The plant hormone jasmonate inhibits leaf growth by delaying the switch from the mitotic cell cycle to the endoreduplication cycle and maintains the cell in a stand-by mode but ready-to-go after the stress.
Abstract
Phytohormones regulate plant growth from cell division to organ development. Jasmonates (JAs) are signaling molecules that have been implicated in stress-induced responses. However, they have also been shown to inhibit plant growth, but the mechanisms are not well understood. The effects of methyl jasmonate (MeJA) on leaf growth regulation were investigated in Arabidopsis (Arabidopsis thaliana) mutants altered in JA synthesis and perception, allene oxide synthase and coi1-16B (for coronatine insensitive1), respectively. We show that MeJA inhibits leaf growth through the JA receptor COI1 by reducing both cell number and size. Further investigations using flow cytometry analyses allowed us to evaluate ploidy levels and to monitor cell cycle progression in leaves and cotyledons of Arabidopsis and/or Nicotiana benthamiana at different stages of development. Additionally, a novel global transcription profiling analysis involving continuous treatment with MeJA was carried out to identify the molecular players whose expression is regulated during leaf development by this hormone and COI1. The results of these studies revealed that MeJA delays the switch from the mitotic cell cycle to the endoreduplication cycle, which accompanies cell expansion, in a COI1-dependent manner and inhibits the mitotic cycle itself, arresting cells in G1 phase prior to the S-phase transition. Significantly, we show that MeJA activates critical regulators of endoreduplication and affects the expression of key determinants of DNA replication. Our discoveries also suggest that MeJA may contribute to the maintenance of a cellular “stand-by mode” by keeping the expression of ribosomal genes at an elevated level. Finally, we propose a novel model for MeJA-regulated COI1-dependent leaf growth inhibition.
In plants, jasmonates (JAs) form a group of oxylipins (oxygenated fatty acids) produced via the octadecanoid pathway (for review, see Balbi and Devoto, 2008; Schaller and Stintzi, 2009; Wasternack and Kombrink, 2010). The volatile phytohormone methyl jasmonate (MeJA), produced through the activity of JA carboxyl methyltransferase (Seo et al., 2001), is an easy-to-handle JA conjugate extensively used in biological assays (Staswick et al., 1992; Pauwels et al., 2008). The roles of these lipid-derived hormones (signaling molecules) have been traditionally studied in the context of plant biotic and abiotic stress-mediated responses such as pathogen attack, wounding, or drought (Reymond et al., 2004; Devoto et al., 2005; Browse, 2009; Yoshida et al., 2009; Avanci et al., 2010; Harb et al., 2010). They are also implicated in plant development by influencing processes like senescence, secondary metabolism, and reproduction (Devoto et al., 2002; Avanci et al., 2010; De Geyter et al., 2012). More recently, evidence has emerged for a role for JA-mediated responses in cell cycle progression. JA blocks synchronized tobacco (Nicotiana tabacum) Bright Yellow-2 cell cultures in both G1 and G2 phases (Świątek et al., 2002), and the G2 arrest is combined with a reduced accumulation of cyclinB1;1 and the cyclin-dependent kinase CDK-B (Świątek et al., 2004). Similarly, in Arabidopsis (Arabidopsis thaliana) cell cultures, MeJA-mediated arrest of the cell cycle in G2 is associated with the repressed expression of mitotic phase genes (Pauwels et al., 2008). Although JAs have been implicated in plant growth inhibition and mitosis (Yan et al., 2007; Zhang and Turner, 2008; Brioudes et al., 2009; Chen et al., 2011), little is known about the molecular mechanisms through which this class of hormones regulates the dynamic process of plant organ development and growth.
Plant organ development is the result of strict spatial and temporal genetic control and the coordination of cell division, growth, and differentiation (Beemster et al., 2003; Tsukaya, 2005; Harashima and Schnittger, 2010; Gonzalez et al., 2012). However, the role of cell cycle regulation and cell division in plant growth and organ development is controversial. Some experimental data are most easily interpreted from the “cellular perspective,” where cell division drives growth, whereas other observations are more consistent with the “organismal perspective,” according to which cell division is merely a consequence of organ growth and to a large extent facultative (Massonnet et al., 2010). In the plant Arabidopsis, two types of cell cycle have been identified: the mitotic cell cycle and the endocycle (or endoreduplication cycle). The process of endoreduplication, which is an increase of ploidy levels via rereplication of DNA in the absence of mitosis (M phase), has been associated with cell expansion, where an increased cell volume is driven by internal turgor pressure (Sugimoto-Shirasu and Roberts, 2003). However, increased ploidy level by endoreduplication appears not to be the unique determinant of plant cell size (Massonnet et al., 2010). For example, some Arabidopsis dwarf mutants defective in hormone signaling or cell wall biosynthesis retain similar ploidy levels to the wild type but exhibit reduced cell size (Caño-Delgado et al., 2000; Ishida et al., 2009). Conversely, some Arabidopsis or tobacco mutant lines overexpressing key cell cycle regulatory genes have altered ploidy levels but do not differ in cell size (De Veylder et al., 2001; Jasinski et al., 2002; Hu et al., 2006). In leaf development, once founder cells are recruited into leaf primordia, cells actively proliferate and the growth of the first true leaves correlates with a switch for most cells to begin endoreduplication with a basipetal polarity (Donnelly et al., 1999; Beemster et al., 2005).
Recently, the occurrence of endoreduplication has been associated with an increased potential for further cell functions (De Veylder et al., 2011), such as the capacity for future cellular growth (Breuer et al., 2010), the regulation of responses against plant pathogen attacks (Chandran et al., 2010; Wildermuth, 2010), or a role in the maintenance of cell fate (Bramsiepe et al., 2010). In terms of cellular machinery, although basic plant cell cycle and growth regulators have become increasingly well characterized during Arabidopsis development (Gutierrez, 2009; Boruc et al., 2010; Van Leene et al., 2010, 2011), little is known about their precise regulation/coordination. It has been shown that cell cycle progression and the switch from the mitotic to the endoreduplication cycle result from the accurate and sequential activity of a conserved core set of regulators implicated in DNA replication and mitosis (Inzé and De Veylder, 2006; Gutierrez, 2009; De Veylder et al., 2011). In animals, E2F transcription factors are major components of this pathway. At the end of the G1 phase, CDK-cyclin complexes phosphorylate the retinoblastoma protein bound to E2F factors, allowing them to activate the expression of genes required for the onset of the synthesis (S) phase, such as Cell Division Control6 (CDC6), CDC10-dependent transcript (CDT1), Origin Recognition Complex (ORCs), and Mini Chromosome Maintenance proteins (MCMs; Attwooll et al., 2004). In plants, two activating E2F proteins, E2Fa (De Veylder et al., 2002; Stevens et al., 2002) and E2Fb (Magyar et al., 2005), have been characterized and likely function according to this model (Bosco, 2010). The transcriptional down-regulation of premitotic/mitotic regulators to inhibit mitosis (Wuarin et al., 2002; Inzé and De Veylder, 2006), combined with maintaining G1-S CDK activity levels to continue DNA replication, contributes to the switch from the mitotic cycle to the endocycle (for review, see De Veylder et al., 2011). Following DNA replication, a key decision within the G2 phase of the cell cycle is whether to commit to mitosis or engage in cycles of repeated DNA synthesis. In plants, endocycle onset involves the selective inactivation of M phase-promoting factors, such as the B1-type CDK (CDKB1;1), through proteolytic destruction of its cyclin partner, CYCA2;3 (Boudolf et al., 2009). The cell cycle regulation machinery is tightly coordinated temporally and spatially during plant development by the action of phytohormones (Gutierrez, 2009; Gonzalez et al., 2010; Dudits et al., 2011). Several studies involving plant hormones such as auxin (Perrot-Rechenmann, 2010), abscisic acid (Świątek et al., 2002; Skirycz and Inzé, 2010), brassinosteroids (Clouse, 2011), GAs (Achard et al., 2009), or cytokinins (Haberer and Kieber, 2002) show how hormonal signaling networks are able to modulate cell division parameters to impact plant growth and development.
JA-synthesis and JA-perception mutants have proven to be efficient tools to dissect JA-mediated plant responses (Berger, 2002; Devoto and Turner, 2003; Lorenzo and Solano, 2005; Browse, 2009). In our analysis, we use the JA-biosynthesis mutant aos, in which the single Allene Oxide Synthase (AOS) gene in Arabidopsis has been knocked out (Park et al., 2002) and the production of JA and its precursors is impaired (von Malek et al., 2002). In the aos mutant, JA downstream signaling can be rescued by exogenous JAs. The second key mutant analyzed here is coi1-16 (for coronatine insensitive1), which displays a JA-insensitive phenotype intermediate between that of the wild type and other coi1 alleles; this mutant has been the system of choice in our study, as in several others, because it is conditionally male fertile and can be maintained as a homozygous line (Ellis and Turner, 2002). The identification and study of coi1 mutants (Feys et al., 1994) led to the discovery that COI1 plays a crucial role in the JA signaling pathway (Xie et al., 1998; Yan et al., 2009). The COI1 gene encodes a 66-kD F-box protein that is part of an E3 ubiquitin-ligase complex of the SKP1/CUL1/F-box type (SCF; Devoto et al., 2002; Xu et al., 2002). In the SCFCOI1 complex, COI1, the JA-Ile receptor (Chini et al., 2007; Thines et al., 2007; Fonseca et al., 2009; Yan et al., 2009), targets repressor components of transcriptional responses to JAs, the jasmonate ZIM-domain (JAZ) proteins, for ubiquitination and subsequent degradation by the 26S proteasome system (Chini et al., 2007; Thines et al., 2007; Gfeller et al., 2010).
While MeJA has been proven to generally inhibit plant growth and to negatively affect mitosis, how this signaling molecule affects specific aspects of plant development remains unclear. In this study, we assess the role of JA signaling in cell cycle control and evaluate the contributions of COI1, AOS, and MeJA in Arabidopsis plants to these processes. Our studies have provided new insights into the cellular basis of the MeJA-induced stunting of plant growth and how MeJA interferes with the switch from proliferative growth to differentiation. First, the results of our studies reveal that MeJA delays the switch from the mitotic cell cycle to the endoreduplication cycle in a COI1-dependent manner as well as inhibits the mitotic cycle itself, arresting the cell cycle in G1 prior to the S transition. These data allow us to propose a novel model for MeJA-regulated COI1-dependent leaf growth inhibition. Second, a novel global transcriptional profiling of Arabidopsis seedlings grown in medium containing MeJA was carried out to identify the molecular players whose expression is regulated during leaf development by this hormone via COI1. Significantly, we show that MeJA activates critical regulators of endoreduplication and affects the expression of key determinants of DNA replication. Together, our discoveries also suggest that MeJA may contribute to the maintenance of a cellular “stand-by mode” by maintaining a high level of expression of ribosomal genes.
RESULTS
MeJA Negatively Affects Cell Cycle Progression during Leaf Development in Arabidopsis
We set out to understand the mechanisms through which JAs affect growth and development in the Arabidopsis leaf by studying the mutants coi1-16B and aos and their corresponding genetic background plant Col glabrous1 (gl1). At 9 d after stratification (DAS), leaves 3 and 4 began to emerge for all the untreated seedlings (Fig. 1), and the aos mutant appeared to be the largest of these followed by coi1-16B. When grown in the presence of MeJA, all seedlings appeared less developed in comparison with their untreated counterparts. However, this did not have a major effect on leaf emergence. In any case, the development of coi1-16B-treated seedlings was not affected as dramatically as were the Col gl1 and aos seedlings.
Figure 1.
Rosette phenotype of MeJA-treated (+) and untreated (−) wild-type and mutant Arabidopsis plants at different stages of development. Plants were grown in vitro and photographed at 9, 13, and 19 DAS. Bar = 5 mm.
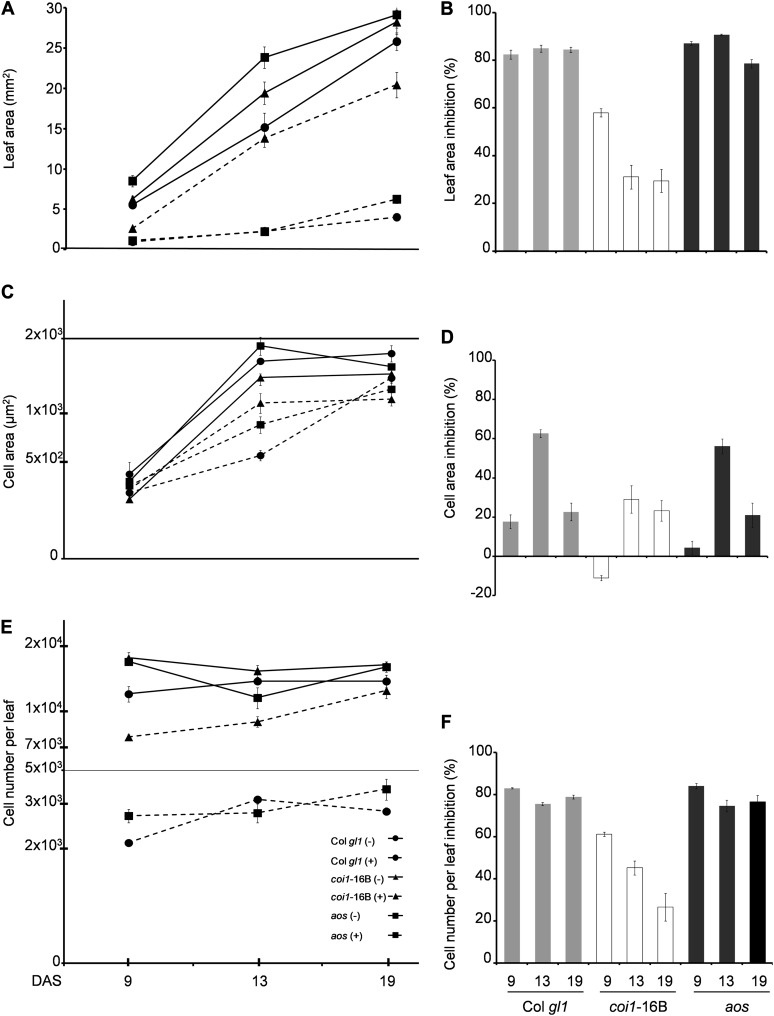
We measured the leaf area of in vitro-grown Col gl1, coi1-16B, and aos plants, harvesting leaves 1 and 2 at 9, 13, and 19 DAS. This kinematic analysis showed that leaf growth was dramatically inhibited by MeJA treatment in Col gl1 and aos plants (i.e. about 80% of leaf area growth inhibition) consistently at all stages of development (Fig. 2, A and B; Supplemental Table S1). The inhibition is evident also for the coi1-16B mutant, although to a much lesser extent than for Col gl1 and aos, and in a more pronounced manner at 9 DAS.
Figure 2.
MeJA alters the evolution of the leaf area by affecting cell size and cell number differentially and in a COI1-dependent manner. Kinematic analysis of leaf growth of Col gl1, coi1-16B, and aos throughout their development is shown. Plants were grown in vitro in the absence (−) or presence (+) of 50 µm MeJA. Leaves 1 and 2 were harvested at 9, 13, and 19 DAS to measure the leaf area (A), the epidermal cell area (C), and the number of epidermal cells per leaf (E). MeJA inhibition (%) has been calculated for the leaf area (B), the epidermal cell area (D), and the number of epidermal cells per leaf (F). Values denote averages ± se of at least three biological replicates (n = 4–21, where n is the number of plants analyzed per biological replicate). In A, C, and E, continuous and dashed lines indicate untreated (−) and MeJA-treated (+) samples, respectively. C and E use a log10 scale.
At the same time points, the epidermal cell areas were determined. It has been shown previously that the development of the epidermal cells reflects that of other cell types in the leaf (Beemster et al., 2005) and that the epidermis is the tissue driving organ growth (Savaldi-Goldstein et al., 2007; Marcotrigiano, 2010; Hacham et al., 2011; Skirycz et al., 2011). Early during development in leaf dividing cells, minor inhibitory effects in Col gl1 and aos, and no inhibitory effect in coi1-16B, were seen. At 13 DAS, MeJA clearly inhibited the expansion of the cell area in Col gl1 (62%) and aos (56%), and this inhibition was obviously reduced in coi1-16B and comparable to that occurring at 19 DAS in the three tested genotypes (Fig. 2, C and D; Supplemental Table S1). The number of cells in Col gl1 and aos was greatly reduced in the presence of MeJA at all stages of development (i.e. about 75%). This cell number MeJA-induced reduction was really restrained in coi1-16B at 13 and 19 DAS (Fig. 2, E and F; Supplemental Table S1). These data, taken together, suggest that the negative effect on leaf growth triggered by MeJA is the result of the dramatic reduction in cell number and of an evident (even if less pronounced) reduction in cell size. Both effects are largely COI1 dependent. The reduction in cell number is more evident during proliferation, while cell size is particularly reduced when cells start expanding.
MeJA Alters Nuclear DNA Content by Delaying the Onset of Endoreduplication in a COI1-Dependent Manner
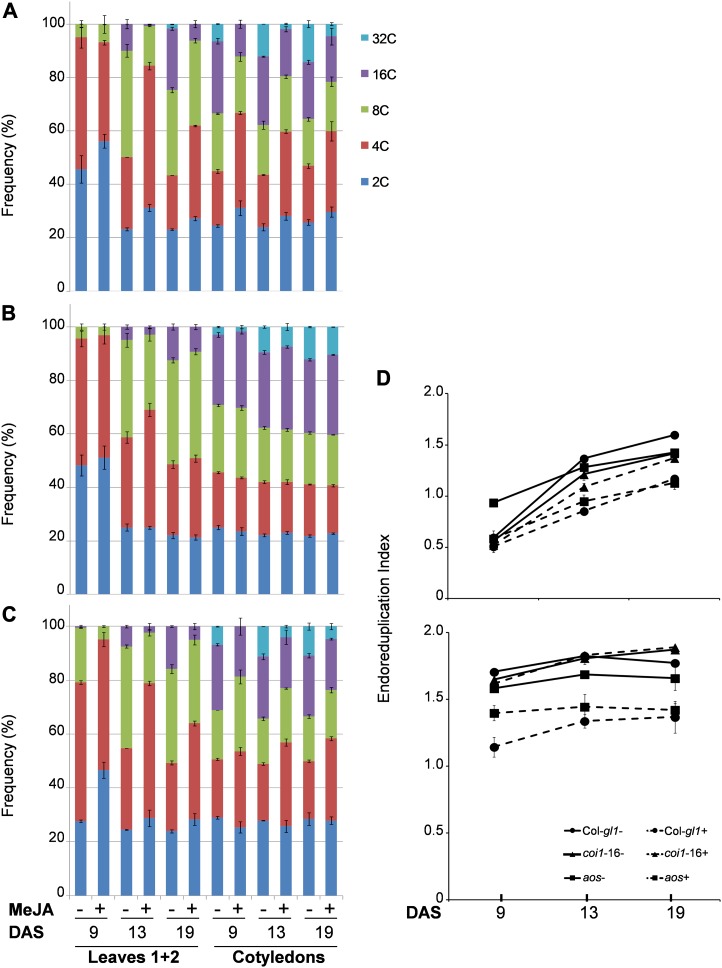
It has been shown that in Arabidopsis, leaves initiate at the flank of the meristem and that their initial growth is driven by cell proliferation (Donnelly et al., 1999). At a later stage, starting from the tip onward, cells will exit the mitotic cycle and begin to expand. This transition is indicated by the onset of endoreduplication, resulting in ploidy levels higher than 4C (Beemster et al., 2005). To better understand the effect of MeJA treatment on the cell cycle and differentiation, the ploidy distribution was examined by flow cytometry in the first true pair of leaves and cotyledons. At 9 DAS, in untreated and MeJA-treated Col gl1 and coi1-16B, most of the leaf nuclei exhibit a 2C or 4C DNA content (Fig. 3, A and B), consistent with a high mitotic activity at this stage of development (Beemster et al., 2005). Correspondingly, at this stage, the cells entering the endocycle are represented mainly by the 8C fraction. Following growth on MeJA, this fraction is decreased in the aos mutant to the level observed in Col gl1 (Fig. 3C). Notably, MeJA shifts the balance of 2C and 4C toward 2C in Col gl1 and aos, while this is unchanged in coi1-16B. At 13 DAS, when the transition from proliferation to expansion occurs in Arabidopsis leaves (Beemster et al., 2005), the 2C or 4C DNA content frequency is decreased with respect to the earlier time point in all genotypes. Upon MeJA treatment in Col gl1 and aos leaves, both 2C and 4C fractions are increased, while in coi1-16B, they remain largely unchanged. A very similar pattern is observed in 19-DAS leaves. Again, upon MeJA treatment in Col gl1 and aos leaves, both 2C and 4C fractions are increased, while they remain unaffected in coi1-16B. In cotyledons, in the absence of MeJA treatment, and at every time point in Col gl1 and coi1-16B seedlings, we observed a higher or similar percentage of nuclei for cells entering into the endocycle, whereas in aos seedlings, we observed similar percentages (8C–32C) compared with those with high mitotic activity (2C and 4C). At all time points, while MeJA causes an increase of the 2C or 4C fraction and a corresponding decrease of the 8C to 32C fractions in Col gl1 and aos (Table I), remarkably, the percentages of nuclei in these two collective fractions remain unchanged by MeJA in coi1-16B. Examination of the ploidy distribution by flow cytometry is a measure of cellular differentiation and can be expressed as an endoreduplication index (EI; Fig. 3D). The EI represents the average number of endocycles undergone by a given nucleus. This parameter typically increases during leaf development, while cotyledons, where divisions cease before the end of embryo ontogeny and growth depends mainly on cell expansion, show precociously high EI values (Mansfield and Briarty, 1992; Tsukaya, 1994). Plotting the EI obtained from our flow cytometry data (Fig. 3; Table I; Supplemental Table S2) revealed that the average number of endocycles in Col gl1 and aos is reduced by MeJA treatment in both leaves and cotyledons. The effect of MeJA is particularly evident in leaves at 13 and 19 DAS. The EI of coi1-16B upon MeJA treatment does not change at any time point for both leaves and cotyledons.
Figure 3.
MeJA alters nuclear DNA content and delays the onset of endoreduplication in a COI1-dependent manner during leaf and cotyledon development. Quantitative analysis is shown for flow cytometry data performed on the first true pair of leaves (1+2) or cotyledons at 9, 13, and 19 DAS of Col gl1 (A), coi1-16B (B), and aos (C) mutants. The values represent average frequencies of the observed ploidy (or C) levels of three independent biological replicates ± se. D, EI calculated from the flow cytometry data on the first true pair of leaves (top panel) or cotyledons (bottom panel). EI represents the average number of endocycles undergone by a typical nucleus {EI = [(0*%2C) + (1*%4C) + (2*%8C) + (3*%16C) + (4*%32C)]/100}. Continuous and dashed lines indicate untreated and treated samples, respectively. For all panels, seedlings were grown in vitro on medium in the absence (−) or presence (+) of 50 μm MeJA. The analyses were performed on at least 20,000 nuclei isolated from 10 to 15 pairs of leaves/cotyledons for each ploidy measurement. Flow cytometry experiments were repeated at least three times for each genotype using independent biological replicates. Error bars indicate se.
Table I. Frequency of nuclei exhibiting 2C to 4C or 8C to 32C DNA content in first true leaves and cotyledons of Arabidopsis Col gl1, coi1-16B, and aos in the absence (−) or presence (+) of MeJA.
The values represent sums of the corresponding 2C + 4C and 8C + 16C + 32C level fractions illustrated in Figure 3, A to C. The analyses were performed on at least 20,000 nuclei isolated from 10 to 15 pairs of leaves/cotyledons for each ploidy measurement. Flow cytometry experiments were repeated at least three times for each genotype using independent biological replicates.
| Plant | Variable | Leaves 1 + 2 |
Cotyledons |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 9 DAS |
13 DAS |
19 DAS |
9 DAS |
13 DAS |
19 DAS |
||||||||
| − | + | − | + | − | + | − | + | − | + | − | + | ||
| Col gl1 | % 2C + 4C | 95.0 | 93.2 | 50.2 | 84.4 | 43.4 | 61.9 | 44.9 | 66.8 | 43.5 | 59.8 | 46.9 | 59.8 |
| % 8C + 16C + 32C | 5.0 | 6.8 | 49.8 | 15.6 | 56.6 | 38.1 | 55.1 | 33.2 | 56.5 | 40.2 | 53.1 | 40.2 | |
| coi1-16B | % 2C + 4C | 95.6 | 96.9 | 58.8 | 69.0 | 48.7 | 50.9 | 45.6 | 43.6 | 42.1 | 42.0 | 41.1 | 40.7 |
| % 8C + 16C + 32C | 4.4 | 3.1 | 41.2 | 31.0 | 51.4 | 49.1 | 54.4 | 56.4 | 57.9 | 58.0 | 58.9 | 59.3 | |
| aos | % 2C + 4C | 79.2 | 95.1 | 54.8 | 78.9 | 49.3 | 64.0 | 50.6 | 53.6 | 48.9 | 56.9 | 49.9 | 58.3 |
| % 8C + 16C + 32C | 20.8 | 4.9 | 45.2 | 21.1 | 50.7 | 36.0 | 49.4 | 46.4 | 51.1 | 43.1 | 50.1 | 41.7 | |
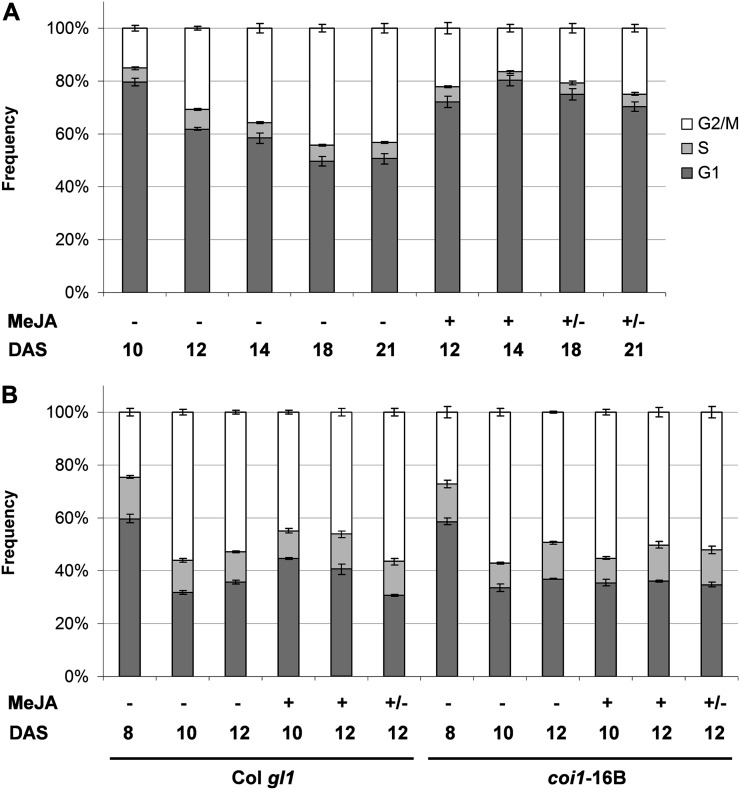
To investigate the dynamics of ploidy changes following MeJA treatment, we extended the analysis of the flow cytometry data by monitoring cell cycle progression in the treated and untreated Arabidopsis first leaf pair as well as in Nicotiana benthamiana cotyledons. We analyzed the cell cycle progression of 9-DAS Arabidopsis seedlings grown continuously in vitro in the presence or absence of MeJA (Supplemental Fig. S2) and Arabidopsis seedlings grown in the absence of treatment up to 8 DAS and then transferred to MeJA-containing medium and analyzed 48 and 96 h after treatment (i.e. at 10 and 12 DAS, respectively). We also tested the ability of leaves to recover from the treatment 48 h after retransfer on medium without MeJA (Fig. 4B). Similar experiments were performed on N. benthamiana seedlings during development from 10 to 21 DAS (Fig. 4A). In Col gl1 at 48 h after MeJA treatment, we observed similar alterations of DNA content to those caused by continuous treatment. Again, the ploidy balance was unchanged in coi1-16B (Supplemental Fig. S1). In both Arabidopsis and N. benthamiana, MeJA treatment causes a significantly increased frequency of nuclei remaining at the G1 phase of the cell cycle after 48 h. The population of nuclei in the G2/M fraction consequently decreases in comparison with the control. The G2/M fraction progressively decreases after prolonged treatment, indicating that the cell cycle is arrested in G1 prior to the S transition. Such a decrease is particularly evident in N. benthamiana throughout the time course, while in Arabidopsis, this is clearer at the earlier time points before the onset of endoreduplication. The effect of MeJA appears to be reversible once the seedlings are transferred back to hormone-free medium (Fig. 4).
Figure 4.
MeJA alters cell cycle progression during leaf and cotyledon development. Cell cycle analysis of flow cytometry data was performed on the cotyledons at 10, 12, 14, 18, and 21 DAS in N. benthamiana (A) or on the first true pair of leaves (1+2) at 8, 10, and 12 DAS of Col gl1 and coi1-16B Arabidopsis plantlets (B). Seedlings were vertically grown in vitro on standard medium for 10 (A) or 8 (B) DAS and transferred to medium containing (+) or not containing (−) 50 μm MeJA. Seedlings growing on MeJA-containing medium were retransferred to MeJA-free medium (+/−) at 14 (A) or 10 (B) DAS. The values represent average frequencies of the observed cell cycle phases (G1, S, or G2/M) of at least five individual seedlings on independent biological replicates. Error bars indicate se.
MeJA Treatment Down-Regulates the Expression and Spatial Distribution of the Core Cell Cycle Marker CYCB1;1:Dbox-GUS Early after Treatment
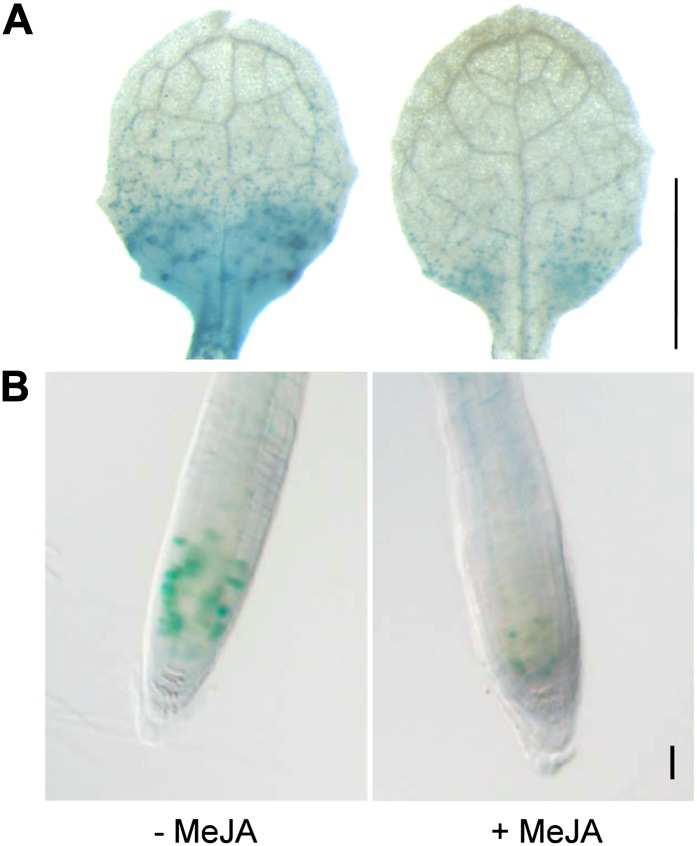
To further test the negative effect of MeJA on cell proliferation, we analyzed in leaf and root organs the mitotic activity of pCYCB1;1:Dbox-GUS transgenic lines, which allowed us to visualize cells at the G2-to-M transition of the cell cycle, reflecting mitotic activity in a very sensitive manner (Colón-Carmona et al., 1999). Arabidopsis CYCB1;1:Dbox-GUS seedlings were grown on control medium for 10 DAS until the third true leaf was fully proliferating (Skirycz et al., 2011). Seedlings were subsequently transferred to control or 50 µm MeJA-containing medium for 48 h. The overall GUS activity was much weaker in the treated leaves, in agreement with Zhang and Turner (2008). Significantly, this reduction indicates a spatial reduction in the number of active mitotic cells once the hormone is taken up from the medium (Fig. 5A). While CYCB1;1-GUS signals are normally detected in a punctate pattern across the root meristem, both the intensity and the number of CYCB1;1-GUS-positive cells dropped substantially in MeJA-treated roots, mirroring the results obtained in leaves (Fig. 5B). Supporting evidence from propidium iodide staining and confocal microscopy showed that the size or cellular organization of the root meristem were not reduced or altered, respectively, by treatment (Supplemental Fig. S3). Taken together, these results strongly suggest that the transcriptional and/or posttranscriptional repression of the CYCB1;1 cell cycle regulator is a part of the cellular responses triggered by MeJA to block cell proliferation.
Figure 5.
MeJA treatment negatively affects cell proliferation in leaves and roots by down-regulating the expression of CYCB1;1. Leaf 3 at 12 DAS (A; bar = 1 mm) and root tip at 6 DAS (B; bar = 50 µm) of Arabidopsis seedlings carrying pCYCB1;1:Dbox-GUS were transferred for 48 h on medium without (−MeJA) or with (+MeJA) 50 µm MeJA. Blue staining indicates mitotic activity. Representative images of numerous independent biological replicates are shown.
In the Primary Root Organ, MeJA Reduces Cell Length and the Size of Mitotic Nuclei But Does Not Affect the Size of the Meristem
The effects of MeJA were also scrutinized in the root growth context, and our studies confirmed that in the primary roots, MeJA reduces the length of mature cells (Supplemental Fig. S4; Supplemental Table S3). While the total root length of untreated coi1-16B is comparable to that of Col gl1 and aos, as expected (Staswick et al., 1992), MeJA dramatically inhibited the total growth of the root in Col gl1 and aos, whereas coi1-16B’s reduced sensitivity to MeJA resulted in a much lower inhibition. Differential interference contrast (DIC) microscopy was used to determine the cell length between fully expanded root hairs on the same cell file of the root in the epidermic mature zone in both MeJA-treated and untreated plants. MeJA inhibits the elongation of the cells in Col gl1 and aos (63% and 67%, respectively), although to a lower extent with respect to the entire root organ. In coi1-16B, the elongation of the cell was inhibited about 36%, in agreement with previous reports (Adams et al., 2010). While the root hair distribution was not obviously affected by the mutations analyzed or by MeJA treatment, the length of fully elongated root hairs in the mature region was also negatively affected by MeJA treatment (Supplemental Fig. S4; Supplemental Table S3). In addition, the cellular organization of the root meristem was verified, and the distance between the quiescent center (QC) and the first visible elongating cell was measured in 6-d-old wild-type roots untreated or continuously treated with MeJA. Upon MeJA treatment, no significant effect on the length of the meristematic zone was observed between the three genotypes (Supplemental Fig. S3; Supplemental Table S4).
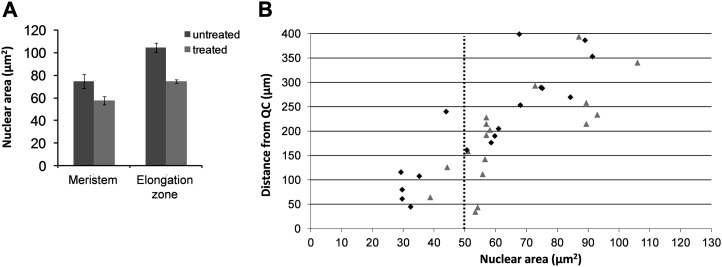
Finally, quantification of nuclear surface area visualized by histone 2B-yellow fluorescent protein (H2B-YFP; Campilho et al., 2006) fusion proteins using confocal microscopy was performed according to Ishida et al. (2010) in 6-d-old wild-type roots untreated or continuously treated with MeJA. Following exposure to MeJA, the average size of the nuclei in Arabidopsis root cells of the meristematic and elongation zones was reduced (Fig. 6A). The analysis of the distribution of the nuclear size along the root meristem demonstrated that in the presence of MeJA, the number of smaller, premitotic nuclei was higher with respect to the untreated meristem (Fig. 6B). In fact, the average percentage of nuclei with size above 50 µm was 87% and 39% in untreated and treated samples, respectively. In addition, in treated root meristems, a higher number of smaller nuclei was distributed along a distance of about 250 µm (largely corresponding to the extent of the meristem) from the QC when compared with the untreated samples. These data, clearly supporting the data obtained in leaves, indicate that MeJA reduces the number of endocycling nuclei in roots as well, and the enrichment in smaller nuclei following MeJA treatment in proximity to the QC is suggestive of a cell cycle arrest in the G1 phase.
Figure 6.
MeJA triggers a reduction of nucleus size in Arabidopsis root cells. Quantification of nuclear surface area visualized by H2B-YFP fusion proteins in 6-d-old roots untreated and treated with 50 µm MeJA was performed according to Ishida et al. (2010). A, Average nuclear area, defined as total pixel intensities of H2B-YFP signals per individual nucleus, was quantified within cells of the same epidermal layers in the meristem and the elongation zone of untreated and MeJA-treated roots. B, Nuclear area plotted against the distance from the QC. The dotted line indicates the average size at the time of the first endocycles. Representative results are shown for 6-d-old untreated (triangles) and MeJA-treated (diamonds) wild-type roots. Data were obtained from at least three biological replicates (n = 18–47).
Molecular Understanding of the Action of MeJA on Cell Cycle Regulation during Leaf Development Using Full-Genome Transcript Profiling
Publicly available microarray studies conducted on whole seedlings and single time points designed mainly to analyze the role of JAs with regard to their function during stress and defense are unsuitable to dissect the MeJA-dependent signaling underlying the physiological responses we have observed in planta. Hence, we have undertaken a novel full-genome transcription profiling to dissect the effect of MeJA on the cell cycle and differentiation using the well-established Arabidopsis leaf as a model system. We present here new data about MeJA inducibility, developmental regulation, as well as COI1 and AOS dependency at the same time.
Plant microarray studies have revealed that JA signaling alters gene expression and that genes induced by MeJA generally required COI1 in Arabidopsis (for review, see Balbi and Devoto, 2008; Acosta and Farmer, 2010; Avanci et al., 2010). We dissected here MeJA-dependent global transcriptional regulation during leaf development using the Arabidopsis ATH1 full-genome DNA arrays (Affymetrix; NASCARRAY-568/573). We performed global expression profiling of the first true leaves of Col gl1 and coi1-16B at the three developmental stages described and of aos at 13 DAS in the presence and absence of MeJA. The MeJA-treated and control samples were compared pairwise at the three time points for each genotype. Global statistical analysis in Col gl1 samples revealed differential regulation by MeJA of an increasing number of genes over time: 695, 1,886, and 2,261 up-regulated and 726, 1,433, and 2,239 down-regulated transcripts at 9, 13, and 19 DAS, respectively, under MeJA growth conditions. In the aos mutant at 13 DAS, 2,486 and 2,281 genes are up- or down-regulated, respectively, and about 80% of the genes differentially regulated in Col gl1 are also differentially regulated in this mutant. On average, about 90% of the MeJA differentially regulated genes in Col gl1 were COI1 dependent. This finding is largely in agreement with previous studies (Devoto et al., 2005; Chini et al., 2007; Thines et al., 2007). In treated Col gl1 plants, about 66% and 53% of the genes up- and down-regulated, respectively, at 9 DAS were similarly regulated at later time points (13 and 19 DAS). In addition, we observed that in untreated Col gl1, 656 and 2,143 transcripts were up-regulated and 1,596 and 3,012 transcripts were down-regulated at 13 and 19 DAS, respectively, when compared with 9 DAS. Globally, about 30% of the genes that are differentially regulated in Col gl1 during leaf development are COI1 dependent. Interestingly, about 60% of the developmentally regulated COI1-dependent genes are not differentially regulated by MeJA. This could be suggestive of a role for COI1 in the development of the leaf that is irrespective of MeJA (Supplemental Table S5). The differentially regulated transcripts in Col gl1 grown in the presence of MeJA were subjected to Gene Ontology (GO) analysis. As expected, among the main GO terms overrepresented for the genes up-regulated at 9 DAS are those associated with response to wounding and JA stimulus. At 13 and 19 DAS, we observed an enrichment of GO categories associated with the ribonucleoprotein complex, ribosome, and ribosome biogenesis (Supplemental Table S6).
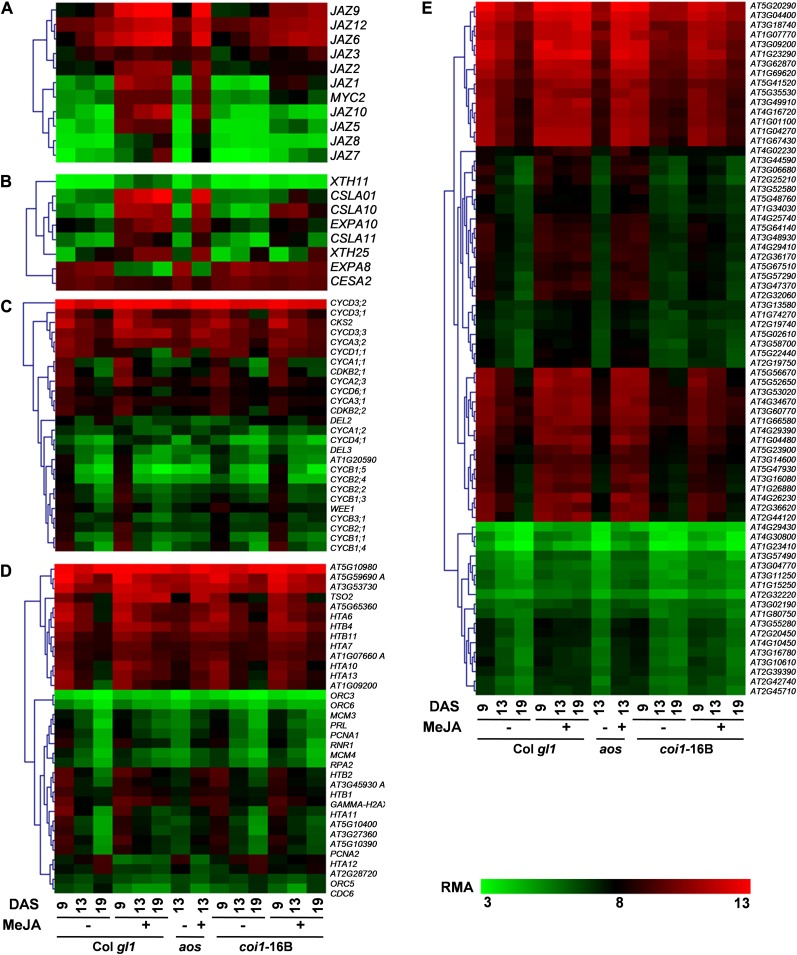
To validate the effectiveness of the continuous MeJA treatment used for microarray experimental conditions, the expression levels of transcripts known to be of JA-responsive genes were confirmed. Noteworthy and previously unreported, the expression of several JAZ genes as well as their ability to be induced by MeJA change during leaf development in both Col gl1 and aos (Fig. 7; Supplemental Fig. S5; Supplemental Table S5).
Figure 7.
Clustering display of classes of selected genes differentially regulated by MeJA and/or during development in Col gl1, aos, and coi1-16B at 9, 13, and 19 DAS. A, MeJA-responsive transcription factors. B, Cell expansion. C, Cell proliferation. D, S phase. E, Ribosomal. RMA-normalized values are displayed.
Several reports have suggested that JAs play a role in cell wall synthesis (Koda, 1997; Caño-Delgado et al., 2000; Ellis and Turner, 2001; Ellis et al., 2002), although the association between JAs and cell expansion is limited (Brioudes et al., 2009). Therefore, we monitored the expression of genes associated with cell expansion and loosening (López-Juez et al., 2008). Examples of significantly up-regulated and COI1-dependent genes are EXPA10, CSLA01, CSLA11, CSLA15, XTH11, and XTH25, and among the down-regulated genes are EXPA8 and CESA2 (Fig. 7; Supplemental Fig. S5; Supplemental Table S5).
Previous studies on the effect of MeJA on the cell cycle have been carried out on actively dividing and synchronized cell cultures (Świątek et al., 2002, 2004; Pauwels et al., 2008). Our study, in planta, uses, to our knowledge for the first time, the leaf system representing an asynchronous population of cells at various stages of the cell cycle to study the effect of JAs on the cell cycle. To further validate our data with a focus of gaining a better understanding of the action of MeJA on cell proliferation, we verified the expression of genes identified as differentially regulated in previously published experiments using the first leaf pair at similar time points during development (Beemster et al., 2005). Reassuringly, in our analysis, 93% of the known cell cycle genes fall into the described “proliferation” category, being consistently down-regulated during development (Fig. 7; Supplemental Fig. S5; Supplemental Table S5).
MeJA Activates Critical Regulators of Endoreduplication and Affects the Expression of Key Determinants of DNA Replication
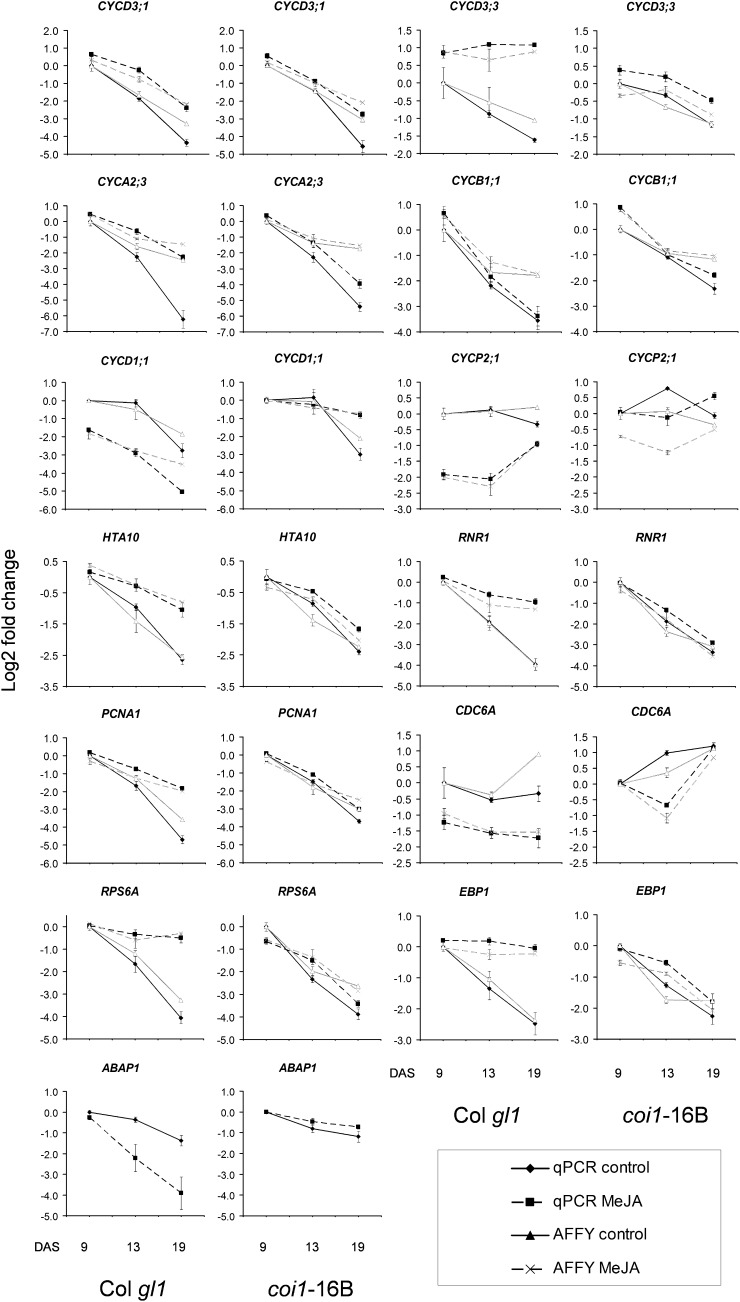
Based on our observations showing the MeJA-mediated negative effects on leaf development and reduced cell proliferation, our molecular analysis focused first on transcripts of core cell cycle regulators previously identified in a global analysis by Menges et al. (2005) and genome-wide analyses by Vandepoele et al. (2002) and López-Juez et al. (2008). We focused our attention on analyzing MeJA-dependent changes in gene expression, and we present here robust multiarray average (RMA) expression levels as hierarchical clustered heat maps of Col gl1, coi1-16B, and aos samples (Fig. 7; Supplemental Fig. S5). In addition, quantitative real-time (QRT)-PCR was used to monitor the expression of selected genes differentially regulated upon MeJA treatment in our microarray analysis. Namely, the results related to S-phase gene markers and DNA replication licensing genes (i.e. CDC6A, PCNA1, RNR1, and HTA10), regulators of cell division (i.e. CYCD1;1), markers with a role in the switch to endoreduplication (i.e. CYCA2;3, CYCD3;1, and CYCD3;3), M-phase gene markers (i.e. CYCB1;1 and CYCP2;1), and ribosomal genes (i.e. RPS6A and EBP1) are presented in Figure 8 for Col gl1 and coi1-16B and in Supplemental Figure S6 for aos for all three time points in addition to that analyzed for this mutant in the microarrays. Overall, for all genes tested, the QRT-PCR and microarray data showed extremely consistent expression patterns, indicating excellent reliability of the array results.
Figure 8.
QRT-PCR validation of microarray-detected changes in MeJA-responsive genes in Col gl1 and coi1-16B. Single RNA samples from each time point tested for microarray analysis were used in triplicate for relative QRT-PCR (as a ratio against the geometric mean of two internal control genes; Vandesompele et al., 2002). For details, see “Materials and Methods.” The genes were selected as representative of those differentially regulated in the first leaf pair of Col gl1 in comparison with coi1-16B during development upon continuous MeJA treatment. The black and gray lines represent the expression levels from QRT-PCR and from RMA-normalized microarray values from Affymetrix ATH1, respectively; continuous and dashed lines represent untreated and treated samples, respectively. Both QRT-PCR and microarray expression levels are shown as log2 fold change. Data are averages ± se of three independent biological replicates, two of which are independent from the microarray experiments. For each genotype, data from the 9-DAS untreated sample are used as the baseline expression value (calibrator).
In order to investigate whether a particular phase of the cell cycle or cellular process associated with it was affected in our experiments, we searched our data set for (1) the expression of 39 M-phase genes identified by Pauwels et al. (2008; cluster 5 therein) and described as being down-regulated by MeJA; (2) 76 genes associated with the DNA synthesis phase (S phase) and encoding prereplication complex (pre-RC) components selected primarily from previous studies (Masuda et al., 2004; Menges et al., 2005); as well as (3) histones (http://chromdb.org/; López-Juez et al., 2008). Our analysis shows, in agreement with previous studies, that many core cell cycle regulators such as cyclins are expressed at higher levels early during leaf development and that transcript levels decrease at later time points; however, the expression of the majority of these genes was not affected by MeJA (Supplemental Table S5). This is the case in our analysis for the expression of CYCB1;1, an M-phase marker gene. On the contrary, but still in agreement with Beemster et al. (2005), the gene CYCP2;1 is not developmentally regulated in untreated leaves irrespective of the genotype, whereas upon MeJA treatment, the expression of this other M-phase marker is down-regulated in Col gl1 and aos (Figs. 7 and 8; Supplemental Figs. S5 and S6). Our microarray study does not show consistent down-regulation of the M-phase genes as identified by Pauwels et al. (2008; Supplemental Table S5).
Notably, the transcripts of D3-type cyclins, negative regulators of endoreduplication such as CYCD3;1, CYCD3;2, and CYCD3;3 (Dewitte et al., 2007), are up-regulated in Col gl1 at the later time points when grown on MeJA-containing medium. The expression of CYCD3;1, and CYCD3;3 decreases during development irrespective of the genotype tested, in agreement with Beemster et al. (2005). These genes have been shown to repress the switch of the mitotic cell cycle toward endocycling (Dewitte et al., 2007), and strikingly, upon MeJA treatment, their levels of expression are increased consistently in Col gl1 and aos but to a greatly reduced extent in coi1-16B (Figs. 7 and 8; Supplemental Fig. S6; Supplemental Table S5). Such change may contribute to inhibiting the endoreduplication mechanisms triggered by MeJA (Fig. 3). Our QRT-PCR analysis confirmed the expression pattern of another negative regulator of endoreduplication, CYCA2;3 (At1g15570; Imai et al., 2006), as being similar to the above genes in Col gl1. With the exception of CYCD3;2 (At1g47210; Yu et al., 2003), all the above genes are significantly up-regulated also in the aos mutant (Fig. 8; Supplemental Fig. S6), providing independent evidence for the reproducibility of our results. Moreover, the cell cycle inhibitor KRP7 is also up-regulated in Col gl1 and the aos mutant at the later time points when grown on MeJA-containing medium. On the contrary, genes such as CYCD1;1, and CDK8/CDKE;1 are down-regulated by MeJA in a COI1-dependent manner in both Col gl1 and aos.
A fundamental regulatory mechanism controlling cell division is licensing DNA for replication at late G1 phase, allowing cells to progress into S phase (Inzé, 2005). This process is regulated by the assembly of ORC subunits, CDC6, CDT1, and MCMs onto replication origins, forming the pre-RC (Castellano et al., 2004; Blow and Dutta, 2005; Diaz-Trivino et al., 2005; Masuda et al., 2008; Costas et al., 2011). Out of 73 S-phase genes analyzed (López-Juez et al., 2008), 34 were differentially regulated by MeJA in Col gl1, and of these, 29 were up-regulated. Of these, 23 were also up-regulated in aos at 13 DAS in a COI1-dependent manner (Figs. 7 and 8; Supplemental Figs. S5 and S6). This category includes MCM3, MCM4, and MCM7, ORC3 and ORC6, PROLIFERATING CELL NUCLEAR ANTIGEN1 (PCNA1) and PCNA2, as well as RIBONUCLEOTIDE REDUCTASE1 (RNR1)/CRINKLY LEAVES8. These genes have been identified as putative target genes for the transcription factor E2FA in a study evaluating the transcriptional reprogramming caused by the overexpression of the proliferation-promoting, differentiation-inhibiting pair E2FA/DPa (Vandepoele et al., 2005). We analyzed the expression of 193 positively or negatively regulated putative E2FA-DPa target genes described by Vandepoele et al. (2005) and classified by López-Juez et al. (2008) and found that several of them are also differentially regulated in the presence of MeJA (Supplemental Fig. S5; Supplemental Table S5). We observed that MeJA increases the expression of some “E2F up” genes and at the same time delays or fails to induce that of “E2F down” genes. Regarding CDC6A, another known E2F target gene in Arabidopsis, it has been shown previously that its overexpression stimulated cell proliferation and triggered extra endocycles in leaf in a cell type-specific manner (Castellano et al., 2001, 2004). Our analysis shows that in untreated Col gl1, the expression of CDC6/CDC6A (At2g29680) does not change during development in either Col gl1 or aos, while CDC6A is down-regulated by MeJA in both genotypes at all time points and in coi1-16B at 13 DAS (Figs. 7 and 8). The regulation of CDC6A is consistent with the reduction of cell proliferation (Fig. 2) and the repression of endoreduplication at 13 and 19 DAS (Fig. 3) in MeJA-treated leaves.
We show here as well that a number of class H2A and H2B histones required for the onset of the S phase (for review, see Desvoyes et al., 2010) are up-regulated in the presence of MeJA in both Col gl1 and aos. For example, the expression of the H2A class histone HTA10, which is down-regulated during development, is induced by MeJA in Col gl1 and aos at the later time points in a COI1-dependent manner (Figs. 7 and 8; Supplemental Fig. S5). The PCNA functions as a sliding clamp for DNA polymerase and is a key factor in DNA replication as well as DNA repair and maintenance of heterochromatic regions, cell cycle regulation, and programmed cell death (Tsurimoto, 1998; Blow and Dutta, 2005; Raynaud et al., 2006). We found that the expression of PCNA1 and PCNA2 is induced by MeJA in Col gl1 and aos at 13 and 19 DAS but not in coi1-16B. Finally, the RNR, comprising two large (R1) and two small (R2) subunits, catalyzes a rate-limiting step in the production of deoxyribonucleotides needed for DNA replication and repair (Wang and Liu, 2006). In Arabidopsis, the large RNR1 subunit is encoded by a single-copy gene and the small RNR2 subunit by three genes (Philipps et al., 1995; Wang and Liu, 2006). In our experiments, the expression of RNR1 (At2g21790) is down-regulated at later stages during development, and its expression is induced by MeJA in Col gl1 and aos at 13 and 19 DAS but not in coi1-16B. RNR2A (At3g23580) is also up-regulated by MeJA at all time points in Col gl1 and aos, and this is COI1 dependent (Figs. 7 and 8; Supplemental Fig. S5; Supplemental Table S5). Altogether, while growth conditions in the presence of MeJA result in plant development retardation, these differential S-phase gene regulations suggest that all the machinery necessary to initiate DNA synthesis prerequisite for further cell proliferation is maintained or prepared in the cells as a stand-by “ready-to-go” status waiting for favorable growth conditions.
MeJA Maintains High Expression of Ribosomal Genes
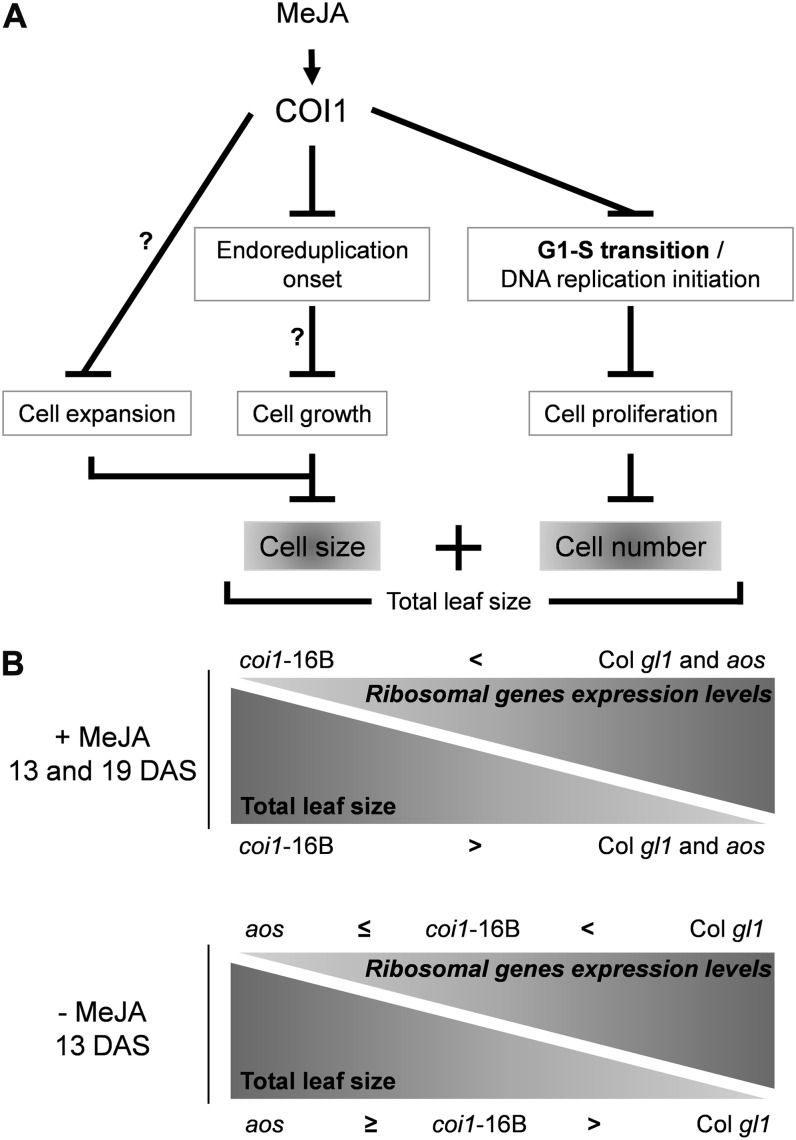
The rate of protein synthesis is key to cellular growth, and stimulation of the expression of genes encoding components of the translational apparatus occurs in cycling cells and upon the addition of auxin in plant cells (Trémousaygue et al., 2003; Gao et al., 2008). In multicellular organisms, it is debated whether the factors determining growth impose their influence on organs as a whole or whether they regulate growth and proliferation at the cellular level (Tsukaya and Beemster, 2006). Therefore, we also analyzed genes coding for ribosomal proteins (a nonredundant list of 166 genes as classified by López-Juez et al., 2008) to evaluate whether the translational capacity of the leaf cells was affected in the presence of MeJA. In our experiment, most ribosomal genes are down-regulated during leaf development in a COI1-independent manner, with the highest expression levels being at the earliest time point (9 DAS) in Col gl1. In the untreated leaves, the global expression level of these ribosomal genes shows an inverse correlation with leaf size: while it is evident that the size of aos and coi1-16B leaves at 13 DAS is bigger than Col gl1 (Fig. 2), the expression levels of the ribosomal genes in the two mutants are significantly lower at this time point (where the expression levels are aos ≤ coi1-16B < Col gl1; Fig; 7; Supplemental Fig. S5). Remarkably, upon MeJA treatment, the expression levels of 140 ribosomal genes are significantly up-regulated by MeJA at one time point at least, and the levels of expression of 72 of them are maintained as high as in the 9-DAS untreated samples throughout the development of the leaf in Col gl1. All these genes are also up-regulated by MeJA at 13 DAS in aos (Fig. 7; Supplemental Fig. S5), and the majority of them are differentially regulated by MeJA and COI1 dependent. A MeJA-dependent reduction in leaf size of aos and Col gl1 is associated with higher global gene expression levels (Fig. 7; Supplemental Fig. S5). The expression analysis of RIBOSOMAL PROTEIN S6A (RPS6A; At4g31700), for example, is down-regulated at later stages during development, and MeJA induces the expression of RPS6A in Col gl1 and aos at 13 and 19 DAS in a COI1-dependent manner (Figs. 7 and 8). ERBB-3-BINDING PROTEIN1 (EBP1) is a possible link between ribosome biosynthesis and cell proliferation, and its stability may influence plant growth via auxin signaling (Squatrito et al., 2004; Horváth et al., 2006). As the expression of EBP1 (At3g51800) was consistent with that of ribosomal genes up-regulated by MeJA, we checked the expression of the 103 EBP1 coregulated genes (Horváth et al., 2006). Of these, 94 are up-regulated by MeJA at one time point at least and are COI1 dependent (Supplemental Fig. S5).
DISCUSSION
Cell division, endoreduplication, and expansion are alternative strategies contributing to plant organ growth and size (Massonnet et al., 2010). JAs have been long associated with the inhibition of plant growth (Creelman and Mullet, 1997) and more recently have been described as negatively affecting cell cycle progression in cell cultures (Świątek et al., 2002, 2004; Pauwels et al., 2008) and generally mitosis in plants (Zhang and Turner, 2008). In this study, using Arabidopsis seedlings at 9, 13, and 19 DAS as a developmental model, a physiological characterization of the JA mutants coi1-16B and aos combined with global transcription profiling allowed us to dissect the link between JA signaling, cell cycle regulation, and leaf growth control and to provide novel insights in planta on the effect on the cellular machinery with respect to previous studies.
MeJA Reduces Both Cell Division and Cell Expansion and Delays the Onset of Endoreduplication during Leaf and Root Development
Our findings indicate that MeJA treatment strongly affects leaf area size in both Col gl1 and aos seedlings. The kinematic analysis (Fig. 2) revealed that MeJA reduces cell number and size in a COI1-dependent manner. These changes are associated with the reduction of cell proliferation and, during the transition phase (13 DAS), when the cell division rates decrease faster than the expansion rates (Donnelly et al., 1999), also with the negative effect of MeJA on cell expansion.
The determination of nuclear DNA content of whole first true leaves and cotyledons showed that the general pattern in the untreated samples was similar to that defined by Beemster et al. (2005; Fig. 3). In the aos mutant, leaves 1 and 2 exhibit a fraction of 8C DNA that is much higher than in Col gl1, suggesting that a lack of endogenous JAs may trigger an increase in the endocycling cell fraction, in agreement with the larger cell size observed at this stage (Figs. 2 and 3). In later leaf development stages of Col gl1 and aos, MeJA treatment increased the proportion of cells with lower ploidy levels. These findings indicate that the seedlings grown in the presence of MeJA had undergone fewer endoreduplication cycles, although without compensating with extra mitosis that is also temporarily arrested, and that COI1 mediates these responses. Altogether, these observations suggest that MeJA treatment negatively affects the switch from the mitotic cycle to the endocycle. The lack of endogenous JAs in the aos mutant may promote an earlier onset of endoreduplication. At the same time, this observation strengthens the direct role of MeJA in the delayed timing of the endocycle onset. Therefore, we hypothesize that plants continuously subjected to MeJA treatment might be kept in stand-by mode, while arresting the mitotic cycle, to exit it later during development. A delay in the onset of endoreduplication was observed already in Arabidopsis plants after 48 h of treatment (Supplemental Fig. S1). The cell cycle analysis (Fig. 4; Supplemental Fig. S2) revealed that removal from MeJA rapidly results in cellular proliferation recovery, possibly as a result of the stand-by mode induction. It is remarkable that within the 48-h time frame, the seedlings are capable of flushing out MeJA, cease to respond, and show cell cycle changes. Therefore, more than an exit from proliferation, we observe inhibition and entry in stand-by mode; as a result, both the rate of division and entry into endoreduplication are negatively affected. Interestingly, several analyses report cross talk between JAs and GAs (Navarro et al., 2008; Cheng et al., 2009; Hong et al., 2012; Yang et al., 2012). This other class of plant-specific hormones plays crucial roles in the mechanisms promoting plant growth and development, notably by triggering the degradation of plant growth repressors, the DELLA proteins (for review, see Harberd et al., 2009). Reinforcing the role of DELLAs as integrators of plant hormone and environmental change responses, recent studies have shown the interaction of DELLAs with JAZ proteins (Hou et al., 2010; Wild et al., 2012; Yang et al., 2012) as well as the MYC2 transcription factor (Hong et al., 2012; Wild et al., 2012). In addition, DELLAs negatively affect plant organ growth by reducing both cell proliferation and cell expansion (Achard et al., 2009). It is then tempting to speculate that JA-triggered growth inhibition could be fine tuned by binding competition among JAZ, DELLA, and MYC2 proteins.
Due to the greater reduction observed in the root length (80%) rather than the cell length, the data suggest that exogenous MeJA inhibits both root-cell proliferation and root-cell elongation, the latter to a greater extent. These observations are in agreement with those showing that MeJA reduces cell size in leaves and that this process is COI1 dependent. The process of endoreduplication normally occurs in expanding root hairs (Sugimoto-Shirasu and Roberts, 2003; Sugimoto-Shirasu et al., 2005; Guimil and Dunand, 2007). The comparison of the effect of MeJA and the role of COI1 in root cells and root hair cells would suggest that COI1 has a differential role in mediating endoreduplication and/or cell expansion. This is also consistent with data showing that COI1 was required for 1-aminocyclopropane-1-carboxylic acid-induced root growth inhibition but was not required for 1-aminocyclopropane-1-carboxylic acid-induced root hair elongation (Adams et al., 2010). The quantification of nuclear surface area with H2B-YFP showed that the endocycle is initiated farther away from the QC (Fig. 6). These observations strengthen the role of MeJA in inhibiting proliferation in G1/S as well as in delaying the onset of endoreduplication in root cells as in leaves. Moreover, the lack of a MeJA effect on the cellular organization of the root meristem (Supplemental Fig. S3) highlights further that a block of endoreduplication is not necessarily indicative of cell cycle arrest.
Effects of MeJA on the Expression of the Core Cell Cycle Regulator CYCB1;1
The repression of pCYCB1;1:Dbox-GUS after 48 h of exposure to MeJA (Fig. 5) indicates that MeJA reduces the number of cells that are actively dividing, contributing to the reduction of the number of cells forming the leaf. It is at the G2/M transition that the cell can choose to switch to the endocycle rather than proceed through mitosis (Zhiponova et al., 2006). Recently, Skirycz et al. (2011) distinguished different mechanisms of cell cycle inhibition depending on the duration of continuous osmotic stress during early leaf development in Arabidopsis. Previously, Skirycz and Inzé (2010) showed that within 72 h of stress imposition, cell proliferation in leaves was again identical to the control conditions and suggested that leaves adapted to a restrictive environment established a new steady state, referred to as the adaptive growth response. In our analysis, it is also possible, therefore, that the 48-h MeJA treatment induces different responses compared with the continuous treatment applied elsewhere in our study. Thus, it is possible that we are monitoring acute responses after 48 h compared with adaptive responses upon continuous treatment. The inhibition of the G1/S transition mirrored by an increase in G1 following MeJA treatment (Fig. 4) could not only explain a decreased mitotic activity but at the same time cause a delayed onset of endoreduplication (Fig. 3). This could also explain the lack of a compensation effect where increased cell size would make up for reduced cell numbers. Interestingly, Massonnet et al. (2010) postulate that leaf growth functions as a hub driving cellular processes where leaf growth is necessary to drive endoreduplication and a direct compensation mechanism between cell division and endoreduplication does not necessarily occur. Our data show that MeJA treatment reduced the expression of pCYCB1;1:Dbox-GUS as well as the relative size of the cell proliferation zone in both leaf and root after 48 h. In addition, in leaves, MeJA delays the onset of endoreduplication as well as blocking G1 prior to S transition. While it is not debatable that a reduction in CYCB1;1 expression can be associated with a decrease in G2/M, it cannot be excluded that this observation is actually mirroring the effect of the inhibition of the G1/S transition.
The Transcriptional Regulation of Specific Clusters of Cell Cycle Genes Supports a Role for MeJA in the Switch from Mitotic Cycle to Endocycle while Preparing for Recovery
Recently, targeted proteomics has uncovered and mapped several interactions of core cell cycle genes (Boruc et al., 2010; Van Leene et al., 2010, 2011). While our study has been informed by previous ones carried out with JA and MeJA in synchronized, actively dividing Bright Yellow-2 or Arabidopsis cell cultures (Świątek et al., 2002, 2004; Pauwels et al., 2008), it has revealed several differences with important physiological relevance for the whole plant system. These previous results suggested that the effect of JA is dependent on the cell cycle stage of the cells when treated. We recognized, therefore, the importance of analyzing the effect of MeJA in the leaf system, which contains cells at different phases of the cell cycle and is also undergoing differentiation to highlight specific differentially expressed regulators. We successfully verified in our study that the expression of genes associated with cell proliferation in leaf is down-regulated during development, but we showed that the majority of these were not differentially regulated by MeJA (Fig. 7). In our experiment, the obvious absence of expression blockage of genes associated with proliferation or M phase, shown by Pauwels et al. (2008) in cell cultures, could be explained by the inherent properties of the leaf system containing actively dividing and differentiating cells in varying ratios according to the developmental stage analyzed. This observation further strengthens the relevance of our analysis for plant development. Actually, the system certainly proved successful in highlighting specific clusters of genes with physiological relevance to our observations.
The null mutation cyca2;3 correlates with a faster endocycle onset, and its overexpression results in a cellular phenotype similar to that of the Arabidopsis overexpressing tobacco CYCA3;2, showing up-regulated S-phase histones but inhibited cell differentiation and endoreduplication (Yu et al., 2003). Moreover, the switch to the endocycle requires that at least the expression of CYCD3;1 is turned off, and the triple mutant cycd3;1-3 showed premature onset of endoreduplication, indicating that the normal function of CYCD3 is to delay this (Dewitte et al., 2003, 2007). According to current models, CYCD3;1 in complex with CDKA;1 regulates cell cycle entry by phosphorylation of RBR1, leading to the release of RBR1-bound E2F transcription factors to drive the expression of genes required for the cell cycle phase transitions (for review, see Magyar, 2008). In accordance, cycd3;1-3 has smaller organs with fewer cells, whereas ectopic expression of CYCD3;1 inhibits organ growth by repressing differentiation, further supporting its role in maintaining the balance between cell proliferation and differentiation (Dewitte et al., 2003, 2007). In our study, we show that the transcripts of CYCD3;1, CYCD3;2, and CYCD3;3 genes are up-regulated at the later time points during development upon MeJA treatment both in Col gl1 and aos in a COI1-dependent manner. A similar up-regulation pattern is associated with CYCA2;3. Taken together, these unforeseen results support our physiological observations that the onset of endoreduplication is delayed upon continuous MeJA treatment (Figs. 7 and 8; Supplemental Table S5). At the same time, this would contribute to the maintenance of the MeJA-induced stand-by mode. Arabidopsis cycd1;1 loss-of-function homozygous mutants show delayed onset of cell division activity and fewer dividing root apical meristem cells, resulting in a lower rate of cell division. On the contrary, the overexpression of early-activated D-type cyclins such as CYCD1;1 promotes the increase of the number of cycling cells within the root apex (Masubelele et al., 2005). Consistent with the above, and given that CYCD1;1 is transcriptionally reduced in Col gl1 and aos upon MeJA treatment, we conclude that MeJA-dependent inhibition of cell division onset in Arabidopsis may also occur through CYCD1;1 and that this mechanism is COI1 dependent.
The activity of CDK-cyclin complexes controlling the cell cycle at different checkpoints is suppressed by negative regulators known as Kip-related proteins (KRPs) in Arabidopsis (De Veylder et al., 2011), a class of CDK inhibitors coordinating cell cycle progression, The expression pattern of the cell cycle inhibitor KRP7 is comparable to those of CYCD3;3 and CYCA2;3 in our analysis (Supplemental Fig. S5; Supplemental Table S5). The reduction of the expression of CYCP2;1 by MeJA adds to the evidence that this hormone may affect the ability of the cell to initiate division (Fig. 8).
The mechanisms regulating growth by cell expansion remain to be elucidated (for review, see Powell and Lenhard, 2012). Our investigation of a number of genes with a recognized role in cell expansion (as classified by López-Juez et al., 2008) revealed a complex picture with about 30% of the genes in this category being up- or down-regulated in a COI1-dependent manner during development, which is certainly worth investigating in future studies.
MeJA Negatively Affects Cell Cycle Progression and the Initiation of DNA Replication, Maintaining the Cell’s Ability to Recover from Treatment
CDC6 and CDT1 are essential factors for DNA replication licensing in eukaryotes because they recruit the MCM DNA helicases that open the replication forks (for review, see De Pamphilis, 2003). Namely, it was observed that overexpression of CDC6/CDC6A (At2g29680) or CDT1a did not affect overall plant development but stimulated endoreduplication (Castellano et al., 2001, 2004), and these two proteins accumulate in limiting amounts for DNA replication licensing. The repression of the expression of CDC6 in our experiments (Figs. 7 and 8) suggests that MeJA may act on this key limiting factor to stall the S phase prior to replication and, consequentially, the G2 phase prior to endoreduplication. These data support the hypothesis that MeJA contributes to inhibiting the initiation of DNA replication, in agreement with the G1 increase upon MeJA treatment shown by our analyses in both Arabidopsis and N. benthamiana (Fig. 4). Other pre-RC components such as MCMs and ORCs, as well as a number of genes encoding class H2A and H2B histones, are transcriptionally up-regulated upon MeJA treatment at later stages of development and are COI1 dependent (Fig. 7; Supplemental Fig. S5). While this may not be crucial for blocking DNA replication/initiation of S phase, it may represent an indicator of the function of MeJA/JAs in preparing the cell for a fast recovery once the inhibition is released. The expression of CDT1A and CDT1B does not seem to be affected by MeJA; nevertheless, it has been shown that Armadillo BTB Protein1 (ABAP1) directly interacts with AtORC1a/b and AtCDT1 homologs and might possibly hamper DNA replication through direct interaction with pre-RC. In AtTCP24-mediated signaling, ABAP1 associates with AtTCP24 and controls AtCDT1a/b homoeostasis by enhancing the transcription repression of AtCDT1A/B (Masuda et al., 2008). The probe for ABAP1 is not located on the Affymetrix ATH1 chip, so we analyzed its expression using QRT-PCR. MeJA negatively affects the expression of ABAP1 in Col gl1 and aos at the later time points in a COI1-dependent manner (Fig. 8; Supplemental Fig. S6). It is intriguing to think that decreased ABAP1 expression upon MeJA treatment could correlate to a decreased complex with TCP24 and that this may result in diminished inhibition of CDT1 expression. Whether MeJA down-regulation of CDC6 could represent the limiting factor for DNA replication remains to be clarified. ABAP1 also could be repressed as part of the stand-by response.
Our experiments show that the expression of the sliding clamps PCNA1 and PCNA2 is induced by MeJA and is COI1 dependent (Fig. 8; Supplemental Fig. S6). PCNA promoters have been shown to contain the cis-regulatory elements “site II motif” and telobox. A conserved topological association between these two elements is observed in the promoter of genes expressed in cycling cells as well as in Arabidopsis genes encoding ribosomal proteins. Interestingly, teloboxes are also observed within the promoters of other plant genes expressed in late G1 such as RNRs, raising the possibility that this element could be involved in a common regulatory process that connects the expression of a set of genes at the G1/S transition (Trémousaygue et al., 2003). RNR2A is up-regulated by MeJA and is COI1 dependent. TSO2 expression is similarly regulated (Fig. 7; Supplemental Fig. S5), TSO2 encodes one of the three ribonucleotide reductase small subunit genes in Arabidopsis, and the double mutant tso2 rnr2a showed extensive programmed cell death, which correlates with accumulating DNA damage. TSO2 is sufficient to provide enough RNR activity to support normal plant development (Wang and Liu, 2006). In Arabidopsis, besides developmental phenotypes, mutation in RNR1 (csl8) does not alter endoreduplication and limited the supply of deoxyribonucleotides (Garton et al., 2007). The association of MeJA with increased levels of RNR1, RNR2A, and TSO2 may highlight a possible role for this hormone in maintaining normal development and genome stability while reacting to the stress.
We found that MeJA treatment up-regulates the expression levels of nucleosome core S-phase histones of classes H2A and H2B (Fig. 7; Supplemental Fig. S5). While it is not known at this stage whether these changes will be accompanied by local changes in the structure of the chromatin and/or have an effect on cell fate, it appears here that MeJA has an active role in ensuring the availability of these histones. This could represent an additional way for the cell to be promptly ready to restore homeostasis once the stress is over. It will be interesting to investigate whether MeJA has an active functional role in regulating these mechanisms also in the light of the previously reported regulation of histone deacetylases by JAs (Devoto et al., 2002; Zhou et al., 2005; Wu et al., 2008).
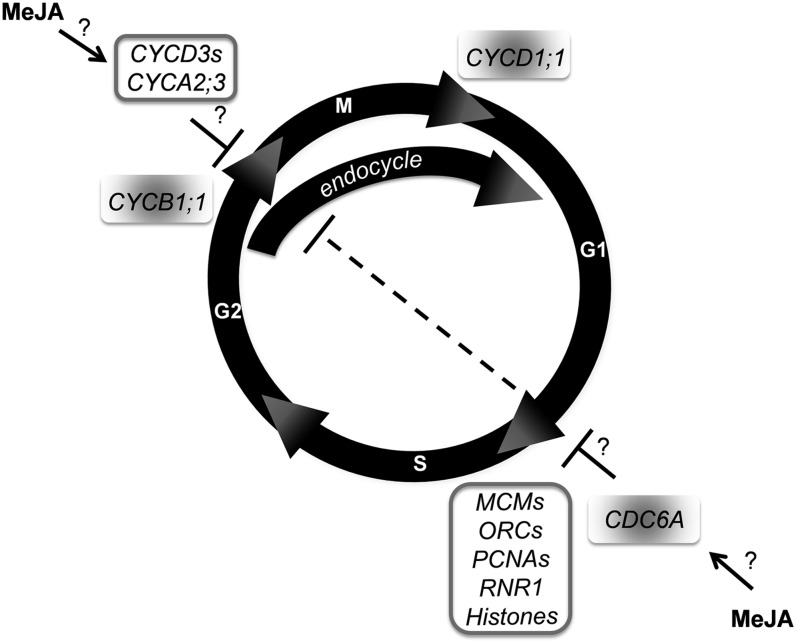
Overall, our data suggest that while MeJA participates in the repression of DNA replication, where MeJA-mediated down-regulation of CDC6 would represent one of the limiting factors for DNA replication, this hormone seems to prepare the cells to progress through the cell cycle once released from the treatment. Altogether, it is clear that MeJA plays a role in these complex mechanisms ultimately regulating growth that will be worth clarifying through future studies. These novel findings are included in our model summarizing the main effects exerted by MeJA on the cell cycle and endocycle mediated by COI1 (Fig. 9). Our flow cytometry analysis showed that MeJA delays the onset of endoreduplication. At the same time, MeJA increases the expression of endoreduplication repressors such as D3-type cyclins and of CYCA2;3 genes as well as down-regulates the expression of the endocycle promoter CDC6A. Supportive evidence is also provided by the reduction in leaf cell size and a reduction of the nuclei size in the treated root, suggesting a delay in differentiation. The down-regulation of CDC6A, a well-known DNA replication licensing gene, might indicate that MeJA could hinder the function of the pre-RC and contribute to inhibiting the G1/S transition. Our cell cycle analysis, showing that an increase in G1 follows MeJA treatment already after 48 h, would support this hypothesis. However, MeJA positively regulates the expression of S-phase genes, several of them encoding pre-RC components, DNA replication, and DNA repair. This could indicate that MeJA maintains a competent environment to guarantee prompt recovery after stress. The inhibitory effect of MeJA on the mitotic activity is suggested by the decrease in the expression of the G2/M-phase marker CYCB1;1:Dbox-GUS in both leaf and root. The reduction in leaf cell number we observed is in agreement with the above. MeJA-dependent repression of the positive regulator of cell division CYCD1;1 might also contribute to inhibiting mitotic activity.
Figure 9.
Main MeJA effects on the cell cycle and endocycle mediated by COI1. Depicted are a normal mitotic cell cycle and an endocycle where cells rereplicate DNA in the absence of intervening mitoses. White and gray-shaded shapes indicate genes up- and down-regulated by MeJA, respectively.
MeJA Maintains a Cellular Stand-By Mode by Keeping High the Expression of Ribosomal Genes
We have observed that upon MeJA treatment, the expression levels of many ribosomal genes are up-regulated by MeJA and maintained at constitutive levels, as in untreated young seedlings (Fig. 7). Among these is RPS6A, one of the two functionally equivalent cytoplasmic RPS6s reported to promote plant growth (Creff et al., 2010). Interestingly, both rps6a and rps6b knockout mutants show a growth-retarded phenotype, and hypomorphic rps6a alleles have been linked to negative effects on cell proliferation (Horiguchi and Tsukaya, 2011). Another example is EBP1, a nucleolar double-stranded RNA-binding protein. Horváth et al. (2006) have shown that, via modulating the levels of EBP1 in potato (Solanum tuberosum) and Arabidopsis, this gene regulates plant organ growth and affects the expression of different cell cycle genes as well as RBR1 protein level. These authors also showed that this candidate cell-autonomous factor stimulates the expression of CYCD3;1. Our observations are consistent with this in showing that MeJA acts positively on the expression levels of these two genes as well as on EBP1 coregulators and additional cyclins involved in endoreduplication (Supplemental Fig. S5). This is also in agreement with Massonnet et al. (2010) suggesting that endoreduplication in leaf cells could be controlled by leaf growth itself. MeJA seems to contribute to the regulation of both by affecting mitosis, delaying endoreduplication and the start of replication, but at the same time establishing a replication-competent environment to guarantee a prompt recovery. The maintenance of high transcript levels of ribosomal proteins as well as histones and pre-RC components by MeJA could put the cell in a stand-by mode ready to go. As a result, to the greatest leaf MeJA-dependent growth inhibition (Fig. 2) correspond the highest levels of ribosomal genes (Fig. 7; Supplemental Fig. S5). A model summarizing some of the key processes revealed to be regulated by continuous MeJA treatment to control leaf size is proposed here (Fig. 10).
Figure 10.
Working model summarizing some of the key processes revealed to be regulated by continuous MeJA treatment to control leaf size. A, The combination of cell size and cell number determines the total leaf size, and this is regulated by MeJA. B, Inverse correlation between leaf size and ribosomal gene expression. To a MeJA-dependent reduction in leaf size of aos and Col gl1 corresponds higher global ribosomal gene expression levels. (This figure is adapted and expanded from Sugimoto-Shirasu and Roberts, 2003.)
CONCLUSION
Our study shows that MeJA contributes to driving two possibly independent strategies to rescheduling the energy between stress responses and growth. Blocking mitosis (G2/M transition) could represent a fast way to block cell division and keep the plant in a ready-to-go state to quickly reinstate homeostasis once the stress signal is released. Alternatively, blocking the cell cycle before the completion of DNA replication (G1/S) could represent a more cost-effective way of slowing down without embarking on the expensive process of DNA replication. In this scenario, MeJA may contribute to holding back DNA replication by negatively affecting the assembly of the pre-RC and at the same time maintaining appropriate cell conditions to ensure a fast recovery. Our results in planta demonstrate that the definition of “JA-mediated inhibition of cell proliferation” accepted so far is rather simplistic, and we have identified a much more complex regulatory network, containing novel elements balancing MeJA-regulated trade off between growth ability and stress tolerance, that will be exciting to study further. Our research sheds light on a rather unprecedented MeJA-regulated COI1-dependent leaf growth control mechanism and supports the existence of this mechanism in Arabidopsis as well as in N. benthamiana, opening the doors to uncoupling MeJA stress to increase yield and to increase the production of defense-related secondary metabolites.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
The Arabidopsis (Arabidopsis thaliana) lines coi1-16 (At2g39940; Ellis and Turner, 2002), subjected to backcrossing to eliminate a pen2 mutation (Westphal et al., 2008), aos (At5g42650; N6149), H2B-YFP (Campilho et al., 2006), and pCYCB1;1:Dbox-GUS (Colón-Carmona et al., 1999) have been described. For clarity, we refer to the coi1-16 line used here as coi1-16B (for backcrossed). The mutant lines coi1-16B and aos are in the Col gl1 background that is referred to as the wild type in this study. Because environmental conditions during seed formation as well as seed storage duration can affect seed vigor, all the analyses on coi1-16B, aos, and Col gl1 were performed on synchronized seeds (i.e. harvested at the same time and in the same light conditions). Germination tests were also performed to confirm that in our conditions, no significant difference in germination time was observed between the lines used. Col gl1, aos, coi1-16B, and Nicotiana benthamiana seeds were sterilized in 100% ethanol for 2 min. Ethanol was removed, and 20% sodium hypochloride (12%, w/v) and 0.1% Tween 20 were added to seeds and mixed for 10 min. Seeds were repeatedly washed with sterile deionized water before sowing on buffered (1% MES, pH 5.8) 0.5× Murashige and Skoog medium (Duchefa) supplemented with 1% Suc and 0.8% phytoagar (Duchefa), containing 50 µm MeJA (Bedoukian Research) when indicated. For in vitro culture, plants were grown at a density of one plant per 3 cm2. After cold treatment in the dark for 3 d (stratification), seeds were incubated under 16-h/8-h light/dark cycle conditions at 22°C with a light intensity of 100 to 120 µmol m−2 s−1.
Leaf Measurements
First true leaves of Arabidopsis seedlings grown on horizontal plates as described above were collected at 9, 13, and 19 DAS, cleared in 100% ethanol, mounted in dl-lactic acid on microscope slides, and photographed. Leaf abaxial epidermal cells (40–100 cells) were drawn for four to 21 leaves with a DMLB microscope (Leica) using a DIC objective. Photographs of leaves and drawings were used to measure the leaf area and calculate the average cell area and number with ImageJ software (http://rsb.info.nih.gov/ij/). Leaf and cell areas were used to calculate cell numbers.
Root Measurements
In both MeJA-treated and untreated plants, primary root, cell, and root hair lengths were measured in 6-d-old seedlings vertically grown (75° to the horizontal). The length of a cell was determined between two fully expanded root hairs on the same cell file via high-resolution images using DIC microscopy and ImageJ software. This was done for six to 10 cells per plant. The length of fully grown root hairs was also measured. A minimum of eight seedlings per genotype were analyzed, and three independent biological replicates were performed. Percentage inhibition in the MeJA-treated seedlings was calculated. As described by Ishida et al. (2009), root meristem organization was visualized using seedlings stained with 5 µg mL−1 propidium iodide solution, and root nuclei were visualized using transgenic plants harboring H2B-YFP fusion protein constructs. Samples were observed using a Carl Zeiss LSM510 META confocal laser microscope. To measure nucleus size in the root meristem, three to six confocal optical sections, collected at approximately 1-µm intervals using identical confocal settings, were merged. Surface areas of individual nuclei within the same epidermal cell files were measured using ImageJ software.
GUS Staining Analysis
Arabidopsis pCYCB1;1:Dbox-GUS seedlings were germinated and vertically grown on control medium for 10 DAS and subsequently transferred to control or 50 µm MeJA-containing medium for 48 h. Histochemical GUS staining was performed as described by Ishida et al. (2009) and imaged using DIC microscopy for the roots or a dissecting microscope (Nikon) for the leaves.
Flow Cytometry Experiments and Ploidy Measurement
Ploidy levels were measured using the Cystain UV Precise P high-resolution DNA staining kit (Partec) according to the manufacturer’s instructions and adapting a procedure from Dolezel et al. (2007). Briefly, nuclei from leaves or cotyledons of seedlings grown under in vitro conditions were released in nuclei extraction buffer (Partec) by lightly chopping the leaves with a razor blade, stained with 4′,6-diamidino-2-phenylindole buffer, and filtered through a Celltrics 30-μm mesh (Partec). At least 20,000 nuclei isolated from approximately 10 to 15 pairs of leaves/cotyledons were used for each ploidy measurement using a Partec PAS flow cytometer and FloMax software (Partec) recording the relative fluorescence intensities. The FloMax software was also used to perform cell cycle analysis. Flow cytometry experiments were repeated at least three times for each genotype using independent biological replicates.
Microarray Experiments and Data Analysis
The first true leaves of Col gl1 and coi1-16B at 9, 13, and 19 DAS grown horizontally on Murashige and Skoog medium as described above with and without 50 µm MeJA were collected, snap frozen immediately, and stored at −80°C until further RNA extraction. aos samples were collected at 13 DAS only. Total RNA was isolated using Tri Reagent according to the manufacturer’s instructions (Sigma-Aldrich). Isolated RNA was further purified using RNeasy mini columns (Qiagen) according to the manufacturer’s instructions. After quality controls, copy RNA, labeling, hybridization, and scanning of the high-density oligonucleotide microarrays (Arabidopsis ATH1 genome array; Affymetrix) were performed at the Nottingham Arabidopsis Stock Centre microarray platform (http://affymetrix.arabidopsis.info/). Data were generated from two independent biological replicates for all samples but the 19-DAS time point, for which only one replicate was available (Affymetrix; NASCARRAY-568/573). ATH1 Expression Profiling and Data Analysis RNA samples were hybridized to single Affymetrix ATH1 Genome arrays at the Nottingham Arabidopsis Stock Centre.
Expression data were processed with RMA (background correction, normalization, and summarization) as implemented in BioConductor (Irizarry et al., 2003a, 2003b; Gentleman et al., 2004). The microarray data were checked for anomalous RNA degradation by ordering and grouping the probes for the ATH1 array according to their 5′ to 3′ position. In order to check the reproducibility of the biological replicates, (1) dendrograms using hierarchical clustering of the correlation between any two pairs of CEL files based on the normalized expression of all their probe sets and (2) scatterplots of normalized expression values for individual genes between replicates and samples were drawn (Supplemental Fig. S7, A and B). The BioConductor package Limma, which employs a hierarchical model to estimate the t statistics, was used to identify differentially expressed genes (Smyth, 2004). The default settings from the front-end package affylmGUI were employed (Wettenhall et al., 2006). The hierarchical model applied by Limma allows computing Student’s t tests for samples where only one replicate was available. False discovery rate-corrected P < 0.05 was used as a cutoff (Benjamini and Hochberg, 1995). Finally, any genes that did not display at least a 1.5-fold change between any two genotypes or time points or between treated versus untreated samples within a genotype were removed. The remaining list was considered a robust set of differentially expressed genes. Venn diagrams with the numbers of genes up-regulated or down-regulated at each time point were used to compare the results between the three genotypes (Oliveros, 2007). We performed GO analysis using the DAVID package (Huang et al., 2007a, 2007b) of the differentially regulated transcripts in Col gl1. Clustered heat maps and similarity trees were produced with The Institute for Genomic Research Multi Experiment Viewer (www.tm4.org/mev/).
QRT-PCR Analysis
Using plant material grown as described above for the microarray experiments, purification of total RNA from plant material was performed as described using the RNeasy plant mini kit (Qiagen). Analysis of total RNA yield was performed on a Nanodrop spectrophotometer (Labtech). One microgram of total RNA from each sample was used for complementary DNA (cDNA) preparation. cDNA preparation was performed using the QuantiTect_Reverse Transcription kit (Qiagen) that includes a genomic DNA removal step. Real-time amplification in the presence of SYBR Green was performed using Precision Mastermix as per the manufacturer’s instructions. Accordingly, no more than one-tenth of the final PCR volume derived from the finished reverse transcription reaction was used. Standard amplification protocols were used and carried out on a Rotor-gene Q real-time machine (Qiagen; Corbett Research RG-6000). All reactions took place in triplicate. Melting curves were analyzed to check primer specificity. We assessed the primer reaction efficiencies using the Comparative Quantitation Report for all genes of interest and the reference genes. All primer reaction efficiencies were comparable around 90%. Levels of each transcript as a ratio against the geometric mean (Vandesompele et al., 2002) of the two internal control genes At5g55840 and At5g17510, identified using the Arabidopsis geNorm reference gene selection kit (Primerdesign) and the qBase software package (Hellemans et al., 2007), were quantified using the comparative threshold cycle calculation. For each genotype, data from the 9-DAS untreated sample were used as the baseline expression value (calibrator). Primers were designed using QuantPrime (http://quantprime.mpimp-golm.mpg.de/; Arvidsson et al., 2008) and settings to maximize the cDNA and single primer specificity. The primers used in this study are listed in Supplemental Table S7.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Alteration of nuclear DNA content and delay of the onset of endoreduplication after 48 h of MeJA treatment.
Supplemental Figure S2. Continuous MeJA treatment alters the cell cycle progression during leaf development.
Supplemental Figure S3. MeJA treatment does not significantly affect the size of the root meristem.
Supplemental Figure S4. MeJA inhibition of growth and cell elongation in roots.
Supplemental Figure S5. RMA expression levels and clustering display of main gene categories analyzed.
Supplemental Figure S6. QRT-PCR validation of microarray-detected changes of MeJA-responsive genes in aos.
Supplemental Figure S7. Quality control of microarray data.
Supplemental Table S1. MeJA-mediated inhibition of the leaf growth of Arabidopsis mutants.
Supplemental Table S2. Flow cytometric analysis of first true leaves or cotyledons of Arabidopsis Col gl1, coi1-16B, and aos in absence or presence of MeJA.
Supplemental Table S3. MeJA effects on root growth.
Supplemental Table S4. Distance of the quiescent center from the first visible elongating cell.
Supplemental Table S5. Genes affected by MeJA treatment during leaf development.
Supplemental Table S6. Main GO categories for genes up-regulated by MeJA.
Supplemental Table S7. Primers used for QRT-PCR.
Acknowledgments
We thank Amar Patel and Danielle Minns for technical help and Fred Ausubel, Lazslo Bögre, Enrique Lopez-Juez, and Fran Robson for critical comments. A.D. designed the research; A.D., S.N., B.M., N.T., T.I., T.-L.T., and V.B. performed the research; A.D., S.N., B.M., N.T., T.I., and H.S. analyzed the data; and A.D., S.N., B.M., and K.S. wrote and edited the paper.
Glossary
- JA
jasmonate
- MeJA
methyl jasmonate
- JAZ
jasmonate ZIM-domain
- DAS
d after stratification
- EI
endoreduplication index
- DIC
differential interference contrast
- QC
quiescent center
- H2B-YFP
histone 2B-yellow fluorescent protein
- GO
Gene Ontology
- RMA
robust multiarray average
- QRT
quantitative real-time
- pre-RC
prereplication complex
- cDNA
complementary DNA
References
- Achard P, Gusti A, Cheminant S, Alioua M, Dhondt S, Coppens F, Beemster GT, Genschik P. (2009) Gibberellin signaling controls cell proliferation rate in Arabidopsis. Curr Biol 19: 1188–1193 [DOI] [PubMed] [Google Scholar]
- Acosta IF, Farmer EE. (2010) Jasmonates. The Arabidopsis Book 8: e0129, doi/10.1199/tab.0129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams E, Devoto A, Turner J. (2010) COI1, a jasmonate receptor, is involved in ethylene-induced inhibition of Arabidopsis root growth in the light. J Exp Bot 61: 4373–4386; erratum Adams E, Devoto A, Turner J (2011) J Exp Bot 62: 5735–5736 [Google Scholar]
- Arvidsson S, Kwasniewski M, Riaño-Pachón DM, Mueller-Roeber B. (2008) QuantPrime: a flexible tool for reliable high-throughput primer design for quantitative PCR. BMC Bioinformatics 9: 465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attwooll C, Lazzerini Denchi E, Helin K. (2004) The E2F family: specific functions and overlapping interests. EMBO J 23: 4709–4716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avanci NC, Luche DD, Goldman GH, Goldman MH. (2010) Jasmonates are phytohormones with multiple functions, including plant defense and reproduction. Genet Mol Res 9: 484–505 [DOI] [PubMed] [Google Scholar]
- Balbi V, Devoto A. (2008) Jasmonate signalling network in Arabidopsis thaliana: crucial regulatory nodes and new physiological scenarios. New Phytol 177: 301–318 [DOI] [PubMed] [Google Scholar]
- Beemster GT, De Veylder L, Vercruysse S, West G, Rombaut D, Van Hummelen P, Galichet A, Gruissem W, Inzé D, Vuylsteke M. (2005) Genome-wide analysis of gene expression profiles associated with cell cycle transitions in growing organs of Arabidopsis. Plant Physiol 138: 734–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beemster GT, Fiorani F, Inzé D. (2003) Cell cycle: the key to plant growth control? Trends Plant Sci 8: 154–158 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. (1995) Controlling the false positive rate: a practical and powerful approach to multiple testing. J R Stat Soc B 57: 289–300 [Google Scholar]
- Berger S. (2002) Jasmonate-related mutants of Arabidopsis as tools for studying stress signaling. Planta 214: 497–504 [DOI] [PubMed] [Google Scholar]
- Blow JJ, Dutta A. (2005) Preventing re-replication of chromosomal DNA. Nat Rev Mol Cell Biol 6: 476–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boruc J, Van den Daele H, Hollunder J, Rombauts S, Mylle E, Hilson P, Inzé D, De Veylder L, Russinova E. (2010) Functional modules in the Arabidopsis core cell cycle binary protein-protein interaction network. Plant Cell 22: 1264–1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosco G. (2010) Cell cycle: retinoblastoma, a trip organizer. Nature 466: 1051–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudolf V, Lammens T, Boruc J, Van Leene J, Van Den Daele H, Maes S, Van Isterdael G, Russinova E, Kondorosi E, Witters E, et al. (2009) CDKB1;1 forms a functional complex with CYCA2;3 to suppress endocycle onset. Plant Physiol 150: 1482–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramsiepe J, Wester K, Weinl C, Roodbarkelari F, Kasili R, Larkin JC, Hülskamp M, Schnittger A. (2010) Endoreplication controls cell fate maintenance. PLoS Genet 6: e1000996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuer C, Ishida T, Sugimoto K. (2010) Developmental control of endocycles and cell growth in plants. Curr Opin Plant Biol 13: 654–660 [DOI] [PubMed] [Google Scholar]
- Brioudes F, Joly C, Szécsi J, Varaud E, Leroux J, Bellvert F, Bertrand C, Bendahmane M. (2009) Jasmonate controls late development stages of petal growth in Arabidopsis thaliana. Plant J 60: 1070–1080 [DOI] [PubMed] [Google Scholar]
- Browse J. (2009) The power of mutants for investigating jasmonate biosynthesis and signaling. Phytochemistry 70: 1539–1546 [DOI] [PubMed] [Google Scholar]
- Campilho A, Garcia B, Toorn HV, Wijk HV, Campilho A, Scheres B. (2006) Time-lapse analysis of stem-cell divisions in the Arabidopsis thaliana root meristem. Plant J 48: 619–627 [DOI] [PubMed] [Google Scholar]
- Caño-Delgado AI, Metzlaff K, Bevan MW. (2000) The eli1 mutation reveals a link between cell expansion and secondary cell wall formation in Arabidopsis thaliana. Development 127: 3395–3405 [DOI] [PubMed] [Google Scholar]
- Castellano MM, Boniotti MB, Caro E, Schnittger A, Gutierrez C. (2004) DNA replication licensing affects cell proliferation or endoreplication in a cell type-specific manner. Plant Cell 16: 2380–2393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano MM, del Pozo JC, Ramirez-Parra E, Brown S, Gutierrez C. (2001) Expression and stability of Arabidopsis CDC6 are associated with endoreplication. Plant Cell 13: 2671–2686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandran D, Inada N, Hather G, Kleindt CK, Wildermuth MC. (2010) Laser microdissection of Arabidopsis cells at the powdery mildew infection site reveals site-specific processes and regulators. Proc Natl Acad Sci USA 107: 460–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Sun J, Zhai Q, Zhou W, Qi L, Xu L, Wang B, Chen R, Jiang H, Qi J, et al. (2011) The basic helix-loop-helix transcription factor MYC2 directly represses PLETHORA expression during jasmonate-mediated modulation of the root stem cell niche in Arabidopsis. Plant Cell 23: 3335–3352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Song S, Xiao L, Soo HM, Cheng Z, Xie D, Peng J. (2009) Gibberellin acts through jasmonate to control the expression of MYB21, MYB24, and MYB57 to promote stamen filament growth in Arabidopsis. PLoS Genet 5: e1000440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini A, Fonseca S, Fernández G, Adie B, Chico JM, Lorenzo O, García-Casado G, López-Vidriero I, Lozano FM, Ponce MR, et al. (2007) The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448: 666–671 [DOI] [PubMed] [Google Scholar]
- Clouse SD. (2011) Brassinosteroid signal transduction: from receptor kinase activation to transcriptional networks regulating plant development. Plant Cell 23: 1219–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colón-Carmona A, You R, Haimovitch-Gal T, Doerner P. (1999) Spatio-temporal analysis of mitotic activity with a labile cyclin-GUS fusion protein. Plant J 20: 503–508 [DOI] [PubMed] [Google Scholar]
- Costas C, de la Paz Sanchez M, Stroud H, Yu Y, Oliveros JC, Feng S, Benguria A, López-Vidriero I, Zhang X, Solano R, et al. (2011) Genome-wide mapping of Arabidopsis thaliana origins of DNA replication and their associated epigenetic marks. Nat Struct Mol Biol 18: 395–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creelman RA, Mullet JE. (1997) Biosynthesis and action of jasmonates in plants. Annu Rev Plant Physiol Plant Mol Biol 48: 355–381 [DOI] [PubMed] [Google Scholar]
- Creff A, Sormani R, Desnos T. (2010) The two Arabidopsis RPS6 genes, encoding for cytoplasmic ribosomal proteins S6, are functionally equivalent. Plant Mol Biol 73: 533–546 [DOI] [PubMed] [Google Scholar]
- De Geyter N, Gholami A, Goormachtig S, Goossens A. (2012) Transcriptional machineries in jasmonate-elicited plant secondary metabolism. Trends Plant Sci 17: 349–359 [DOI] [PubMed] [Google Scholar]
- DePamphilis ML. (2003) Eukaryotic DNA replication origins: reconciling disparate data. Cell 114: 274–275 [DOI] [PubMed] [Google Scholar]
- Desvoyes B, Sanchez MP, Ramirez-Parra E, Gutierrez C. (2010) Impact of nucleosome dynamics and histone modifications on cell proliferation during Arabidopsis development. Heredity (Edinb) 105: 80–91 [DOI] [PubMed] [Google Scholar]
- De Veylder L, Beeckman T, Beemster GT, de Almeida Engler J, Ormenese S, Maes S, Naudts M, Van Der Schueren E, Jacqmard A, Engler G, et al. (2002) Control of proliferation, endoreduplication and differentiation by the Arabidopsis E2Fa-DPa transcription factor. EMBO J 21: 1360–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder L, Beeckman T, Beemster GT, Krols L, Terras F, Landrieu I, van der Schueren E, Maes S, Naudts M, Inzé D. (2001) Functional analysis of cyclin-dependent kinase inhibitors of Arabidopsis. Plant Cell 13: 1653–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder L, Larkin JC, Schnittger A. (2011) Molecular control and function of endoreplication in development and physiology. Trends Plant Sci 16: 624–634 [DOI] [PubMed] [Google Scholar]
- Devoto A, Ellis C, Magusin A, Chang HS, Chilcott C, Zhu T, Turner JG. (2005) Expression profiling reveals COI1 to be a key regulator of genes involved in wound- and methyl jasmonate-induced secondary metabolism, defence, and hormone interactions. Plant Mol Biol 58: 497–513 [DOI] [PubMed] [Google Scholar]
- Devoto A, Nieto-Rostro M, Xie D, Ellis C, Harmston R, Patrick E, Davis J, Sherratt L, Coleman M, Turner JG. (2002) COI1 links jasmonate signalling and fertility to the SCF ubiquitin-ligase complex in Arabidopsis. Plant J 32: 457–466 [DOI] [PubMed] [Google Scholar]
- Devoto A, Turner JG. (2003) Regulation of jasmonate-mediated plant responses. Ann Bot (Lond) 92: 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewitte W, Riou-Khamlichi C, Scofield S, Healy JM, Jacqmard A, Kilby NJ, Murray JA. (2003) Altered cell cycle distribution, hyperplasia, and inhibited differentiation in Arabidopsis caused by the D-type cyclin CYCD3. Plant Cell 15: 79–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewitte W, Scofield S, Alcasabas AA, Maughan SC, Menges M, Braun N, Collins C, Nieuwland J, Prinsen E, Sundaresan V, et al. (2007) Arabidopsis CYCD3 D-type cyclins link cell proliferation and endocycles and are rate-limiting for cytokinin responses. Proc Natl Acad Sci USA 104: 14537–14542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Trivino S, del Mar Castellano M, de la Paz Sanchez M, Ramirez-Parra E, Desvoyes B, Gutierrez C. (2005) The genes encoding Arabidopsis ORC subunits are E2F targets and the two ORC1 genes are differently expressed in proliferating and endoreplicating cells. Nucleic Acids Res 33: 5404–5414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolezel J, Greilhuber J, Suda J. (2007) Estimation of nuclear DNA content in plants using flow cytometry. Nat Protoc 2: 2233–2244 [DOI] [PubMed] [Google Scholar]
- Donnelly PM, Bonetta D, Tsukaya H, Dengler RE, Dengler NG. (1999) Cell cycling and cell enlargement in developing leaves of Arabidopsis. Dev Biol 215: 407–419 [DOI] [PubMed] [Google Scholar]
- Dudits D, Abrahám E, Miskolczi P, Ayaydin F, Bilgin M, Horváth GV. (2011) Cell-cycle control as a target for calcium, hormonal and developmental signals: the role of phosphorylation in the retinoblastoma-centred pathway. Ann Bot (Lond) 107: 1193–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis C, Karafyllidis I, Wasternack C, Turner JG. (2002) The Arabidopsis mutant cev1 links cell wall signaling to jasmonate and ethylene responses. Plant Cell 14: 1557–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis C, Turner JG. (2001) The Arabidopsis mutant cev1 has constitutively active jasmonate and ethylene signal pathways and enhanced resistance to pathogens. Plant Cell 13: 1025–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis C, Turner JG. (2002) A conditionally fertile coi1 allele indicates cross-talk between plant hormone signalling pathways in Arabidopsis thaliana seeds and young seedlings. Planta 215: 549–556 [DOI] [PubMed] [Google Scholar]
- Feys B, Benedetti CE, Penfold CN, Turner JG. (1994) Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell 6: 751–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca S, Chini A, Hamberg M, Adie B, Porzel A, Kramell R, Miersch O, Wasternack C, Solano R. (2009) (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nat Chem Biol 5: 344–350 [DOI] [PubMed] [Google Scholar]
- Gao X, Nagawa S, Wang G, Yang Z. (2008) Cell polarity signaling: focus on polar auxin transport. Mol Plant 1: 899–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garton S, Knight H, Warren GJ, Knight MR, Thorlby GJ. (2007) crinkled leaves 8—a mutation in the large subunit of ribonucleotide reductase—leads to defects in leaf development and chloroplast division in Arabidopsis thaliana. Plant J 50: 118–127 [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gfeller A, Dubugnon L, Liechti R, Farmer EE. (2010) Jasmonate biochemical pathway. Sci Signal 3: cm3. [DOI] [PubMed] [Google Scholar]
- Gonzalez N, De Bodt S, Sulpice R, Jikumaru Y, Chae E, Dhondt S, Van Daele T, De Milde L, Weigel D, Kamiya Y, et al. (2010) Increased leaf size: different means to an end. Plant Physiol 153: 1261–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez N, Vanhaeren H, Inzé D. (2012) Leaf size control: complex coordination of cell division and expansion. Trends Plant Sci 17: 332–340 [DOI] [PubMed] [Google Scholar]
- Guimil S, Dunand C. (2007) Cell growth and differentiation in Arabidopsis epidermal cells. J Exp Bot 58: 3829–3840 [DOI] [PubMed] [Google Scholar]
- Gutierrez C. (2009) The Arabidopsis cell division cycle. The Arabidopsis Book 7: e0120, doi/10.1199/tab.0120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberer G, Kieber JJ. (2002) Cytokinins: new insights into a classic phytohormone. Plant Physiol 128: 354–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacham Y, Holland N, Butterfield C, Ubeda-Tomas S, Bennett MJ, Chory J, Savaldi-Goldstein S. (2011) Brassinosteroid perception in the epidermis controls root meristem size. Development 138: 839–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harashima H, Schnittger A. (2010) The integration of cell division, growth and differentiation. Curr Opin Plant Biol 13: 66–74 [DOI] [PubMed] [Google Scholar]
- Harb A, Krishnan A, Ambavaram MM, Pereira A. (2010) Molecular and physiological analysis of drought stress in Arabidopsis reveals early responses leading to acclimation in plant growth. Plant Physiol 154: 1254–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harberd NP, Belfield E, Yasumura Y. (2009) The angiosperm gibberellin-GID1-DELLA growth regulatory mechanism: how an “inhibitor of an inhibitor” enables flexible response to fluctuating environments. Plant Cell 21: 1328–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J. (2007) qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol 8: R19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong GJ, Xue XY, Mao YB, Wang LJ, Chen XY. (2012) Arabidopsis MYC2 interacts with DELLA proteins in regulating sesquiterpene synthase gene expression. Plant Cell 24: 2635–2648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiguchi G, Tsukaya H. (2011) Organ size regulation in plants: insights from compensation. Front Plant Sci 2: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horváth BM, Magyar Z, Zhang Y, Hamburger AW, Bakó L, Visser RG, Bachem CW, Bögre L. (2006) EBP1 regulates organ size through cell growth and proliferation in plants. EMBO J 25: 4909–4920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X, Lee LY, Xia K, Yan Y, Yu H. (2010) DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev Cell 19: 884–894 [DOI] [PubMed] [Google Scholar]
- Hu Y, Poh HM, Chua NH. (2006) The Arabidopsis ARGOS-LIKE gene regulates cell expansion during organ growth. Plant J 47: 1–9 [DOI] [PubMed] [Google Scholar]
- Huang W, Sherman BT, Tan Q, Collins JR, Alvord WG, Roayaei J, Stephens R, Baseler MW, Lane HC, Lempicki RA. (2007a) The DAVID Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol 8: R183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Sherman BT, Tan Q, Kir J, Liu D, Bryant D, Guo Y, Stephens R, Baseler MW, Lane HC, et al. (2007b) DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res 35: W169–W175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai KK, Ohashi Y, Tsuge T, Yoshizumi T, Matsui M, Oka A, Aoyama T. (2006) The A-type cyclin CYCA2;3 is a key regulator of ploidy levels in Arabidopsis endoreduplication. Plant Cell 18: 382–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inzé D. (2005) Green light for the cell cycle. EMBO J 24: 657–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inzé D, De Veylder L. (2006) Cell cycle regulation in plant development. Annu Rev Genet 40: 77–105 [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. (2003a) Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res 31: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. (2003b) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264 [DOI] [PubMed] [Google Scholar]
- Ishida T, Adachi S, Yoshimura M, Shimizu K, Umeda M, Sugimoto K. (2010) Auxin modulates the transition from the mitotic cycle to the endocycle in Arabidopsis. Development 137: 63–71 [DOI] [PubMed] [Google Scholar]
- Ishida T, Fujiwara S, Miura K, Stacey N, Yoshimura M, Schneider K, Adachi S, Minamisawa K, Umeda M, Sugimoto K. (2009) SUMO E3 ligase HIGH PLOIDY2 regulates endocycle onset and meristem maintenance in Arabidopsis. Plant Cell 21: 2284–2297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasinski S, Riou-Khamlichi C, Roche O, Perennes C, Bergounioux C, Glab N. (2002) The CDK inhibitor NtKIS1a is involved in plant development, endoreduplication and restores normal development of cyclin D3;1-overexpressing plants. J Cell Sci 115: 973–982 [DOI] [PubMed] [Google Scholar]
- Koda Y. (1997) Possible involvement of jasmonates in various morphogenic events. Physiol Plant 100: 639–646 [Google Scholar]
- López-Juez E, Dillon E, Magyar Z, Khan S, Hazeldine S, de Jager SM, Murray JA, Beemster GT, Bögre L, Shanahan H. (2008) Distinct light-initiated gene expression and cell cycle programs in the shoot apex and cotyledons of Arabidopsis. Plant Cell 20: 947–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O, Solano R. (2005) Molecular players regulating the jasmonate signalling network. Curr Opin Plant Biol 8: 532–540 [DOI] [PubMed] [Google Scholar]
- Magyar Z (2008) Keeping the balance between proliferation and differentiation by the E2F transcriptional regulatory network is central to plant growth and development. In L Bogre, GT Beemster, eds, Plant Growth Signaling. Springer, Berlin, pp 89–105 [Google Scholar]
- Magyar Z, De Veylder L, Atanassova A, Bakó L, Inzé D, Bögre L. (2005) The role of the Arabidopsis E2FB transcription factor in regulating auxin-dependent cell division. Plant Cell 17: 2527–2541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield SG, Briarty LG. (1992) Cotyledon cell development in Arabidopsis thaliana during reserve deposition. Can J Bot 70: 151–164 [Google Scholar]
- Marcotrigiano M. (2010) A role for leaf epidermis in the control of leaf size and the rate and extent of mesophyll cell division. Am J Bot 97: 224–233 [DOI] [PubMed] [Google Scholar]
- Massonnet C, Vile D, Fabre J, Hannah MA, Caldana C, Lisec J, Beemster GT, Meyer RC, Messerli G, Gronlund JT, et al. (2010) Probing the reproducibility of leaf growth and molecular phenotypes: a comparison of three Arabidopsis accessions cultivated in ten laboratories. Plant Physiol 152: 2142–2157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masubelele NH, Dewitte W, Menges M, Maughan S, Collins C, Huntley R, Nieuwland J, Scofield S, Murray JA. (2005) D-type cyclins activate division in the root apex to promote seed germination in Arabidopsis. Proc Natl Acad Sci USA 102: 15694–15699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda HP, Cabral LM, De Veylder L, Tanurdzic M, de Almeida Engler J, Geelen D, Inzé D, Martienssen RA, Ferreira PC, Hemerly AS. (2008) ABAP1 is a novel plant Armadillo BTB protein involved in DNA replication and transcription. EMBO J 27: 2746–2756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda HP, Ramos GB, de Almeida-Engler J, Cabral LM, Coqueiro VM, Macrini CM, Ferreira PC, Hemerly AS. (2004) Genome based identification and analysis of the pre-replicative complex of Arabidopsis thaliana. FEBS Lett 574: 192–202 [DOI] [PubMed] [Google Scholar]
- Menges M, de Jager SM, Gruissem W, Murray JA. (2005) Global analysis of the core cell cycle regulators of Arabidopsis identifies novel genes, reveals multiple and highly specific profiles of expression and provides a coherent model for plant cell cycle control. Plant J 41: 546–566 [DOI] [PubMed] [Google Scholar]
- Navarro L, Bari R, Achard P, Lisón P, Nemri A, Harberd NP, Jones JD. (2008) DELLAs control plant immune responses by modulating the balance of jasmonic acid and salicylic acid signaling. Curr Biol 18: 650–655 [DOI] [PubMed] [Google Scholar]
- Oliveros JC (2007) VENNY: an interactive tool for comparing lists with Venn diagrams. http://bioinfogp.cnb.csic.es/tools/venny/index.html (December 15, 2012)
- Park JH, Halitschke R, Kim HB, Baldwin IT, Feldmann KA, Feyereisen R. (2002) A knock-out mutation in allene oxide synthase results in male sterility and defective wound signal transduction in Arabidopsis due to a block in jasmonic acid biosynthesis. Plant J 31: 1–12 [DOI] [PubMed] [Google Scholar]
- Pauwels L, Morreel K, De Witte E, Lammertyn F, Van Montagu M, Boerjan W, Inzé D, Goossens A. (2008) Mapping methyl jasmonate-mediated transcriptional reprogramming of metabolism and cell cycle progression in cultured Arabidopsis cells. Proc Natl Acad Sci USA 105: 1380–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrot-Rechenmann C. (2010) Cellular responses to auxin: division versus expansion. Cold Spring Harb Perspect Biol 2: a001446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philipps G, Clément B, Gigot C. (1995) Molecular characterization and cell cycle-regulated expression of a cDNA clone from Arabidopsis thaliana homologous to the small subunit of ribonucleotide reductase. FEBS Lett 358: 67–70 [DOI] [PubMed] [Google Scholar]
- Powell AE, Lenhard M. (2012) Control of organ size in plants. Curr Biol 22: R360–R367 [DOI] [PubMed] [Google Scholar]
- Raynaud C, Sozzani R, Glab N, Domenichini S, Perennes C, Cella R, Kondorosi E, Bergounioux C. (2006) Two cell-cycle regulated SET-domain proteins interact with proliferating cell nuclear antigen (PCNA) in Arabidopsis. Plant J 47: 395–407 [DOI] [PubMed] [Google Scholar]
- Reymond P, Bodenhausen N, Van Poecke RM, Krishnamurthy V, Dicke M, Farmer EE. (2004) A conserved transcript pattern in response to a specialist and a generalist herbivore. Plant Cell 16: 3132–3147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savaldi-Goldstein S, Peto C, Chory J. (2007) The epidermis both drives and restricts plant shoot growth. Nature 446: 199–202 [DOI] [PubMed] [Google Scholar]
- Schaller A, Stintzi A. (2009) Enzymes in jasmonate biosynthesis: structure, function, regulation. Phytochemistry 70: 1532–1538 [DOI] [PubMed] [Google Scholar]
- Seo HS, Song JT, Cheong JJ, Lee YH, Lee YW, Hwang I, Lee JS, Choi YD. (2001) Jasmonic acid carboxyl methyltransferase: a key enzyme for jasmonate-regulated plant responses. Proc Natl Acad Sci USA 98: 4788–4793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skirycz A, Claeys H, De Bodt S, Oikawa A, Shinoda S, Andriankaja M, Maleux K, Eloy NB, Coppens F, Yoo SD, et al. (2011) Pause-and-stop: the effects of osmotic stress on cell proliferation during early leaf development in Arabidopsis and a role for ethylene signaling in cell cycle arrest. Plant Cell 23: 1876–1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skirycz A, Inzé D. (2010) More from less: plant growth under limited water. Curr Opin Biotechnol 21: 197–203 [DOI] [PubMed] [Google Scholar]
- Smyth GK. (2004) Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: 1544–6115 [DOI] [PubMed] [Google Scholar]
- Squatrito M, Mancino M, Donzelli M, Areces LB, Draetta GF. (2004) EBP1 is a nucleolar growth-regulating protein that is part of pre-ribosomal ribonucleoprotein complexes. Oncogene 23: 4454–4465 [DOI] [PubMed] [Google Scholar]
- Staswick PE, Su W, Howell SH. (1992) Methyl jasmonate inhibition of root growth and induction of a leaf protein are decreased in an Arabidopsis thaliana mutant. Proc Natl Acad Sci USA 89: 6837–6840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens R, Mariconti L, Rossignol P, Perennes C, Cella R, Bergounioux C. (2002) Two E2F sites in the Arabidopsis MCM3 promoter have different roles in cell cycle activation and meristematic expression. J Biol Chem 277: 32978–32984 [DOI] [PubMed] [Google Scholar]
- Sugimoto-Shirasu K, Roberts GR, Stacey NJ, McCann MC, Maxwell A, Roberts K. (2005) RHL1 is an essential component of the plant DNA topoisomerase VI complex and is required for ploidy-dependent cell growth. Proc Natl Acad Sci USA 102: 18736–18741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto-Shirasu K, Roberts K. (2003) “Big it up”: endoreduplication and cell-size control in plants. Curr Opin Plant Biol 6: 544–553 [DOI] [PubMed] [Google Scholar]
- Świątek A, Azmi A, Stals H, Inzé D, Van Onckelen H. (2004) Jasmonic acid prevents the accumulation of cyclin B1;1 and CDK-B in synchronized tobacco BY-2 cells. FEBS Lett 572: 118–122 [DOI] [PubMed] [Google Scholar]
- Świątek A, Lenjou M, Van Bockstaele D, Inzé D, Van Onckelen H. (2002) Differential effect of jasmonic acid and abscisic acid on cell cycle progression in tobacco BY-2 cells. Plant Physiol 128: 201–211 [PMC free article] [PubMed] [Google Scholar]
- Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu G, Nomura K, He SY, Howe GA, Browse J. (2007) JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448: 661–665 [DOI] [PubMed] [Google Scholar]
- Trémousaygue D, Garnier L, Bardet C, Dabos P, Hervé C, Lescure B. (2003) Internal telomeric repeats and ‘TCP domain’ protein-binding sites co-operate to regulate gene expression in Arabidopsis thaliana cycling cells. Plant J 33: 957–966 [DOI] [PubMed] [Google Scholar]
- Tsukaya H. (1994) Genetics of shoot morphogenesis. Tanpakushitsu Kakusan Koso 39: 2580–2590 [PubMed] [Google Scholar]
- Tsukaya H. (2005) Leaf shape: genetic controls and environmental factors. Int J Dev Biol 49: 547–555 [DOI] [PubMed] [Google Scholar]
- Tsukaya H, Beemster GT. (2006) Genetics, cell cycle and cell expansion in organogenesis in plants. J Plant Res 119: 1–4 [DOI] [PubMed] [Google Scholar]
- Tsurimoto T. (1998) PCNA, a multifunctional ring on DNA. Biochim Biophys Acta 1443: 23–39 [DOI] [PubMed] [Google Scholar]
- Vandepoele K, Raes J, De Veylder L, Rouzé P, Rombauts S, Inzé D. (2002) Genome-wide analysis of core cell cycle genes in Arabidopsis. Plant Cell 14: 903–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandepoele K, Vlieghe K, Florquin K, Hennig L, Beemster GT, Gruissem W, Van de Peer Y, Inzé D, De Veylder L. (2005) Genome-wide identification of potential plant E2F target genes. Plant Physiol 139: 316–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3: RESEARCH0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Leene J, Boruc J, De Jaeger G, Russinova E, De Veylder L. (2011) A kaleidoscopic view of the Arabidopsis core cell cycle interactome. Trends Plant Sci 16: 141–150 [DOI] [PubMed] [Google Scholar]
- Van Leene J, Hollunder J, Eeckhout D, Persiau G, Van De Slijke E, Stals H, Van Isterdael G, Verkest A, Neirynck S, Buffel Y, et al. (2010) Targeted interactomics reveals a complex core cell cycle machinery in Arabidopsis thaliana. Mol Syst Biol 6: 397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Malek B, van der Graaff E, Schneitz K, Keller B. (2002) The Arabidopsis male-sterile mutant dde2-2 is defective in the ALLENE OXIDE SYNTHASE gene encoding one of the key enzymes of the jasmonic acid biosynthesis pathway. Planta 216: 187–192 [DOI] [PubMed] [Google Scholar]
- Wang C, Liu Z. (2006) Arabidopsis ribonucleotide reductases are critical for cell cycle progression, DNA damage repair, and plant development. Plant Cell 18: 350–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack C, Kombrink E. (2010) Jasmonates: structural requirements for lipid-derived signals active in plant stress responses and development. ACS Chem Biol 5: 63–77 [DOI] [PubMed] [Google Scholar]
- Westphal L, Scheel D, Rosahl S. (2008) The coi1-16 mutant harbors a second site mutation rendering PEN2 nonfunctional. Plant Cell 20: 824–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wettenhall JM, Simpson KM, Satterley K, Smyth GK. (2006) affylmGUI: a graphical user interface for linear modeling of single channel microarray data. Bioinformatics 22: 897–899 [DOI] [PubMed] [Google Scholar]
- Wild M, Davière JM, Cheminant S, Regnault T, Baumberger N, Heintz D, Baltz R, Genschik P, Achard P. (2012) The Arabidopsis DELLA RGA-LIKE3 is a direct target of MYC2 and modulates jasmonate signaling responses. Plant Cell 24: 3307–3319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildermuth MC. (2010) Modulation of host nuclear ploidy: a common plant biotroph mechanism. Curr Opin Plant Biol 13: 449–458 [DOI] [PubMed] [Google Scholar]
- Wu K, Zhang L, Zhou C, Yu CW, Chaikam V. (2008) HDA6 is required for jasmonate response, senescence and flowering in Arabidopsis. J Exp Bot 59: 225–234 [DOI] [PubMed] [Google Scholar]
- Wuarin J, Buck V, Nurse P, Millar JB. (2002) Stable association of mitotic cyclin B/Cdc2 to replication origins prevents endoreduplication. Cell 111: 419–431 [DOI] [PubMed] [Google Scholar]
- Xie DX, Feys BF, James S, Nieto-Rostro M, Turner JG. (1998) COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280: 1091–1094 [DOI] [PubMed] [Google Scholar]
- Xu L, Liu F, Lechner E, Genschik P, Crosby WL, Ma H, Peng W, Huang D, Xie D. (2002) The SCF(COI1) ubiquitin-ligase complexes are required for jasmonate response in Arabidopsis. Plant Cell 14: 1919–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Zhang C, Gu M, Bai Z, Zhang W, Qi T, Cheng Z, Peng W, Luo H, Nan F, et al. (2009) The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor. Plant Cell 21: 2220–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y, Stolz S, Chételat A, Reymond P, Pagni M, Dubugnon L, Farmer EE. (2007) A downstream mediator in the growth repression limb of the jasmonate pathway. Plant Cell 19: 2470–2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang DL, Yao J, Mei CS, Tong XH, Zeng LJ, Li Q, Xiao LT, Sun TP, Li J, Deng XW, et al. (2012) Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc Natl Acad Sci USA 109: E1192–E1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida Y, Sano R, Wada T, Takabayashi J, Okada K. (2009) Jasmonic acid control of GLABRA3 links inducible defense and trichome patterning in Arabidopsis. Development 136: 1039–1048 [DOI] [PubMed] [Google Scholar]
- Yu Y, Steinmetz A, Meyer D, Brown S, Shen WH. (2003) The tobacco A-type cyclin, Nicta;CYCA3;2, at the nexus of cell division and differentiation. Plant Cell 15: 2763–2777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Turner JG. (2008) Wound-induced endogenous jasmonates stunt plant growth by inhibiting mitosis. PLoS ONE 3: e3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhiponova MK, Pettkó-Szandtner A, Stelkovics E, Neer Z, Bottka S, Krenács T, Dudits D, Fehér A, Szilák L. (2006) Mitosis-specific promoter of the alfalfa cyclin-dependent kinase gene (Medsa;CDKB2;1) is activated by wounding and ethylene in a non-cell division-dependent manner. Plant Physiol 140: 693–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Zhang L, Duan J, Miki B, Wu K. (2005) HISTONE DEACETYLASE19 is involved in jasmonic acid and ethylene signaling of pathogen response in Arabidopsis. Plant Cell 17: 1196–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]