Integration of transcriptional and signaling pathways promote rice floret meristem specification and organ development.
Abstract
SEPALLATA (SEP) MADS box transcription factors mediate floral development in association with other regulators. Mutants in five rice (Oryza sativa) SEP genes suggest both redundant and unique functions in panicle branching and floret development. LEAFY HULL STERILE1/OsMADS1, from a grass-specific subgroup of LOFSEP genes, is required for specifying a single floret on the spikelet meristem and for floret organ development, but its downstream mechanisms are unknown. Here, key pathways and directly modulated targets of OsMADS1 were deduced from expression analysis after its knockdown and induction in developing florets and by studying its chromatin occupancy at downstream genes. The negative regulation of OsMADS34, another LOFSEP gene, and activation of OsMADS55, a SHORT VEGETATIVE PHASE-like floret meristem identity gene, show its role in facilitating the spikelet-to-floret meristem transition. Direct regulation of other transcription factor genes like OsHB4 (a class III homeodomain Leu zipper member), OsBLH1 (a BEL1-like homeodomain member), OsKANADI2, OsKANADI4, and OsETTIN2 show its role in meristem maintenance, determinacy, and lateral organ development. We found that the OsMADS1 targets OsETTIN1 and OsETTIN2 redundantly ensure carpel differentiation. The multiple effects of OsMADS1 in promoting auxin transport, signaling, and auxin-dependent expression and its direct repression of three cytokinin A-type response regulators show its role in balancing meristem growth, lateral organ differentiation, and determinacy. Overall, we show that OsMADS1 integrates transcriptional and signaling pathways to promote rice floret specification and development.
Patterning an angiosperm flower requires the combined and individual functions of class A, B, C, D, and E MADS box transcription factors (Krizek and Fletcher, 2005; Thompson and Hake, 2009). In Arabidopsis (Arabidopsis thaliana), the class E activity is conferred by four redundant proteins, SEPALLATA1 (SEP1), SEP2, SEP3, and SEP4, that are cofactors in complexes with other MADS box factors that determine floral organ identities and meristem determinacy (Pelaz et al., 2000). Studies on loss- and gain-of-function mutants in SEP genes together with protein interaction analyses point to their pivotal role in mediating interactions among other floral organ-patterning genes (Honma and Goto, 2001; Ditta et al., 2004; Immink et al., 2009). The largely shared functions of Arabidopsis SEP genes differ from observations that homologs in other plants often have discrete roles in floral development (Malcomber and Kellogg, 2005), but the molecular mechanism underlying their species-specific roles is not well studied. In addition to the ABCDE class of organ fate regulators, a number of hormone signaling pathways influence floral transition, organ numbers, fertility, and floral meristem (FM) determinacy. Dynamic interactions are reported between transcription factors and specific hormone signaling factors during the establishment of Arabidopsis FMs (Sessions et al., 1997; Leibfried et al., 2005; Shani et al., 2006), but links between floral organ patterning and hormone signaling are not entirely clear.
In the ancient LOFSEP clade, grass-specific genes constitute a subgroup diversified from other monocot and eudicot family members (Prasad et al., 2001; Malcomber and Kellogg, 2005). The rice (Oryza sativa) inflorescence (also called a panicle), a model for grass species, is branched and produces many kinds of lateral meristems. A short-branch meristem called the spikelet produces a pair of rudimentary glumes and then a pair of empty glumes before its transition to a single FM (Bommert et al., 2005). Various studies show that five rice SEP genes have both redundant and nonredundant roles. They contribute to panicle morphology, spikelet and FM specification, floral organ differentiation, and meristem determinacy (Jeon et al., 2000; Prasad et al., 2005; Cui et al., 2010; Gao et al., 2010; Kobayashi et al., 2010). Rice LEAFY HULL STERILE (LHS1)/OsMADS1, a member of the grass subgroup of LOFSEP genes, referred to henceforth as OsMADS1, is expressed in FMs, functions during FM establishment and floret organ patterning, and contributes to meristem termination (Jeon et al., 2000; Prasad et al., 2001, 2005; Agrawal et al., 2005; Ohmori et al., 2009; Cui et al., 2010; Gao et al., 2010; Li et al., 2010; Wang et al., 2010). In addition to studies on mutants in rice SEP factors, their protein-protein interactions are reported from yeast two-hybrid analyses. Yet, despite this extensive knowledge, the downstream genes and pathways regulated by rice SEP factors are uncharacterized. Here, we have studied the mechanistic basis of OsMADS1 function by investigating its downstream targets and pathways in developing rice panicles. Our investigations show its global effects on transcriptional networks and hormonal signaling pathways. We demonstrate its role in modulating the expression of various shoot meristem and FM transcription factors and of auxin- and cytokinin-mediated signaling events. We find that some directly regulated transcription factors and signaling targets of OsMADS1 are distinct from those of Arabidopsis SEP3. Interestingly, our data also show, in instances, inverse gene expression outcomes for some targets common to OsMADS1 and SEP3 (Kaufmann et al., 2009). Thus, these findings provide insights on how OsMADS1 complexes modulate other transcription factors and the balance between auxin and cytokinin signaling to promote the determinate development of a rice FM.
RESULTS
Global Profile of Genes Regulated by OsMADS1 in Developing Rice Panicles
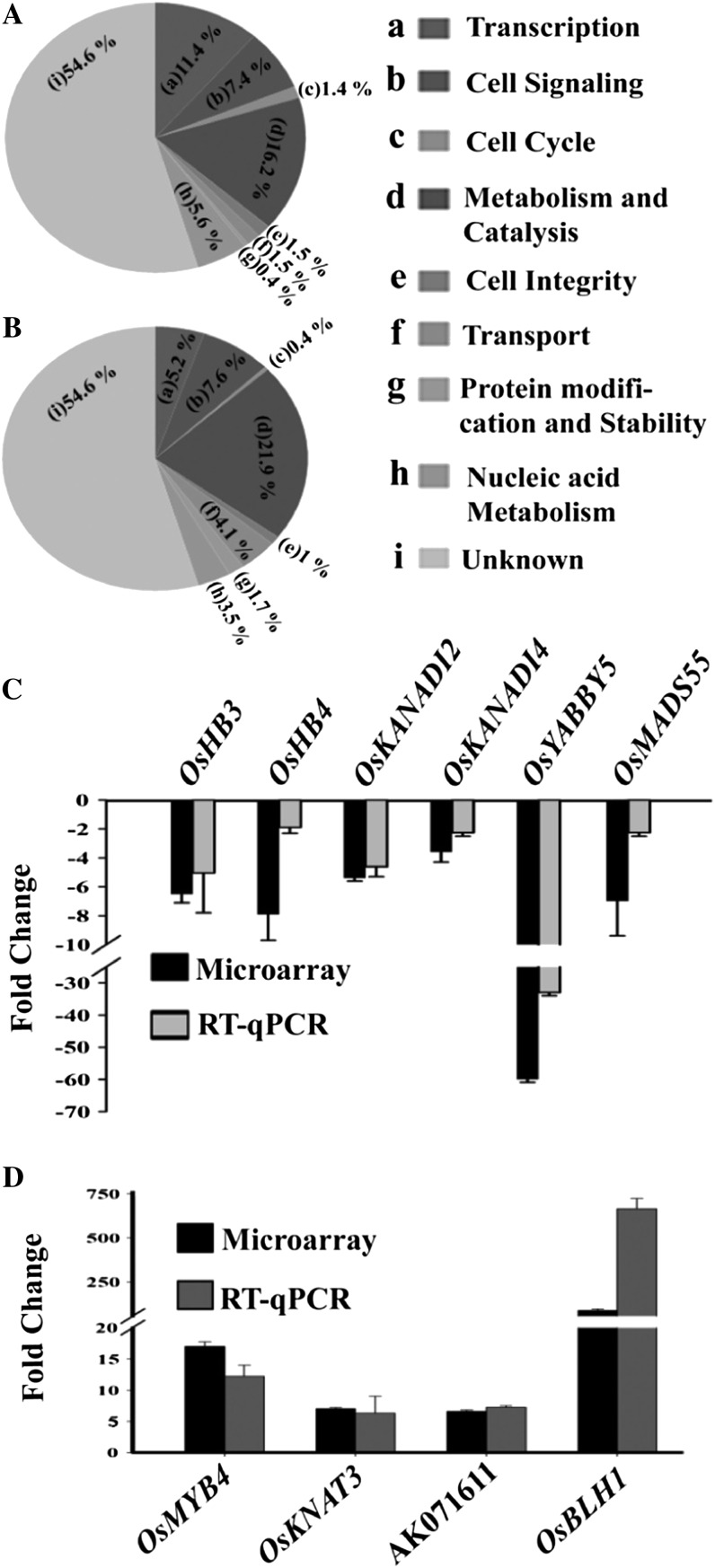
To identify target genes controlled by OsMADS1 during rice floret development, we employed multiple complementary approaches. Rice Affymetrix Gene Chips were used to find general patterns in global expression profiles in young developing panicles with strong knockdown of OsMADS1 by RNA interference (OsMADS1-RNAi) as compared with similarly staged wild-type tissues (Supplemental Data Set S1). On functional classification of deregulated genes with 3-fold or greater expression level change, we found a significantly high proportion to be implicated in transcription and signaling (Fig. 1, A and B; Supplemental Data Set S2). These data gave a gross overview of genes and pathways downstream to OsMADS1. Primary verification of these data was done by reverse transcription-quantitative PCR (RT-qPCR) analyses of transcript levels for 17 transcription factors (Supplemental Table S1) in RNA samples used in microarray analysis and in an independently generated pool of wild-type and OsMADS1-RNAi panicles (Fig. 1, C and D; Supplemental Fig. S1). These factors have varied DNA-binding motifs (Supplemental Table S2), suggesting that OsMADS1-containing complexes are regulators of many other transcription factors during floret development.
Figure 1.
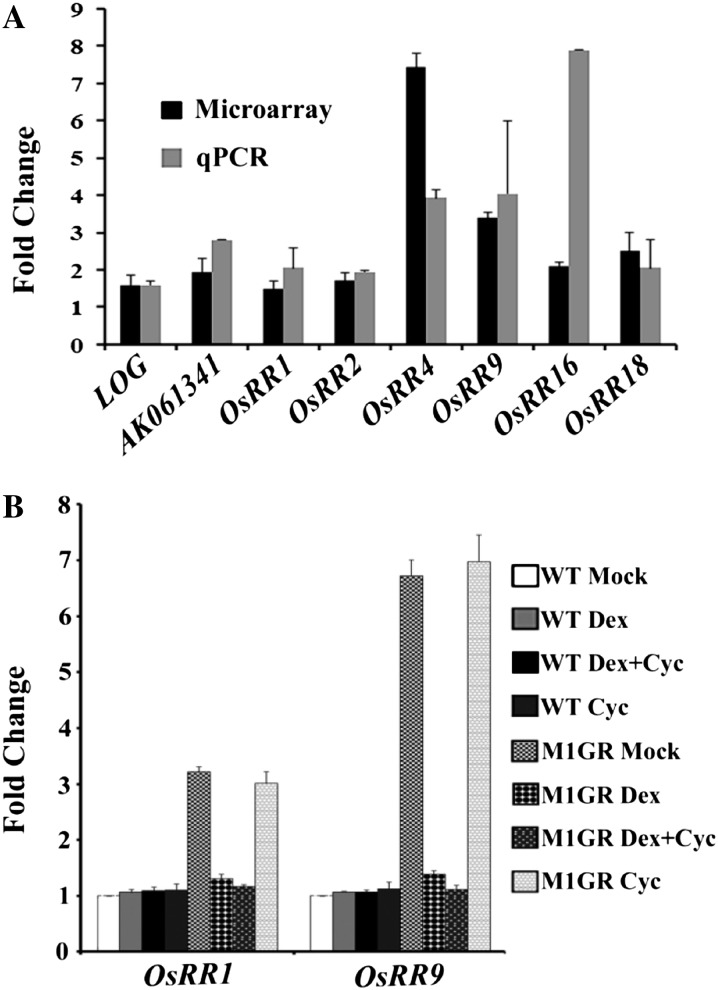
Global profile of genes and pathways deregulated in OsMADS1 knockdown panicles. A and B, Functional categorization of deregulated transcriptome (3-fold or more; P < 0.05) in developing panicles of OsMADS1-RNAi knockdown lines as compared with the wild type. Down-regulated transcripts are classified in A and up-regulated transcripts in B. Categorization was based on their predicted functional domains as annotated in various databases. C and D, RT-qPCR analyses of transcript levels for representative transcription factor genes down-regulated (C) and up-regulated (D). The fold change, with se bars, is calculated for the normalized transcript level in OsMADS1 knockdown versus wild-type panicles. RT-qPCR data are compared with data from microarray analysis.
Induction of OsMADS1-ΔGR Complements OsMADS1 Loss-of-Function Phenotypes
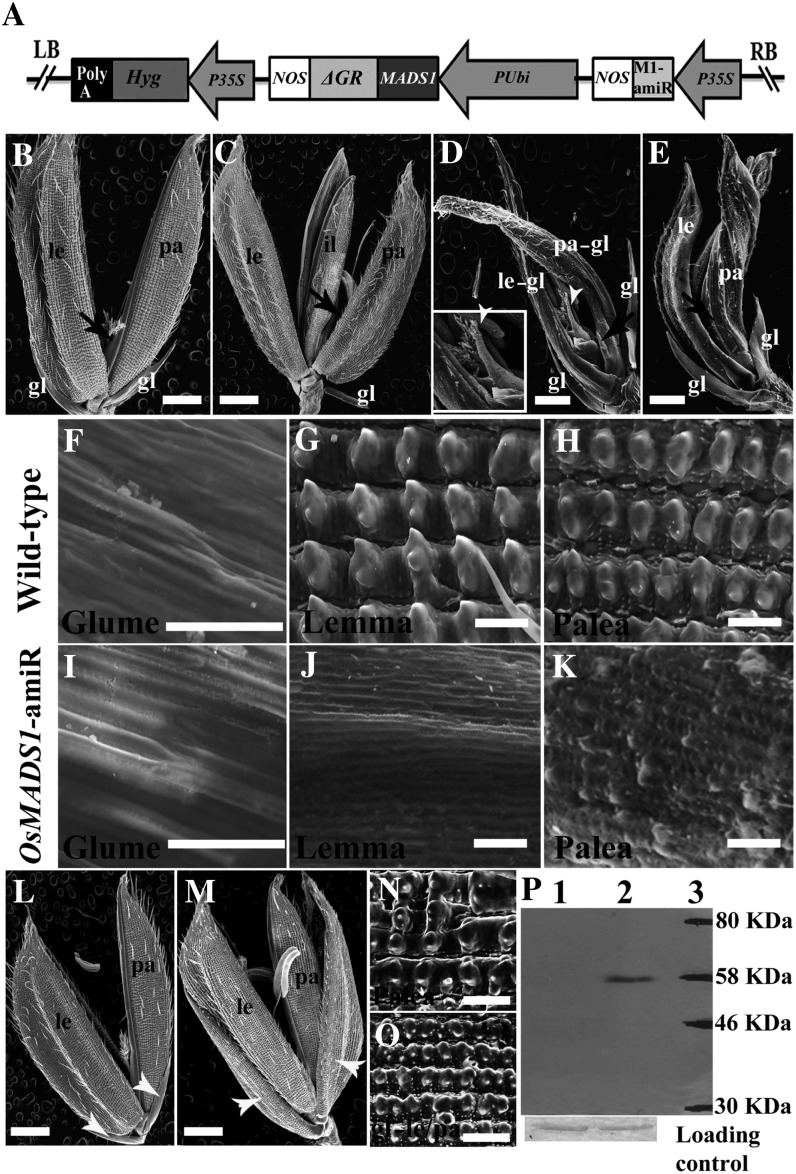
To gather evidence for the regulatory effects of OsMADS1 on its downstream pathways and to identify direct targets, we developed an artificial microRNA (amiRNA)-based knockdown system. The amiRNA-triggered knockdown was coupled with the expression of a chemically inducible OsMADS1-ΔGR fusion protein (Fig. 2A). The microRNA was designed against the 3′ untranslated region (UTR) of OsMADS1 to target the specific down-regulation of endogenous OsMADS1 transcripts. Since the transgenic copy of OsMADS1 complementary DNA (cDNA) is fused to ΔGR at its C-terminal end and is devoid of the 3′ UTR, it is not a target for suppression by the amiRNA. From 35 PUbi:OsMADS1-ΔGR; P35S:OsMADS1amiR transgenics, eight lines with single-copy insertions and good levels of amiRNAs (Supplemental Fig. S2, A and B) were further analyzed. In these plants, the developmental defects caused due to knockdown of endogenous OsMADS1 should be complemented on induction of the OsMADS1-ΔGR protein. Under noninductive conditions, when OsMADS1-ΔGR is sequestered in the cytoplasm, we observe that florets recapitulate strong loss-of-function OsMADS1 mutant phenotypes (Fig. 2, B–E; Jeon et al., 2000; Agrawal et al., 2005; Prasad et al., 2005). The outer floret organs, lemma and palea, were narrow, poorly developed, and failed to enclose the inner organs (Fig. 2, B–D). The underdeveloped palea lacked the characteristic marginal tissue (Fig. 2, C–E, black arrows), and typical epidermal features of the lemma and palea were also lost (Fig. 2, F–K). Furthermore, lodicules and stamens were converted to multiple lemma/glume-like organs (Fig. 2, C–E). This, coupled with the multiple carpels seen in some florets (Fig. 2D, white arrowheads), indicates meristem indeterminacy. Thus, the down-regulation of OsMADS1 was efficient, and OsMADS1-ΔGR protein was not leaky when not induced. In young OsMADS1-amiRNA knockdown panicles, the extent of OsMADS1 down-regulation and the transcript levels for OsMADS5 and OsMADS34, genes from the LOFSEP subclade, and for OsMADS7, a rice SEP3 gene family member (Supplemental Fig. S2C), were determined. OsMADS1 expression was much reduced, OsMADS5 transcripts were very marginally increased, and OsMADS34 expression was significantly increased. OsMADS7 expression status was not altered (Supplemental Fig. S2C). We deduce that the amiRNA-directed knockdown of OsMADS1 was relatively specific.
Figure 2.
The OsMADS1-ΔGR fusion protein is functional on induction in transgenic rice. A, Transfer DNA segment in the construct PUbi:OsMADS1-ΔGR; P35S:OsMADS1amiR. The 35S promoter drives transcription of an amiRNA targeting the 3′ UTR of endogenous OsMADS1 transcripts. The maize ubiquitin cis-elements drive the expression of OsMADS1 translational fusion with the partial rat glucocorticoid receptor (ΔGR) domain. LB, Left border; RB, right border. B, A wild-type floret. C to E, Floret phenotypes in PUbi:OsMADS1-ΔGR; P35S:OsMADS1amiR transgenics grown in noninductive mock conditions. F to K, Scanning electron microscopy of epidermal cellular features in glumes, lemma, and palea in wild-type and mock-treated PUbi:OsMADS1-ΔGR; P35S:OsMADS1amiR lines. L and M, Florets of PUbi:OsMADS1-ΔGR; P35S:OsMADS1amiR plants grown in inductive conditions (dexamethasone treated). All floret organ phenotypes are rescued. N, Cellular features of rescued palea shown in L. O, Conversion of empty glumes to a lemma/palea-like organ in M, an overexpression phenotype seen in occasional spikelets. gl, Empty glumele; il, internal lemma; le, lemma; pa, palea. Bars = 1 mm in B to E and L to M and 100 µm in F to K and N to O. P, Western blot with nuclear extracts from leaves of PUbi:OsMADS1-ΔGR; P35S:OsMADS1amiR lines to detect OsMADS1-ΔGR protein before induction (lane 1) and after dexamethasone induction (lane 2).
To investigate the complementation of the knockdown phenotypes of PUbi:OsMADS1-ΔGR; P35S:OsMADS1amiR transgenics, we treated these plants with the inducer dexamethasone or subjected them to mock treatment. Treatments were given from just prior to floral transition until the fully developed panicle emerged. In inducer-treated plants, the lemma, marginal tissue of palea, lodicules, stamens, and carpels developed normally (Fig. 2, H, L, and N; Supplemental Fig. S2, D and E). This proved that the induced OsMADS1-ΔGR fusion protein is fully functional in planta and regulates downstream events leading to organ development and floret determinacy. By western-blot analyses, we ascertained that OsMADS1-ΔGR protein is nucleus localized only in inducer-treated tissues (Fig. 2P). This inducible OsMADS1-ΔGR system was used to identify its putative direct target genes in panicles with developing florets, as summarized in Table I and described in following sections.
Table I. Genes directly regulated by OsMADS1 during rice floret development.
+ indicates that the gene is positively regulated (activated), and − denotes that it is negatively regulated (repressed) by OsMADS1.
| Gene | Function/Predicted Function | Regulation by OsMADS1 |
|---|---|---|
| OsMADS34 | Spikelet meristem specification, organ development | − |
| OsHB4 | Meristem specification, termination | + |
| OsMADS55 | Meristem identity | + |
| OsKANADI2 | Organ polarity and development | + |
| OsKANADI4 | Organ polarity and development | + |
| OsBLH1 | Meristem specification, organ development | − |
| OsARF-GAP | Auxin transport | − |
| OsPIN1 | Auxin transport | + |
| OsETTIN2 | Auxin response, organ development | + |
| OsARF9 | Auxin response | + |
| OsRR1 | Cytokinin response | − |
| OsRR4 | Cytokinin response | − |
| OsRR9 | Cytokinin response | − |
Regulatory Effects of OsMADS1 on Other MADS Factor Genes to Control FM Fate
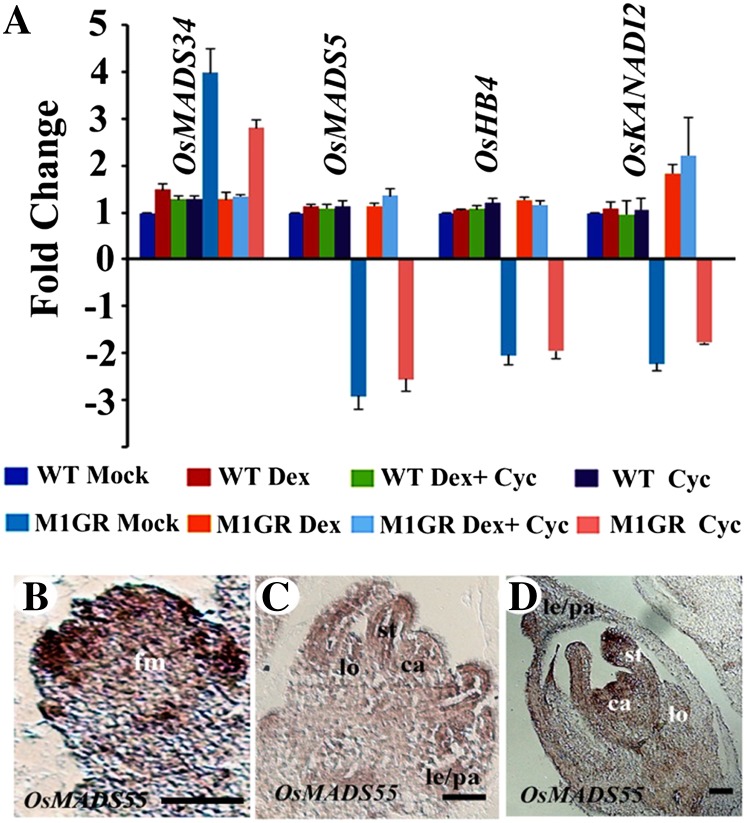
MADS box factors control flowering time, floral organ identity, and FM determinacy. OsMADS34, a rice LOFSEP member, regulates inflorescence branching and promotes spikelet meristem fate (Gao et al., 2010; Kobayashi et al., 2010, 2012) and thus is suggested to act temporally earlier than OsMADS1. In developing panicles, spikelets, and florets, these two genes have partly exclusive and some overlapping expression patterns (Kobayashi et al., 2010). OsMADS34 transcripts occur at high levels in inflorescence branch meristems, spikelet meristems, and rudimentary glumes and empty glumes (also called sterile lemmas) that are spikelet lateral organs. OsMADS34 transcripts are at reduced levels in the young FM. In contrast, OsMADS1 transcripts are abundant in the incipient FM and nearly undetectable in empty glumes (Prasad et al., 2001; Gao et al., 2010; Kobayashi et al., 2010, 2012). We found 4-fold increased transcript levels for OsMADS34 in PUbi:OsMADS1-ΔGR; P35S:OsMADS1amiR panicles (Fig. 3A). Interestingly, this abnormal OsMADS34 expression can be restored to wild-type levels by dexamethasone-based induction of OsMADS1-ΔGR even in the presence of cycloheximide (Fig. 3A). These data suggest that OsMADS1 can directly modulate the expression of OsMADS34 in young spikelet/FMs.
Figure 3.
Direct transcriptional regulation, by OsMADS1, of transcription factor genes with meristem functions. A, Quantitation of normalized OsMADS34, OsMADS55, OsHB4, and OsKANADI2 transcript levels in wild-type (WT) and PUbi:OsMADS1-ΔGR; P35S:OsMADS1amiR panicles by RT-qPCR. Plants of both genotypes were mock treated or were given dexamethasone, cycloheximide, or a combination of both chemicals. B to D, Spatial localization of OsMADS55 transcripts in wild-type florets. FM with initiating lemma and palea primordia (B), floret with differentiating inner organs (C), and a nearly mature floret (D) are shown. ca, Carpel; fm, FM; le, lemma; lo, lodicule; pa, palea; st, stamen. Bars = 50 μm.
The rice genome encodes three SHORT VEGETATIVE PHASE (SVP)-like genes: OsMADS22, OsMADS55, and OsMADS47 (Arora et al., 2007; Lee et al., 2008). Among these, we detect decreased expression of OsMADS22 and OsMADS55 but not of OsMADS47 in OsMADS1-RNAi panicles (Fig. 1C; Supplemental Table S2). Since OsMADS55 has two consensus binding sites (CArG) for MADS domain proteins in its proximal promoter, it was a more probable candidate target of OsMADS1; hence, we validated its down-regulation in OsMADS1-amiRNA panicles by RT-qPCR (Fig. 3A) and carefully examined its expression status on induction of OsMADS1-ΔGR. The reduced OsMADS55 expression in the panicles of mock-treated PUbi:OsMADS1-ΔGR; P35S:OsMADS1amiR plants was restored to wild-type levels in dexamethasone-treated plants. Notably, this effect was maintained in the presence of cycloheximide and dexamethasone (Fig. 3A), indicating that OsMADS1 can directly modulate OsMADS55 expression status. The overlap in the expression domains of OsMADS1 and OsMADS55 in young florets with initiating lemma and palea (Prasad et al., 2005; Fig. 3B) supports the suggested regulatory relationship between these genes. In slightly older florets, both transcripts occur in the lemma, palea, and carpel, but OsMADS55 alone is detected in developing lodicules and stamens (Prasad et al., 2005; Fig. 3, C and D). Together, the data show that OsMADS1 can promote the spikelet-to-floret transition by simultaneously activating positive regulators of FM fate (i.e. OsMADS55) and repressing spikelet meristem factors like OsMADS34.
Regulation of Homeodomain Transcription Factor Genes by OsMADS1
While functions for rice class III homeodomain Leu zipper (HD-Zip III) genes in shoot apical meristem initiation, its maintenance, and their role in leaf polarity are inferred from their ectopic overexpression phenotypes (Nagasaki et al., 2007; Itoh et al., 2008), their roles in FMs and floral organ development are not known. Recent data reveal an unexpected but crucial contribution of HD-Zip III factors in the precise regulation of Arabidopsis floral stem cells, which suggests additional functions for these factors (Ji et al., 2011). Our microarray data showed that four rice HD-Zip III genes (OsHB1–OsHB4) of the REVOLUTA (REV)/RBV clade have reduced transcript levels in OsMADS1-RNAi panicles (Supplemental Fig. S3A). RT-qPCR confirmed these effects for two of these genes (Fig. 1C). We chose OsHB4 to interrogate its direct regulation by OsMADS1, as its highest expression levels are in developing panicles (Itoh et al., 2008). In mock-treated PUbi:OsMADS1-ΔGR; P35S:OsMADS1amiR plants, with early and differentiating florets, OsHB4 was down-regulated (Fig. 3A). This defect was rescued to near wild-type levels in panicles treated with dexamethasone alone or when a combination of dexamethasone and cycloheximide was used (Fig. 3A). OsHB4 transcriptional activation was also observed on ectopic induction of OsMADS1-ΔGR in nonflowering vegetative shoot apical meristems (Supplemental Fig. S3B). These analyses indicate that OsMADS1 can directly regulate the transcription of OsHB4. In addition to HD-Zip III genes, several BEL1-like homeodomain genes (BLH) are misexpressed in OsMADS1-RNAi panicles (Fig. 1D; Supplemental Table S2). Thus, multiple members of the homeodomain class of shoot meristem regulators are regulated by OsMADS1 in developing florets.
OsMADS1 Regulates OsKANADI2 and OsKANADI4 Expression
Arabidopsis KANADI genes promote abaxial polarity during meristem specification and organogenesis (Kerstetter et al., 2001; Emery et al., 2003). Interestingly, two rice KANADI genes, related to Arabidopsis ATS and KANADI2, were down-regulated in knockdown panicles (Fig. 1C; Supplemental Tables S1 and S2). Since the upstream sequences of these genes have multiple CArG elements, we investigated the possibility of their direct regulation by OsMADS1. In panicles of mock-treated PUbi:OsMADS1-ΔGR; P35S:OsMADS1amiR plants, OsKANADI2 and OsKANADI4 transcript levels were low (Fig. 3A; Supplemental Fig. S3C). This deregulation was rescued on OsMADS1-ΔGR induction, and abundant transcription occurred even in the presence of cycloheximide (Fig. 3A; Supplemental Fig. S3C). We interpret that both OsKANADI2 and OsKANADI4 are candidate direct targets of OsMADS1. Overall, the data show that transcription regulators implicated in early events of FM establishment, its regulated maintenance, and lateral organ development are deregulated on OsMADS1 knockdown.
Auxin Signaling and the Effects of OsMADS1 on OsPIN1, OsETTIN2, and OsARF-GAP
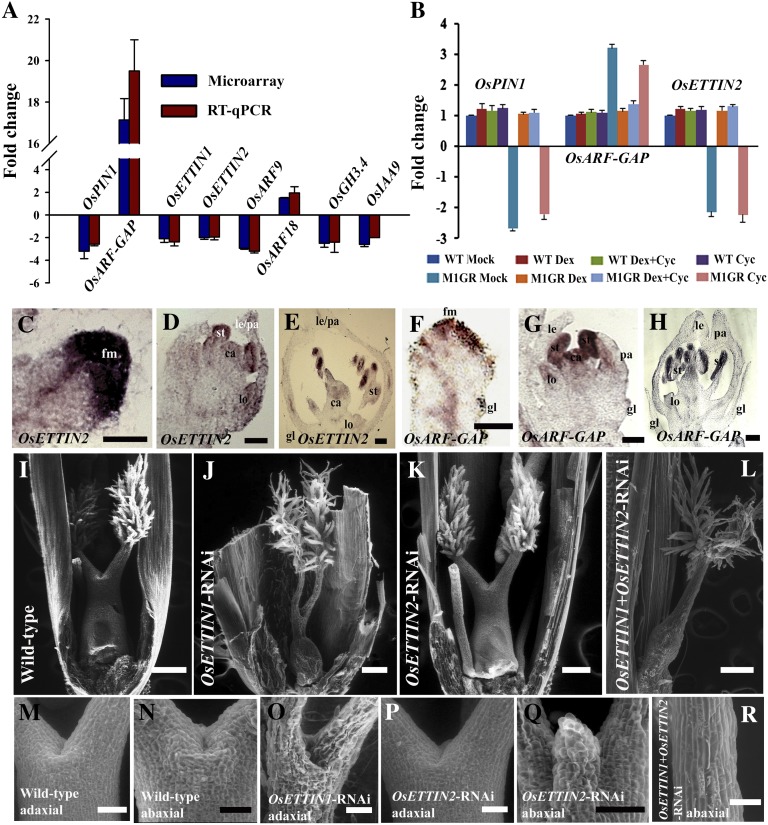
Mutants in rice and maize (Zea mays) coorthologs for Arabidopsis auxin biosynthesis factors, auxin transport, and signaling factors display defects in inflorescence axillary meristem development, spikelet and FM initiation, and organ numbers and organ development (McSteen et al., 2007; Morita and Kyozuka, 2007; McSteen, 2010; Phillips et al., 2011). Our microarray data showed that the second largest functional category of affected genes encodes signaling factors, and we note global effects on auxin metabolism and response (Supplemental Table S3; Supplemental Fig. S4, A and B). For example, several auxin response factor (ARF) genes (OsETTIN1, OsETTIN2, OsARF9, OsARF12, OsARF15, OsARF16, OsARF18, and OsARF25), a number of small auxin up RNA and GH3 auxin response genes, and polar auxin transport and signaling factors such as OsPINOID (Morita and Kyozuka, 2007) and rice PIN FORMED1 (Supplemental Fig. S4, A and B; Supplemental Table S3) were affected. This was paralleled by elevated expression of a predicted C2 domain-containing OsARF-GAP (an ADP ribosylation GTPase activating protein) and of several Aux/IAA (for Auxin/indole-3-acetic acid) repressors of auxin-dependent gene expression (Jain et al., 2006; Supplemental Fig. S4, A and B; Supplemental Table S3). The deregulated expression of OsARF9, OsARF18, two ETTIN-like genes, OsETTIN1 and OsETTIN2, and two key signaling factors, OsPIN1 and OsARF-GAP, was validated by RT-qPCR (Fig. 4A).
Figure 4.
Regulation of the auxin signaling pathway by OsMADS1. A, Fold change determined by RT-qPCR in the normalized expression of OsPIN1, OsARF-GAP, four ARFs (OsETTIN1, OsETTIN2, OsARF9, and OsARF18), and two auxin-responsive genes (OsGH3.4 and OsIAA9) in OsMADS1 knockdown florets. The fold change detected by microarray analysis is also shown. B, Expression levels for OsPIN1, OsARF-GAP, and OsETTIN2 in the panicles of wild-type (WT) and PUbi:OsMADS1-ΔGR; P35S:OsMADS1amiR plants treated individually with dexamethasone and cycloheximide and also in combination. The effects of these treatments were compared with that of wild-type plants. C to H, Spatial distribution of OsETTIN2 (C–E) and OsARF-GAP (F–H) transcripts in developing wild-type florets. C and F, Young FMs before organ initiation. D and G, FMs with differentiating floret organs. E and H, Florets with mature organs. Meristems and organs are labeled as in Figure 3. Bars = 50 µm in C, D, F, and G and 100 µm in E and H. I to L, Functional characterization of OsETTIN1 and OsETTIN2. Carpels are shown in the wild type (I), OsETTIN1-RNAi (J), OsETTIN2-RNAi (K), and OsETTIN1+OsETTIN2-RNAi (L). M to O, Abaxial and adaxial views of the style region above the ovary. The wild-type (M and N), OsETTIN1-RNAi (O), OsETTIN2-RNAi (P and Q), and OsETTIN1+OsETTIN2-RNAi (R) are shown. Bars = 200 µm in I to L and 50 µm in M to R.
We tested the possibility of direct regulation, by OsMADS1, of OsETTIN2, OsPIN1, and OsARF-GAP genes using panicles from PUbi:OsMADS1-ΔGR; P35S:OsMADS1amiR lines. As seen in OsMADS1-RNAi panicles, in mock-treated plants, OsPIN1 and OsETTIN2 expression was down-regulated while OsARF-GAP expression was enhanced (Fig. 4B). These abnormal expression levels were rescued to near wild-type levels in dexamethasone-treated panicles (Fig. 4B) and in panicles treated with both dexamethasone and cycloheximide. These data indicate that OsMADS1 directly modulates the expression of these genes in developing florets. The regulation of OsETTIN2 by OsMADS1 is further supported by a complete overlap in the spatial distribution of OsMADS1 and OsETTIN2 transcripts in early FMs and in emerging lemma/palea primordia (Fig. 4, C and D; Prasad et al., 2001, 2005). At later developmental stages, OsETTIN2 expression is mainly confined to inner floret organs (Fig. 4E). The distribution of OsARF-GAP transcripts in developing florets has some patterns distinct from OsMADS1 (Fig. 4, F–H). For example, in very early FMs, OsARF-GAP transcripts are largely restricted to the apical and central regions, unlike the uniform distribution of OsMADS1 in florets at this stage (Fig. 4F; Prasad et al., 2001). Differentiating lemma and palea do not express OsARF-GAP, while abundant OsMADS1 transcripts occur in these organs (Fig. 4, G and H; Prasad et al., 2001, 2005). Taken together, we deduce that OsMADS1 has a direct role in positively regulating auxin signaling and response during floret development.
OsETTIN1 and OsETTIN2 Redundantly Regulate Carpel Development
The ETTIN class of ARFs lack the carboxy terminal domain required for Aux/IAA interaction and are not influenced by auxin-mediated proteolysis; therefore, independent mechanisms regulate their function (Guilfoyle and Hagen, 2007). Arabidopsis ETTIN plays an important role in patterning FM and reproductive organs (Sessions et al., 1997). Our evidence for direct transcriptional regulation of OsETTIN2 by OsMADS1 prompted us to functionally characterize the closely related rice OsETTIN1 and OsETTIN2 genes by creating individual and double knockdown lines (Supplemental Fig. S5, A–C). Seventeen, 22, and 12 transgenic lines, several with near complete knockdown, were raised for OsETTIN1-RNAi, OsETTIN2-RNAi, and OsETTIN1+OsETTIN2-RNAi, respectively (Supplemental Fig. S5, D–I). While knockdown of OsETTIN1 or OsETTIN2 conferred mild floral phenotypes, simultaneous knockdown of both the genes in OsETTIN1+OsETTIN2-RNAi transgenics caused severe carpel defects that resembled Arabidopsis ettin mutants (Fig. 4, I–L; Sessions and Zambryski, 1995). The carpels in OsETTIN1-RNAi knockdown florets have a small increase in the style and stigma length, with a minor compromise in ovary size (Fig. 4, I and J). In OsETTIN2-RNAi florets, enhanced growth of the transmitting tract region from the abaxial (palea) face of the carpel was seen (Fig. 4, N and Q). Carpels in the florets of OsETTIN1+OsETTIN2-RNAi have severely reduced ovaries and undivided elongated styles (Fig. 4, L and R). Thus, rice OsETTIN1 and OsETTIN2 are redundant for ovary development and contribute to coordinate carpel tissue differentiation.
OsMADS1 Negatively Regulates Cytokinin Signaling with Direct Effects on A-Type Response Regulators
Interactions between cytokinin signaling and homeodomain transcription factors govern meristem maintenance in Arabidopsis, maize, and rice (Gordon et al., 2009; Tsuda et al., 2011; Bolduc et al., 2012). In OsMADS1 knockdown panicles, we detected an overall increased activity of cytokinin pathway genes, examples being LONELY GUY (LOG) family members, a predicted receptor, HKL1/CRL4, and several response regulator (RR) genes (Supplemental Table S4). Many of these expression effects were validated by RT-qPCR (Fig. 5A; Supplemental Table S1). Also, several of these genes were negatively modulated on ectopic expression of OsMADS1-ΔGR in vegetative meristems (Supplemental Fig. S6A).
Figure 5.
Negative regulation of components in cytokinin signaling by OsMADS1. A, Fold increase in the normalized transcript levels for two cytokinin biosynthetic genes (LOG and AK061341), four A-type response regulators (OsRR1, OsRR2, OsRR4, and OsRR9), and two B-type response regulators (OsRR16 and OsRR18) in OsMADS1 knockdown panicles versus wild-type panicles. The data from RT-qPCR are compared with those from microarray analysis. B, Transcript levels for OsRR1 and OsRR9 in wild-type (WT) and PUbi:OsMADS1-ΔGR; P35S:OsMADS1amiR plants treated with dexamethasone and cycloheximide individually and those given both chemicals compared with levels in mock-treated plants.
The repressive effects of OsMADS1 on cytokinin signaling in developing florets were further examined to determine the directly regulated genes from this pathway. Expression levels were studied for two type A response regulators, OsRR1 and OsRR9, in panicles from mock- and dexamethasone-treated PUbi:OsMADS1-ΔGR; P35S:OsMADS1amiR lines. We observed increased expression of OsRR1 and OsRR9 in mock-treated panicles, while in panicles of dexamethasone-treated plants, this was reduced to near wild-type levels (Fig. 5B). This reversal of expression status also occurred in the presence of cycloheximide, suggesting that OsMADS1 can repress the expression of these genes in wild-type panicles. To summarize, OsMADS1 may directly target A-type response regulators to negatively modulate cytokinin signaling during rice floret development.
The repressive effect of OsMADS1 on the expression of cytokinin pathway genes implied that their transcripts should be present at low levels in tissues with abundant OsMADS1. We analyzed the spatial distribution of OsRR1 and OsRR9 RNAs in developing wild-type florets. Their uniform expression in very young FMs overlapped with OsMADS1 (Fig. 6, A and D; Prasad et al., 2001, 2005). In differentiating floret organs, these rice A-type response regulators were expressed in lodicules, stamens, and carpel (Fig. 6, B, C, E, and F), wherein OsMADS1 transcripts are undetectable or at low levels. In the lemma and palea with high levels of OsMADS1 (Prasad et al., 2001, 2005), these A-type response regulator genes were not expressed. These data are consistent with their negative regulation by OsMADS1 in florets at later stages of development. We presume that the overall up-regulation of the cytokinin signaling pathway in OsMADS1-RNAi panicles may be an indirect effect of increased cytokinin biosynthesis. This notion is supported by the ectopic overexpression of LOG, a biosynthetic gene, in OsMADS1-RNAi florets (Supplemental Fig. S6, B–G).
Figure 6.
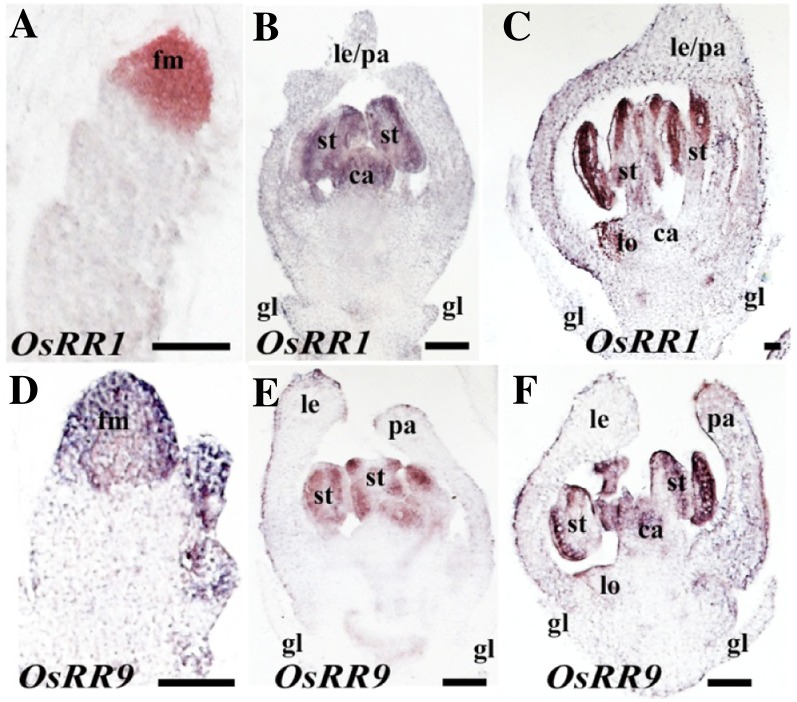
Spatial distribution of OsRR1 and OsRR9 transcripts in developing wild-type florets. A to C, Expression pattern for OsRR1. D to F, Expression pattern for OsRR9. A and D, FMs with emerging lemma/palea primordia. B and E, Florets at the early stages of inner floret organ development. C and F, Florets with nearly mature organs. Organs are labeled as in Figure 3. Bars = 50 µm.
Chromatin Association of OsMADS1 at CArG cis-Elements of Target Loci
Our data on the gene expression effects triggered by the induction of OsMADS1-ΔGR protein in panicles of OsMADS1 knockdown plants identified some key directly modulated target genes. These include genes for meristem regulators (OsMADS34, OsMADS55, OsHB4, OsKANADI2, OsKANADI4, and OsBLH1) and components in auxin (OsPIN1, OsARF-GAP, OsETTIN2, and OsARF9) and cytokinin (OsRR1, OsRR4, and OsRR9) signaling. Using young wild-type florets, we examined the in vivo association of OsMADS1 protein with CArG elements at these genomic loci. Chromatin immunoprecipitation (ChIP) analysis was done with chromatin from panicles (0.2–2 cm) with florets at early stages of meristem and organ development using affinity-purified polyclonal antibodies (Supplemental Fig. S7A). The positions of CArG elements in the DNA fragments taken for ChIP analysis are indicated in Figure 7 and in Supplemental Table S5. As controls, for each locus, we tested the OsMADS1 association with a chromatin fragment with no binding sites. We observed high levels of OsMADS1 occupancy at OsMADS34, OsMADS55, OsHB4, OsKANADI2, KANADI4, and OsBLH1 loci (Fig. 7, A–E and M; Supplemental Fig. S3, D and E). These data strongly correlate with the effects of OsMADS1 in modulating the expression status of these genes with known or predicted functions in spikelet/FMs. Among OsMADS1 targets from the auxin pathway, we detect significant OsMADS1 enrichment at OsETTIN2, OsPIN1, and OsARF-GAP loci and moderate occupancy at the OsARF9 locus (Fig. 7, F–I and N).
Figure 7.
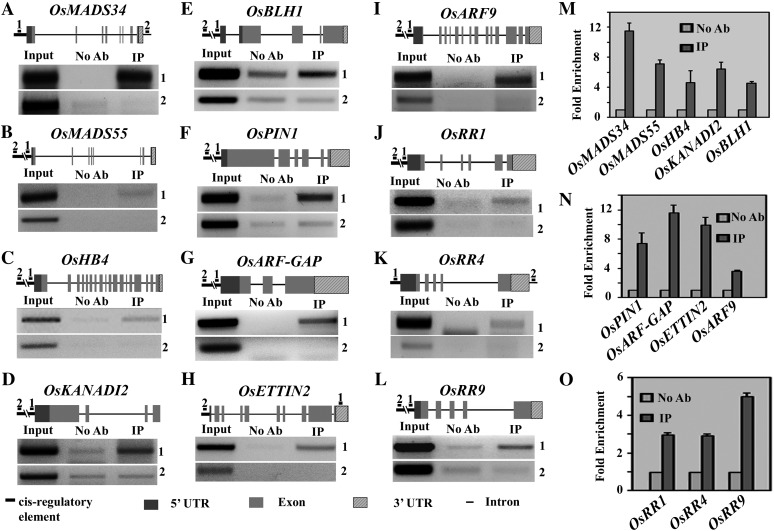
ChIP assay for OsMADS1 occupancy at genomic loci of target genes. Schematic diagrams show exons (light gray box), introns (thin black line), 5′ UTRs (dark gray box), 3′ UTRs (diagonally striped box), and upstream sequences (thick black line) for each locus analyzed. A short bar numbered 1 indicates the position of the DNA fragment with CArG elements that was taken for PCRs shown in the top panel for each locus. The region numbered 2 in the schematic shows the position of the PCR amplicon for a CArG-less nonspecific control DNA segment shown in the bottom panel for each locus. A to E, ChIP analysis for selected target genes encoding meristem regulatory transcription factors. PCR amplicons are shown for OsMADS34 (A), OsMADS55 (B), OsHB4 (C), OsKANADI2 (D), and BLH1 (E). In each case, the Input lane is a control PCR with sheared chromatin, the No Ab lane is PCR on chromatin mock precipitated without antibodies, and the IP lane is PCR on immunoprecipitated chromatin. F to I, Enrichment of OsMADS1 at cis-elements of genes in the auxin pathway: OsPIN1 (F), OsARF-GAP (G), OsETTIN2 (H), and OsARF9 (I). J to L, OsMADS1 occupancy on cytokinin A-type response regulators OsRR1 (J), OsRR4 (K), and OsRR9 (L). M to O, Quantitative representation of the fold enrichment for PCR amplicons from ChIP as compared with the no-antibody control in each case. The average enrichment is shown for two biological replicates, each analyzed in triplicate reactions.
To ascertain the modulation of cytokinin signaling pathway genes, we tested OsMADS1 occupancy at the cis-elements from loci for response regulators (A and B type) and at LOG, a cytokinin biosynthetic gene. The OsRR2 locus does not have any CArG motifs, so its promoter fragment also served as a nonspecific negative control. Specific OsMADS1 enrichment at OsRR1, OsRR4, and OsRR9 loci was observed (Fig. 7, J–L and O). OsMADS1 was not associated with OsRR2 genomic sequences or at loci for the B-type response regulators (OsRR16 and OsRR18) or the LOG locus (Supplemental Fig. S7, E–H). We conclude that some A-type response regulators are directly regulated by OsMADS1 in developing rice panicles. As expected, at each locus, genomic sequences that do not contain CArG elements were not enriched for OsMADS1 (denoted as fragment 2 in Fig. 7, A–L). In summary, OsMADS1 complexes bind and directly modulate the expression status of genes for transcription factors and hormone signaling components to promote the formation of a single determinate fertile floret on a spikelet meristem.
DISCUSSION
OsMADS1 Is a Regulator of Genetic Networks That Control the Spikelet-to-Floret Transition
Grass inflorescences (panicles) generate multiple kinds of branch meristems before forming a short branch called the spikelet that bears a variable number of florets. A rice spikelet has only a single floret subtended by two sterile lemmas (empty glumes). The latter represent vestiges of lower florets found in other grass spikelets (Bommert et al., 2005). Of the five SEP-like rice genes, OsMADS7 and OsMADS8 are members of SEP3 clade and have retained conserved functions, as they regulate organ development in inner floret whorls (Cui et al., 2010). But members of a grass-specific subgroup in the LOFSEP clade, OsMADS1, OsMADS5, and OsMADS34, have acquired novel functions (Malcomber and Kellogg, 2005; Gao et al., 2010; Kobayashi et al., 2010). OsMADS34 regulates panicle branching and promotes spikelet meristem and empty glume fate (Gao et al., 2010; Kobayashi et al., 2010, 2012). OsMADS1 is expressed developmentally slightly later and is restricted to the incipient FM region of the spikelet and controls its fate (Jeon et al., 2000; Prasad et al., 2005). Our data are consistent with an early OsMADS1 function in promoting the spikelet-to-floret transition by directly reducing OsMADS34 expression levels in the young spikelet. This interpretation is consistent with the phenotypes of osmads1 and osmads34 mutants. OsMADS1 loss-of-function mutants have indeterminate FM with misspecified multiple glume/lemma-like organs (Jeon et al., 2000; Prasad et al., 2005; Gao et al., 2010). Conversely, its ectopic overexpression is sufficient to initiate lemma formation, a floret-specific developmental program, in the spikelet empty glumes (Prasad et al., 2001). Interestingly, loss-of-function osmads34 mutant spikelets also show the conversion of empty glumes to lemma-like organs (Gao et al., 2010). We provide here, to our knowledge, the first molecular evidence of the regulatory relationship between these rice LOFSEP factors and the role of OsMADS1 in the spikelet-to-floret transition.
OsMADS1 Promotes FM Identity and Its Maintenance
After ensuring the transition of a spikelet meristem to a fertile FM, OsMADS1 contributes to the maintenance of FM identity and its determinate development. Genetic and physical interactions of OsMADS1 suggest that in promoting FM identity, it acts in conjunction with a SEP sister clade (AGL6-like) member, MOSAIC FLORAL ORGANS1/OsMADS6 (Ohmori et al., 2009; Li et al., 2010), and with OsMADS15/DEP, an APETALA1/FRUITFULL-like homolog (Supplemental Fig. S8). The interaction between OsMADS15 and OsMADS1 is particularly important for the maintenance of FMs, as they revert to shoot meristems in a mutant with loss-of-function alleles in both these genes (Wang et al., 2010). Together, these data suggest that OsMADS1 has both nonredundant and redundant roles in promoting FM development.
Arabidopsis SVP family genes, AGL24 and SVP, regulate floral transition and FM identity (Hartmann et al., 2000; Michaels et al., 2003; Gregis et al., 2008). Interestingly, rice SVP genes are not important for flowering time but have a conserved role in meristem identity and have additional functions in controlling the response to brassinosteroids (Lee et al., 2008). We provide here evidence for the divergence in the regulatory relationship between SVP and SEP genes of rice and Arabidopsis. Arabidopsis SEP3 directly represses AGL24 and SVP expression (Kaufmann et al., 2009). In contrast, we find that OsMADS1 can directly activate OsMADS55 in panicles with developing florets, consistent with the coexpression of these genes in developing panicles (Arora et al., 2007). Furthermore, overexpression phenotypes for OsMADS1, OsMADS22, and OsMADS55 share some characteristics like short panicle branches and spikelets with empty glumes transformed to lemma-like organs (Prasad et al., 2001; Sentoku et al., 2005; Lee et al., 2008). Thus, our data on the positive regulatory relationship between OsMADS1 and OsMADS55 are functionally supported. These regulatory patterns indicate some divergence between rice OsMADS1 and Arabidopsis SEP3 during FM establishment (i.e. OsMADS1 is a regulator of meristem identity early during its formation, while SEP3 plays a major role in promoting floral organ development).
Interactions among MADS Factors during Floret Organogenesis and Meristem Termination
OsMADS1, OsMADS6, and OsMADS15/DEP together control FM specification, as discussed above, yet they have unequal roles during organ differentiation, as suggested by differences in their expression domains concomitant with the emergence of lateral organs (Kyozuka et al., 2000; Prasad et al., 2005; Ohmori et al., 2009; Supplemental Fig. S8). Our data imply a role for OsMADS1 during growth of the newly initiated FM, perhaps to aid its transient indeterminacy. We speculate that this may occur by delaying organogenesis by repressing organ identity class B (OsMADS2 and OsMADS16) and class C (OsMADS3 and OsMADS58) genes in these very early FMs. This supposition agrees with the elevated expression levels of these organ identity genes on OsMADS1 knockdown (Supplemental Fig. S1). At slightly later developmental stages, OsMADS6 could participate in activating class B, C, and E (OsMADS7 and OsMADS8) floret organ identity genes. This postulate is consistent with their reduced expression in osmads6 mutants and with the expression domain of OsMADS6 in FMs, palea, lodicule, and carpel primordia (Ohmori et al., 2009; Li et al., 2011a; Supplemental Fig. S8). These regulatory relationships between LOFSEP, AGL6-like gene family members, and organ-patterning genes point to functional divergence between rice and Arabidopsis SEP factors.
In rice florets, the meristem terminates once the ovule primordium is generated as the final lateral organ (Yamaki et al., 2011). OsMADS1, OsMADS6, other SEP genes, and class C and D genes (OsMADS3, OsMADS58, OsMADS13, OsMADS21, OsMADS7, and OsMADS8) contribute to rice FM termination (Yamaguchi et al., 2006; Ohmori et al., 2009; Li et al., 2010, 2011a; Supplemental Fig. S8). Mutants in these factors share the phenotype of abnormal extra carpels (Fig. 2D; Yamaguchi et al., 2006; Dreni et al., 2007; Ohmori et al., 2009; Cui et al., 2010). Protein interactions of OsMADS1 with OsMADS13 (a class D gene), OsMADS7, and OsMADS8 (Favaro et al., 2002; Cui et al., 2010) and its genetic interactions with OsMADS6, OsMADS3, OsMADS58, and OsMADS13 (Dreni et al., 2011; Li et al., 2011a, 2011b; Supplemental Fig. S8) suggest that higher order complexes of these factors may promote meristem termination and ovule development.
OsMADS1 Directly Regulates Several Other Transcription Factors Implicated in FM Specification, Maintenance, and Determinacy
A balance between meristem indeterminacy and lateral organ emergence is crucial during early stages of FM development. The genetic interactions among FM identity, organ identity, and meristem maintenance genes (e.g. between WUSCHEL (WUS), AGAMOUS (AG), AGL24, and SVP; Liu et al., 2009; Grandi et al., 2012) and the combinatorial activity of three BLH proteins and SHOOT MERISTEMLESS (STM), a KNOX HD protein, regulate Arabidopsis FM initiation and maintenance (Rutjens et al., 2009). Recently, the miR165/166-targeted HD-Zip III members PHAVOLUTA and PHABULOSA were found to have an unexpected crucial role in Arabidopsis FM stem cell regulation (Ji et al., 2011), aside from their role in specifying organ polarity. Our data suggest that OsMADS1 aids rice FM development and determinacy possibly by regulating genes like OsHB4/HD-Zip III and OsYABBY5/TONGARI-BOUSHI1 (TOB1). A recent study demonstrated a role for TOB1 (OsYABBY5) in rice FM maintenance and determinacy (Tanaka et al., 2012). As loss-of-function mutants in all rice HD-Zip III genes are not known, a complete understanding of their functions is awaited. Regardless, as discussed by Itoh et al. (2008), the regulatory interactions between auxin and HD-Zip III factors in rice shoot meristems, where miR165/166 are not expressed (Nagasaki et al., 2007), may permit differential actions for HD-Zip III in meristems versus lateral organs. This conceptually allows auxin-regulated, and perhaps OsMADS1-regulated, functions for HD-Zip III transcription factors in early FM. OsMADS1 also links meristem identity with local auxin signaling by regulating genes for several ARFs (OsETTIN2, OsARF9, and OsARF12) and for other components in auxin signaling (Fig. 8). As homologs of some of these genes in auxin signaling are also direct targets of Arabidopsis SEP3 (Kaufmann et al., 2009; Supplemental Table S7) and we find that OsPIN1 and OsETTIN2 are directly modulated by OsMADS1, we infer that SEP factors have a conserved role in regulating auxin signaling during flower development.
Figure 8.
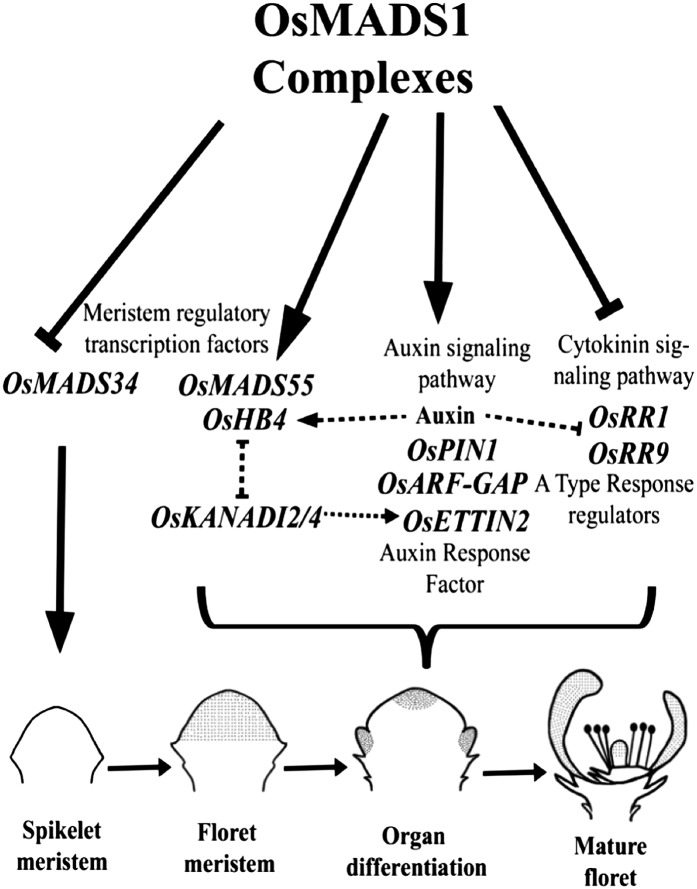
Model depicting the likely mechanism of OsMADS1 action in regulating meristem specification, determinacy, and organ differentiation through different stages of floret development. Gray dots depict the spatial domains of OsMADS1 expression in a young spikelet with an incipient FM, in a young floret at the early stages of lateral organ patterning, and in a differentiated floret. OsMADS1 and its partners in various complexes directly repress OsMADS34 to promote the transition from spikelet to FM. It activates other meristem regulators and genes in the auxin signaling pathway. These events could regulate early events in meristem specification, its floral fate, and lateral primordia differentiation. OsMADS1-containing complexes are also negative regulators of the cytokinin pathway that can contribute to meristem determinacy. These pathways and the cross talk between them in developing florets culminate in a determinate meristem, with its typically differentiated organs.
Regulation of KANADI and ETTIN Genes Contributes to Organ Development and Polarity
KANADI genes promote abaxial identity and laminar growth of lateral organs. ett arf4 double mutants have several lateral organ defects, such as short sepals with abaxial protrusions, narrow petals, sterile stamens, and gynoecia with distal abaxial and adaxial ovules, thus showing that these genes act in concert. Interestingly, these phenotypes are also seen in kan1 kan2 double mutants (Pekker et al., 2005). Recently, physical interactions between KANADI4 (KAN4/ATS) and ETTIN were shown to coordinate ovule and integument development (Kelley et al., 2012). Our finding that gene homologs of Arabidopsis KANADI2, ATS, and ETTIN (Supplemental Table S1) are directly regulated by OsMADS1 in rice florets brings new light on how OsMADS1 can contribute to lateral organ development. Derangements in lateral organs, such as narrow malformed lemma and palea with loss of abaxial features and decreased stamen numbers, observed in loss-of-function OsMADS1 mutants (Fig. 2; Jeon et al., 2000; Prasad et al., 2005), are broadly similar to the organ defects of Arabidopsis ett arf4 or kan1 kan2 mutants. Our studies on OsETTIN2 and OsETTIN1, a direct and indirect target regulated by OsMADS1, show their role in ensuring normal carpel differentiation. Interestingly the defects in style and stigma development and the abaxial overgrowth of the transmitting tract seen in OsETTIN1+OsETTIN2-RNAi knockdown carpels resemble ett phenotypes (Sessions and Zambryski, 1995). However, as we do not detect a complete failure of ovary formation in OsETTIN1+OsETTIN2-RNAi carpels, the possibility of redundancy with two other rice ETTIN-like genes cannot be ruled out. The contribution of other abaxial fate determinants such as OsKANADI2 and OsKANADI4 for floret organ development remains to be investigated.
OsMADS1 Has Regulatory Effects on Multiple Auxin Signaling Events
Auxin is often attributed morphogen-like properties in developmental processes like meristem specification and lateral organ formation (Benková et al., 2003). The auxin response is mediated by auxin response transcription factors (ARFs), which are repressed by Aux/IAA proteins in the absence of auxin (Guilfoyle and Hagen, 2007). OsMADS1 enhances the auxin response by simultaneously activating genes for ARFs with activation domains (e.g. OsARF9, OsARF12, OsARF16, and OsARF25) and repressing genes for ARFs with repression domains (e.g. OsARF18). Aside from these effects, we note that genes for auxin biosynthesis, homeostasis factors, polar distribution, and signaling factors were affected in OsMADS1 knockdown florets. We find that OsMADS1 directly modulates the expression of a predicted rice OsARF-GAP gene and of OsPIN1, encoding a key factor for polar auxin distribution. These data show that multiple shared elements in auxin signaling are regulated by OsMADS1 and SEP3 (Supplemental Table S7; Kaufmann et al., 2009).
OsMADS1 Negatively Regulates Cytokinin Signaling in Florets with Direct Effects on A-Type Response Regulators
The two major signaling-transcription factor networks that control meristem size and maintenance involve WUS and KNOX homeodomain factors and cytokinin signaling. Cytokinin induces WUS expression by multiple pathways (Gordon et al., 2009; Chickarmane et al., 2012). WUS in turn directly represses the expression of some type A cytokinin response regulators in a feedback loop that regulates stem cell number in Arabidopsis shoot meristem (Leibfried et al., 2005; Gordon et al., 2009). The importance of cytokinin levels for the size and determinacy of rice FMs is clearly illustrated by the phenotypes of log and ckx2 mutants in cytokinin metabolism (Ashikari et al., 2005; Kurakawa et al., 2007). OsMADS1 expression in the meristem center of florets undergoing organ differentiation and the indeterminate phenotype of OsMADS1 knockdown florets made it plausible that OsMADS1 modulates the cytokinin pathway. Our global expression analyses showed an overall increase in cytokinin levels and/or response in OsMADS1 knockdown florets. However, no significant changes in the expression of OsWOX1 (orthologous to WUS) or OsH1 (orthologous to STM) were observed, which hints that OsMADS1-mediated regulation of FM determinacy may occur independently. Our expression analyses after OsMADS1 induction and data from ChIP show that it can repress OsRR1, OsRR4, and OsRR9 A-type response regulator genes, while B-type response regulators and the biosynthetic gene LOG are indirectly affected. We cannot exclude the effects of auxin signaling on cytokinin pathway genes, as the promoter elements for several cytokinin pathway genes contain auxin-responsive elements (data not shown) and also because cross talk between auxin and cytokinin pathways are known (Swarup et al., 2002).
Collectively, our findings indicate that OsMADS1 complexes regulate the critical balance between these hormone signaling pathways during the establishment and development of the rice FM (Fig. 8). Importantly, the data illustrate that OsMADS1 complexes can either directly activate (e.g. meristem regulatory transcription factors and auxin signaling components) or repress (e.g. cytokinin pathway components) its target genes. We speculate that its interactions with partners in higher order complexes, and their interactions with chromatin modifiers, may determine its effect on gene expression, as is known for other transcription factor complexes (Sridhar et al., 2006; Liu et al., 2007; Smaczniak et al., 2012). The cumulative effects of these complexes on net target gene expression status culminates in a rice spikelet with a determinate floret with differentiated organs (Fig. 8).
MATERIALS AND METHODS
Plant Materials and Chemical Treatments
Rice (Oryza sativa var japonica) TP309 embryogenic calli were used to raise transgenics as described by Prasad et al. (2001). For global expression analysis, 0.2- to 2-cm panicles from wild-type and OsMADS1-RNAi plants were used. The ratio of panicles in different stages was 50% (0.2–0.5 cm), 30% (0.5–1.0 cm), and 20% (1.0–2.0 cm). For OsMADS1-ΔGR induction in PUbi:OsMADS1-ΔGR; P35S:OsMADS1amiR knockdown plants, these transgenics and wild-type plants were treated with one-half-strength Murashige and Skoog salts supplemented with 0.1% ethanol (mock), 10 µm dexamethasone (Sigma), 10 µm cycloheximide (Sigma), or 10 µm dexamethasone and 10 µm cycloheximide for 3 h. The developmental stages of panicle sizes and the ratios used to generate RNA pools for these experiments were the same as used in experiments with OsMADS1-RNAi panicles.
Microarray Analysis
Total RNAs from two biological pools of wild-type and OsMADS1-RNAi panicles were prepared as described (Yadav et al., 2007). The manufacturer’s instructions were followed for complementary RNA labeling and hybridization to 57K Affymetrix GeneChip Rice Genome Arrays, and the arrays were scanned using the Gene-Chip Scanner 7000. The *.CEL files generated by GeneChip Operating Software were analyzed by Avadis (version 4.2; currently GeneSpring). High correlation values of 0.9718 and 0.9981 between two biological replicates of mutant and wild-type samples, respectively, ensured the quality of our microarray experiments. The data were normalized by gene chip robust multiarray analysis algorithm and log2 transformed. Following standard one-way Student’s t test, the genes with 2-fold or greater change in expression values at P < 0.05 in mutant panicles as compared with wild-type panicles were identified as differentially expressed. To further avoid any false positives, we excluded very-low-expressing genes whose normalized signal intensity values were less than 10 in wild-type samples for down-regulated genes and less than 10 in OsMADS1-RNAi samples for up-regulated genes.
RT-qPCR
Oligo(dT)-primed cDNAs were synthesized using SuperScript III (Invitrogen) as described (Yadav et al., 2007). A total of 25 ng of cDNA was used in quantitative PCRs with 250 nm gene-specific primers and DyNAmo SYBR Green (Finnzymes) in an ABI 7900 HT system. For each deregulated gene, the difference in cycle threshold value between OsMADS1-RNAi and wild-type RNAs for the UBQ5-normalized transcript levels (ΔΔCt) was used to calculate fold change, with se being calculated from six replicates derived from two biological samples. For determining transcript levels in PUbi:OsMADS1-ΔGR; P35S:OsMADS1amiR transgenics and control wild-type plants before and after chemical treatments, the following approach was taken. The normalized ΔΔCt values, for each transcript, in all treatments were calculated for the wild type and transgenics individually. The actual fold change was computed after subtracting the normalized comparative threshold cycle values obtained in wild-type mock-treated samples from the comparative threshold cycle obtained for all other treatments. Primers used are listed in Supplemental Table S6.
In Situ Hybridization
Antisense RNA probes for OsMADS55, OsETTIN2, and OsRR9 were transcribed with T7 RNA polymerase from pBluescript KS+ clones of gene-specific cDNA fragments. T3 RNA polymerase transcriptions on these recombinants gave sense RNA probes. Antisense probes for OsRR1 and OsARF-GAP were prepared with T7 RNA polymerase and sense probes with SP6 RNA polymerase. Hybridizations on sections of developing spikelets and florets were done as described by Prasad et al. (2005), and signals were visualized using anti-digoxygenin-alkaline phosphatase conjugated antibodies (Roche).
Plasmid Construction, Protein Expression, and Antibody Production
A gene-specific OsMADS1 cDNA fragment encoding C-terminal amino acids 160 to 257 was cloned in pET32a, and the thioredoxin-His-tagged protein was expressed in bacterial cultures grown at 37°C for 3 h with 0.5 mm isopropylthio-β-galactoside. The C-terminal partial OsMADS1 protein was purified by nickel-nitrilotriacetic acid agarose chromatography, the thioredoxin-His tags were removed by thrombin cleavage. The purified protein was used to raise rabbit polyclonal antibodies. The latter were antigen affinity purified, and specificity was established by western blotting of panicle lysates (Supplemental Fig. S5A). Rice nuclear extracts were prepared as described by Busk and Pages (1997). amiRNA against OsMADS1 was designed according to Warthmann et al. (2008) using the Web tool http://wmd2.weigelworld.org/cgi-bin/mirnatools.pl, and this was cloned for expression from the 35S promoter in a binary vector (Supplemental Materials and Methods S1). amiRNA targets nucleotides 1,071 to 1,091 of the cDNA, 230 bp downstream of the stop codon in the 3′ UTR. For generating RNAi constructs for ETTIN genes, 316- and 395-bp gene-specific cDNA regions for OsETTIN1 and OsETTIN2, respectively, were amplified and cloned in the sense and antisense orientations in pBluescript KS+ separated by a 270-bp linker fragment. These assembled constructs for RNAi of these genes were cloned in pUN vector (Prasad et al., 2001) to yield PUbi:OsETTIN1-RNAi, PUbi:OsETTIN2-RNAi, and PUbi:OsETTIN1+OsETTIN2-RNAi plasmids (Supplemental Fig. S5). Primer sequences are listed in Supplemental Table S6.
ChIP Analysis
ChIP was carried out as described (Liu et al., 2008) with some modifications. Briefly, about 250 mg of 1% formaldehyde-fixed young wild-type rice panicles (0.2–2 cm) was ground in liquid nitrogen and then sonicated to yield chromatin with DNA of less than 500 bp. Immunoprecipitation was carried out at 4°C for 6 h in a buffer containing 150 mm NaCl, 25 mm Tris-HCl, pH 7.5, 5 mm EDTA, 1% Triton X-100, 0.1% SDS, 0.5% sodium deoxycholate, 1 mm phenylmethylsulfonyl fluoride, and protease inhibitors (Roche) with affinity-purified anti-OsMADS1 antibody and protein G-Sepharose beads (GE Healthcare). Bead-bound complexes were eluted in 2% SDS, 0.1 m NaHCO3, and 10 mm dithiothreitol at 37°C, and the cross links were reversed. A total of 25 ng of ethanol-precipitated DNA quantified on a NanoDrop ND-1000 spectrophotometer (Thermo Scientific) was used in PCR for 36 or 40 cycles. The primers used are listed in Supplemental Table S6.
The accession numbers for the cDNA sequences analyzed in this article are as follows: OsMADS1 (L34271), OsMADS5 (AK064184), OsMADS7 (AK100263), OsMADS16 (AK069317), OsMADS34 (AK100227), OsMADS55 (AK111859), OsMADS58 (AK111723), OsHB3 (AK102183), OsHB4 (AK103284), OsYABBY5 (AK070205), OsERTF4 (AK062882), OsKANADI2 (Os02g46940), OsKANADI4 (AK108408), OsDOF (AK119803), OsBLH1 (AK120844), OsKNAT3 (AK241438), OsMYB4 (AK109125), OsbHLH (AK107626), OsPIN1 (AK102343), OsETTIN1 (AK099793), OsETTIN2 (AK103776), OsARF9 (AK064925), OsARF18 (AK100322), OsRR1 (AK072736), OsRR2 (AK070645), OsRR4 (AK101721), OsRR9 (AK058585), OsRR16 (AK062250), OsRR18 (AK065276), OsBIM2 (AK071315), OsLOB37 (AK071624), OsGH3.4 (AK101932), LOG (AK071695), OsIAA9 (AK073365), and OsARF-GAP (AK099299).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Deregulation of transcription factor genes in OsMADS1-RNAi panicles.
Supplemental Figure S2. Characterization of PUbi:OsMADS1-ΔGR; P35S:OsMADS1amiR transgenic lines.
Supplemental Figure S3. Regulation of HD-Zip III and OsKANADI4 expression by OsMADS1.
Supplemental Figure S4. Regulation of components in the auxin signaling pathway by OsMADS1.
Supplemental Figure S5. Characterization of OsETTIN1 and OsETTIN2 knockdown plants.
Supplemental Figure S6. Regulation of cytokinin response regulators by OsMADS1-ΔGR.
Supplemental Figure S7. Specificity of rabbit anti-OsMADS1 antibodies.
Supplemental Figure S8. Model summarizing the genetic and physical interactions of OsMADS1.
Supplemental Table S1. List of genes studied in this article with their predicted Arabidopsis orthologs.
Supplemental Table S2. List of transcription factor genes from various classes with deregulated expression on OsMADS1 knockdown.
Supplemental Table S3. List of genes from the auxin signaling pathway deregulated in OsMADS1-RNAi.
Supplemental Table S4. List of genes from the cytokinin signaling pathway deregulated in OsMADS1-RNAi.
Supplemental Table S5. CArG elements at loci taken for ChIP analysis.
Supplemental Table S6. List of primers used in this study.
Supplemental Table S7. List of some unique and common regulated targets of rice OsMADS1 and Arabidopsis SEP3.
Supplemental Data Set S1. List of genes deregulated 2-fold or more (P < 0.05) when OsMADS1-RNAi panicles are compared with the wild type.
Supplemental Data Set S2. List of genes used for functional categorization.
Acknowledgments
We thank Prof. Sanjay Kapoor (University of Delhi) for assistance with the statistical analysis of microarray experiments and Dr. Kalika Prasad (Indian Institute of Science Education and Research, Thiruvanathapuram) for his critical comments. Grace Lhaineikim (Indian Institute of Science) is acknowledged for generating some P35S:OsMADS1amiR lines.
Glossary
- FM
floral meristem
- RNAi
RNA interference
- RT-qPCR
reverse transcription-quantitative PCR
- amiRNA
artificial microRNA
- UTR
untranslated region
- ChIP
chromatin immunoprecipitation
- cDNA
complementary DNA
References
- Agrawal GK, Abe K, Yamazaki M, Miyao A, Hirochika H. (2005) Conservation of the E-function for floral organ identity in rice revealed by the analysis of tissue culture-induced loss-of-function mutants of the OsMADS1 gene. Plant Mol Biol 59: 125–135 [DOI] [PubMed] [Google Scholar]
- Arora R, Agarwal P, Ray S, Singh AK, Singh VP, Tyagi AK, Kapoor S. (2007) MADS-box gene family in rice: genome-wide identification, organization and expression profiling during reproductive development and stress. BMC Genomics 8: 242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashikari M, Sakakibara H, Lin S, Yamamoto T, Takashi T, Nishimura A, Angeles ER, Qian Q, Kitano H, Matsuoka M. (2005) Cytokinin oxidase regulates rice grain production. Science 309: 741–745 [DOI] [PubMed] [Google Scholar]
- Benková E, Michniewicz M, Sauer M, Teichmann T, Seifertová D, Jürgens G, Friml J. (2003) Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602 [DOI] [PubMed] [Google Scholar]
- Bolduc N, Yilmaz A, Mejia-Guerra MK, Morohashi K, O’Connor D, Grotewold E, Hake S. (2012) Unraveling the KNOTTED1 regulatory network in maize meristems. Genes Dev 26: 1685–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bommert P, Satoh-Nagasawa N, Jackson D, Hirano HY. (2005) Genetics and evolution of inflorescence and flower development in grasses. Plant Cell Physiol 46: 69–78 [DOI] [PubMed] [Google Scholar]
- Busk PK, Pages M. (1997) Microextraction of nuclear proteins from single maize embryos. Plant Mol Biol Rep 15: 371–376 [Google Scholar]
- Chickarmane VS, Gordon SP, Tarr PT, Heisler MG, Meyerowitz EM. (2012) Cytokinin signaling as a positional cue for patterning the apical-basal axis of the growing Arabidopsis shoot meristem. Proc Natl Acad Sci USA 109: 4002–4007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui R, Han J, Zhao S, Su K, Wu F, Du X, Xu Q, Chong K, Theissen G, Meng Z. (2010) Functional conservation and diversification of class E floral homeotic genes in rice (Oryza sativa). Plant J 61: 767–781 [DOI] [PubMed] [Google Scholar]
- Ditta G, Pinyopich A, Robles P, Pelaz S, Yanofsky MF. (2004) The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Curr Biol 14: 1935–1940 [DOI] [PubMed] [Google Scholar]
- Dreni L, Jacchia S, Fornara F, Fornari M, Ouwerkerk PB, An G, Colombo L, Kater MM. (2007) The D-lineage MADS-box gene OsMADS13 controls ovule identity in rice. Plant J 52: 690–699 [DOI] [PubMed] [Google Scholar]
- Dreni L, Pilatone A, Yun D, Erreni S, Pajoro A, Caporali E, Zhang D, Kater MM. (2011) Functional analysis of all AGAMOUS subfamily members in rice reveals their roles in reproductive organ identity determination and meristem determinacy. Plant Cell 23: 2850–2863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery JF, Floyd SK, Alvarez J, Eshed Y, Hawker NP, Izhaki A, Baum SF, Bowman JL. (2003) Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr Biol 13: 1768–1774 [DOI] [PubMed] [Google Scholar]
- Favaro R, Immink RG, Ferioli V, Bernasconi B, Byzova M, Angenent GC, Kater M, Colombo L. (2002) Ovule-specific MADS-box proteins have conserved protein-protein interactions in monocot and dicot plants. Mol Genet Genomics 268: 152–159 [DOI] [PubMed] [Google Scholar]
- Gao X, Liang W, Yin C, Ji S, Wang H, Su X, Guo C, Kong H, Xue H, Zhang D. (2010) The SEPALLATA-like gene OsMADS34 is required for rice inflorescence and spikelet development. Plant Physiol 153: 728–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon SP, Chickarmane VS, Ohno C, Meyerowitz EM. (2009) Multiple feedback loops through cytokinin signaling control stem cell number within the Arabidopsis shoot meristem. Proc Natl Acad Sci USA 106: 16529–16534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi V, Gregis V, Kater MM. (2012) Uncovering genetic and molecular interactions among floral meristem identity genes in Arabidopsis thaliana. Plant J 69: 881–893 [DOI] [PubMed] [Google Scholar]
- Gregis V, Sessa A, Colombo L, Kater MM. (2008) AGAMOUS-LIKE24 and SHORT VEGETATIVE PHASE determine floral meristem identity in Arabidopsis. Plant J 56: 891–902 [DOI] [PubMed] [Google Scholar]
- Guilfoyle TJ, Hagen G. (2007) Auxin response factors. Curr Opin Plant Biol 10: 453–460 [DOI] [PubMed] [Google Scholar]
- Hartmann U, Höhmann S, Nettesheim K, Wisman E, Saedler H, Huijser P. (2000) Molecular cloning of SVP: a negative regulator of the floral transition in Arabidopsis. Plant J 21: 351–360 [DOI] [PubMed] [Google Scholar]
- Honma T, Goto K. (2001) Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 409: 525–529 [DOI] [PubMed] [Google Scholar]
- Immink RG, Tonaco IA, de Folter S, Shchennikova A, van Dijk AD, Busscher-Lange J, Borst JW, Angenent GC. (2009) SEPALLATA3: the ‘glue’ for MADS box transcription factor complex formation. Genome Biol 10: R24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh J, Hibara K, Sato Y, Nagato Y. (2008) Developmental role and auxin responsiveness of class III homeodomain leucine zipper gene family members in rice. Plant Physiol 147: 1960–1975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M, Kaur N, Garg R, Thakur JK, Tyagi AK, Khurana JP. (2006) Structure and expression analysis of early auxin-responsive Aux/IAA gene family in rice (Oryza sativa). Funct Integr Genomics 6: 47–59 [DOI] [PubMed] [Google Scholar]
- Jeon JS, Jang S, Lee S, Nam J, Kim C, Lee SH, Chung YY, Kim SR, Lee YH, Cho YG, et al (2000) leafy hull sterile1 is a homeotic mutation in a rice MADS box gene affecting rice flower development. Plant Cell 12: 871–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji L, Liu X, Yan J, Wang W, Yumul RE, Kim YJ, Dinh TT, Liu J, Cui X, Zheng B, et al. (2011) ARGONAUTE10 and ARGONAUTE1 regulate the termination of floral stem cells through two microRNAs in Arabidopsis. PLoS Genet 7: e1001358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann K, Muiño JM, Jauregui R, Airoldi CA, Smaczniak C, Krajewski P, Angenent GC. (2009) Target genes of the MADS transcription factor SEPALLATA3: integration of developmental and hormonal pathways in the Arabidopsis flower. PLoS Biol 7: e1000090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley DR, Arreola A, Gallagher TL, Gasser CS. (2012) ETTIN (ARF3) physically interacts with KANADI proteins to form a functional complex essential for integument development and polarity determination in Arabidopsis. Development 139: 1105–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerstetter RA, Bollman K, Taylor RA, Bomblies K, Poethig RS. (2001) KANADI regulates organ polarity in Arabidopsis. Nature 411: 706–709 [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Maekawa M, Miyao A, Hirochika H, Kyozuka J. (2010) PANICLE PHYTOMER2 (PAP2), encoding a SEPALLATA subfamily MADS-box protein, positively controls spikelet meristem identity in rice. Plant Cell Physiol 51: 47–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Yasuno N, Sato Y, Yoda M, Yamazaki R, Kimizu M, Yoshida H, Nagamura Y, Kyozuka J. (2012) Inflorescence meristem identity in rice is specified by overlapping functions of three AP1/FUL-like MADS box genes and PAP2, a SEPALLATA MADS box gene. Plant Cell 24: 1848–1859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizek BA, Fletcher JC. (2005) Molecular mechanisms of flower development: an armchair guide. Nat Rev Genet 6: 688–698 [DOI] [PubMed] [Google Scholar]
- Kurakawa T, Ueda N, Maekawa M, Kobayashi K, Kojima M, Nagato Y, Sakakibara H, Kyozuka J. (2007) Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature 445: 652–655 [DOI] [PubMed] [Google Scholar]
- Kyozuka J, Kobayashi T, Morita M, Shimamoto K. (2000) Spatially and temporally regulated expression of rice MADS box genes with similarity to Arabidopsis class A, B and C genes. Plant Cell Physiol 41: 710–718 [DOI] [PubMed] [Google Scholar]
- Lee S, Choi SC, An G. (2008) Rice SVP-group MADS-box proteins, OsMADS22 and OsMADS55, are negative regulators of brassinosteroid responses. Plant J 54: 93–105 [DOI] [PubMed] [Google Scholar]
- Leibfried A, To JP, Busch W, Stehling S, Kehle A, Demar M, Kieber JJ, Lohmann JU. (2005) WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature 438: 1172–1175 [DOI] [PubMed] [Google Scholar]
- Li H, Liang W, Hu Y, Zhu L, Yin C, Xu J, Dreni L, Kater MM, Zhang D. (2011a) Rice MADS6 interacts with the floral homeotic genes SUPERWOMAN1, MADS3, MADS58, MADS13, and DROOPING LEAF in specifying floral organ identities and meristem fate. Plant Cell 23: 2536–2552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Liang W, Jia R, Yin C, Zong J, Kong H, Zhang D. (2010) The AGL6-like gene OsMADS6 regulates floral organ and meristem identities in rice. Cell Res 20: 299–313 [DOI] [PubMed] [Google Scholar]
- Li H, Liang W, Yin C, Zhu L, Zhang D. (2011b) Genetic interaction of OsMADS3, DROOPING LEAF, and OsMADS13 in specifying rice floral organ identities and meristem determinacy. Plant Physiol 156: 263–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Chen H, Er HL, Soo HM, Kumar PP, Han JH, Liou YC, Yu H. (2008) Direct interaction of AGL24 and SOC1 integrates flowering signals in Arabidopsis. Development 135: 1481–1491 [DOI] [PubMed] [Google Scholar]
- Liu C, Thong Z, Yu H. (2009) Coming into bloom: the specification of floral meristems. Development 136: 3379–3391 [DOI] [PubMed] [Google Scholar]
- Liu C, Zhou J, Bracha-Drori K, Yalovsky S, Ito T, Yu H. (2007) Specification of Arabidopsis floral meristem identity by repression of flowering time genes. Development 134: 1901–1910 [DOI] [PubMed] [Google Scholar]
- Malcomber ST, Kellogg EA. (2005) SEPALLATA gene diversification: brave new whorls. Trends Plant Sci 10: 427–435 [DOI] [PubMed] [Google Scholar]
- McSteen P. (2010) Auxin and monocot development. Cold Spring Harb Perspect Biol 2: a001479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McSteen P, Malcomber S, Skirpan A, Lunde C, Wu X, Kellogg E, Hake S. (2007) barren inflorescence2 encodes a co-ortholog of the PINOID serine/threonine kinase and is required for organogenesis during inflorescence and vegetative development in maize. Plant Physiol 144: 1000–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, Ditta G, Gustafson-Brown C, Pelaz S, Yanofsky M, Amasino RM. (2003) AGL24 acts as a promoter of flowering in Arabidopsis and is positively regulated by vernalization. Plant J 33: 867–874 [DOI] [PubMed] [Google Scholar]
- Morita Y, Kyozuka J. (2007) Characterization of OsPID, the rice ortholog of PINOID, and its possible involvement in the control of polar auxin transport. Plant Cell Physiol 48: 540–549 [DOI] [PubMed] [Google Scholar]
- Nagasaki H, Itoh J, Hayashi K, Hibara K, Satoh-Nagasawa N, Nosaka M, Mukouhata M, Ashikari M, Kitano H, Matsuoka M, et al (2007) The small interfering RNA production pathway is required for shoot meristem initiation in rice. Proc Natl Acad Sci USA 104: 14867–14871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmori S, Kimizu M, Sugita M, Miyao A, Hirochika H, Uchida E, Nagato Y, Yoshida H. (2009) MOSAIC FLORAL ORGANS1, an AGL6-like MADS box gene, regulates floral organ identity and meristem fate in rice. Plant Cell 21: 3008–3025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekker I, Alvarez JP, Eshed Y. (2005) Auxin response factors mediate Arabidopsis organ asymmetry via modulation of KANADI activity. Plant Cell 17: 2899–2910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelaz S, Ditta GS, Baumann E, Wisman E, Yanofsky MF. (2000) B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405: 200–203 [DOI] [PubMed] [Google Scholar]
- Phillips KA, Skirpan AL, Liu X, Christensen A, Slewinski TL, Hudson C, Barazesh S, Cohen JD, Malcomber S, McSteen P. (2011) vanishing tassel2 encodes a grass-specific tryptophan aminotransferase required for vegetative and reproductive development in maize. Plant Cell 23: 550–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad K, Parameswaran S, Vijayraghavan U. (2005) OsMADS1, a rice MADS-box factor, controls differentiation of specific cell types in the lemma and palea and is an early-acting regulator of inner floral organs. Plant J 43: 915–928 [DOI] [PubMed] [Google Scholar]
- Prasad K, Sriram P, Kumar CS, Kushalappa K, Vijayraghavan U. (2001) Ectopic expression of rice OsMADS1 reveals a role in specifying the lemma and palea, grass floral organs analogous to sepals. Dev Genes Evol 211: 281–290 [DOI] [PubMed] [Google Scholar]
- Rutjens B, Bao D, van Eck-Stouten E, Brand M, Smeekens S, Proveniers M. (2009) Shoot apical meristem function in Arabidopsis requires the combined activities of three BEL1-like homeodomain proteins. Plant J 58: 641–654 [DOI] [PubMed] [Google Scholar]
- Sentoku N, Kato H, Kitano H, Imai R. (2005) OsMADS22, an STMADS11-like MADS-box gene of rice, is expressed in non-vegetative tissues and its ectopic expression induces spikelet meristem indeterminacy. Mol Genet Genomics 273: 1–9 [DOI] [PubMed] [Google Scholar]
- Sessions A, Nemhauser JL, McColl A, Roe JL, Feldmann KA, Zambryski PC. (1997) ETTIN patterns the Arabidopsis floral meristem and reproductive organs. Development 124: 4481–4491 [DOI] [PubMed] [Google Scholar]
- Sessions RA, Zambryski PC. (1995) Arabidopsis gynoecium structure in the wild and in ettin mutants. Development 121: 1519–1532 [DOI] [PubMed] [Google Scholar]
- Shani E, Yanai O, Ori N. (2006) The role of hormones in shoot apical meristem function. Curr Opin Plant Biol 9: 484–489 [DOI] [PubMed] [Google Scholar]
- Smaczniak C, Immink RG, Muiño JM, Blanvillain R, Busscher M, Busscher-Lange J, Dinh QD, Liu S, Westphal AH, Boeren S, et al. (2012) Characterization of MADS-domain transcription factor complexes in Arabidopsis flower development. Proc Natl Acad Sci USA 109: 1560–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridhar VV, Surendrarao A, Liu Z. (2006) APETALA1 and SEPALLATA3 interact with SEUSS to mediate transcription repression during flower development. Development 133: 3159–3166 [DOI] [PubMed] [Google Scholar]
- Swarup R, Parry G, Graham N, Allen T, Bennett M. (2002) Auxin cross-talk: integration of signalling pathways to control plant development. Plant Mol Biol 49: 411–426 [DOI] [PubMed] [Google Scholar]
- Tanaka W, Toriba T, Ohmori Y, Yoshida A, Kawai A, Mayama-Tsuchida T, Ichikawa H, Mitsuda N, Ohme-Takagi M, Hirano HY. (2012) The YABBY gene TONGARI-BOUSHI1 is involved in lateral organ development and maintenance of meristem organization in the rice spikelet. Plant Cell 24: 80–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson BE, Hake S. (2009) Translational biology: from Arabidopsis flowers to grass inflorescence architecture. Plant Physiol 149: 38–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda K, Ito Y, Sato Y, Kurata N. (2011) Positive autoregulation of a KNOX gene is essential for shoot apical meristem maintenance in rice. Plant Cell 23: 4368–4381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Tang D, Hong L, Xu W, Huang J, Li M, Gu M, Xue Y, Cheng Z. (2010) DEP and AFO regulate reproductive habit in rice. PLoS Genet 6: e1000818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warthmann N, Chen H, Ossowski S, Weigel D, Herve P. (2008) Highly specific gene silencing by artificial miRNAs in rice. PLoS ONE 3: e1829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav SR, Prasad K, Vijayraghavan U. (2007) Divergent regulatory OsMADS2 functions control size, shape and differentiation of the highly derived rice floret second-whorl organ. Genetics 176: 283–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T, Lee DY, Miyao A, Hirochika H, An G, Hirano HY. (2006) Functional diversification of the two C-class MADS box genes OSMADS3 and OSMADS58 in Oryza sativa. Plant Cell 18: 15–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaki S, Nagato Y, Kurata N, Nonomura K. (2011) Ovule is a lateral organ finally differentiated from the terminating floral meristem in rice. Dev Biol 351: 208–216 [DOI] [PubMed] [Google Scholar]