Functional transfer of a chloroplast gene gene to the nucleus of Campanulaceae species is a remarkable example of the processes underpinning endosymbiotic evolution.
Abstract
Eukaryotic cells originated when an ancestor of the nucleated cell engulfed bacterial endosymbionts that gradually evolved into the mitochondrion and the chloroplast. Soon after these endosymbiotic events, thousands of ancestral prokaryotic genes were functionally transferred from the endosymbionts to the nucleus. This process of functional gene relocation, now rare in eukaryotes, continues in angiosperms. In this article, we show that the chloroplastic acetyl-CoA carboxylase subunit (accD) gene that is present in the plastome of most angiosperms has been functionally relocated to the nucleus in the Campanulaceae. Surprisingly, the nucleus-encoded accD transcript is considerably smaller than the plastidic version, consisting of little more than the carboxylase domain of the plastidic accD gene fused to a coding region encoding a plastid targeting peptide. We verified experimentally the presence of a chloroplastic transit peptide by showing that the product of the nuclear accD fused to green fluorescent protein was imported in the chloroplasts. The nuclear gene regulatory elements that enabled the erstwhile plastidic gene to become functional in the nuclear genome were identified, and the evolution of the intronic and exonic sequences in the nucleus is described. Relocation and truncation of the accD gene is a remarkable example of the processes underpinning endosymbiotic evolution.
Photosynthetic eukaryotes arose more than a billion years ago through the endosymbiotic association of an α proteobacterium (Margulis, 1970; Gray et al., 1999) and a cyanobacterium with the progenitor of the nucleated cell (Mereschkowsky, 1905; Goksoyr, 1967; Deusch et al., 2008). These proteobacterial and cyanobacterial endosymbionts subsequently evolved into mitochondria and chloroplasts, respectively. This transition from endosymbionts to integrated cytoplasmic organelles involved the loss of nonessential or redundant bacterial genes, the creation of protein import machinery, and extensive functional relocation of genes from the organelle ancestors to the nuclear genome. As a consequence, modern cytoplasmic organellar genomes are much smaller in size compared with their prokaryotic ancestors, even though the spectrum of proteins required for function and biogenesis is not substantially different (Timmis et al., 2004). As an example, the human mitochondrial genome encodes only 37 genes, and most flowering plant plastomes encode only approximately 120 genes compared with several thousand genes in the proposed extant relatives of their bacterial ancestors (Timmis et al., 2004).
The merging of two genomes from different lineages through endosymbiosis not only permitted the functional relocation of ancestral organellar genes to the nucleus but also significantly contributed to eukaryote evolution and adaptation to new ecological niches by combining the different biochemical capabilities encoded by each genome and by providing, through endosymbiotic DNA transfer, a continuous rich source of genetic diversity, new genes, exons, introns, and gene regulatory elements (Martin et al., 2002; Martin and Koonin, 2006; Noutsos et al., 2007). These transfers of DNA from the organelles to the nucleus continue to occur at a surprisingly high frequency (Thorsness and Fox, 1990; Huang et al., 2003; Stegemann et al., 2003; Sheppard et al., 2008). The nuclear copies of extant organelle DNA are referred to as norgs (for nuclear integrants of organelle DNA; Leister, 2005), and they can be further classified by their cytoplasmic organelle origin as either numts (for nuclear integrants of mitochondrial DNA; Lopez et al., 1994) or nupts (for nuclear integrants of plastid DNA; Timmis et al., 2004). The fate of nupts is variable, with some being lost within a single generation (Sheppard and Timmis, 2009) while others remain in the nucleus for millions of years and evolve neutrally in a nucleus specific manner (Rousseau-Gueutin et al., 2011a, 2012).
Organellar genes are usually nonfunctional after transfer to the nuclear environment, as they require the acquisition of nuclear gene regulatory elements to become active and a target peptide-encoding sequence if the protein is to be targeted back to the organelle. This process, which has occurred over long evolutionary time periods, has been partially reconstructed experimentally, and some of the molecular mechanisms responsible for these rare events have been described (Stegemann and Bock, 2006; Lloyd and Timmis, 2011; Fuentes et al., 2012). Presumably the activity of organellar genes functionally transferred to the nucleus will be duplicated for a certain period of time with functional genes existing in both genetic compartments of the cell, until one becomes defunct by chance mutation (Adams et al., 1999). In the case of the functional transfer of a chloroplastic gene to the nucleus, the retention of the plastidic copy is usually favored (Rousseau-Gueutin et al., 2012). However, functional gene transfer can occur repeatedly, and eventually the loss of functionality of the plastidic copy results in permanent nuclear residence, since no reciprocal exchange of genetic material between the plastome and the nucleus has ever been observed. This one-way mechanism is referred to as a “gene ratchet” (Doolittle, 1998).
In animals, the functional relocation of mitochondrial genes to the nucleus appears to have stopped but it is still occurring in plants, particularly in the angiosperms, where the molecular mechanisms of activation and further evolution are uniquely amenable to study. Most of the discovered recent functional gene relocations in angiosperms have involved transfer of mitochondrial genes to the nucleus (Liu et al., 2009), with only a few plastid examples reported (Gantt et al., 1991; Millen et al., 2001; Cusack and Wolfe, 2007; Ueda et al., 2007; Magee et al., 2010; Rousseau-Gueutin et al., 2011b). However, with the recent availability of more than a hundred angiosperm plastome sequences, it has become apparent that several genes have been lost recently in various fully photosynthetically competent lineages (Magee et al., 2010), suggesting their functional relocation to the nucleus. Included in this relocation process is the acetyl-CoA carboxylase subunit (accD), which has been lost independently from the plastomes of some angiosperm families: Acoraceae (Goremykin et al., 2005), Campanulaceae (Haberle et al., 2008), Fabaceae (Magee et al., 2010), Geraniaceae (Guisinger et al., 2008), and Poaceae (Konishi and Sasaki, 1994; Martin et al., 1998). In those species, it is expected that an alternative version of accD of eukaryotic or prokaryotic origin will exist in their nuclear genomes to carry out fatty acid biosynthesis in the chloroplast since knockout experiments of plastid accD (pt-accD) in tobacco (Nicotiana tabacum) showed that it is an essential gene (Kode et al., 2005). In addition, several lines of evidence suggest that expression of accD is indispensable during embryo development in Arabidopsis (Arabidopsis thaliana; Bryant et al., 2011). In the Campanulaceae, which lack accD from their plastome, it was observed that prokaryotic acetyl-CoA carboxylase (ACCase) proteins were nevertheless still present in protein extracts from chloroplasts, as in all flowering plants except the Poaceae family (Konishi et al., 1996). These results indicate that chloroplastic accD must have been functionally transferred from the chloroplast to the nucleus in that family.
Here, we report the identification in Trachelium caeruleum (Campanulaceae) of a chimeric nuclear accD (n-accD) of chloroplast origin that encodes an abridged version of the protein. The entire n-accD transcript encodes only a target peptide fused to the carboxylase domain of the plastidic accD gene. Evidence is provided to show that this nuclear gene has functionally replaced the plastidic gene in T. caeruleum. We also provide substantial insights into the acquisition of functionality of the plastidic gene in the nuclear genome and on its subsequent nuclear evolution in this new genetic compartment. Finally, we discuss the genetic changes that may have facilitated its loss from the plastome and its functional relocation to the nucleus in a few plant families during angiosperm evolution.
RESULTS
accD Has Been Functionally Transferred from the Chloroplast to the Nucleus in T. caeruleum by Acquiring Nuclear Gene Regulatory Elements
A comparison of approximately a hundred angiosperm plastome sequences showed that accD was defunct and often completely missing in species belonging to the Acoraceae, Campanulaceae, Fabaceae, Geraniaceae (two independent losses), and Poaceae, suggesting at least six independent losses of the plastidic accD gene, consistent with previous reports (Jansen et al., 2007). Since knockout experiments in tobacco have shown that it is an essential gene (Kode et al., 2005), it is likely that accD has been functionally replaced by a eukaryotic or prokaryotic-like version in the nuclear genome. In a study of the presence and absence of a prokaryotic type and a eukaryotic type of ACCase in 28 plant families (including Campanulaceae), it was observed that all plant families (with the exception of Poaceae) contained a prokaryotic ACCase in the protein extracts of plastids (Konishi et al., 1996). However, from the comparison of 23 Asterid plastome sequences, it was observed that the plastidic accD gene was missing in the Campanulaceae T. caeruleum (Haberle et al., 2008). Its absence in the plastome of a Campanulaceae species (T. caeruleum) and its presence in the closely related Asteraceae species (Guizotia abyssinica, Helianthus annuus, and Lactuca sativa) suggest a relatively recent loss of this plastid gene following the divergence of these two families. The presence of a prokaryotic type ACCase in the protein extracts of Campanulaceae plastids (Konishi et al., 1996) further indicates that the accD gene must have been functionally transferred from the chloroplast to the nucleus in that family.
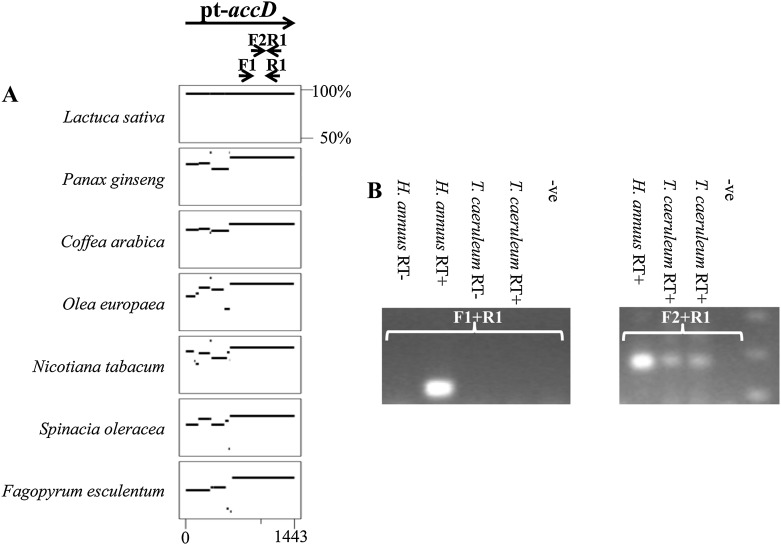
Comparison of pt-accD sequences (Fig. 1A) indicated that the last 250 amino acids encoded by this gene are highly conserved among Asterids. This C-terminal region encodes a carboxylase domain, which is the only known functional domain of the accD protein (Zhang et al., 2003). One of the primer pairs designed to part of this conserved region amplified a transcribed sequence using poly(A) primed complementary DNA (cDNA) from T. caeruleum (Fig. 1). The sequence of this 128-bp product does not correspond to any region of T. caeruleum plastome and shows 83% to 84% nucleotide identity to a region of the carboxylase domain of Asteraceae pt-accD genes. These sequence data suggest that a nupt encoding part of accD gene is actively transcribed and polyadenylated in T. caeruleum. The entire sequence of this putative n-accD transcript was obtained by RACE-PCR. The putative n-accD gene encodes a protein of 331 amino acids, compared with the approximately 500 amino acids encoded by the accD gene in the plastomes of Asterids (Fig. 2). The paucity of nuclear sequence data for Asterids precludes unequivocal characterization of the border between plastid-like and preexisting nuclear sequence in this transcript. However, we found that 75 residues at the N terminus of the nuclear-encoded protein show low similarity (e-value approximately equal to 10−6; 40% amino acid sequence similarity) to the middle of the intron-less 3-ketoacyl-acyl carrier protein synthase I (KAS I) gene from Vitis vinifera and that the 235 amino acids encoded at the 3′ end of the T. caeruleum n-accD transcript are 69% similar to the accD carboxylase domain encoded by Asteraceae pt-accD genes.
Figure 1.
Amplification of a partial nuclear accD (n-accD) transcript that encodes a conserved region of the pt-accD gene in T. caeruleum. A, Multiple percentage identity plots of pt-accD gene sequences from Asterid species compared with an H. annuus pt-accD reference sequence. The percentage identity (50% to 100% scale to right of L. sativa plot) of each gap-free aligning sequence is indicated. Two primer pairs (Accd-Aster-F1 and Accd-Aster-R1; AccD-Aster-F2 and AccD-Aster-R1) specific for the conserved 3′ end of the pt-accD gene were used for amplification of n-accD transcripts from T. caeruleum. B, Ethidium bromide-stained agarose gels showing RT-PCR amplification products from cDNA of T. caeruleum. An n-accD transcript was amplified from T. caeruleum cDNA using a primer pair specific for the conserved 3′ end of Asterid pt-accD genes (F2 and R1), but not by a second primer pair (F1 and R1) specific for a larger region of pt-accD sequence. Annealing sites of these primers are indicated (A). H. annuus cDNA was used as a positive control for each primer pair.
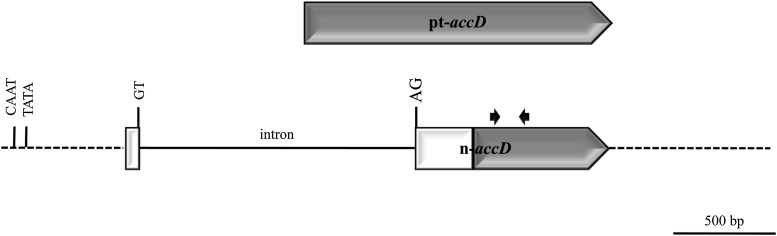
Figure 2.
Comparison of the plastidic accD gene present in most angiosperms with the nuclear accD gene from T. caeruleum. Protein-coding and noncoding regions are indicated by bars and lines, respectively. A region of the T. caeruleum n-accD gene with homology to the pt-accD ORF is shown as a shaded region in the n-accD sequence. Coding regions of n-accD gene of presumed nuclear origin are shown as white boxes. The intron present in n-accD and its consensus splice site sequences are indicated. The positions of putative CAAT and TATA boxes are also indicated. Annealing sites of primers accD-Aster-F1 and accD-Aster-R1 are represented by arrows. The scale bar indicates 500 bp.
To become functional in the nucleus, the chloroplast-derived accD gene must acquire a promoter, a transit peptide-encoding sequence to import the cytoplasmic protein back into the chloroplast and appropriate RNA processing motifs (Bock and Timmis, 2008). Thermal asymmetric interlaced (TAIL)-PCR allowed the characterization of 584 bp of genomic DNA (gDNA) 5′ of the translation initiation site of the transcript and 793 bp 3′ of the stop codon (Fig. 2). Putative eukaryotic TATA and CAAT boxes were identified 431 bp and 493 bp 5′ of the predicted translation start site of the gene, respectively. The sequence downstream of the open reading frame (ORF) showed no plastid sequence similarity, indicating that it is likely nuclear in origin. A chloroplastic target peptide-encoding sequence was predicted at the 5′ end of the nuclear protein (Supplemental Table S1) by using five different softwares.
To detect the presence of possible intronic sequences within T. caeruleum n-accD, the gene was amplified from gDNA. A single intron of 1.4 kb, which did not present any similarity to any Kas1 sequences, was identified. It bisects the 64th and 65th bp of the ORF and interrupts the target peptide-encoding sequence. It contains splice site sequences and a branch point (YNYYRAY) near the 3′ end of the intron, consistent with efficient splicing (Konarska et al., 1985; Roy and Gilbert, 2006).
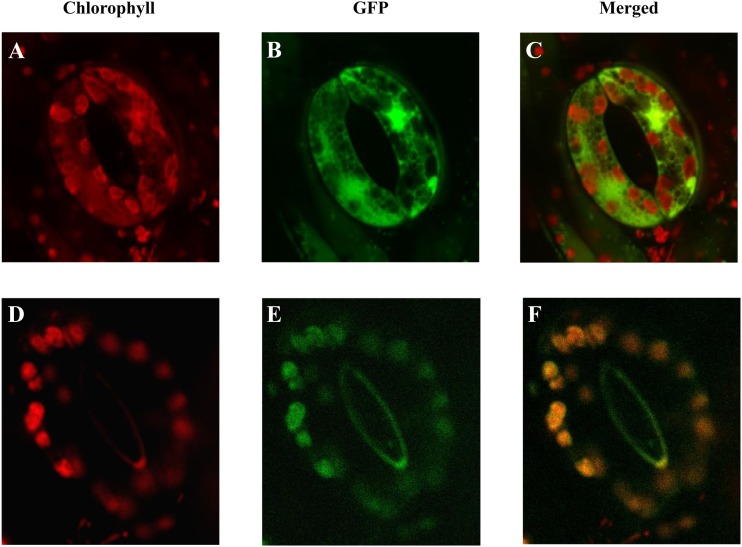
To verify experimentally the existence of the predicted transit peptide, the sequence encoding the majority of the n-accD protein was fused in frame to a GFP gene (GFP) and the subcellular location of the n-accD-GFP proteins was determined. A control construct without any accD sequence (P35S:GFP) was used to verify that no vector sequences adjacent to the GFP gene could encode a cryptic target peptide. Stable tobacco lines transformed with the control construct showed GFP fluorescence in the cytoplasm of leaf guard cells, whereas transgenic plants expressing the fusion construct showed clear plastid localized GFP fluorescence (Fig. 3). These results confirm that the amino acid sequence at the N terminus of n-ACCD acts as a chloroplastic target peptide.
Figure 3.
The T. caeruleum n-accD encodes a plastid target peptide-encoding sequence. Confocal laser scanning microscopy of guard cells from tobacco plants transformed with either a GFP gene (A, B, and C: P35S:GFP construct) or with part of the n-accD ORF (first 222 amino acids of the ORF) fused in frame to the 5′ end of the GFP gene (D, E, and F: pTc-accD:GFP construct). A and D, Chlorophyll fluorescence. B and E, GFP fluorescence. C and F, Merged images of A + B and D + E, respectively. These images demonstrate that the n-ACCD-GFP fusion proteins are targeted to the plastids contained in these tobacco guard cells.
accD Has Been Functionally Transferred to the Nucleus in Campanulaceae Species
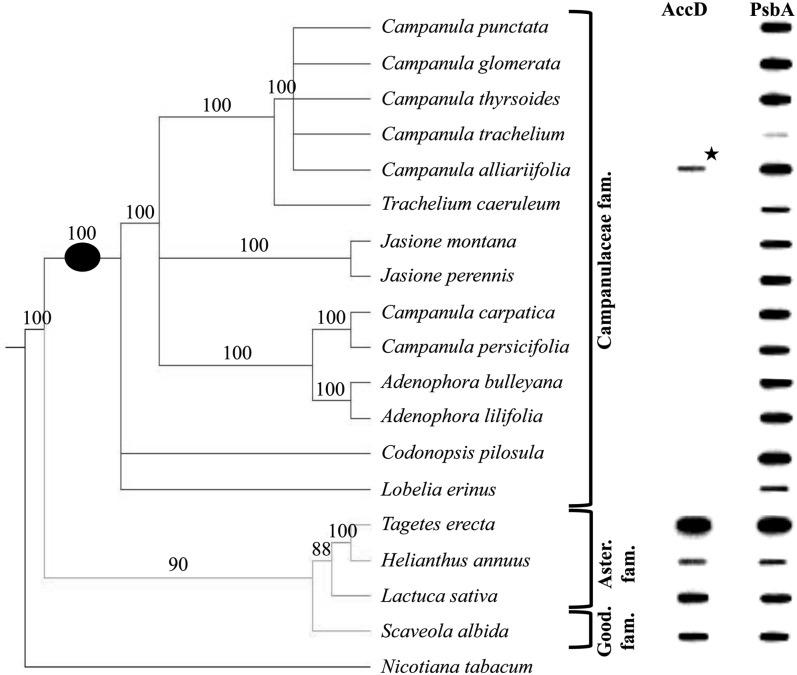
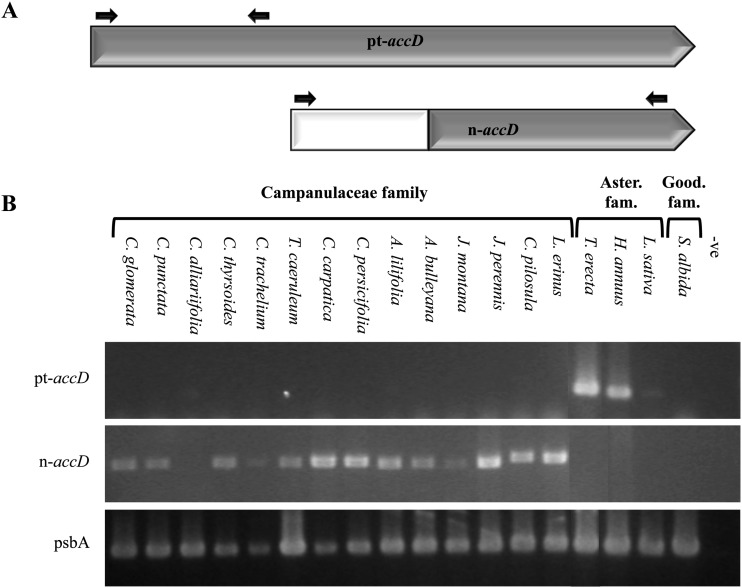
Plastome sequencing studies have revealed that pt-accD is present in three Asteraceae species (G. abyssinica, H. annuus, and L. sativa) but absent in T. caeruleum (Campanulaceae) plastome due to DNA rearrangements (Timme et al., 2007; Haberle et al., 2008; Dempewolf et al., 2010). To estimate more precisely the evolutionary timing of the loss of this pt-accD gene, slot-blot hybridization of total cellular DNA was undertaken from 18 Asterales species (Fig. 4). This method allows inference of gene location because of the high copy number of plastomes compared with the low copy number of nuclear genomes per cell. Thus a strong signal is obtained if the gene is located in the chloroplast, whereas no (or weak) signal is obtained if the gene is in the nucleus. The psbA (ubiquitous plastidic gene) and pt-accD probes produced similar level of hybridization for both probes in all the Goodeniaceae and Asteraceae species investigated, whereas little hybridization of the pt-accD gene compared with psbA was detected in any Campanulaceae tested. These data suggest that pt-accD was lost from the plastome of these latter species near the time of divergence of the Asteraceae and Campanulaceae. A very weak but still significant pt-accD hybridization (10% of hybridization signal) was observed in Campanula alliariifolia. However, its origin was not further investigated because it was considered as a pseudogene.
Figure 4.
Absence of accD in Campanulaceae plastomes. Total DNA from 14 Campanulaceae, three Asteraceae, and one Goodeniaceae species was subjected to slot-blot DNA hybridization using an accD probe and a plastidic psbA probe (control) obtained using H. annuus gDNA as template. The phylogenetic relationships of the species used in this analysis, based on ribosomal RNA ITS1 and ITS2 sequences and a Neighbor Joining analysis, are shown on the left. The absence of strong hybridization in Campanulaceae suggests that this gene is no longer present in the high copy number plastomes of these species. The only exception is in C. alliariifolia (highlighted with an asterisk) where a weak accD hybridization (10%) was observed. A black circle on the phylogenetic tree indicates the evolutionary time point when the putative pt-accD deletion event occurred. A single sample of triplicate loadings is presented.
The loss of pt-accD in the Campanulaceae implies the presence of a functional nuclear copy since the gene is essential (Kode et al., 2005; Bryant et al., 2011). Among Asterales species that still contain a functional pt-accD, it is possible that some may also express a functional n-accD if evolutionary relocation is at an intermediate stage. To determine if some species present the plastidic as well as the nuclear copy, reverse transcription (RT)-PCR with primer pairs specific to each copy were produced (Fig. 5A). RT-PCR was undertaken on leaf mRNA from the same 18 Asterales species (14 Campanulaceae, three Asteraceae, and one Goodeniaceae) that were used in the slot blot (Fig. 4). All species that lacked pt-accD produced nuclear transcripts (Fig. 5B), apart from C. alliariifolia. In all of the Campanulaceae species, consistent with the slot-blot results, no pt-accD transcript was identified, confirming the loss of the plastidic gene in all these species. A nuclear transcript was subsequently amplified from C. alliariifolia using an alternative primer pair, and the rearrangements that caused the loss of pt-accD in T. caeruleum (Haberle et al., 2008) were also confirmed in C. alliarifolia.
Figure 5.
Detection of plastidic (pt-accD) or nuclear (n-accD) transcripts in 18 Asterales by RT-PCR. A, Positions of the primer pairs that allowed the specific amplification of the plastidic accD (pt-accD: approximately 350 bp) or of the nuclear accD (n-accD: approximately 970 bp) transcripts. B, RT-PCR results showing the amplification of pt-accD only from Asteraceae species while n-accD was only amplified from Campanulaceae species. C. alliarifolia was the only Campanulaceae species for which no n-accD amplification product was obtained, but an n-accD transcript could be amplified in that species using another primer pair. -ve indicates the absence of template. A primer pair specific for the plastidic psbA gene was used as a positive amplification control for all species.
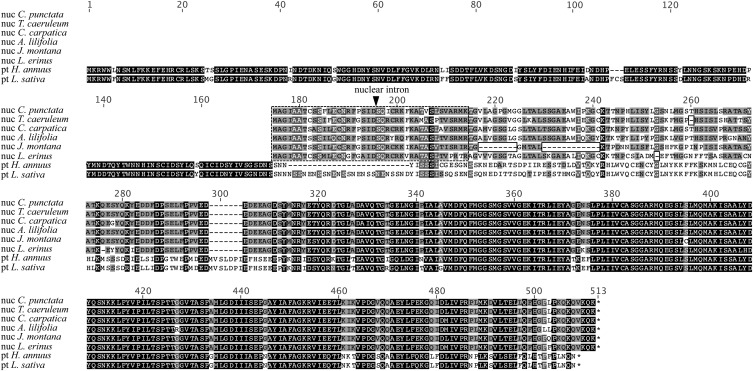
n-accD transcripts from 10 Campanulaceae species (Adenophora bulleyana, Adenophora lilifolia, Campanula carpatica, Campanula punctata, Campanula thyrsoides, Campanula trachelium, Jasione montana, Jasione perennis, Lobelia erinus, and T. caeruleum) were sequenced and all contained an intact ORF. Seven species had an n-accD transcript of an identical size, while deletions of three, six (two deletion events), and 60 bp (two deletion events) were present in T. caeruleum, L. erinus, and the two Jasione species (J. montana and J. perennis), respectively. These deletions occurred toward the 5′ end of n-accD transcript, outside the plastid-derived sequence (Fig. 6). The greatest conservation of these n-accD transcript sequences was found in the accD enzymatic domain encoded by the nupt sequence, emulating conservation of the 3′ region of pt-accD that also encodes the carboxylase functional domain.
Figure 6.
Multiple alignment of the predicted nuclear and plastidic accD proteins from eight Asterales. Nucleus-encoded accD predicted protein sequences from six Campanulaceae species (C. punctata, T. caeruleum, C. carpatica, A. lilifolia, J. montana, and L. erinus) and plastid-encoded accD protein sequences from two Asteraceae species (H. annuus and L. sativa) are aligned. The predicted plastidic transit peptide encoding-sequence present at the amino-terminus of the n-accD protein is boxed with hatched lines. The black boxes indicate the deletion events that occurred in the n-accD ORF of some Campanulaceae species. The location of the intron in the n-accD gene is indicated by a black triangle.
Pairwise analyses of nonsynonymous and synonymous nucleotide substitution rates (Ka and Ks, respectively) were undertaken to determine whether n-accD was under positive selection. In the Campanulaceae species analyzed, nuclear accD showed a Ka ranging from 0.13 to 0.15 and a Ks ranging from 1.01 to 1.55. These values are in accordance with the observation that the time of functional transfer of accD was close to the formation of the Campanulaceae. Pairwise Ka/Ks ratios suggested that the n-accD gene was not positively selected, as is nearly always observed for genes functionally transferred from an organelle to the nucleus (Liu et al., 2009). Indeed, from the study of more than a hundred organellar genes transferred to the nucleus in various angiosperms (including the multiple transfers of 11 genes in several lineages) it was observed that only 1% of genes in pairwise comparisons showed evidence of positive selection (Liu et al., 2009).
The Asteraceae and Goodeniaceae species investigated in this study that were shown to possess a pt-accD gene by slot-blot hybridization were subsequently shown to produce pt-accD transcripts (Fig. 5B). The only exception was Scaveola albida, for which the absence of a transcript is presumed to be due to sequence divergence with the primer pairs since the plastidic gene was shown to be present on the slot blot. In Tagetes erecta, the functionality of the gene was further verified by sequencing pt-accD and by observing the presence of an intact ORF. None of the Asteraceae species used in this study contained both plastidic and nuclear transcripts and thus were not at an intermediate stage of functional relocation of the accD gene to the nucleus. Numerous attempts to amplify a whole or partial n-accD in those species were unsuccessful.
Independent Functional Relocation of pt-accD in a Few Angiosperm Families
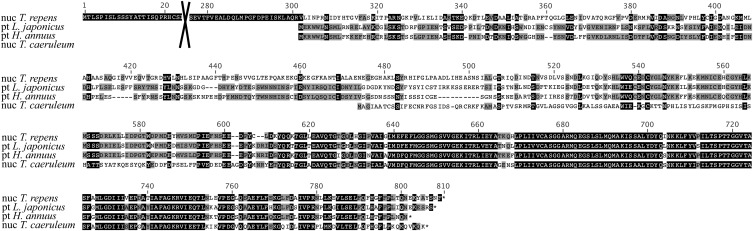
Currently available plastome data reveal the loss of a functional pt-accD in species belonging to five different angiosperm families. In the Campanulaceae family and more specifically in T. caeruleum, our work shows that pt-accD appears to have been functionally transferred to the nucleus and replaced by a nucleus-encoded version of prokaryotic origin. The n-accD ORF identified in T. caeruleum encodes approximately 200 amino acids of plastid origin, whereas pt-accD encodes a 500-amino acid protein in the closely related Asteraceae species. Recently ESTs corresponding to another putative n-accD gene of plastid origin have been discovered in Trifolium repens (Fabaceae; Magee et al., 2010). Interestingly, both T. caeruleum and T. repens n-accD genes each encode only the 3′-end region of the plastidic gene, which corresponds to the carboxyl transferase domain of the ACCD protein (Fig. 7). This is the only known functional domain present in this protein (Zhang et al., 2003). Despite this common feature, these two n-accD genes have distinct gene structures and have acquired different nuclear regions to promote the nuclear expression and chloroplast targeting of the gene product, implying the occurrence of two different functional accD transfer events in the Fabaceae and Campanulaceae.
Figure 7.
T. caeruleum and T. repens n-accD protein sequences only present similarity for plastid derived sequences. Multiple alignment of n-accD predicted protein sequences from T. repens (Fabaceae) and T. caeruleum (Asterales) with the pt-accD protein sequences from Lathyrus japonicus (Fabaceae) and H. annuus (Asterales). Sequence conservation between all four peptides is restricted to the carboxy terminus corresponding to the carboxylase domain of the plastidic protein. Amino acids 24-275 encoded by the n-accD transcript of T. repens are not shown.
The Intronic Region in the n-accD Is Highly Variable
Introns sometimes play a role in the regulation of gene expression (Sheldon et al., 2002; Schauer et al., 2009). To determine if the 1.4-kb intron present in the T. caeruleum n-accD transcript may have a role in the gene expression, we sequenced the intronic region from three other Campanulaceae species (C. punctata, C. thyrsoides, and J. perennis). The intron was present at an identical position in this gene in all four species examined, suggesting that it was relocated early in the Campanulaceae lineage. However, its size is variable, ranging from 1,357 bp in T. caeruleum to 2,430 bp in C. punctata. None of these intronic sequences are highly conserved between the four species (Supplemental Fig. S1), suggesting an absence of conserved regulatory elements.
DISCUSSION
ACCases catalyze the formation of malonyl-CoA from acetyl-CoA and are required for de novo fatty acid synthesis (Konishi and Sasaki, 1994; Sasaki and Nagano, 2004). Dicotyledonous plants possess two forms of ACCase: a eukaryotic form composed of a single multifunctional polypeptide located in the cytosol and a prokaryotic ACCase composed of several subunits in the stroma of plastids (Sasaki et al., 1995; Konishi et al., 1996). The prokaryote-type ACCase is composed of four subunits (Gornicki et al., 1997): the α-carboxyltransferase subunit (accA), the biotin carboxyl carrier (accB), and the biotin carboxylase (accC), which are all nucleus encoded, and the β-carboxyltransferase subunit (accD) that is encoded in the plastome (even in the reduced plastomes of parasitic and nonphotosynthetic plants; Wolfe et al., 1992; Bungard, 2004; Delannoy et al., 2011). From a study of 28 plant families, it was observed that, apart from Poaceae, all flowering plants (including Campanulaceae) contain prokaryotic-like ACCase proteins in their chloroplasts (Konishi and Sasaki, 1994; Konishi et al., 1996; Gornicki et al., 1997). However, species belonging to the Acoraceae (Goremykin et al., 2005), Campanulaceae (Haberle et al., 2008), Fabaceae (Magee et al., 2010), Geraniaceae (Guisinger et al., 2008), and Poaceae (Konishi and Sasaki, 1994; Martin et al., 1998) were shown to have lost chloroplastic accD from their plastomes. In this study, we provide evidence that members of the Campanulaceae family have functionally transferred the chloroplastic accD gene to the nucleus prior to its loss from the plastome.
It has been hypothesized that accD is retained in the plastome to allow each plastid to control ACCase activity according to its needs (Bungard 2004) because this enzyme is a limiting factor in fatty acid biosynthesis (Madoka et al., 2002). However, the absence of a plastidic version in T. caeruleum (and other angiosperms) and the identification of a functional accD gene (of prokaryotic origin) encoded in the nucleus of that species is not consistent with a plastid autonomous regulatory role for this subunit. Remarkably, this n-accD gene encodes a protein that is about one-half the size of the plastidic version and shows similarity to only one-third of the entire plastidic protein. The region of n-ACCD protein encoded by nuclear sequences is very short (approximately 100 amino acids) and consists of little more than a target peptide-encoding sequence.
Several lines of evidence strongly indicate that the minimal n-accD gene of prokaryotic origin present in the Campanulaceae is functionally equivalent to the plastid gene found in most flowering plant species. First, the plastid-derived region of the nuclear transcript encodes the carboxyltransferase domain that is the only known functional domain of the n-accD polypeptide. Within the Campanulaceae, the 3′ end of n-accD that corresponds to the accD carboxylase domain is highly conserved compared with the 5′ end of the gene, consistent with it also encoding a functional domain in this nuclear gene. Second, the carboxyltransferase domain of the nuclear protein maintains the “PLIIVCASGGARMQE” motif that is considered to be the accD putative catalytic site (Lee et al., 2004). Third, we demonstrated that this nuclear transcript encodes a transit peptide that targets the accD precursor proteins to the chloroplast. Fourth, we were able to isolate a similar nuclear transcript from all the Campanulaceae species that have lost pt-accD, whereas it was missing in related species where a pt-accD gene is retained in the plastome. Finally, and despite testing many primer pairs designed in various regions of the plastidic or nuclear accD gene, we were not able to amplify or sequence any other candidate accD transcripts in Campanulaceae. All of these results are in accordance with the functional replacement of the plastidic accD with this minimal nuclear version of accD in the Campanulaceae.
It has been inferred by sequence comparisons that the cytochrome c oxydase subunit1 (cox1), cox2, ribosomal proteinL2 (rpl2), and iron-sulfur subunit (sdhB) mitochondrial genes have functionally relocated to the nucleus in two pieces (gene fission; Adams et al., 2001; Funes et al., 2002; Gawryluk and Gray, 2009, 2010). In the Campanulaceae, as well as in the Fabaceae (Magee et al., 2010), functional transfer of plastidic accD by gene fission seems to be unlikely because the identified n-accD lacks only the N terminus of the plastidic accD that has no functional domain and is very variable in size and sequence among angiosperms, suggesting that this region of the protein is nonessential. Moreover, experiments to amplify the accD 5′ region from any T. caeruleum transcripts were unsuccessful despite testing many primer pairs. Similarly, in T. repens only n-accD ESTs that are also restricted to the functional domain of the plastidic accD gene were found, while no sequences corresponding to the 5′ end of plastidic accD gene were reported among the 700,000 ESTs sequenced (Magee et al., 2010).
How did pt-accD become functional in the nuclear genome within the Campanulaceae? To retain enzyme activity in the chloroplasts after transposition to the nucleus, pt-accD required the acquisition of several nuclear gene regulatory elements, including a promoter, a target peptide-encoding sequence, and a polyadenylation signal (Timmis et al., 2004). The complexity of these activation events can vary, since some plastid promoters can function in the nucleus (Cornelissen and Vandewiele, 1989; Lloyd and Timmis, 2011), some organellar genes already contain the genetic information for protein targeting into the organelles (Ueda et al., 2008), and sequences in the 3′ untranslated region of some plastid genes may fortuitously encode polyadenylation sites (Stegemann and Bock, 2006; Lloyd and Timmis, 2011). To determine the number of nuclear regulatory elements that accD acquired to become functional in the nucleus, we attempted to identify the origin of the different regions of the gene. Apart from the chloroplastic target peptide that may derive from KAS I, the nuclear regulatory sequences acquired by the n-accD gene show no sequence similarity to any other sequences available in the public databases and therefore the promoter and the polyadenylation signal most presumably derive from preexisting nuclear sequences rather than plastidic sequences.
How has the plastidic accD gene been functionally replaced in a few angiosperms? From the complete plastome sequences currently available in public databases, at least six instances of loss of pt-accD can be inferred during angiosperm evolution. In the Poaceae, the plastidic accD gene has been replaced by a nuclear encoded homomeric ACCase of eukaryotic origin that acquired a plastidic transit peptide encoding-sequence (Konishi et al., 1996). However, in Campanulaceae, the plastidic gene has been functionally transferred and relocated in the nucleus. In T. repens (Fabaceae), similar functional gene transfer from the chloroplast has been inferred (Magee et al., 2010). This putative n-accD gene from T. repens also contains only the sequence of the functional domain of the plastidic gene. Since different nuclear sequences are encoded by n-accD and regulate expression of this gene in T. repens and Campanulaceae species, these two genes derive from separate plastid gene transfer events.
In Arabidopsis, an acc gene of eukaryotic origin has been tandemly duplicated on chromosome 1 (Yanai et al., 1995) and one of the duplicated copies (acc2) encodes a plastid-targeted product. The ACC2 protein has a similar function to the plastidic ACCD protein (Babiychuk et al., 2011) that is still functional in Arabidopsis plastome (Konishi et al., 1996; Babiychuk et al., 2011). Among the 22 eudicot nuclear genomes fully sequenced to date, only Brassicaceae species (Arabidopsis lyrata, Arabidopsis, Brassica rapa, and Capsella rubella) encode an additional plastid-targeted eukaryotic ACCase, whereas the remaining species only encode cytosolic proteins (Supplemental Table S2). This plastid-targeted eukaryotic-type ACCase, first reported in Brassica napus (Schulte et al., 1997), appeared more than 16 million years ago before the divergence of the Brassica and Arabidopsis genera (Supplemental Fig. S2; Hedges et al., 2006). All Brassicaceae plastomes sequenced to date (Aethionema cordifolium, Aethionema grandiflorum, Arabidopsis, Arabis hirsuta, B. napus, B. rapa, Barbarea verna, Capsella bursa-pastoris, Crucihimalaya wallichii, Draba nemorosa, Lepidium virginicum, Lobularia maritima, Nasturtium officinale, Olimarabidopsis pumila) encode a functional plastidic accD gene, in spite of the presence of a plastid targeted nuclear acc2 gene in some of these species. In Arabidopsis, mutation of this nuclear acc2 gene produced no phenotype, suggesting that the plastid encoded gene product provides most ACCase activity in this species (Babiychuk et al., 2011). Although a prokaryotic ACCase activity has been detected in plastid protein extracts of all eudicots tested (Konishi et al., 1996), it is possible that some other eudicot species may also contain a plastid-targeted ACCase of eukaryotic origin, as observed for some Brassicaceae members (Babiychuk et al., 2011).
Following activation of accD in the nucleus, both nuclear and chloroplastic copies presumably are functional for a period of time. Coexpression of an organellar gene in two different cellular genetic compartments has only been reported for a few mitochondrial genes (cox2, rpl5, sdh4) in land plants (Adams et al., 1999; Sandoval et al., 2004; Choi et al., 2006). In our study, none of the 18 species tested showed coexpression of n-accD and pt-accD. We were unable to find any vestige of n-accD (using multiple primer pairs) in the Goodeniaceae and Asteraceae species that possess a functional pt-accD, suggesting that the relocation of the gene occurred soon after the functional gene transfer, most likely either prior to or immediately after the emergence of the Campanulaceae lineage.
Are there reasons for the relocation of pt-accD to the nucleus in certain angiosperm families? Plastome organization, gene content, and gene order are generally well conserved among flowering plants. However, recent complete sequencing revealed that the plastomes in a few plant families, such as the Campanulaceae, Fabaceae, and Geraniaceae, are highly rearranged, causing the disruption of operons and loss of genes (Cosner et al., 1997; Cai et al., 2008; Haberle et al., 2008; Guisinger et al., 2011). It has been suggested that a higher number of repeat sequences present in these rearranged plastomes (Guisinger et al., 2008, 2011) can promote illegitimate homologous recombination and cause these multiple structural changes (Maréchal and Brisson, 2010). Recent studies also showed that nucleotide substitution rates have been accelerated in some angiosperm families such as the Geraniaceae (Guisinger et al., 2008, 2011) and Fabaceae (Magee et al., 2010), possibly from a less efficient chloroplastic DNA repair mechanism (Guisinger et al., 2008, 2011). In Fabaceae, each of the four consecutive plastidic genes “ycf4-psaI-accD-rps16” that are present in a hypermutable region has been lost from the plastome of at least one member of that family (Magee et al., 2010). Even though the molecular mechanisms at the origin of this unusual and faster plastome evolution are still unknown, they most presumably have contributed to the functional relocation of plastidic genes, such as accD, to the nucleus.
These results provide an example of the evolutionary processes leading to the functional relocation of a chloroplastic gene to the nucleus. The plastidic accD gene, which has been lost independently in diverse glaucophyte, diatom, protist, or land plant species (Martin et al., 1998), has been functionally replaced by an abridged nuclear-encoded gene of prokaryotic origin in the Campanulaceae. This transfer has involved the acquisition of additional nuclear sequence that provides both gene expression and protein targeting back to the plastid. This functional relocation has been accompanied and probably facilitated by extensive rearrangements of the plastome in Campanulaceae species.
MATERIALS AND METHODS
Plant Material and Plant Growth Conditions
Eighteen species belonging to the Asterales were investigated: two Adenophora species (Adenophora bulleyana and Adenophora lilifolia), seven Campanula species (Campanula alliariifolia, Campanula carpatica, Campanula glomerata, Campanula persicifolia, Campanula punctata, Campanula trachelium, and Campanula thyrsoides), Codonopsis pilosula, Helianthus annuus, two Jasione species (Jasione montana and Jasione perennis), Lactuca sativa, Lobelia erinus, Scaveola albida, Tagetes erecta, and Trachelium caeruleum. These plants were grown in soil in a controlled environment chamber under 14-h-light/25°C and 10-h-dark/18°C conditions.
Isolation of Nucleic Acids and cDNA Synthesis
gDNA was isolated from 100 mg of fresh leaf tissue using the DNeasy Plant Mini kit (Qiagen). Total RNA was prepared using an RNeasy Plant Mini kit (Qiagen) and gDNA was removed using a TURBO DNA free kit (Ambion). RT was then performed using an Advantage RT-for-PCR kit (Clontech) with oligo(dT) primers. All kits were used in accordance with the manufacturers’ instructions.
PCR and RT-PCR Amplification
Amplifications were performed using KapaTaq polymerase (Kapa Biosystems) or Phusion High-Fidelity polymerase (Finnzymes) following the manufacturers’ instructions. For PCR reactions using KapaTaq, gDNA or cDNA was denatured at 95°C for 2 min and amplified using 35 cycles of 95°C for 30 s, 50°C to 60°C for 30 s, and 72°C for 1 min. For PCR reactions using the Phusion High-Fidelity polymerase, gDNA or cDNA was denatured at 95°C for 2 min and amplified using 35 cycles of 98°C for 10 s, 60°C for 15 s, and 72°C for 1 min.
Primers Used for PCR Amplifications
All primers used for amplifications (PCR, RT-PCR, RNA ligase-mediated [RLM]-RACE, and TAIL-PCR) and sequencing are listed in Supplemental Table S3. To amplify part of the nuclear accD transcript in T. caeruleum, RT-PCR of T. caeruleum cDNA was undertaken using primers accD-Aster-F2 and accD-Aster-R1. To determine the presence of an intron in the n-accD gene of the different species, gDNA was PCR amplified using primers nuc-accD-intron-F and nuc-accD-intron-R. To detect the presence of n-accD or pt-accD transcripts in each species, RT-PCR was undertaken using primers specific to the nuclear (nuc-accD-F and nuc-accD-stop-R) or plastidic copy (pt-accD-F and pt-accD-R). The ribosomal DNA (rDNA)-internal transcribed spacer (ITS) regions of each species was amplified from gDNA using the primer pairs rDNA18S-F and rDNA28S-R or ITS-F and ITS-R.
DNA Sequencing Reactions
PCR products were cleaned using the QIAquick PCR purification kit (Qiagen), cloned into pGEM-T vector (pGEM-T Vector System 1, Promega) after adenosine addition according to the manufacturers’ instructions. Positive and independent clones were purified using the GenElute Plasmid Miniprep kit (Sigma-Aldrich) and sequenced using universal primers or randomly designed primers.
RLM-RACE
A FirstChoice RLM-RACE kit (Ambion) was used for 5′ and 3′ RACE of T. caeruleum cDNA template. For 5′ RACE of n-accD cDNA, nested primers accD-RLM-R1 and accD-RLM-R2 were used sequentially in two rounds of PCR. For 3′ RACE, primers accD-RLM-F1 and accD-RLM-F2 were used in two sequential rounds of PCR.
TAIL-PCR
TAIL-PCR was performed as described (Liu and Whittier, 1995) using degenerate primer AD6 (Sessions et al., 2002) and gene specific primers accD-TAIL-R1, accD-TAIL-R2 and accD-TAIL-R3 to obtain the sequences at the 5′ end of the n-accD gene and the degenerate primer AD6 and gene specific primers accD-TAIL-F1, accD-TAIL-F2 and accD-TAIL-F3 for the 3′ end. T. caeruleum gDNA was used as template for the PCR reactions.
DNA Blot Analysis
For DNA slot blotting, 350 ng of total cellular DNA per slot was fixed to Amersham Hybond-N+ membrane (GE Healthcare) using a SRC 072/0 Minifold II apparatus (Scleicher and Schuell). Membranes were hybridized with [32P]dATP-labeled probe (accD or psbA). The probes were generated by PCR using primers accD963F2 and accD1593R (accD) or cp957F and cp1469R (psbA) from H. annuus gDNA as template. Detection and quantification was performed using a Typhoon Trio Imaging system and ImageQuant TL software (GE Healthcare).
Transit Peptide Prediction
Transit peptide predictions were made using BaCelLo (Pierleoni et al., 2006), MultiLoc (Höglund et al., 2006), Predotar (Small et al., 2004), Protein Prowler (Hawkins and Bodén, 2006), and TargetP (Emanuelsson et al., 2000) software programs.
Vector Construction
Expression cassettes pTc-accD, P35S:GFP, and pTc-accD:GFP were created using the pGreen system of binary vectors (Hellens et al., 2000). pGreen0029 contains the neomycin phosphotransferaseII gene flanked by the nopaline synthase promoter and terminator. The 35S terminator and promoter from pPRVIIIA:neoSTLS2 (Huang et al., 2003) were cloned into pGreen0029 using HindIII/BamHI and NotI/XbaI, respectively, to generate P35SPT. To create the pTc-accD construct that contains the n-accD transcript under the regulatory control of the 35S promoter and terminator, the n-accD transcript was amplified from T. caeruleum cDNA with nuc-accD-XbaIF and nuc-accD-XbaIR primers (both with 5′ XbaI sites). The PCR product was digested with XbaI and cloned into P35SPT. To create the P35S:GFP construct (control), the GFP gene was amplified from the ptGW vector using GFPXbaIF and GFPXbaIR primers (each primer contained an XbaI site at its 5′ end). The PCR product was digested with XbaI and cloned into P35SPT. To generate the pTc-accD:GFP construct that contains the target peptide-encoding sequence of the T. caeruleum n-accD gene in frame with the GFP gene and between the 35S promoter and terminator, the GFP gene was amplified from the ptGW vector using GFP-AfeIF and GFP-SpeIR primers (primers contain AfeI and SpeI sites at their 5′ ends, respectively). The PCR product was digested with AfeI and SpeI and cloned into pTc-accD. In this contruct, the first 668 nucleotides of the n-accD transcript (containing the target peptide) were fused in frame with the GFP ORF. All these amplifications were performed using the Phusion High-Fidelity polymerase (Finnzymes), and each expression cassette was sequenced prior to use.
Production of Stable Transgenic Tobacco Lines and Expression Analysis of Fluorescent Proteins by Microscopy
An Agrobacterium tumefaciens strain containing pSoup (Hellens et al., 2000) was transformed with p35S:GFP or pTc-accD:GFP using the “freeze-thaw” method (An et al., 1988). These strains were used to transform tobacco (Nicotiana tabacum) using a standard leaf disc method and kanamycin selection (300 mg L–1; Mathis and Hinchee, 1994). Leaf material from explants showing high GFP expression were cut into small sections, submerged in 10 μg mL–1 propidium iodide for 10 min, mounted in water, and viewed by confocal laser scanning microscopy using a TCS NT/SP microscope (Leica).
Pairwise Analyses of Nucleotide Substitution Rates
Ka and Ks were calculated using Mega5 software (Tamura et al., 2011) and the Nei-Gojobori method (Jukes-Cantor). For pairwise analysis, the chloroplastic accD sequence from Ranunculus macranthus (basal eudicot: NC_006796) was used as a reference. A codon-based Z test of selection was then used to determine the type of selection acting on the n-accD gene in Campanulaceae species.
Molecular Phylogenetic Analyses
The ITS1 and ITS2 regions from each species were sequenced to verify the good identification of the accessions. A sequence matrix of 19 species was obtained by multiple alignment using Geneious (Drummond et al., 2010) and by adjusting the resulting alignment manually. The data matrix was analyzed using PHYML (Guindon and Gascuel, 2003) and the General Time Reversible model (Tavaré, 1986) or the Neighbor Joining method, with tobacco as outgroup. Bootstrap analyses were performed with 10,000 replicates (Felsenstein, 1985). Phylogenetic trees were drawn and edited using Archaeoptryx (http://www.phylosoft.org/archaeopteryx). The positions of the species within the phylogenetic tree were congruent with previous phylogenetic studies (Albach et al., 2001; Eddie et al., 2003).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers JQ693016 to JQ693033.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Multiple percentage identity plots of the n-accD gene in Campanulaceae species using MultiPipMaker.
Supplemental Figure S2. Maximum likelihood tree obtained with Brassicaceae ACC transcript sequences showing the duplication of the eukaryotic acc gene before the divergence of the Brassica and Arabidopsis genera.
Supplemental Table S1. Prediction of the presence of a transit peptide-encoding sequence in the nuclear accD transcript of T. caeruleum using five different softwares.
Supplemental Table S2. Test of the putative presence of a transit peptide-encoding sequence in the eukaryotic (homomeric) acc transcripts of eudicots having their nuclear genome fully sequenced.
Supplemental Table S3. Oligonucleotide primer list.
Acknowledgments
We thank Dr. Stuart Roy for his help with the confocal laser scanning microscopy.
Glossary
- ORF
open reading frame
- ACCase
acetyl-CoA carboxylase
- gDNA
genomic DNA
- cDNA
complementary DNA
- RT
reverse transcription
- TAIL
thermal assymetric interlaced
- RLM
RNA ligase-mediated
- Ka
nonsynonymous nucleotide substitution rate
- Ks
synonymous nucleotide substitution rate
References
- Adams KL, Ong HC, Palmer JD. (2001) Mitochondrial gene transfer in pieces: fission of the ribosomal protein gene rpl2 and partial or complete gene transfer to the nucleus. Mol Biol Evol 18: 2289–2297 [DOI] [PubMed] [Google Scholar]
- Adams KL, Song K, Roessler PG, Nugent JM, Doyle JL, Doyle JJ, Palmer JD. (1999) Intracellular gene transfer in action: dual transcription and multiple silencings of nuclear and mitochondrial cox2 genes in legumes. Proc Natl Acad Sci USA 96: 13863–13868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albach DC, Soltis PS, Soltis DE, Olmstead RG. (2001) Phylogenetic analysis of asterids based on sequences of four genes. Ann Mo Bot Gard 88: 163–212 [Google Scholar]
- An G, Ebert P, Mitra A, Ha S (1988) Binary vectors. In SB Gelvin, RA Schilperoort, eds, Plant Molecular Biology Manual, Vol A3. Kluwer Academic Publishers, Boston, pp 1–19 [Google Scholar]
- Babiychuk E, Vandepoele K, Wissing J, Garcia-Diaz M, De Rycke R, Akbari H, Joubès J, Beeckman T, Jänsch L, Frentzen M, et al. (2011) Plastid gene expression and plant development require a plastidic protein of the mitochondrial transcription termination factor family. Proc Natl Acad Sci USA 108: 6674–6679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock R, Timmis JN. (2008) Reconstructing evolution: gene transfer from plastids to the nucleus. Bioessays 30: 556–566 [DOI] [PubMed] [Google Scholar]
- Bryant N, Lloyd J, Sweeney C, Myouga F, Meinke D. (2011) Identification of nuclear genes encoding chloroplast-localized proteins required for embryo development in Arabidopsis. Plant Physiol 155: 1678–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bungard RA. (2004) Photosynthetic evolution in parasitic plants: insight from the chloroplast genome. Bioessays 26: 235–247 [DOI] [PubMed] [Google Scholar]
- Cai Z, Guisinger M, Kim HG, Ruck E, Blazier JC, McMurtry V, Kuehl JV, Boore J, Jansen RK. (2008) Extensive reorganization of the plastid genome of Trifolium subterraneum (Fabaceae) is associated with numerous repeated sequences and novel DNA insertions. J Mol Evol 67: 696–704 [DOI] [PubMed] [Google Scholar]
- Choi C, Liu Z, Adams KL. (2006) Evolutionary transfers of mitochondrial genes to the nucleus in the Populus lineage and coexpression of nuclear and mitochondrial Sdh4 genes. New Phytol 172: 429–439 [DOI] [PubMed] [Google Scholar]
- Cornelissen M, Vandewiele M. (1989) Nuclear transcriptional activity of the tobacco plastid psbA promoter. Nucleic Acids Res 17: 19–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosner ME, Jansen RK, Palmer JD, Downie SR. (1997) The highly rearranged chloroplast genome of Trachelium caeruleum (Campanulaceae): multiple inversions, inverted repeat expansion and contraction, transposition, insertions/deletions, and several repeat families. Curr Genet 31: 419–429 [DOI] [PubMed] [Google Scholar]
- Cusack BP, Wolfe KH. (2007) When gene marriages don’t work out: divorce by subfunctionalization. Trends Genet 23: 270–272 [DOI] [PubMed] [Google Scholar]
- Delannoy E, Fujii S, Colas des Francs-Small C, Brundrett M, Small I. (2011) Rampant gene loss in the underground orchid Rhizanthella gardneri highlights evolutionary constraints on plastid genomes. Mol Biol Evol 28: 2077–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempewolf H, Kane NC, Ostevik KL, Geleta M, Barker MS, Lai Z, Stewart ML, Bekele E, Engels JM, Cronk QC, et al. (2010) Establishing genomic tools and resources for Guizotia abyssinica (L.f.) Cass.—the development of a library of expressed sequence tags, microsatellite loci, and the sequencing of its chloroplast genome. Mol Ecol Res 10: 1048–1058 [DOI] [PubMed] [Google Scholar]
- Deusch O, Landan G, Roettger M, Gruenheit N, Kowallik KV, Allen JF, Martin W, Dagan T. (2008) Genes of cyanobacterial origin in plant nuclear genomes point to a heterocyst-forming plastid ancestor. Mol Biol Evol 25: 748–761 [DOI] [PubMed] [Google Scholar]
- Doolittle WF. (1998) You are what you eat: A gene transfer ratchet could account for bacterial genes in eukaryotic nuclear genomes. Trends Genet 14: 307–311 [DOI] [PubMed] [Google Scholar]
- Drummond AJ, Ashton B, Buxton S, Cheung M, Cooper A, Heled J, Kearse M, Moir R, Stones-Havas S, Sturrock S, et al (2010) Geneious v5.1. http://www.geneious.com (February 10, 2013)
- Eddie W, Shulkina T, Gaskin J, Haberle R, Jansen R. (2003) Phylogeny of campanulaceae s. str. inferred from its sequences of nuclear ribosomal DNA. Ann Mo Bot Gard 90: 554–575 [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G. (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Felsenstein J. (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39: 783–791 [DOI] [PubMed] [Google Scholar]
- Fuentes I, Karcher D, Bock R. (2012) Experimental reconstruction of the functional transfer of intron-containing plastid genes to the nucleus. Curr Biol 22: 763–771 [DOI] [PubMed] [Google Scholar]
- Funes S, Davidson E, Reyes-Prieto A, Magallón S, Herion P, King MP, González-Halphen D. (2002) A green algal apicoplast ancestor. Science 298: 2155. [DOI] [PubMed] [Google Scholar]
- Gantt JS, Baldauf SL, Calie PJ, Weeden NF, Palmer JD. (1991) Transfer of rpl22 to the nucleus greatly preceded its loss from the chloroplast and involved the gain of an intron. EMBO J 10: 3073–3078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gawryluk RM, Gray MW. (2009) A split and rearranged nuclear gene encoding the iron-sulfur subunit of mitochondrial succinate dehydrogenase in Euglenozoa. BMC Res Notes 2: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gawryluk RM, Gray MW. (2010) An ancient fission of mitochondrial Cox1. Mol Biol Evol 27: 7–10 [DOI] [PubMed] [Google Scholar]
- Goksoyr J. (1967) Evolution of eucaryotic cells. Nature 214: 1161. [DOI] [PubMed] [Google Scholar]
- Goremykin VV, Holland B, Hirsch-Ernst KI, Hellwig FH. (2005) Analysis of Acorus calamus chloroplast genome and its phylogenetic implications. Mol Biol Evol 22: 1813–1822 [DOI] [PubMed] [Google Scholar]
- Gornicki P, Faris J, King I, Podkowinski J, Gill B, Haselkorn R. (1997) Plastid-localized acetyl-CoA carboxylase of bread wheat is encoded by a single gene on each of the three ancestral chromosome sets. Proc Natl Acad Sci USA 94: 14179–14184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray MW, Burger G, Lang BF. (1999) Mitochondrial evolution. Science 283: 1476–1481 [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52: 696–704 [DOI] [PubMed] [Google Scholar]
- Guisinger MM, Kuehl JV, Boore JL, Jansen RK. (2008) Genome-wide analyses of Geraniaceae plastid DNA reveal unprecedented patterns of increased nucleotide substitutions. Proc Natl Acad Sci USA 105: 18424–18429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guisinger MM, Kuehl JV, Boore JL, Jansen RK. (2011) Extreme reconfiguration of plastid genomes in the angiosperm family Geraniaceae: rearrangements, repeats, and codon usage. Mol Biol Evol 28: 583–600 [DOI] [PubMed] [Google Scholar]
- Haberle RC, Fourcade HM, Boore JL, Jansen RK. (2008) Extensive rearrangements in the chloroplast genome of Trachelium caeruleum are associated with repeats and tRNA genes. J Mol Evol 66: 350–361 [DOI] [PubMed] [Google Scholar]
- Hawkins J, Bodén M. (2006) Detecting and sorting targeting peptides with neural networks and support vector machines. J Bioinform Comput Biol 4: 1–18 [DOI] [PubMed] [Google Scholar]
- Hedges SB, Dudley J, Kumar S. (2006) TimeTree: a public knowledge-base of divergence times among organisms. Bioinformatics 22: 2971–2972 [DOI] [PubMed] [Google Scholar]
- Hellens RP, Edwards EA, Leyland NR, Bean S, Mullineaux PM. (2000) pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol Biol 42: 819–832 [DOI] [PubMed] [Google Scholar]
- Höglund A, Dönnes P, Blum T, Adolph HW, Kohlbacher O. (2006) MultiLoc: prediction of protein subcellular localization using N-terminal targeting sequences, sequence motifs and amino acid composition. Bioinformatics 22: 1158–1165 [DOI] [PubMed] [Google Scholar]
- Huang CY, Ayliffe MA, Timmis JN. (2003) Direct measurement of the transfer rate of chloroplast DNA into the nucleus. Nature 422: 72–76 [DOI] [PubMed] [Google Scholar]
- Jansen RK, Cai Z, Raubeson LA, Daniell H, Depamphilis CW, Leebens-Mack J, Müller KF, Guisinger-Bellian M, Haberle RC, Hansen AK, et al. (2007) Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc Natl Acad Sci USA 104: 19369–19374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kode V, Mudd EA, Iamtham S, Day A. (2005) The tobacco plastid accD gene is essential and is required for leaf development. Plant J 44: 237–244 [DOI] [PubMed] [Google Scholar]
- Konarska MM, Grabowski PJ, Padgett RA, Sharp PA. (1985) Characterization of the branch site in lariat RNAs produced by splicing of mRNA precursors. Nature 313: 552–557 [DOI] [PubMed] [Google Scholar]
- Konishi T, Sasaki Y. (1994) Compartmentalization of two forms of acetyl-CoA carboxylase in plants and the origin of their tolerance toward herbicides. Proc Natl Acad Sci USA 91: 3598–3601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi T, Shinohara K, Yamada K, Sasaki Y. (1996) Acetyl-CoA carboxylase in higher plants: most plants other than gramineae have both the prokaryotic and the eukaryotic forms of this enzyme. Plant Cell Physiol 37: 117–122 [DOI] [PubMed] [Google Scholar]
- Lee SS, Jeong WJ, Bae JM, Bang JW, Liu JR, Harn CH. (2004) Characterization of the plastid-encoded carboxyltransferase subunit (accD) gene of potato. Mol Cells 17: 422–429 [PubMed] [Google Scholar]
- Leister D. (2005) Origin, evolution and genetic effects of nuclear insertions of organelle DNA. Trends Genet 21: 655–663 [DOI] [PubMed] [Google Scholar]
- Liu SL, Zhuang Y, Zhang P, Adams KL. (2009) Comparative analysis of structural diversity and sequence evolution in plant mitochondrial genes transferred to the nucleus. Mol Biol Evol 26: 875–891 [DOI] [PubMed] [Google Scholar]
- Liu YG, Whittier RF. (1995) Thermal asymmetric interlaced PCR: automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking. Genomics 25: 674–681 [DOI] [PubMed] [Google Scholar]
- Lloyd AH, Timmis JN. (2011) The origin and characterization of new nuclear genes originating from a cytoplasmic organellar genome. Mol Biol Evol 28: 2019–2028 [DOI] [PubMed] [Google Scholar]
- Lopez JV, Yuhki N, Masuda R, Modi W, O’Brien SJ. (1994) Numt, a recent transfer and tandem amplification of mitochondrial DNA to the nuclear genome of the domestic cat. J Mol Evol 39: 174–190 [DOI] [PubMed] [Google Scholar]
- Madoka Y, Tomizawa K, Mizoi J, Nishida I, Nagano Y, Sasaki Y. (2002) Chloroplast transformation with modified accD operon increases acetyl-CoA carboxylase and causes extension of leaf longevity and increase in seed yield in tobacco. Plant Cell Physiol 43: 1518–1525 [DOI] [PubMed] [Google Scholar]
- Magee AM, Aspinall S, Rice DW, Cusack BP, Sémon M, Perry AS, Stefanović S, Milbourne D, Barth S, Palmer JD, et al. (2010) Localized hypermutation and associated gene losses in legume chloroplast genomes. Genome Res 20: 1700–1710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maréchal A, Brisson N. (2010) Recombination and the maintenance of plant organelle genome stability. New Phytol 186: 299–317 [DOI] [PubMed] [Google Scholar]
- Margulis L (1970) The Origin of the Eukaryotic Cell. Yale University Press, New Haven, CT, and London [Google Scholar]
- Martin W, Koonin EV. (2006) Introns and the origin of nucleus-cytosol compartmentalization. Nature 440: 41–45 [DOI] [PubMed] [Google Scholar]
- Martin W, Rujan T, Richly E, Hansen A, Cornelsen S, Lins T, Leister D, Stoebe B, Hasegawa M, Penny D. (2002) Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proc Natl Acad Sci USA 99: 12246–12251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin W, Stoebe B, Goremykin V, Hapsmann S, Hasegawa M, Kowallik KV. (1998) Gene transfer to the nucleus and the evolution of chloroplasts. Nature 393: 162–165 [DOI] [PubMed] [Google Scholar]
- Mathis NL, Hinchee AW (1994) Agrobacterium inoculation techniques for plant tissues. In SB Gelvin, RA Schilperoort, eds, Plant Molecular Biology Manual, Ed 2. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 1–9 [Google Scholar]
- Mereschkowsky C. (1905) Über natur und ursprung der chromatophoren im pflanzenreiche. Biologisches Centralblatt 25: 593–604 [Google Scholar]
- Millen RS, Olmstead RG, Adams KL, Palmer JD, Lao NT, Heggie L, Kavanagh TA, Hibberd JM, Gray JC, Morden CW, et al. (2001) Many parallel losses of infA from chloroplast DNA during angiosperm evolution with multiple independent transfers to the nucleus. Plant Cell 13: 645–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noutsos C, Kleine T, Armbruster U, DalCorso G, Leister D. (2007) Nuclear insertions of organellar DNA can create novel patches of functional exon sequences. Trends Genet 23: 597–601 [DOI] [PubMed] [Google Scholar]
- Pierleoni A, Martelli PL, Fariselli P, Casadio R. (2006) BaCelLo: a balanced subcellular localization predictor. Bioinformatics 22: e408–e416 [DOI] [PubMed] [Google Scholar]
- Rousseau-Gueutin M, Ayliffe MA, Timmis JN. (2011a) Conservation of plastid sequences in the plant nuclear genome for millions of years facilitates endosymbiotic evolution. Plant Physiol 157: 2181–2193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseau-Gueutin M, Ayliffe MA, Timmis JN. (2012) Plastid DNA in the nucleus: new genes for old. Plant Signal Behav 7: 269–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseau-Gueutin M, Lloyd AH, Sheppard AE, Timmis JN (2011b) Gene transfer to the nucleus. In C Bullerwell, ed, Organelle Genetics: Evolution of Organelle Genomes and Gene Expression. Springer, Berlin, pp 147–171 [Google Scholar]
- Roy SW, Gilbert W. (2006) The evolution of spliceosomal introns: patterns, puzzles and progress. Nat Rev Genet 7: 211–221 [DOI] [PubMed] [Google Scholar]
- Sandoval P, León G, Gómez I, Carmona R, Figueroa P, Holuigue L, Araya A, Jordana X. (2004) Transfer of RPS14 and RPL5 from the mitochondrion to the nucleus in grasses. Gene 324: 139–147 [DOI] [PubMed] [Google Scholar]
- Sasaki Y, Konishi T, Nagano Y. (1995) The compartmentation of acetyl-coenzyme A carboxylase in plants. Plant Physiol 108: 445–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki Y, Nagano Y. (2004) Plant acetyl-CoA carboxylase: structure, biosynthesis, regulation, and gene manipulation for plant breeding. Biosci Biotechnol Biochem 68: 1175–1184 [DOI] [PubMed] [Google Scholar]
- Schauer SE, Schlüter PM, Baskar R, Gheyselinck J, Bolaños A, Curtis MD, Grossniklaus U. (2009) Intronic regulatory elements determine the divergent expression patterns of AGAMOUS-LIKE6 subfamily members in Arabidopsis. Plant J 59: 987–1000 [DOI] [PubMed] [Google Scholar]
- Schulte W, Töpfer R, Stracke R, Schell J, Martini N. (1997) Multi-functional acetyl-CoA carboxylase from Brassica napus is encoded by a multi-gene family: indication for plastidic localization of at least one isoform. Proc Natl Acad Sci USA 94: 3465–3470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions A, Burke E, Presting G, Aux G, McElver J, Patton D, Dietrich B, Ho P, Bacwaden J, Ko C, et al. (2002) A high-throughput Arabidopsis reverse genetics system. Plant Cell 14: 2985–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon CC, Conn AB, Dennis ES, Peacock WJ. (2002) Different regulatory regions are required for the vernalization-induced repression of FLOWERING LOCUS C and for the epigenetic maintenance of repression. Plant Cell 14: 2527–2537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard AE, Ayliffe MA, Blatch L, Day A, Delaney SK, Khairul-Fahmy N, Li Y, Madesis P, Pryor AJ, Timmis JN. (2008) Transfer of plastid DNA to the nucleus is elevated during male gametogenesis in tobacco. Plant Physiol 148: 328–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard AE, Timmis JN. (2009) Instability of plastid DNA in the nuclear genome. PLoS Genet 5: e1000323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small I, Peeters N, Legeai F, Lurin C. (2004) Predotar: A tool for rapidly screening proteomes for N-terminal targeting sequences. Proteomics 4: 1581–1590 [DOI] [PubMed] [Google Scholar]
- Stegemann S, Bock R. (2006) Experimental reconstruction of functional gene transfer from the tobacco plastid genome to the nucleus. Plant Cell 18: 2869–2878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegemann S, Hartmann S, Ruf S, Bock R. (2003) High-frequency gene transfer from the chloroplast genome to the nucleus. Proc Natl Acad Sci USA 100: 8828–8833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavaré S. (1986) Some Probabilistic and Statistical Problems in the Analysis of DNA Sequences. Lect Math Life Sci 17: 57–86 (American Mathematical Society) [Google Scholar]
- Thorsness PE, Fox TD. (1990) Escape of DNA from mitochondria to the nucleus in Saccharomyces cerevisiae. Nature 346: 376–379 [DOI] [PubMed] [Google Scholar]
- Timme RE, Kuehl JV, Boore JL, Jansen RK. (2007) A comparative analysis of the Lactuca and Helianthus (Asteraceae) plastid genomes: identification of divergent regions and categorization of shared repeats. Am J Bot 94: 302–312 [DOI] [PubMed] [Google Scholar]
- Timmis JN, Ayliffe MA, Huang CY, Martin W. (2004) Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat Rev Genet 5: 123–135 [DOI] [PubMed] [Google Scholar]
- Ueda M, Fujimoto M, Arimura S, Murata J, Tsutsumi N, Kadowaki K. (2007) Loss of the rpl32 gene from the chloroplast genome and subsequent acquisition of a preexisting transit peptide within the nuclear gene in Populus. Gene 402: 51–56 [DOI] [PubMed] [Google Scholar]
- Ueda M, Fujimoto M, Arimura SI, Tsutsumi N, Kadowaki KI. (2008) Presence of a latent mitochondrial targeting signal in gene on mitochondrial genome. Mol Biol Evol 25: 1791–1793 [DOI] [PubMed] [Google Scholar]
- Wolfe KH, Morden CW, Palmer JD. (1992) Function and evolution of a minimal plastid genome from a nonphotosynthetic parasitic plant. Proc Natl Acad Sci USA 89: 10648–10652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai Y, Kawasaki T, Shimada H, Wurtele ES, Nikolau BJ, Ichikawa N. (1995) Genomic organization of 251 kDa acetyl-CoA carboxylase genes in Arabidopsis: tandem gene duplication has made two differentially expressed isozymes. Plant Cell Physiol 36: 779–787 [DOI] [PubMed] [Google Scholar]
- Zhang H, Yang Z, Shen Y, Tong L. (2003) Crystal structure of the carboxyltransferase domain of acetyl-coenzyme A carboxylase. Science 299: 2064–2067 [DOI] [PubMed] [Google Scholar]