDOG1 protein and can be used for analyzing LepaDOG1 protein expression in the spatiotemporal gradient of seed development, maturation, and dormancy induction of the L. papillosum infructescence.
Analysis of L. papillosum DOG1 Protein Expression during Seed Maturation and Dormancy Induction
DOG1 transcript expression in Arabidopsis has been described to peak during seed maturation (Bentsink et al., 2006) and is regulated by environmental conditions in association with primary dormancy induction (Kendall et al., 2011). Also, AtDOG1 proteins have recently been shown to accumulate during Arabidopsis seed maturation (Nakabayashi et al., 2012). Using an α-DOG1 polyclonal antibody raised against AtDOG1 (Schwab, 2008), we detected an antigen with the apparent molecular mass of 34 kD in seeds of type III L. papillosum infructescences in the FO prior to dormancy induction (FO8; Fig. 6B), with seeds being able to germinate. We did not detect this band in old octets after dormancy induction, where the fruits were brown and dry and dormancy had been induced (FO1; Fig. 6B).
Figure 6.
Analysis of LepaDOG1 protein expression within the L. papillosum infructescence. A, Type III L. papillosum infructescence exhibiting the seed maturation gradient and the experimental dissection into FOs. B, Top panel, FO-specific germination time courses to determine the developmental switch to seed dormancy; bottom panel, FO-specific immunoblot analysis of LepaDOG1 protein (arrow) expression using a polyclonal antibody (α-DOG11) raised against AtDOG1. C, Control experiments (left panel, Ponceau-stained blotting membrane; middle and right panels, immunoblot analyses) demonstrating that two different antibodies raised against AtDOG1 (α-DOG11 and α-DOG12) detect a recombinant 37-kD LesaDOG1* antigen that corresponds to a 37-kD protein purified from an extract of E. coli expressing GST-LesaDOG1 fusion protein (DOG1; for constructs and purification procedure, see Supplemental Figs. S3 and S4). Note that in the E. coli DOG1 extract, the α-DOG1 antibodies detect the strongest 37-kD protein band (Ponceau) as LesaDOG1* (blue arrows), while this antigen is not detected by the α-GST antibody. The asterisk indicates the presence of an approximately 3-kD residual linker peptide in the recombinant LesaDOG1* protein left after GST cleavage (Supplemental Fig. S3). Therefore, an apparent molecular mass of 34 kD is expected for LesaDOG1. Both α-DOG1 antibodies detect 34-kD LepaDOG1 antigens in a L. papillosum protein extract from type I FO prior to dormancy induction (red arrows). Note that the two α-DOG1 antibodies differ in sensitivity and that the highly similar 34-kD LepaDOG1a and LepaDOG1c proteins cannot be distinguished. kD indicates the protein molecular mass marker ladder. [See online article for color version of this figure.]
To confirm the identity of the detected band and to show that Lepidium spp. DOG1 proteins are detectable with antibodies raised against AtDOG1, we expressed and purified recombinant LesaDOG1 fused to glutathione S-transferase (GST) in Escherichia coli (Supplemental Figs. S3 and S4). This fusion protein includes a linker peptide containing a tobacco etch virus (TEV) protease cleavage site between LesaDOG1 and GST. After protein purification and proteolytic cleavage, the E. coli protein extract contained as the strongest band a protein with the apparent molecular mass of 37 kD (Ponceau staining in Fig. 6C; Supplemental Fig. S4). This 37-kD protein is detected by the α-DOG1 polyclonal antibody but not by the α-GST polyclonal antibody (Fig. 6C); as expected, the residual GST-LesaDOG1 fusion as well as free GST protein were detected by the α-GST polyclonal antibody (Fig. 6C). We refer to this α-DOG1 antigen as the 37-kD LesaDOG1* protein, which consists of an approximately 3-kD linker peptide fused to the LesaDOG1 protein (Supplemental Fig. S3). After subtracting the size of the linker peptide from the 37-kD LesaDOG1*, the expected apparent molecular mass of the LesaDOG1 protein is 34 kD. As the sequence similarities between the predicted LesaDOG1, LepaDOG1a, and LepaDOG1c proteins are very high (Fig. 5), we conclude that the α-DOG1 antigens with the apparent molecular mass of 34 kD detected in the L. papillosum protein extracts from FOs (Figs. 6 and 7) are the LepaDOG1 proteins.
Figure 7.
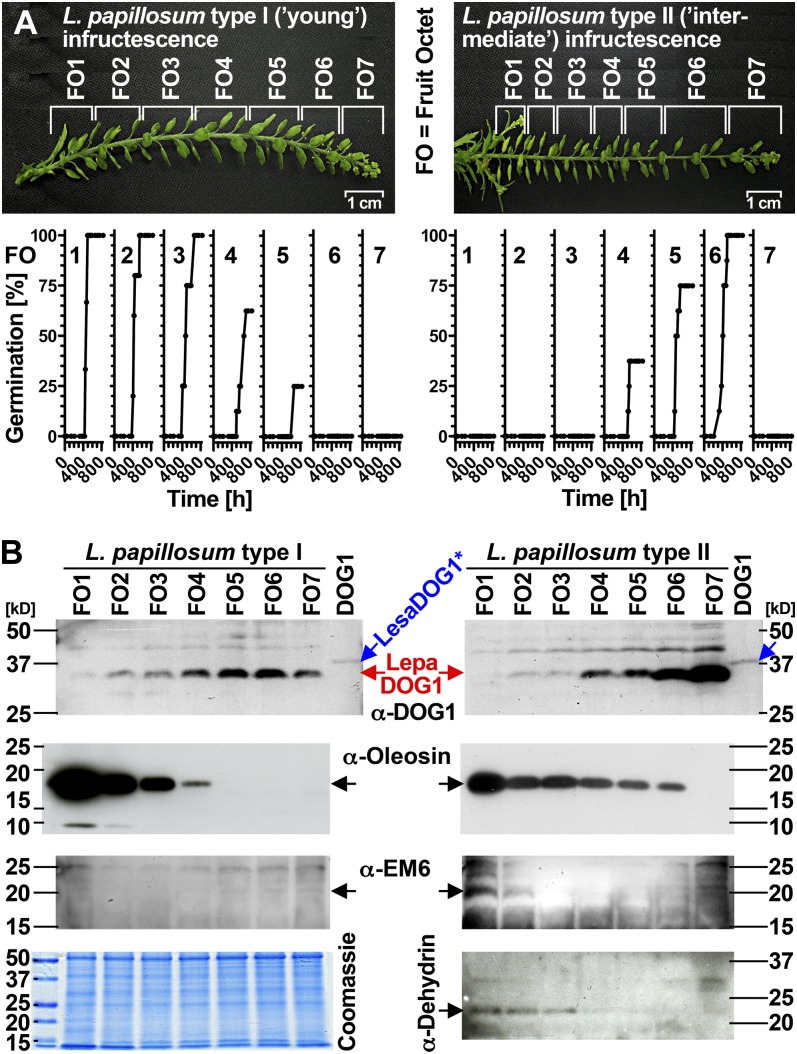
Spatial analysis of LepaDOG1 protein expression within the L. papillosum infructescence. A, Type I (left) and type II (right) L. papillosum infructescences, their experimental dissection into FO (each containing eight fruits and thereby 16 seeds), and their FO-specific germination time courses. Note that nongerminating seeds from top FOs are immature, while nongerminating seeds from bottom FOs are dormant. B, FO-specific immunoblot analysis of type I (left) and type II (right) infructescences shown in A displaying the protein expression patterns for LepaDOG1, oleosin (expressed during early maturation, indicative for seed filling), EM6, and dehydrin (expressed during late maturation, indicative for seed maturation and desiccation tolerance) by using corresponding antibodies (α-DOG12, α-oleosin, α-EM6, α-dehydrin). A Coomassie blue-stained gel showing the FO protein extract banding patterns is seen at bottom left. kD indicates the protein molecular mass marker ladder. [See online article for color version of this figure.]
Further support for our conclusion that the L. papillosum 34-kD antigens in Figures 6 and 7 are indeed the LepaDOG1 proteins comes from an interesting observation of Nakabayashi et al. (2012). Those authors demonstrated that the AtDOG1 proteins with a calculated molecular mass of approximately 32 kD exhibit significantly slower SDS-PAGE migration (i.e. they run at an apparent molecular mass of approximately 40 kD). AtDOG1, LesaDOG1, LepaDOG1a, and LepaDOG1c are all acidic proteins with calculated pI values between 5.1 and 5.7, and the acidity of the AtDOG1 proteins was proposed to be the reason for the slower migration (Nakabayashi et al., 2012). Like the Arabidopsis DOG1 proteins, the Lepidium spp. DOG1 proteins exhibit significantly slower SDS-PAGE migration: LesaDOG1, LepaDOG1a, and LepaDOG1c all have calculated molecular masses of approximately 31.6 kD, but their apparent molecular masses during SDS-PAGE is approximately 34 kD. Discrepancies between calculated and apparent molecular mass seem to be a general phenomenon, as shown by a global proteome analysis of the gel mobility of proteins indicating that more than 40% of the proteins did not migrate within 10% of the predicted position (Shirai et al., 2008). It is interesting that the DOG1 proteins of all three species share a common acidity feature, since Nakabayashi et al. (2012) suggested that alteration of the AtDOG1 protein pI value may lead to altered DOG1 function.
Figure 7 (left panels) shows that the 34-kD LepaDOG1 proteins are abundant in seeds from type I (young) infructescence FO1 to FO6; all these FOs contain seeds prior to dormancy induction. The onset of LepaDOG1 protein expression between FO7 and FO6 is associated with immature seeds prior to their ability to germinate. In agreement with this observation, in type II (intermediate) infructescences, the highest LepaDOG1 abundance is in FOs with immature (FO7) and fully germinating (FO6) seeds, and the LepaDOG1 abundance decreases prior to dormancy induction and remains low in primary dormant seeds (Fig. 7, right panels). To further assign these protein patterns of LepaDOG1 expression to developmental stages within the L. papillosum infructescence, we used monoclonal antibodies directed against oleosin and polyclonal antibodies against EM6 (for Early Met labeled) and dehydrin; the expression abundance of these three proteins is indicative for seed storage accumulation, maturation, and the induction of desiccation tolerance, respectively (Close et al., 1993; Crowe et al., 2000; Boudet et al., 2006; Siloto et al., 2006). In support of the finding that LepaDOG1 is already abundant in immature FOs, abundant LepaDOG1 expression preceded oleosin expression and, therefore, occurs early during seed development already at the filling stage (Fig. 6B). In contrast to these early phases of seed development, LepaDOG1 abundance was low in the late phases of seed development characterized by abundant EM6 (maturation) and dehydrin (desiccation tolerance) proteins (Fig. 6B, right panels). In conclusion, utilization of the well-structured infructescences of L. papillosum demonstrated that LepaDOG1 protein is most abundant in immature and fully germinating seeds and that its abundance decreases prior to the induction of primary seed dormancy.
DISCUSSION
Spatiotemporal Maturation Patterns in L. papillosum Infructescences to Study the Induction of Seed Dormancy in the Mother Plant Environment
The timing of flowering and germination are the most significant developmental transitions for local adaptation to environments. Several recent publications clearly demonstrate that in Arabidopsis, QTLs with large effects on fitness colocalize with QTLs for field germination timing and seed dormancy (Huang et al., 2010; Moyers and Kane, 2010; Chiang et al., 2011; Barua et al., 2012; Kronholm et al., 2012). Germination phenology is under intense, geographically variable, natural selection in Arabidopsis, since it is the major determinant of fitness during the earliest stages of colonization (Huang et al., 2010; Moyers and Kane, 2010; Chiang et al., 2011; Barua et al., 2012; Kronholm et al., 2012). It is also evident that QTLs including DOG1 determine the phenotypic plasticity of seed germination/dormancy responses already on the mother plant in interaction with environmental factors such as seed maturation photoperiod and temperature. The phenomenon that the maternal environment during seed development and maturation determines the germination phenology of Arabidopsis and other species is well-established knowledge (Kugler, 1951; Gutterman, 1980/1981; Munir et al., 2001). However, as a major advance, we now know from ecological work with Arabidopsis (Huang et al., 2010; Moyers and Kane, 2010; Chiang et al., 2011; Barua et al., 2012; Kronholm et al., 2012) that natural genetic variation at the DOG1 locus is a major determinant for the local adaptation to novel environments, and we also know that the maternal environment controls DOG1 gene expression during seed maturation. For example, low temperature during seed maturation causes increased DOG1 gene expression and deeper primary dormancy of mature seeds (Chiang et al., 2011; Kendall et al., 2011; Nakabayashi et al., 2012). Arabidopsis, therefore, is a very good model also for addressing intraspecific questions in ecological, evolutionary, and developmental biology. Since species can differ considerably, this species cannot be used to address all questions of interest, but Arabidopsis tools and knowledge are of particular advantage for moving beyond this species to other Brassicaceae spp. (Tonsor et al., 2005; Schranz et al., 2007; Oyama et al., 2008; Franzke et al., 2011; Graeber et al., 2010). The large Brassicaceae genus Lepidium (cress) is highly suited for comparative cross-species work, and we show here that L. papillosum provides an insight into dormancy induction during seed maturation in a developmental (infructescence structure and seed physiology) and an evolutionary (Lepidium DOG1 genes) context.
The molecular phylogeny of the Brassicaceae provided evidence that the genus Lepidium represents a monophyletic assemblage consisting of three major lineages (Mummenhoff et al., 2001, 2009). With approximately 250 species, it is one of the largest Brassicaceae genera, and it offers cross-species comparisons regarding unique or common early-life-history traits. Intercontinental dispersal of the mucilaginous seeds adhering to birds led to a relatively rapid worldwide distribution of this genus starting in the Pliocene, which includes adaptation to different environments (Mummenhoff et al., 2001, 2004; Dierschke et al., 2009). Comparative work with Lepidium spp. and Arabidopsis seeds (Linkies et al., 2009; Graeber et al., 2010, 2011; Voegele et al., 2011), fruits (Mummenhoff et al., 2009; Mühlhausen et al., 2010), and flowers (Lee et al., 2002) emphasizes that the genus Lepidium provides a phylogenetically and environmentally defined framework highly suited for evolutionary and developmental research on seed/fruit-related traits. DOG1 gene homologs are present in Lepidium spp. representing all major lineages, subclades, and different geographical ranges. Different Lepidium spp. are characterized by distinct dormancy mechanisms and/or dormancy depths associated either with the covering layers (coat dormancy) or the embryo (Müller et al., 2006; Graeber et al., 2010; this work). We demonstrate here that physiological dormancy is induced during seed maturation of L. papillosum (i.e. on the mother plant), and scarification experiments showed that this primary dormancy is mainly coat imposed, as is the case for Arabidopsis. But in contrast to Arabidopsis, neither cold stratification nor after-ripening storage alone is sufficient to completely release the coat dormancy of L. papillosum, which can only be achieved by a combination of both or by scarification.
The L. papillosum infructescence is a well-ordered raceme with a seed maturation gradient from the bottom (mature) to the top (immature) fruits. The typical Lepidium spp. inflorescence/infructescence is an elongated raceme in which the pedicels (stalks) of the individual flowers/fruits are arranged along an unbranched axis, but neither in L. sativum nor in many other Lepidium species or in Arabidopsis is the anatomy of the infructescence so well ordered as in L. papillosum. This enabled us to study the pattern of seed germinability and dormancy induction with a high spatiotemporal resolution (Figs. 2 and 7). Premature germination can be induced by GAs during Arabidopsis seed development when embryo development has been completed (at approximately 10 DAP), and subsequent seed maturation (approximately 10–20 DAP) includes the induction of primary dormancy (Ooms et al., 1993; Raz et al., 2001; Le et al., 2010; van Zanten et al., 2011). In contrast to Arabidopsis, premature seed germination prior to dormancy induction does not require any hormonal addition in L. papillosum (this work), as is also known from Brassica spp. (Fernandez, 1997; Ren and Bewley, 1999) or cereal grains (Rodríguez et al., 2012). However, L. papillosum premature seeds need approximately four times longer to complete germination as compared with mature seeds in which dormancy has been broken (compare Fig. 3 with Figs. 6 and 7), and even though no hormonal addition is needed, they require light (data not shown). Nitrate is known to break the coat dormancy of Arabidopsis by decreasing seed abscisic acid contents (Alboresi et al., 2005; Finch-Savage and Leubner-Metzger, 2006; Matakiadis et al., 2009), and it is also known to synchronize the germination of various seeds by enhancing GA effects (Hilhorst et al., 1986; Leubner-Metzger 2002). Our germination medium contains 4 mm nitrate, but as for the coat dormancy of tobacco seeds (Leubner-Metzger, 2002, and refs. therein), this concentration did not release the coat dormancy induced during seed development of L. papillosum seeds (Figs. 2 and 3).
In our experiments with L. papillosum (Figs. 2 and 7), we captured in type I infructescences stages prior to and at the onset of seed germinability until full premature germination (prior to dormancy induction), and in types II and III infructescences we captured all stages prior to, during, and after the induction of primary dormancy, including the full premature germination stage. To assign these stages to events during seed development and maturation on the molecular level, we analyzed the expression patterns of well-known marker proteins including oleosin (seed storage accumulation), EM6 (seed maturation), and dehydrin (desiccation tolerance; Close et al., 1993; Huang, 1996; Bies et al., 1998; Crowe et al., 2000; Ruuska et al., 2002; Boudet et al., 2006; Siloto et al., 2006; Leprince and Buitink, 2010). Oleosin accumulation during L. papillosum seed development was enhanced with the onset of premature germinability and early maturation in type I and II infructescences. EM6 was not evident in type I but only during late maturation in type II infructescences, when dehydrins also are expressed.
The division of the L. papillosum infructescence into FOs in combination with the large seed size facilitated physiological (germination assay) and molecular (e.g. immunoblot analysis of protein expression patterns) analyses simultaneously from the same infructescence. This resulted in a comprehensive and highly resolved spatiotemporal analysis of the developmental switches during seed maturation. Each FO contains 16 seeds, of which we used one-half for the germination assays and one-half for protein analyses; however, as each seed is large enough, several further single-seed-based biochemical assays also would be possible. In summary, the genus Lepidium harbors species with scientifically interesting and experimentally advantageous features, such as the nondormant and large-seeded L. sativum that emerged as a Brassicaceae model to study endosperm weakening during seed germination (Linkies et al., 2009; Graeber et al., 2010; Voegele et al., 2012), as well as species with dormant seeds like L. papillosum, for which dormancy induction during seed maturation can be studied with a superior spatiotemporal resolution.
Evolution of the Brassicaceae DOG1 Genes in Diploid and Polyploid Species and Developmentally Regulated DOG1 Protein Expression during L. papillosum Seed Maturation
Here, we provide evidence for the presence of DOG1 gene homologs in nine Lepidium species from all major lineages, subclades, and geographical ranges (Fig. 1). In several of these species, we also obtained evidence for at least two DOG1 genes being present in their genomes (Supplemental Table S1). We found that L. papillosum is a polyploid species with at least four DOG1 homologous genomic sequences (LepaDOG1a to LepaDOG1d; Fig. 5). Furthermore, we detected transcripts of LepaDOG1a and LepaDOG1c in L. papillosum seeds, a transcript origin also evident for the putative Brassicaceae orthologs LesaDOG1, BrDOG1, and AtDOG1 in L. sativum, B. rapa, and Arabidopsis, respectively (Bentsink et al., 2006; Graeber et al., 2010; Footitt et al., 2011; Nakabayashi et al., 2012). The genomic sequences of LepaDOG1a and LesaDOG1 show more than 80% similarity with highly similar exon regions and, due to conserved splice sites, more than 98% putative amino acid sequence similarity despite rather diverging intron sequences. In this respect, the situation is similar to the comparison of the LesaDOG1, BrDOG1, and AtDOG1 exon and intron sequences, which is discussed in relation to speciation by Graeber et al. (2010). The comparison of LepaDOG1a and LesaDOG1 introns revealed two InDels, which could also be variations as a consequence of speciation processes. Interestingly, one of the InDels in the LesaDOG1 intron seems to result from an insertion of a repeat region (Fig. 5). This indicates that the putative orthologs LepaDOG1a and LesaDOG1 diversified rather recently, since only a few other single-nucleotide changes accumulated between the two sequences and the main differences resulted from InDel variations in intron 1. This is in agreement with phylogenetic analyses in Lepidium spp. The low levels of chloroplast DNA and ITS sequence divergence among Lepidium species of lineage II (to which L. papillosum and L. sativum belong) suggest a rapid and recent (Pliocene/Pleistocene; approximately 2.2 million years ago) radiation of Lepidium spp. into lineages of the different continents (Mummenhoff et al., 2001, 2004).
Interestingly, dormancy variation in Arabidopsis populations is associated with a series of functionally distinct alleles showing InDel variations in intron 1 that segregate at the AtDOG1 locus, which probably reflects local environmental adaptation (Kronholm et al., 2012). Although this does not generate any causal relationship, it demonstrates that for the interspecies comparison, a combination between exon sequence conservation (suggesting common function) and intron divergence (suggesting local environmental adaptation and/or speciation) is evident as well. Interestingly, a proximal 150-bp region of the DOG1 gene promoter is conserved in sequences between LesaDOG1 and LepaDOG1d with no exon 1 but only exon 2 homologous sequences being present, indicating a kind of exon skipping by genomic rearrangement (Fig. 5A). A start codon (ATG) at the beginning of this exon 2-similar region in LepaDOG1d in frame with the coding region of LesaDOG1 suggests that a protein could be translated from this gene consisting only of the C-terminal part of a DOG1 protein. We do not know if this gene is expressed, but we have evidence that transcripts of LepaDOG1a and LepaDOG1c are expressed in L. papillosum seeds. In contrast to Arabidopsis, where different transcript splice forms may give rise to different DOG1 protein variants from the single AtDOG1 gene (Bentsink et al., 2006), we have no evidence for alternative splicing of LepaDOG1a, LepaDOG1c, LesaDOG1, or BrDOG1 (Graeber et al., 2010; this work). However, providing that the LepaDOG1a and LepaDOG1c transcripts are translated, at least two similar DOG1 proteins are expressed in L. papillosum seeds. It is tempting to speculate that in L. papillosum, the two LepaDOG1a and LepaDOG1c proteins provide functional roles in dormancy regulation that are fulfilled by different splice variants of the single AtDOG1 gene (Bentsink et al., 2006; Nakabayashi et al., 2012) in the shrunken genome of Arabidopsis, which is characterized by extensive gene loss (Thomas et al., 2006; Oyama et al., 2008). Furthermore, polyploidization can form the basis for neofunctionalization or subfunctionalization of multiplied and retained genes (homeologs; Erdmann et al., 2010; Liu and Adams, 2010; Franzke et al., 2011), providing a mechanism for potentially diversified or more complex functions of the individual LepaDOG1 proteins in a putatively conserved dormancy mechanism. As outlined above, we propose that L. papillosum evolved by autopolyploidization or allopolyploidization from the very closely related L. oxytrichum (or its progenitor). Comparative analysis of the DOG1 genes between the diploid (based on x = 12) L. oxytrichum (2n = 24) and the tetraploid L. papillosum (2n = 48) would thus be an interesting future research project.
Nakabayashi et al. (2012) very recently demonstrated that AtDOG1 proteins accumulate during Arabidopsis seed maturation and dormancy induction. The onset of this accumulation at approximately 12 DAP is followed by a continuous increase until the mature dry-seed state. During subsequent dry seed storage, the AtDOG1 protein content seems to decline. Protein and transcript expression patterns of AtDOG1 differ, since transcript accumulation peaks during maturation and (already) declines toward maturity. The AtDOG1 proteins are localized in the nuclei of the embryo, and their contents in fresh mature dry seeds correlated with the observed effect on dormancy and after-ripening. Furthermore, low temperature during seed maturation caused increased AtDOG1 protein content and deeper dormancy. We show here that LepaDOG1 proteins accumulate during L. papillosum seed maturation. The well-ordered L. papillosum infructescences (types I–III) combined with germinability assays and marker protein expression patterns (see the first section of “Discussion”) allowed us to pinpoint LepaDOG1 protein accumulation and relate it to the effects on germinability and dormancy induction. The onset of LepaDOG1 protein accumulation occurred prior to the onset of germinability of the population (see FO6/FO7 in type I and FO7 in type II infructescences; Fig. 7) and prior to the onset of storage lipid accumulation (oleosin marker protein) during early seed maturation. LepaDOG1 protein strongly accumulated during early seed maturation in seed populations that were in the phase of acquiring their premature germinability (compare LepaDOG1 protein contents in type I [FO5/FO6] and type II [FO6/FO7] infructescences with the patterns in germinability; Fig. 7). An initial decline in LepaDOG1 protein contents occurred prior to the induction of dormancy in FOs that still had full premature germinability (see type I FO1/FO2 and type II FO3/FO4; Fig. 7), and a further decline was associated with dormancy induction, as evident in mature FOs of type II and III infructescences.
Therefore, although the onset of DOG1 protein expression in early seed maturation seems similar in L. papillosum (this work) and Arabidopsis (Nakabayashi et al., 2012), the peak of LepaDOG1 protein is in physiological terms earlier and the decline already during mid maturation is faster, which rather resembles AtDOG1 transcript more than protein accumulation patterns. Most interestingly, while in Arabidopsis the AtDOG1 contents remained high until late seed maturation and were associated with the dormancy level of freshly harvested seeds (Nakabayashi et al., 2012), in L. papillosum (this work) the LepaDOG1 protein peak was associated with the period of highest seed premature germinability, and the content declined prior to the induction of primary dormancy. In agreement with LepaDOG1 being present in seeds with high germinability, it was shown that in Arabidopsis, available AtDOG1 in imbibed mature nondormant seeds does not inhibit germination (Nakabayashi et al., 2012). Therefore, we speculate that, in general, the expression of DOG1 during a defined phase of seed maturation is critical for the subsequent induction of factors mediating primary seed dormancy.
Reasons for the observed species-specific differences in DOG1 protein expression patterns are not known, and we can only speculate here. One important point is that, although both species have physiological primary coat dormancy and the DOG1 proteins may have a conserved role in inducing this dormancy, they are not the only key players in these effects. Brassicaceae spp. differ, for example, in their requirement for GA treatment to induce premature germination, as outlined above. Arabidopsis and L. papillosum also differ in their requirements for seed dormancy release by after-ripening storage and/or cold stratification: here, the combination of both means for L. papillosum is required (this work), whereas either of these means alone releases Arabidopsis seed dormancy (Holdsworth et al., 2008). Although key players like DOG1 may be conserved in their function and downstream mechanisms, the details related to the developmental expression and the relative importance of several factors might differ between ecotypes and species. Like primary dormancy, also secondary dormancy induction and release during dormancy cycling of Arabidopsis in the soil seed bank are associated with the temperature-mediated regulation of DOG1 expression (Footitt et al., 2011). This work shows that AtDOG1 may be involved in a thermal-sensing mechanism that influences the dormancy level by altering the sensitivity to abscisic acid. On the other hand, during seed maturation and imbibition, GAs may be more important in the DOG1-mediated thermal-sensing mechanism (Kendall et al., 2011; Nakabayashi et al., 2012). The presence of the DOG1 genes and the DOG1-associated seed dormancy mechanism(s), therefore, may be evolutionarily conserved among the Brassicaceae, but their evolution during speciation and local environmental adaptation may constitute a variation on the same theme. It seems, therefore, that as with “the class” of physiological dormancy itself, different paths lead to success. Further cross-species work with several other Lepidium and Brassicaceae species will help to elucidate how DOG1-mediated mechanisms regulate the sensitivities of germination timing to environmental factors and contribute to local environmental adaptation.
MATERIALS AND METHODS
Plant Material and Germination Assays
Lepidium spp. seed accessions were obtained from the sources described in Supplemental Table S1. Lepidium papillosum plants were grown in a phytochamber (16/8-h photoperiod, approximately 115 μmol m−2 s−1, and 22°C/18°C) or greenhouse in Freiburg (from July to November with subsequent seed after-ripening on mother plants for 12 months) with no supplemental lighting or temperature control. Seeds were either harvested from completely dried whole infructescences of greenhouse- or phytochamber-grown plants (as indicated) or from developing infructescences categorized into type I to III (see “Results” and figure legends) of phytochamber-grown plants. Developing infructescences were divided into consecutive FOs (see “Results” and figure legends), and each FO was separated into 16 fruit halves (each containing one seed). For germination analysis, isolated seeds were placed onto 1% (w/v) BactoAgar (Becton-Dickinson) with one-tenth-strength (w/v) Murashige and Skoog salts (Duchefa) and 0.1% (v/v) Plant Preservative Mixture (Plant Cell Technology) added in 9-cm petri dishes and were incubated at 24°C in a Sanyo Versatile Environmental Test Chamber (MLR-350) in continuous white light (approximately 75 µmol m−2 s−1). The percentage radicle emergence of the seed populations was scored over time using a binocular microscope (Leica DCF480).
Methods for Species Verification and L. papillosum Genome Analysis
4′,6-Diamino-2-phenylindole-stained mitotic chromosome spreads from L. papillosum flower bud tissue were used for chromosome counts following the protocol of Dierschke et al. (2009). Methods for sequence analysis of ITS of nuclear ribosomal DNA were as described earlier (Bowman et al., 1999; Mummenhoff et al., 2001).
For Southern-blot analysis, a digoxigenin (DIG)-labeled probe covering 353 bp of exon 1 of LesaDOG1 was amplified with primers LesaDOG1a-FP3-wgDNA and GSP-3 (Supplemental Table S2) from 20 pg of pGEM-T Easy vector (Promega) containing the genomic sequence of LesaDOG1 using the PCR DIG Probe Synthesis Kit (Roche) according to the manufacturer’s instructions. The probe was purified through a cellulose acetate syringe filter (Whatman). Seven micrograms of L. papillosum genomic DNA extracted from leaves using the Plant DNeasy Maxi Kit (Qiagen) was digested with EcoRI or XbaI (New England Biolabs) overnight and subsequently run on a 0.8% (w/v) agarose gel together with DIG-labeled DNA Mr marker VII (Roche). DNA was transferred onto Nytran Supercharge membranes (Whatman) using a TurboBlotter (Whatman). DIG-labeled probe (4 µL per mL of hybridization solution DIG Easy Hyb [Roche]) was hybridized to the membrane at 41.5°C for 18 h followed by high-stringency washes in 0.5× SSC + 0.1% (w/v) SDS at 65°C. After incubation with Blocking Reagent (Roche), hybridization signal was detected using alkaline phosphatase-coupled anti-DIG antibody (Roche) and CDP-Star (Roche) chemiluminescent substrate according to the manufacturer’s instructions on ECL Hyperfilm (Amersham).
PCR-Based Cloning of Lepidium DOG1 Genomic DNA and cDNA Fragments
Lepidium spp. DOG1 fragments were PCR amplified from genomic DNA or RNA extracted from seeds. RNA was extracted as described by Graeber et al. (2011) and subsequently cDNA synthesized as described by Graeber et al. (2010), but using random hexamers (Fermentas). DNA extraction from seeds using commercial extraction kits can be difficult due to clogging of membranes by seed storage compounds and mucilage. Therefore, genomic DNA from seeds was extracted by the following protocol: mix 50 mg of seeds ground in liquid N2 with 500 µL of preheated cetyltrimethylammonium bromide buffer (100 mm Tris, pH 8, 20 mm EDTA, 1.4 m NaCl, 2% [w/v] cetyltrimethylammonium bromide, and 1% [w/v] polyvinylpyrrolidone 40000) and incubate 30 min at 65°C. Mix with 750 μL of 12:12:1 phenol:chloroform:isoamyl alcohol, centrifuge 2 min at 11,000 rpm, and transfer upper phase. Mix 750 μL of 24:1 chloroform:isoamyl alcohol with upper phase, centrifuge 2 min at 11,000 rpm, and repeat step. Mix upper phase with 1 µL of RNase A (Invitrogen) and incubate 30 min at 37°C. Add 500 µL of isopropanol, incubate 15 min at room temperature, and centrifuge 15 min at 13,000 rpm. Wash pellet in cold 70% ethanol, dry, and resuspend in 100 µL of water. Primer sequences (Supplemental Tables S1 and S2) used for cloning Lepidium spp. genomic DOG1 fragments were based on Lepidium sativum DOG1 sequence (Graeber et al., 2010) and designed to match conserved positions according to a sequence alignment of AtDOG1, BrDOG1, and LesaDOG1. Primers used for L. papillosum cDNA cloning were based on the obtained genomic L. papillosum DOG1 sequences.
L. sativum DOG1 Recombinant Protein Expression
LesaDOG1 coding sequence was PCR amplified from cloned full-length LesaDOG1 cDNA (Graeber et al., 2010) using LesaDOG1-FP1-GW (containing a Shine-Dalgarno sequence and a Kozac sequence) and LesaDOG1-RP1-GW primers and subsequently cloned into the Gateway-compatible pCR8/GW/TOPO vector (Invitrogen). Using the Gateway cloning system (Invitrogen) following the manufacturer’s instructions, LesaDOG1 coding sequence was introduced into a modified pDEST15 vector (Invitrogen) containing a TEV protease recognition site (ENLYFQG) between the GST coding sequence and the in-frame insertion point. When translated, this results in a predicted 533-amino acid-containing GST-Linker(TEV)-LesaDOG1 fusion protein of 61.1 kD [GST, 25.7 kD; Linker(TEV), 3.9 kD; LesaDOG1; 31.5 kD; Supplemental Fig. S2).
This plasmid was transformed in Escherichia coli BL21-CodonPlus(DE3)-RIL cells (Stratagene). A single positive clone was used to grow a liquid culture, and protein expression was induced with 1 mm isopropylthio-β-galactoside. The recombinant fusion protein was strongly expressed after 3 h (Supplemental Fig. S4A). Cells were pelleted by centrifugation, frozen at −20°C, and thawed in lysis buffer (1.25 m Tris, pH 9, one tablet per 20 mL of Protease Inhibitor Complete Mini [Roche], 53 units mL−1 DNase I [Qiagen], 5 units mL−1 RNase A [Fermentas], 1 mm dithiothreitol, and 1 mm EDTA) two times. The cells were lysed using a French pressure cell press (Aminco) and subsequent sonication three times for 2 min each using Sonopuls GM70 (Bandelin). The lysate was mixed with 1% (v/v) Triton X-100 and centrifuged. From the supernatant, GST-LesaDOG1 fusion protein was purified using Glutathione-Sepharose 4B Matrix (Amersham Bioscience) according to the manufacturer’s instructions in two successive rounds. Therefore, fusion protein was bound to the matrix overnight at 4°C, washed, and the eluted protein from this first purification round was dialyzed in phosphate-buffered saline using Slide-A-Lyzer Dialysis Cassette 20K MWCO (Pierce Protein Research Products, Thermo Fisher Scientific) overnight at 4°C to establish optimal binding conditions for the second purification round. TEV cleavage of the GST-LesaDOG1 fusion protein was performed using AcTEV Protease (Invitrogen) overnight at 16°C, while fusion protein was bound to matrix during the second purification round. To analyze purification and TEV cleavage efficiency, fractions of matrix (i.e. matrix-bound protein) and supernatant (i.e. unbound protein) were analyzed by SDS-PAGE (Supplemental Fig. S4B), and gels were stained with PageBlue Protein Staining Solution (Fermentas).
Sequence Alignments and Molecular Phylogenetic Analyses
The bioinformatic software suite Geneious Pro 5.5.8 (Biomatters) was used for sequence analysis and representation. Pairwise and multiple global DNA sequence alignments were generated using MAFFT version 6.814b (Katoh et al., 2002) using the E-INS-i strategy. Full-length putative amino acid sequences of LepaDOG1 proteins were generated based on corresponding genomic DNA and cDNA fragments (Supplemental Table S1). For phylogenetic analysis, these full-length LepaDOG1 proteins were aligned with DOG1 protein sequences from Arabidopsis (Arabidopsis thaliana) Cape Verde Island (α-splice variant; GenBank accession no. EF028466), Brassica rapa (Bra025050, corresponding to the seed-expressed partial cDNA with GenBank accession no. GQ411194), L. sativum (GenBank accession no. GQ411192), and DOG1-Like1 (At4g18660; the most similar protein to AtDOG1 in Arabidopsis) using MUSCLE (Edgar, 2004) with default settings. The multiple sequence alignment was manually corrected and analyzed using the ProtTest Web server (Abascal et al., 2005) to determine the best-fitting substitution model for evolutionary determination. JTT substitution matrix and allowing rate heterogeneity among sites (+G) was selected as the best-fitting model, and a maximum-likelihood tree was constructed from the alignment using PhyML (Guindon and Gascuel, 2003) with the suggest model JTT+G with four substitution rate categories for the γ distribution and the γ distribution parameter being estimated from the data set and 200 bootstrap replicates. The tree was midpoint rooted. Dot-blot analysis was performed using Emboss dotmatcher software.
Immunoblot Analyses of L. papillosum Protein Extracts
In the case of simultaneous germination and western analysis of developing infructescences, 50% of all fruit halves of each FO was used for germination assays and 50% for protein extraction. Therefore, they were placed into reaction vessels with ceramic beads, frozen in liquid nitrogen, and subsequently homogenized two times for 15 s each at 6,500 rpm using a Precellys Tissue Homogenizer (Peqlab). Extraction buffer (0.1 m Tris, pH 8, 6 m urea, 2 m thiourea, 0.2% [v/v] Triton X-100, 0.2% [v/v] sarcosyl, and 2 mm dithiothreitol), 400 µL per FO, was added to the frozen sample. Thawed samples were incubated by overhead shaking (30 rpm) for 30 min at 4°C. Two times the homogenate was centrifuged (10 min, 13,000 rpm, 4°C), and the supernatant was transferred into a new reaction vessel. The clear extract of soluble proteins was stored at −20°C. Protein contents were determined using the Bio-Rad Protein Assay Kit with bovine γ-globulin as a standard. Generally, 60 µg of L. papillosum protein was separated by SDS-PAGE together with the molecular mass ladder Precision Plus Protein Standard Dual Color (Bio-Rad) using the Mini-Protean 3 Cell System (Bio-Rad) and blotted with Bjerrum transfer buffer (48 mm Tris, pH 9.2, 39 mm Gly, 20% [v/v] methanol, and 0.0375% [w/v] SDS) using a Trans-Blot SD Semi-Dry blotter (Bio-Rad) onto a polyvinylidene difluoride Hybond-P membrane (Amersham) at a constant 25 V for 1 h. Homogenous transfer was validated by staining the membrane with Ponceau S.
Membranes were blocked in 2% (w/v) bovine serum albumin (or 2% [w/v] gelatin in the case of the α-dehydrin antibody) in phosphate-buffered saline, 0.1% (v/v) Tween-20. For immunological detection, the following primary antibodies were used: two different polyclonal antibodies, α-DOG11 (Schwab, 2008) and α-DOG12 (Nakabayashi et al., 2012), directed against AtDOG1; a monoclonal antibody, α-oleosine, directed against Arabidopsis D9 oleosin (Crowe et al., 2000; Siloto et al., 2006); a polyclonal antibody, α-EM6, directed against Medicago truncatula Em6 (Boudet et al., 2006); and a polyclonal antibody, α-dehydrin, directed against a wide range of plant species dehydrins (Close et al., 1993). Anti-rabbit IgG-peroxidase conjugate (Sigma) was used for the detection of α-DOG11, α-DOG12, α-EM6, and α-dehydrin, and anti-mouse IgG-peroxidase conjugate (Promega) was used for the detection of α-oleosine using ECL Plus Western Blotting Detection reagents (Amersham) following the manufacturer’s instructions on ECL Hyperfilm (Amersham), except for the immunoblot shown in Figure 6B, in which nitroblue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate-based detection was used.
Lepidium spp. full-length and partial genomic DOG1 sequence data have been deposited in GenBank with the following accession numbers: JX512181 (Lepidium campestre), JX512177 (Lepidium africanum), JX512179 (Lepidium pseudohyssopifolium), JX512180 (Lepidium ruderale), JX512178 (Lepidium bonariense), JX512176 (Lepidium didymum), JX512182 (Lepidium leptopetalum), JX512183 (L. papillosum DOG1a), JX512184 (L. papillosum DOG1b), JX512185 (L. papillosum DOG1c), JX512186 (L. papillosum DOG1d), as well as partial cDNA sequences JX512187 (L. papillosum DOG1a) and JX512188 (L. papillosum DOG1c).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Comparison of L. oxytrichum and L. papillosum trichome types.
Supplemental Figure S2. ITS sequence comparison from L. oxytrichum, L. papillosum, and the Lepidium spp. accession Mummenhoff 2415 used in this work.
Supplemental Figure S3. Map of the plasmid used for recombinant GST-LesaDOG1 protein expression.
Supplemental Figure S4. SDS-PAGE analysis of recombinant GST-LesaDOG1 fusion protein expression and purification (LesaDOG1*).
Supplemental Table S1. Putative DOG1 orthologs cloned from diverse Lepidium species.
Supplemental Table S2. Primer sequences used for cloning of putative DOG1 orthologs.
Acknowledgments
We thank Sara Mayland-Quellhorst for chromosome counts in L. papillosum, Ulrike Coja, Andreas Mühlhausen, and Lydia Buller for help with DOG1 cloning and sequencing, Maren Siegrist for help with LesaDOG1 recombinant protein expression, and Lucille Schmieding for correcting style and grammar. We thank Wim Soppe and Kazumi Nakabayashi for generously providing the α-DOG1 antibodies, Julia Buitink for the α-EM6 antibody, Timothy Close for the α-dehydrin antibody, and SemBioSys Genetics (www.sembiosys.com) for the α-oleosin antibody.
Glossary
- QTL
quantitative trait locus
- DAP
d after pollination
- ITS
internal transcribed spacers
- FO
fruit octet
- InDel
insertion/deletion
- GST
glutathione S-transferase
- TEV
tobacco etch virus
- DIG
digoxigenin
- cDNA
complementary DNA
References
- Abascal F, Zardoya R, Posada D. (2005) ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21: 2104–2105 [DOI] [PubMed] [Google Scholar]
- Alboresi A, Gestin C, Leydecker M-T, Bedu M, Meyer C, Truong H-N. (2005) Nitrate, a signal relieving seed dormancy in Arabidopsis. Plant Cell Environ 28: 500–512 [DOI] [PubMed] [Google Scholar]
- Alonso-Blanco C, Aarts MG, Bentsink L, Keurentjes JJ, Reymond M, Vreugdenhil D, Koornneef M. (2009) What has natural variation taught us about plant development, physiology, and adaptation? Plant Cell 21: 1877–1896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Shehbaz IA. (1986) The genera of Lepidieae (Cruciferae; Brassicaceae) in the southeastern United States. J Arnold Arbor Harv Univ 67: 265–311 [Google Scholar]
- Barua D, Butler C, Tisdale TE, Donohue K. (2012) Natural variation in germination responses of Arabidopsis to seasonal cues and their associated physiological mechanisms. Ann Bot (Lond) 109: 209–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentsink L, Jowett J, Hanhart CJ, Koornneef M. (2006) Cloning of DOG1, a quantitative trait locus controlling seed dormancy in Arabidopsis. Proc Natl Acad Sci USA 103: 17042–17047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bies N, Aspart L, Carles C, Gallois P, Delseny M. (1998) Accumulation and degradation of Em proteins in Arabidopsis thaliana: evidence for post-transcriptional controls. J Exp Bot 49: 1925–1933 [Google Scholar]
- Boudet J, Buitink J, Hoekstra FA, Rogniaux H, Larré C, Satour P, Leprince O. (2006) Comparative analysis of the heat stable proteome of radicles of Medicago truncatula seeds during germination identifies late embryogenesis abundant proteins associated with desiccation tolerance. Plant Physiol 140: 1418–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman JL, Brüggemann H, Lee JY, Mummenhoff K. (1999) Evolutionary changes in floral structure within Lepidium L. (Brassicaceae). Int J Plant Sci 160: 917–929 [DOI] [PubMed] [Google Scholar]
- Chiang GCK, Bartsch M, Barua D, Nakabayashi K, Debieu M, Kronholm I, Koornneef M, Soppe WJJ, Donohue K, De Meaux J. (2011) DOG1 expression is predicted by the seed-maturation environment and contributes to geographical variation in germination in Arabidopsis thaliana. Mol Ecol 20: 3336–3349 [DOI] [PubMed] [Google Scholar]
- Close TJ, Fenton RD, Moonan F. (1993) A view of plant dehydrins using antibodies specific to the carboxy terminal peptide. Plant Mol Biol 23: 279–286 [DOI] [PubMed] [Google Scholar]
- Crowe AJ, Abenes M, Plant A, Moloney MM. (2000) The seed-specific transactivator, ABI3, induces oleosin gene expression. Plant Sci 151: 171–181 [DOI] [PubMed] [Google Scholar]
- Dierschke T, Mandáková T, Lysak MA, Mummenhoff K. (2009) A bicontinental origin of polyploid Australian/New Zealand Lepidium species (Brassicaceae)? Evidence from genomic in situ hybridization. Ann Bot (Lond) 104: 681–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donohue K, Rubio de Casas R, Burghardt L, Kovach K, Willis CG. (2010) Germination, postgermination adaptation, and species ecological ranges. Annu Rev Ecol Evol Syst 41: 293–319 [Google Scholar]
- Edgar RC. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann R, Gramzow L, Melzer R, Theissen G, Becker A. (2010) GORDITA (AGL63) is a young paralog of the Arabidopsis thaliana B(sister) MADS box gene ABS (TT16) that has undergone neofunctionalization. Plant J 63: 914–924 [DOI] [PubMed] [Google Scholar]
- Fernandez DE. (1997) Developmental basis of homeosis in precociously germinating Brassica napus embryos: phase change at the shoot apex. Development 124: 1149–1157 [DOI] [PubMed] [Google Scholar]
- Ferrándiz C, Pelaz S, Yanofsky MF. (1999) Control of carpel and fruit development in Arabidopsis. Annu Rev Biochem 68: 321–354 [DOI] [PubMed] [Google Scholar]
- Finch-Savage WE, Leubner-Metzger G. (2006) Seed dormancy and the control of germination. New Phytol 171: 501–523 [DOI] [PubMed] [Google Scholar]
- Footitt S, Douterelo-Soler I, Clay H, Finch-Savage WE. (2011) Dormancy cycling in Arabidopsis seeds is controlled by seasonally distinct hormone-signaling pathways. Proc Natl Acad Sci USA 108: 20236–20241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzke A, Lysak MA, Al-Shehbaz IA, Koch MA, Mummenhoff K. (2011) Cabbage family affairs: the evolutionary history of Brassicaceae. Trends Plant Sci 16: 108–116 [DOI] [PubMed] [Google Scholar]
- Gossmann TI, Schmid KJ. (2011) Selection-driven divergence after gene duplication in Arabidopsis thaliana. J Mol Evol 73: 153–165 [DOI] [PubMed] [Google Scholar]
- Graeber K, Linkies A, Müller K, Wunchova A, Rott A, Leubner-Metzger G. (2010) Cross-species approaches to seed dormancy and germination: conservation and biodiversity of ABA-regulated mechanisms and the Brassicaceae DOG1 genes. Plant Mol Biol 73: 67–87 [DOI] [PubMed] [Google Scholar]
- Graeber K, Linkies A, Wood AT, Leubner-Metzger G. (2011) A guideline to family-wide comparative state-of-the-art quantitative RT-PCR analysis exemplified with a Brassicaceae cross-species seed germination case study. Plant Cell 23: 2045–2063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graeber K, Nakabayashi K, Miatton E, Leubner-Metzger G, Soppe WJ. (2012) Molecular mechanisms of seed dormancy. Plant Cell Environ 35: 1769–1786 [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52: 696–704 [DOI] [PubMed] [Google Scholar]
- Guo YL, Todesco M, Hagmann J, Das S, Weigel D. (2012) Independent FLC mutations as causes of flowering-time variation in Arabidopsis thaliana and Capsella rubella. Genetics 192: 729–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutterman Y. (1980/1981) Influences on seed germinability: phenotypic maternal effects during seed maturation. Isr J Bot 29: 105–117 [Google Scholar]
- Hewson H. (1981) The genus Lepidium L. (Brassicaceae) in Australia. Brunonia 4: 217–308 [Google Scholar]
- Hilhorst HWM, Smitt AI, Karssen CM. (1986) Gibberellin-biosynthesis and -sensitivity mediated stimulation of seed germination of Sisymbrium officinale by red light and nitrate. Physiol Plant 67: 285–290 [Google Scholar]
- Holdsworth MJ, Bentsink L, Soppe WJJ. (2008) Molecular networks regulating Arabidopsis seed maturation, after-ripening, dormancy and germination. New Phytol 179: 33–54 [DOI] [PubMed] [Google Scholar]
- Huang AH. (1996) Oleosins and oil bodies in seeds and other organs. Plant Physiol 110: 1055–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Schmitt J, Dorn L, Griffith C, Effgen S, Takao S, Koornneef M, Donohue K. (2010) The earliest stages of adaptation in an experimental plant population: strong selection on QTLS for seed dormancy. Mol Ecol 19: 1335–1351 [DOI] [PubMed] [Google Scholar]
- Hurka H, Mummenhoff K, Schoppmann C. (1992) Chromosome numbers in the genus Lepidium (Brassicaceae). IOPB Newsletter 18/19: 8–9 [Google Scholar]
- Katoh K, Misawa K, Kuma KI, Miyata T. (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30: 3059–3066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendall SL, Hellwege A, Marriot P, Whalley C, Graham IA, Penfield S. (2011) Induction of dormancy in Arabidopsis summer annuals requires parallel regulation of DOG1 and hormone metabolism by low temperature and CBF transcription factors. Plant Cell 23: 2568–2580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Alonso-Blanco C, Peeters AJM, Soppe W. (1998) Genetic control of flowering time in Arabidopsis. Annu Rev Plant Physiol Plant Mol Biol 49: 345–370 [DOI] [PubMed] [Google Scholar]
- Kronholm I, Picó FX, Alonso-Blanco C, Goudet J, de Meaux J. (2012) Genetic basis of adaptation in Arabidopsis thaliana: local adaptation at the seed dormancy QTL DOG1. Evolution 66: 2287–2302 [DOI] [PubMed] [Google Scholar]
- Kugler I. (1951) Untersuchungen über das Keimverhalten einiger Rassen von Arabidopsis thaliana (L.) Heynh: ein Beitrag zum Problem der Lichtkeimung. Beitr Biol Pflanz 28: 211–243 [Google Scholar]
- Le BH, Cheng C, Bui AQ, Wagmaister JA, Henry KF, Pelletier J, Kwong L, Belmonte M, Kirkbride R, Horvath S, et al. (2010) Global analysis of gene activity during Arabidopsis seed development and identification of seed-specific transcription factors. Proc Natl Acad Sci USA 107: 8063–8070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Mummenhoff K, Bowman JL. (2002) Allopolyploidization and evolution of species with reduced floral structures in Lepidium L. (Brassicaceae). Proc Natl Acad Sci USA 99: 16835–16840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leprince O, Buitink J. (2010) Desiccation tolerance: from genomics to the field. Plant Sci 179: 554–564 [Google Scholar]
- Leubner-Metzger G. (2002) Seed after-ripening and over-expression of class I β-1,3-glucanase confer maternal effects on tobacco testa rupture and dormancy release. Planta 215: 959–968 [DOI] [PubMed] [Google Scholar]
- Linkies A, Müller K, Morris K, Turecková V, Wenk M, Cadman CS, Corbineau F, Strnad M, Lynn JR, Finch-Savage WE, et al. (2009) Ethylene interacts with abscisic acid to regulate endosperm rupture during germination: a comparative approach using Lepidium sativum and Arabidopsis thaliana. Plant Cell 21: 3803–3822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SL, Adams KL. (2010) Dramatic change in function and expression pattern of a gene duplicated by polyploidy created a paternal effect gene in the Brassicaceae. Mol Biol Evol 27: 2817–2828 [DOI] [PubMed] [Google Scholar]
- Matakiadis T, Alboresi A, Jikumaru Y, Tatematsu K, Pichon O, Renou JP, Kamiya Y, Nambara E, Truong HN. (2009) The Arabidopsis abscisic acid catabolic gene CYP707A2 plays a key role in nitrate control of seed dormancy. Plant Physiol 149: 949–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyers BT, Kane NC. (2010) The genetics of adaptation to novel environments: selection on germination timing in Arabidopsis thaliana. Mol Ecol 19: 1270–1272 [DOI] [PubMed] [Google Scholar]
- Mühlhausen A, Polster A, Theissen G, Mummenhoff K. (2010) Evolution of fruit dehiscence in Brassicaceae: examples from Aethionema and Lepidium. Acta Hortic 867: 207–219 [Google Scholar]
- Müller K, Tintelnot S, Leubner-Metzger G. (2006) Endosperm-limited Brassicaceae seed germination: abscisic acid inhibits embryo-induced endosperm weakening of Lepidium sativum (cress) and endosperm rupture of cress and Arabidopsis thaliana. Plant Cell Physiol 47: 864–877 [DOI] [PubMed] [Google Scholar]
- Mummenhoff K, Brüggemann H, Bowman JL. (2001) Chloroplast DNA phylogeny and biogeography of Lepidium (Brassicaceae). Am J Bot 88: 2051–2063 [PubMed] [Google Scholar]
- Mummenhoff K, Linder P, Friesen N, Bowman JL, Lee J-Y, Franzke A. (2004) Molecular evidence for bicontinental hybridogenous genomic constitution in Lepidium sensu stricto (Brassicaceae) species from Australia and New Zealand. Am J Bot 91: 254–261 [DOI] [PubMed] [Google Scholar]
- Mummenhoff K, Polster A, Mühlhausen A, Theissen G. (2009) Lepidium as a model system for studying the evolution of fruit development in Brassicaceae. J Exp Bot 60: 1503–1513 [DOI] [PubMed] [Google Scholar]
- Munir J, Dorn LA, Donohue K, Schmitt J. (2001) The effect of maternal photoperiod on seasonal dormancy in Arabidopsis thaliana (Brassicaceae). Am J Bot 88: 1240–1249 [PubMed] [Google Scholar]
- Murín A. (1986) Karyological study of Slovakian flora. VIII. Acta Facultatis Rerum Naturalium Universitatis Comenianae Botanica 33: 1–16 [Google Scholar]
- Nah G, Chen ZJ. (2010) Tandem duplication of the FLC locus and the origin of a new gene in Arabidopsis related species and their functional implications in allopolyploids. New Phytol 186: 228–238 [DOI] [PubMed] [Google Scholar]
- Nakabayashi K, Bartsch M, Xiang Y, Miatton E, Pellengahr S, Yano R, Seo M, Soppe WJ. (2012) The time required for dormancy release in Arabidopsis is determined by DELAY OF GERMINATION1 protein levels in freshly harvested seeds. Plant Cell 24: 2826–2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooms J, Leon-Kloosterziel KM, Bartels D, Koornneef M, Karssen CM. (1993) Acquisition of desiccation tolerance and longevity in seeds of Arabidopsis thaliana: a comparative study using abscisic acid-insensitive abi3 mutants. Plant Physiol 102: 1185–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyama RK, Clauss MJ, Formanová N, Kroymann J, Schmid KJ, Vogel H, Weniger K, Windsor AJ, Mitchell-Olds T. (2008) The shrunken genome of Arabidopsis thaliana. Plant Syst Evol 273: 257–271 [Google Scholar]
- Raz V, Bergervoet JHW, Koornneef M. (2001) Sequential steps for developmental arrest in Arabidopsis seeds. Development 128: 243–252 [DOI] [PubMed] [Google Scholar]
- Ren CW, Bewley JD. (1999) Developmental and germinative events can occur concurrently in precociously germinating Chinese cabbage (Brassica rapa ssp pekinensis) seeds. J Exp Bot 50: 1751–1761 [Google Scholar]
- Rodríguez MV, Mendiondo GM, Cantoro R, Auge GA, Luna V, Masciarelli O, Benech-Arnold RL. (2012) Expression of seed dormancy in grain sorghum lines with contrasting pre-harvest sprouting behavior involves differential regulation of gibberellin metabolism genes. Plant Cell Physiol 53: 64–80 [DOI] [PubMed] [Google Scholar]
- Ruuska SA, Girke T, Benning C, Ohlrogge JB. (2002) Contrapuntal networks of gene expression during Arabidopsis seed filling. Plant Cell 14: 1191–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schranz ME, Osborn TC. (2004) De novo variation in life-history traits and responses to growth conditions of resynthesized polyploid Brassica napus (Brassicaceae). Am J Bot 91: 174–183 [DOI] [PubMed] [Google Scholar]
- Schranz ME, Song BH, Windsor AJ, Mitchell-Olds T. (2007) Comparative genomics in the Brassicaceae: a family-wide perspective. Curr Opin Plant Biol 10: 168–175 [DOI] [PubMed] [Google Scholar]
- Schwab M (2008) Identification of novel seed dormancy mutants in Arabidopsis thaliana and molecular and biochemical characterization of the seed dormancy gene DOG1. PhD thesis. Universität zu Köln, Cologne, Germany
- Shirai A, Matsuyama A, Yashiroda Y, Hashimoto A, Kawamura Y, Arai R, Komatsu Y, Horinouchi S, Yoshida M. (2008) Global analysis of gel mobility of proteins and its use in target identification. J Biol Chem 283: 10745–10752 [DOI] [PubMed] [Google Scholar]
- Siloto RM, Findlay K, Lopez-Villalobos A, Yeung EC, Nykiforuk CL, Moloney MM. (2006) The accumulation of oleosins determines the size of seed oilbodies in Arabidopsis. Plant Cell 18: 1961–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thellung A. (1906) Die Gattung Lepidium (L.) R. Br. Eine Monographische Studie. Neue Denkschriften der Schweizerischen Naturforschenden Gesellschaft 41: 1–304 [Google Scholar]
- Thomas BC, Pedersen B, Freeling M. (2006) Following tetraploidy in an Arabidopsis ancestor, genes were removed preferentially from one homeolog leaving clusters enriched in dose-sensitive genes. Genome Res 16: 934–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonsor SJ, Alonso-Blanco C, Koornneef M. (2005) Gene function beyond a single trait: natural variation, gene effects, and evolutionary ecology in Arabidopsis thaliana. Plant Cell Environ 28: 2–20 [Google Scholar]
- van Zanten M, Koini MA, Geyer R, Liu Y, Brambilla V, Bartels D, Koornneef M, Fransz P, Soppe WJ. (2011) Seed maturation in Arabidopsis thaliana is characterized by nuclear size reduction and increased chromatin condensation. Proc Natl Acad Sci USA 108: 20219–20224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voegele A, Graeber K, Oracz K, Tarkowská D, Jacquemoud D, Turečková V, Urbanová T, Strnad M, Leubner-Metzger G. (2012) Embryo growth, testa permeability, and endosperm weakening are major targets for the environmentally regulated inhibition of Lepidium sativum seed germination by myrigalone A. J Exp Bot 63: 5337–5350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voegele A, Linkies A, Müller K, Leubner-Metzger G. (2011) Members of the gibberellin receptor gene family GID1 (GIBBERELLIN INSENSITIVE DWARF1) play distinct roles during Lepidium sativum and Arabidopsis thaliana seed germination. J Exp Bot 62: 5131–5147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wang X, Tang H, Tan X, Ficklin SP, Feltus FA, Paterson AH. (2011) Modes of gene duplication contribute differently to genetic novelty and redundancy, but show parallels across divergent angiosperms. PLoS ONE 6: e28150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Kulkarni V, Liu N, Del Carpio DP, Bucher J, Bonnema G. (2010) BrFLC2 (FLOWERING LOCUS C) as a candidate gene for a vernalization response QTL in Brassica rapa. J Exp Bot 61: 1817–1825 [DOI] [PMC free article] [PubMed] [Google Scholar]