A novel mitochondrial carrier protein is required for early leaf development in rice.
Abstract
A dual-targeted protein belonging to the mitochondrial carrier family was characterized in rice (Oryza sativa) and designated 3′-Phosphoadenosine 5′-Phosphosulfate Transporter1 (PAPST1). The papst1 mutant plants showed a defect in thylakoid development, resulting in leaf chlorosis at an early leaf developmental stage, while normal leaf development was restored 4 to 6 d after leaf emergence. OsPAPST1 is highly expressed in young leaves and roots, while the expression is reduced in mature leaves, in line with the recovery of chloroplast development seen in the older leaves of papst1 mutant plants. OsPAPST1 is located on the outer mitochondrial membrane and chloroplast envelope. Whole-genome transcriptomic analysis reveals reduced expression of genes encoding photosynthetic components (light reactions) in papst1 mutant plants. In addition, sulfur metabolism is also perturbed in papst1 plants, and it was seen that PAPST1 can act as a nucleotide transporter when expressed in Escherichia coli that can be inhibited significantly by 3′-phosphoadenosine 5′-phosphosulfate. Given these findings, together with the altered phenotype seen only when leaves are first exposed to light, it is proposed that PAPST1 may act as a 3′-phosphoadenosine 5′-phosphosulfate carrier that has been shown to act as a retrograde signal between chloroplasts and the nucleus.
Phosphorous is an essential macronutrient in all cells, where it is an integral component of macromolecules such as DNA, RNA, and phospholipids. It is important in metabolism in that many metabolites exist as phosphorylated forms, and phosphorous is an essential component of the energy currency in cells in the form of ATP. Furthermore, phosphorous plays an important role in signal transduction, and with kinase and phosphatases it plays roles in the posttranslational regulation of a variety of proteins in the cell, including transcription factors (Chiou and Lin, 2011). Plants obtain phosphorous directly from soils, which are often limiting in available phosphate sources and thus require the application of expensive fertilizers to support plant growth. Thus, it is not surprising that the acquisition and regulation of phosphate in plant cells is an intensively studied process (Chiou and Lin, 2011; Chen et al., 2012).
The purine nucleotide ATP is one of the most crucial cellular molecules containing organic phosphate. It is the major energy donor in most metabolic reactions and is also used to generate activated precursors for the synthesis of many macromolecules (Zrenner et al., 2006). In plants, two types of nucleotide transporters have been identified on the molecular level, namely, mitochondrial carrier family-type and plastid nucleotide transporter (NTT)-type carriers (Haferkamp et al., 2011). While initially characterized in mitochondria, mitochondrial carrier family-type ATP carriers are present in almost all organelles, including mitochondria, plastids, peroxisomes, glyoxysomes (Arai et al., 2008; Linka et al., 2008; Palmieri et al., 2011), endoplasmic reticulum (Leroch et al., 2008), and plasma membrane (Rieder and Neuhaus, 2011). The second type of plant ATP carrier, the NTT type, are phylogenetically derived from bacterial homologs in all forms of plastids. In Arabidopsis (Arabidopsis thaliana), two NTT isoforms exist, and both localize to plastids and show tissue-specific expression patterns (Haferkamp et al., 2011).
In nonphotosynthetic plastids, the main function of ATP/ADP transporters is supplying ATP-dependent reactions, such as starch and fatty acid biosynthesis, with cytosolic ATP (Haferkamp et al., 2011). In contrast to nongreen plastids, the function of ATP transport in chloroplasts is not clear. Chloroplasts likely depend on an external supply of ATP during the night for various biosynthetic processes, including the assembly of magnesium chelatase that catalyzes the insertion of an Mg2+ ion into protoporphyrin IX (Reinhold et al., 2007), and analysis of the functions of the two NTT carriers in Arabidopsis suggests that they are required under conditions where photosynthesis is limited; thus, substrate-level phosphorylation cannot supply sufficient ATP (Haferkamp et al., 2011; Weber and Linka, 2011). A thylakoid ATP/ADP carrier (TAAC) encoded by the gene At5g01500 has been characterized in Arabidopsis (Thuswaldner et al., 2007). The lack of the TAAC in an Arabidopsis transfer DNA insertion mutant caused a 30% to 40% reduction in thylakoid ATP transport and metabolism. It is proposed that the TAAC protein supplies ATP for energy-dependent reactions during thylakoid biogenesis and turnover in plants based on the fact that the TAAC is readily expressed in dark-grown Arabidopsis seedlings, and its level remains stable throughout the greening process. Its expression is highest in developing green tissues and in leaves undergoing senescence or abiotic stress (Thuswaldner et al., 2007).
A mitochondrial and chloroplastic dual-targeted ATP/ADP transporter of the mitochondrial carrier family in maize (Zea mays) and Arabidopsis, AtBT1 (for brittle1), has been shown to be localized to the inner plastidial envelope and mitochondria (Kirchberger et al., 2008; Bahaji et al., 2011a). The aberrant growth and sterility phenotype of homozygous Atbt1 transfer DNA mutants was complemented when expressing both the dual-targeted AtBT1 and AtBT1 specifically delivered to mitochondria (Bahaji et al., 2011b). So AtBT1 localized to mitochondria is important for development and growth (Bahaji et al., 2011b). But the function of AtBT1 localized to the inner plastidial envelope is unclear.
Here, we report a novel dual-targeted mitochondrial carrier protein in rice (Oryza sativa) called 3′-Phosphoadenosine 5′-Phosphosulfate Transporter1 (PAPST1; LOC_Os01g16040), which is required for early leaf development in rice. A variety of evidence suggests that it acts at least in part as a 3′-phosphoadenosine 5′-phosphosulfate (PAPS) carrier, revealing that this molecule is associated with chloroplast retrograde signaling (Chen et al., 2011; Estavillo et al., 2011) and also plays a role in chloroplast development in rice.
RESULTS
Isolation and Phenotypic Characterization of the papst1 Mutant
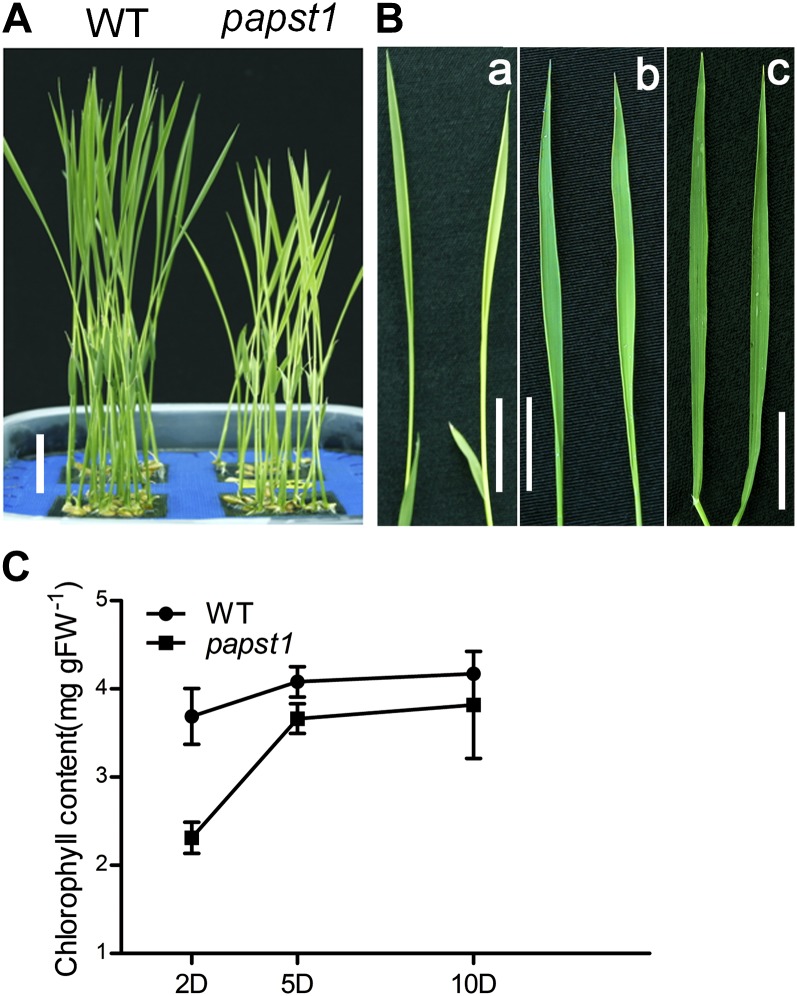
A rice mutant with leaf virescent (Fig. 1A) was isolated from an ethylmethane sulfonate-generated rice mutant library (cv Nipponbare) and was named papst1 after the gene was cloned from the mutant (see below). The newly emerged leaves of the papst1 mutant showed chlorosis and gradually turned green during leaf maturation. Analysis of the third leaf in detail revealed that on day 2 after emerging, chlorosis was evident in papst1 plants when grown under the conditions of 12 h of light (350 mmol m−2 s−1 illumination)/12 h of dark. Over time, the leaves gradually turned green, and 8 d after emergence, they were similar to wild-type leaves (Fig. 1B). papst1 mutant plants were smaller than wild-type plants at all developmental stages (Fig. 1A; Supplemental Fig; S1), although the morphology of roots, and the seed yield of the mutants, were the same as in wild-type plants (Supplemental Fig. S1).
Figure 1.
Phenotypic characterization of papst1 mutant plants. A, Shoots of 7-d-old seedlings. Bar = 2 cm. B, Phenotypes of the third leaf of wild-type (WT; left) and papst1 mutant (right) plants on days 2 (a), 5 (b), and 10 (c) after emerging. Bars = 2 cm. C, Chlorophyll contents of the third leaf at days 2, 5, and 10 after emerging of wild-type and papst1 mutant plants. Values represent means ± sd of 10 seedlings. FW, Fresh weight.
The chlorophyll content of the third leaf 2, 5, and 10 d after emergence was measured. The chlorophyll content of the leaves of papst1 mutants at 2 and 5 d after emergence (2.31 ± 0.17 and 3.66 ± 0.17 mg g−1) was significantly lower than that of wild-type plants (3.65 ± 0.31 and 4.08 ± 0.16 mg g−1). In contrast, the chlorophyll content of the leaves of papst1 mutants at 10 d after emergence was similar to that of the wild type, with papst1 at 3.82 ± 0.6 mg g−1 and wild-type plants at 4.17 ± 0.05 mg g−1 (Fig. 1C).
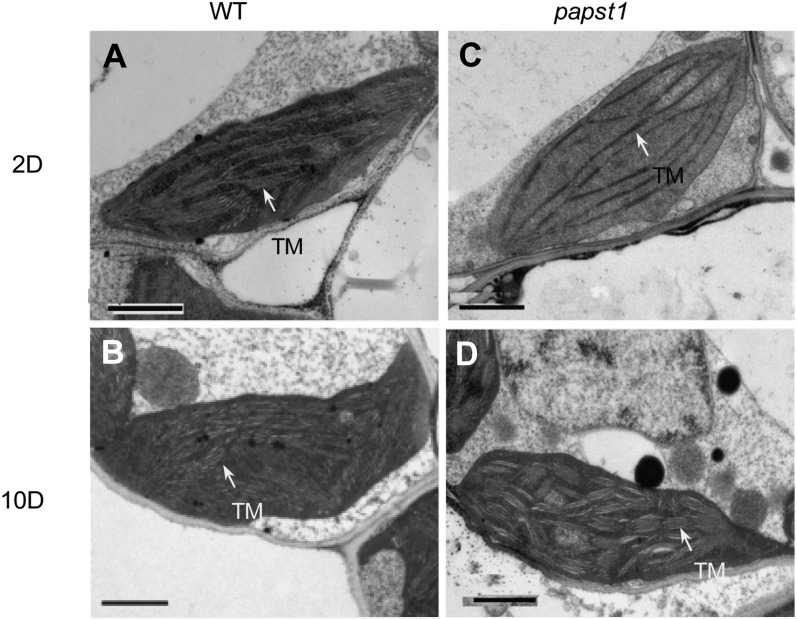
The chloroplast ultrastructure in the third leaf 2 and 10 d after emergence of papst1 and wild-type plants was examined using transmission electron microscopy (TEM). The structure of thylakoid membrane organization was altered in the third leaf of papst1 mutant plants at 2 d after emergence compared with that of wild-type plants. The granal stacks were reduced and less dense and reduced membranes in papst1 compared with the wild type (Fig. 2, A and C). At 10 d after emergence, the chloroplasts of papst1 were normal, like those of the wild type (Fig. 2, B and D). TEM images also showed that mitochondrial morphologies were similar between papst1 mutant and wild-type plants, with electron-dense oval mitochondria evident in both plants with extensive cristae structure (Supplemental Fig. S2).
Figure 2.
TEM images of chloroplast structure in wild-type (WT) and papst1 mutant plants. Electron micrographs show the third leaf at 2 and 10 d after emerging in wild-type (A and B) and papst1 mutant (C and D) plants, respectively. Arrows indicate thylakoid membranes (TM). Bars = 1 µm.
Cloning and Characterization of papst1
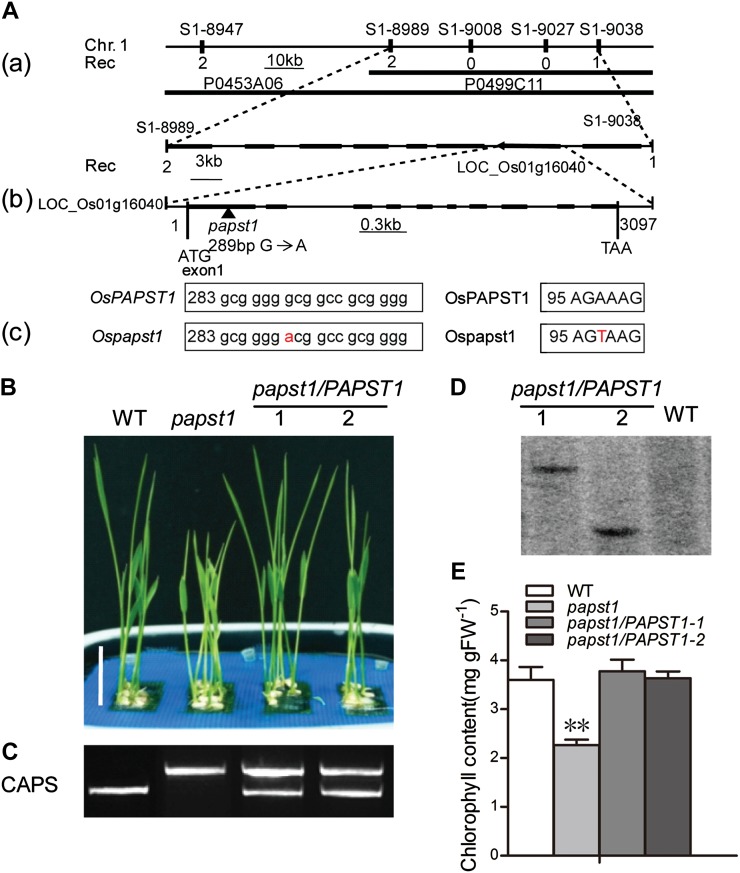
A map-based cloning approach was used to identify the gene from an F2 population derived from a cross between the papst1 mutant and indica rice (var Kasalath). The mutated gene locus was initially mapped between two simple sequence repeat (SSR) markers, S1-8989 and S1-9038, on the short arm of chromosome 1 (Fig. 3A). The region is covered by two plasmid clones, P0453A06 and P0499C11. According to the annotation information (http://rice.plantbiology.msu.edu/), eight putative genes are located in this region. These eight genes were amplified from both papst1 and wild-type plants and sequenced. The results revealed that in exon 1 of LOC_Os01g16040, a single nucleotide change (G289A) in papst1 mutant plants caused a missense mutation resulting in a change of Ala to Thr (A97T). The point mutation in the papst1 allele was visually verified using the cleaved amplified polymorphic sequence (CAPS) marker (Fig. 3C).
Figure 3.
Map-based cloning of OsPAPST1 and complementation test. A, Map-based cloning of OsPAPST1. OsPAPST1 was mapped between two SSR markers, S1-8989 and S1-9038, on the short arm of chromosome 1 (a). The genomic region contains eight genes (black boxes). The sequence surrounding the point mutation (G-to-A transition) in papst1 is shown in b and c. The genomic structure of OsPAPST1, comprising 10 exons and 9 introns, is indicated. B, Phenotypes of 6-d-old plants of wild-type (WT), papst1 mutant, and two transgenic plants transformed with OsPAPST1. Bar = 2 cm. C, PCR analysis using a CAPS marker for wild-type, papst1 mutant, and two transgenic plants transformed with OsPAPST1. D, Southern-blot analysis of the wild type and the two transgenic lines transformed with OsPAPST1, using the hygromycin gene as a probe. E, Chlorophyll contents of the third leaf of 6-d-old wild-type, papst1 mutant, and two transgenic plants transformed with OsPAPST1. Values represent means ± sd of 10 biological replicates. papst1 values that are significantly different from the corresponding wild-type controls are indicated by asterisks (**P < 0.01; Student’s t test). FW, Fresh weight. [See online article for color version of this figure.]
To verify that the mutant phenotype was caused by the point mutation of OsPAPST1, papst1 plants were transformed with the full-length genomic sequence of OsPAPST1 under the control of its native promoter. Two independent transgenic lines were confirmed using the CAPS method (Fig. 3C) and Southern blot (Fig. 3D). Similar phenotypes and leaf chlorophyll content were observed between wild-type and transgenic plants (Fig. 3, B and E). These results confirm that the phenotype in the mutants is caused by mutation of OsPAPST1.
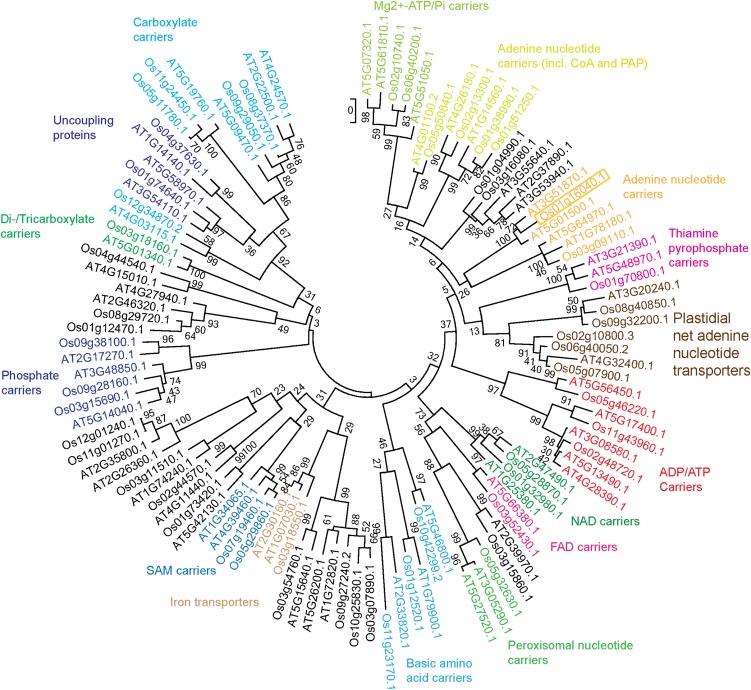
The predicted sequence of OsPAPST1 consists of 381 amino acids that exhibit features of all mitochondrial carrier-type proteins (Supplemental Fig. S3; Palmieri et al., 2011). Several phylogenetic analyses of mitochondrial carrier proteins from plants have been carried out in recent years (Palmieri et al., 2011). Using Arabidopsis as a model, it can be seen that the protein encoded by LOC_Os01g16040 branches closest to two proteins from Arabidopsis, encoded at loci At3g51870 and At5g01500 (Fig. 4). At5g01500 is the TAAC protein and is found in thylakoids (Thuswaldner et al., 2007; Yin et al., 2010); however, the protein encoded by At3g51870 is likely located in the envelopes of plastids according to proteomics-based analysis (Ferro et al., 2003, 2010; Sun et al., 2009).
Figure 4.
Phylogenetic analysis of the rice mitochondrial carrier protein family compared with Arabidopsis. Members of the rice mitochondrial carrier family were identified using orthology to the Arabidopsis members outlined by Haferkamp and Schmitz-Esser (2012). The phylogenetic tree was calculated with MEGA5 using the maximum likelihood tree method and the Jones-Thornton-Taylor model of amino acid substitution. Numbers represent the bootstrapping values of maximum likelihood and maximum parsimony after 1,000 replications. Functional groups were annotated according to Haferkamp and Schmitz-Esser (2012). The gene LOC_Os01g16040 is boxed. [See online article for color version of this figure.]
RNA Interference of OsPAPST1 Mimics the papst1 Mutant Phenotype
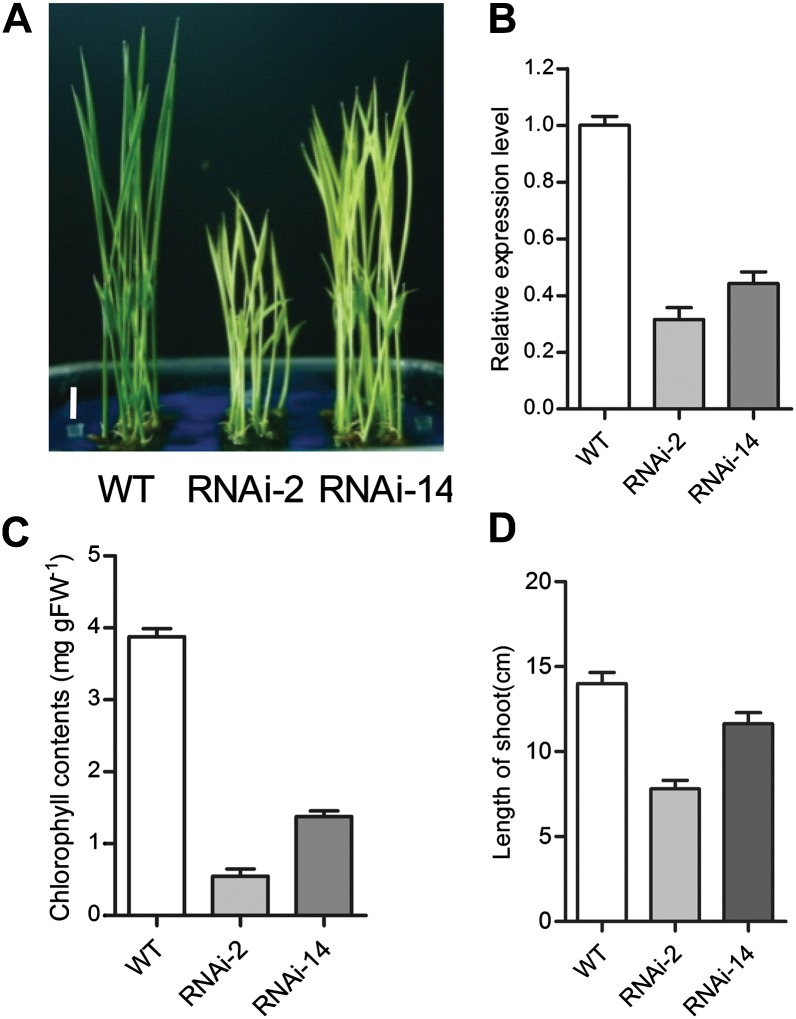
To further confirm that OsPAPST1 was the gene associated with the phenotype observed, transgenic plants were generated in which the endogenous OsPAPST1 gene was suppressed by RNA interference (RNAi). Five RNAi lines with different repression of OsPAPST1 showed the same virescent phenotypes as in the papst1 mutant. Transgenic T2 plants of two lines (RNAi-2 and RNAi-14) with greatly reduced OsPAPST1 transcripts (Fig. 5, A and B) were selected for measurement of chlorophyll content. The chlorophyll content of the newly emerged leaves of the lines was significantly lower (0.55 ± 0.13 and 1.38 ± 0.18 mg g−1, respectively) than that of the wild type (3.89 ± 0.19 mg g−1; Fig. 5C). The growth of RNAi lines was also inhibited (Fig. 5D). These results confirmed that RNAi of OsPAPST1 could mimic the phenotypes of the papst1 mutant.
Figure 5.
Effects of RNAi suppression of OsPAPST1. A, Shoots of 7-d-old seedlings of RNAi lines. WT, Wild type. Bar = 1 cm. B, qRT-PCR analysis of the expression levels of OsPAPST1 in the third leaf of RNAi lines. C, Chlorophyll contents of the third leaf of 7-d-old wild-type and two RNAi plants. Values represent means ± sd of 10 biological replicates. FW, Fresh weight. D, Length of shoots of 7-d-old wild-type and two RNAi plants. Values represent means ± sd of five biological replicates.
Functional Expression of OsPAPST1 in Escherichia coli
The phylogenetic classification suggests that OsPAPST1 is involved in the transport of adenine nucleotides (Fig. 4). To determine if adenine nucleotides are the substrates transported by OsPAPST1, kinetic analysis of adenine nucleotide uptake was performed on heterologously expressed OsPAPST1 and Ospapst1 in E. coli. Recombinant His-6-Xpress-OsPAPST1-FLAG protein and recombinant His-6-Xpress-Ospapst1-FLAG protein were expressed in E. coli cells, as evidenced by western-blot analysis using anti-His antibody (Fig. 6A). Uptake studies with radioactively labeled [α-32P]ATP or [α-32P]ADP into intact bacterial cells harboring OsPAPST1 revealed the time-linear import of both nucleotides for 5 min. In contrast, noninduced E. coli cells with OsPAPST1 imported adenylates at a much lower level (Fig. 6, B and C). For determination of the Km and Vmax values of OsPAPST1 and Ospapst1 to catalyze transport, E. coli cells were incubated with 0 to 900 µm radioactive adenine nucleotides for 1 min. The apparent Km values for ATP and ADP (Table I) of OsPAPST1 were 363.2 ± 63.54 and 753.2 ± 99.73 µm, similar to the Km values of Ospapst1 (258.8 ± 64.5 µm for ATP and 733.7 ± 99.79 µm for ADP). The calculated Vmax values for ATP and ADP uptake of OsPAPST1 were 1.338 and 1.945 nmol mg−1 protein h−1 (Table I), which are higher than those of Ospapst1 (0.67 ± 0.06 nmol mg−1 protein h−1 for ATP and 1.30 ± 0.20 nmol mg−1 protein h−1 for ADP). Both Vmax values were in the same range as those determined for the Arabidopsis mitochondrial ATP/ADP carriers expressed in E. coli (0.18–4.41 nmol mg−1 protein h−1; Haferkamp et al., 2002).
Figure 6.
Analysis of heterologously expressed OsPAPST1 and Ospapst1 in E. coli cells. A, Expression of OsPAPST1, Ospapst1, or vector in E. coli cells induced by IPTG. Western-blot analysis with anti-6-His antibodies was carried out to verify expression. B and C, Kinetics of adenine nucleotide uptake. IPTG-induced E. coli cells harboring the plasmid encoding OsPAPST1 and Ospapst1 were incubated with 50 µm [α-32P]ATP (B) or [α-32P]ADP (C) for up to 15 min. Noninduced E. coli cells transformed with the plasmid encoding OsPAPST1 were used as controls (black circles, OsPAPST1; white circles, Ospapst1; black triangles, noninduced cells harboring OsPAPST1). Values represent means ± sd of three biological replicates. D, Effects of PAPS on [α-32P]ATP transport by OsPAPST1. E, Phylogenetic tree of LOC_Os01g16040 with the yeast ADP/ATP transporter (ScAAC1:NP_013772), the human ADP/ATP transporter (HsANT1:NP_001142), the human PAP transporter (HsPAP:NP_848621), and the two closest Arabidopsis proteins (AtTAAC/PAPST:At5g01500 and AtPAPST1:At5g64970). The tree was drawn in the same manner as in Figure 4.
Table I. Apparent Km and Vmax of OsPAPST1 and Ospapst1 for ATP and ADP.
Each value represents the mean ± sd of three replicates. Data significantly different from the corresponding wild-type controls are indicated by asterisks (**P < 0.01; Student’s t test).
| Substrate | OsPAPST1 | Ospapst1 |
|---|---|---|
| Km (µm) | ||
| ATP | 363.2 ± 63.54 | 258.8 ± 64.45 |
| ADP | 753.2 ± 99.73 | 733.7 ± 99.79 |
| Vmax (nmol mg−1 protein h−1) | ||
| ATP | 1.34 ± 0.09 | 0.67 ± 0.05** |
| ADP | 1.95 ± 0.24 | 1.30 ± 0.20** |
Competition experiments with 27 different potential substrates were performed. Inhibition of [α-32P]ATP uptake catalyzed by OsPAPST1 was observed in the presence of nonlabeled ATP and ADP. These molecules reduced the rate to below 45% of the control values (without effectors). Unlabeled GDP and GMP had relatively lower inhibitory effects on the [α-32P]ATP uptake than nonlabeled ATP and ADP (Table II). The uncoupler compound carbonyl cyanide m-chlorophenyl hydrazone did not inhibit ATP uptake by OsPAPST1 (Table II). While the above data suggest that OsPAPST1 can transport adenine nucleotides, the phylogenetic classification shows that it does not cluster with the classical plastidial transporters (Fig. 4).
Table II. Effects of various metabolites on [α-32P]ATP transport by OsPAPST1.
Uptake in E. coli cells expressing OsPAPST1 was carried out for 1 min and stopped by rapid filtration (see “Materials and Methods”). ATP uptake was measured at a substrate concentration of 50 µm. Metabolic effectors were present in a 10-fold higher concentration than the substrate.
| Effector | Transport Rate |
|---|---|
| % | |
| Control | 100 |
| CTP | 96.8 ± 2.7 |
| UTP | 87.1 ± 4.8 |
| ITP | 90.3 ± 5.2 |
| GTP | 88.8 ± 6.2 |
| ADP | 42.2 ± 2.9 |
| GDP | 48.2 ± 4.5 |
| IDP | 82 ± 2.7 |
| UDP | 83.9 ± 3.8 |
| AMP | 60.3 ± 3.4 |
| CMP | 85 ± 1.2 |
| GMP | 50.3 ± 6.7 |
| IMP | 94.2 ± 2.3 |
| UMP | 89.8 ± 1.6 |
| 3′-AMP | 71.3 ± 6.7 |
| dTTP | 75.5 ± 3.4 |
| dGTP | 71.9 ± 2.9 |
| dCTP | 73.7 ± 9.6 |
| dATP | 65.8 ± 0.4 |
| ADP-Glc | 76.6 ± 4.6 |
| UDP-Glc | 74.3 ± 3.6 |
| NAD | 74.2 ± 6.2 |
| NADH | 74.3 ± 3.4 |
| NADP | 84.2 ± 1.8 |
| NADPH | 103 ± 2.1 |
| FAD | 80.7 ± 5.3 |
| Carbonyl cyanide m-chlorophenyl hydrazone | 78.6 ± 2 |
Given the observed specific phenotype of a delay in chloroplast development and the important roles of PAPS in chloroplast retrograde signaling (Chen et al., 2011; Estavillo et al., 2011), the ability of OsPAPST1 to transport adenine molecules associated with chloroplast development was tested. It was observed that PAPS could effectively compete for the transport of ATP, reducing it to approximately 60% of that observed without any inhibitor (Fig. 6D). A comparison of the protein encoded by LOC_Os01g16040 against a human and yeast ADP/ATP carrier and a mitochondrial carrier protein defined as a PAPS transporter, a mitochondrial carrier family protein from humans (Fiermonte et al., 2009), showed that it branched with the PAPS carrier with high confidence (Fig. 6E), suggesting that it is more related to a PAPS carrier than to an ATP/ADP carrier.
OsPAPST1 Is Targeted to Mitochondria and Chloroplasts
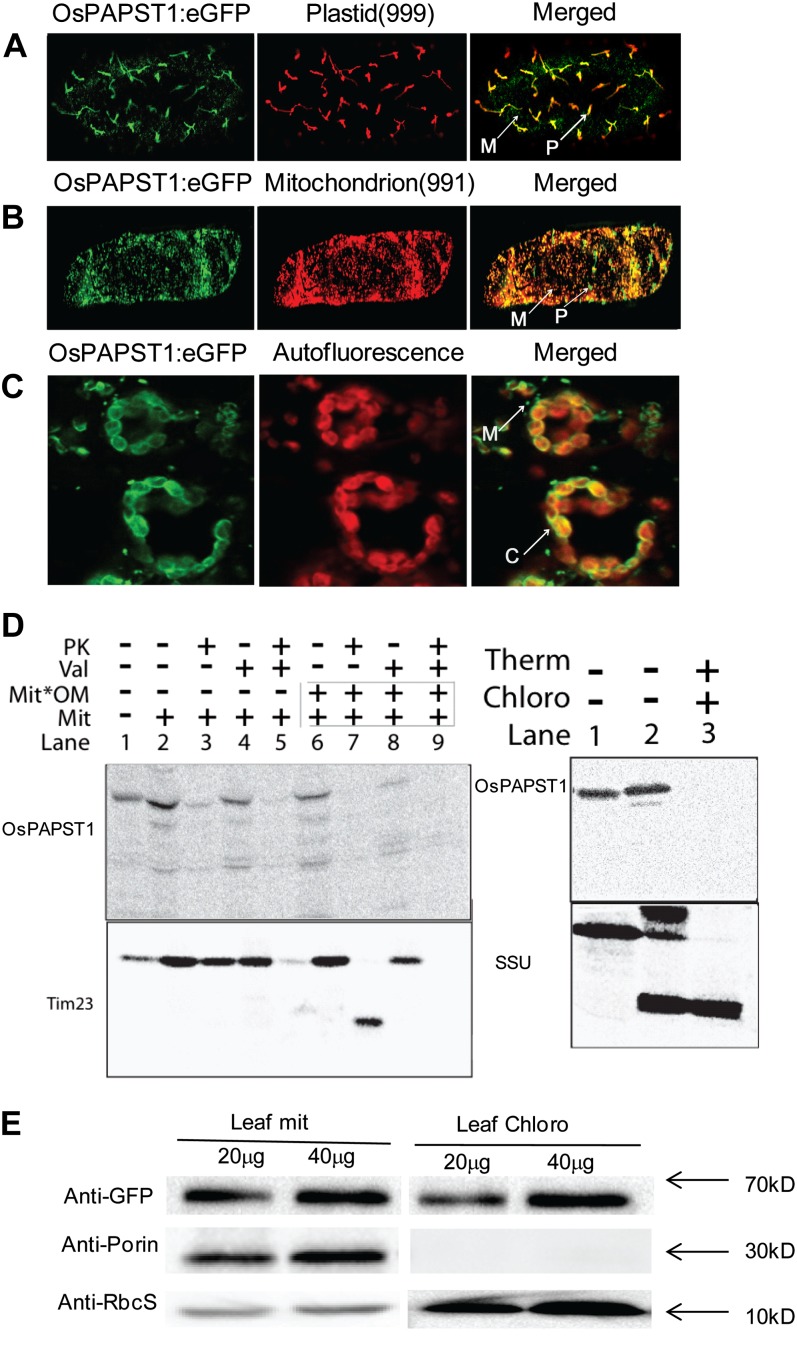
To determine the subcellular localization of the OsPAPST1 protein, OsPAPST1 coding sequence was fused in frame with enhanced GFP (eGFP) and transiently expressed in onion (Allium cepa) and tobacco (Nicotiana tabacum) epidermis. eGFP in onion epidermal cells showed subcellular colocalization of OsPAPST1 to plastids, as determined by overlapping with red fluorescent protein under the targeting signal of the small subunit of ribulose bisphosphate (Carrie et al., 2009), and colocalized to mitochondria, as determined by overlapping with red fluorescent protein directed by the targeting signal of the mitochondrial alternative oxidase protein (Carrie et al., 2009; Fig. 7, A and B). This indicates that OsPAPST1 is a dual-targeted protein. As a closely related protein in Arabidopsis (Fig. 4) has been reported to be a thylakoid protein (Thuswaldner et al., 2007), the analysis of OsPAPST1 targeting in tobacco epidermal cells suggests an envelope localization (Fig. 7C), as only the periphery of the chloroplasts displays GFP fluorescence. The crescent moon-shape-like fluorescent patterns resemble those described from transient expression of outer envelope plastid proteins (Breuers et al., 2012). To investigate the location of OsPAPST1 in both mitochondria and chloroplasts, we carried out in vitro import assays, reasoning that if imported proteins were located on the outer membranes of these organelles, they would be digested by externally added protease. In vitro uptake assays into isolated mitochondria and plastids from rice revealed that while OsPAPST1 bound to these organelles, no protease-protected products were detected (Fig. 7D), compared with the control of Translocase of Inner Membrane23 (Tim23) for mitochondria and the small subunit of ribulose 1,5-bisphosphate for plastids (Fig. 7D). Note that mitochondrial rupture of the outer membrane after import resulted in the loss of most of the bound protein (Fig. 7D, lane 8), but this had no effect on Tim23 (Fig. 7D, lane 8). Notably, no protease-protected products were observed with import into chloroplasts, despite a strong signal with chloroplast without externally added protease (Fig. 7D). This strongly suggests that OsPAPST1 is located on the mitochondrial outer membrane and outer envelope membrane of plastids.
Figure 7.
Subcellular localization of OsPAPST1. A, Localization of the OsPAPST1:eGFP fusion protein in plastid indicated by a plastid marker (999; Nelson et al., 2007) in onion epidermal cells. B, Localization of the OsPAPST1:eGFP fusion protein in mitochondria indicated by a mitochondrial marker (991; Nelson et al., 2007) in onion epidermal cells. C, Crescent-shaped pattern of fluorescence of the OsPAPST1:eGFP fusion protein in chloroplast tobacco epidermal cells. P, Plastid; M, mitochondria; C, chloroplasts. D, In vitro uptake of radiolabeled OsPAPST1 into mitochondria and chloroplasts. Radiolabeled OsPAPST1 was incubated with isolated mitochondria and chloroplasts, and protease was added after 15 min where indicated. Whereas radiolabeled OsPAPST1 bound to mitochondria quite efficiently (mitochondrial panel; lane 2), it was not protected from protease digestion (mitochondrial panel; lane 3), in contrast to the inner membrane protein Tim23. A similar pattern was observed with chloroplasts, where no protease-protected protein was evident (chloroplast panel; lane 3). PK, Protease; Val, valinomycin; Mit, mitochondria; Mit*OM, outer membranes of mitochondria; Therm, thermolysin; Chloro, chloroplasts; SSU, small subunit of ribulose 1,5-bisphosphate carboxylase/oxygenase. E, Immunoblot analysis of OsPAPST1:eGFP-complemented papst1 transgenic plants. The OsPAPST1 protein was detected using an anti-GFP protein antibody. The purity of the chloroplasts and mitochondria was tested using antibodies raised against organelle-specific proteins, the small subunit of 1,5-ribulose bisphosphate for chloroplasts and porin for mitochondria.
To verify the dual-targeting nature of the OsPAPST1 proteins, we performed an immunoblot analysis. As OsPAPST1 is a mitochondrial carrier protein that displays relatively high levels of sequence similarity to other carrier proteins in rice, we generated transgenic plants expressing eGFP fused to the C terminus of the full-length OsPAPST1 protein. The translational fusion protein (OsPAPST1:eGFP) was able to rescue the papst1 mutant phenotype (Supplemental Fig. S4), indicating that the fusion protein was functional. Immunoblot analysis of chloroplasts and mitochondria isolated from OsPAPST1:eGFP-complemented papst1 transgenic plants showed that a protein band with an apparent molecular mass of 70 kD was detected in both chloroplasts and mitochondria (Fig. 7E), indicating that the OsPAPST1 protein was localized in both organelles. Note that the apparent mass of 70 kD is due to the combined size of OsPAPST1 (approximately 40 kD) and GFP (approximately 29 kD). Thus, it was concluded on the basis of these results that OsPAPST1 is a dual-targeted protein located on the outer membrane of mitochondria and plastids.
Plastid Function Is Compromised in the papst1 Mutant
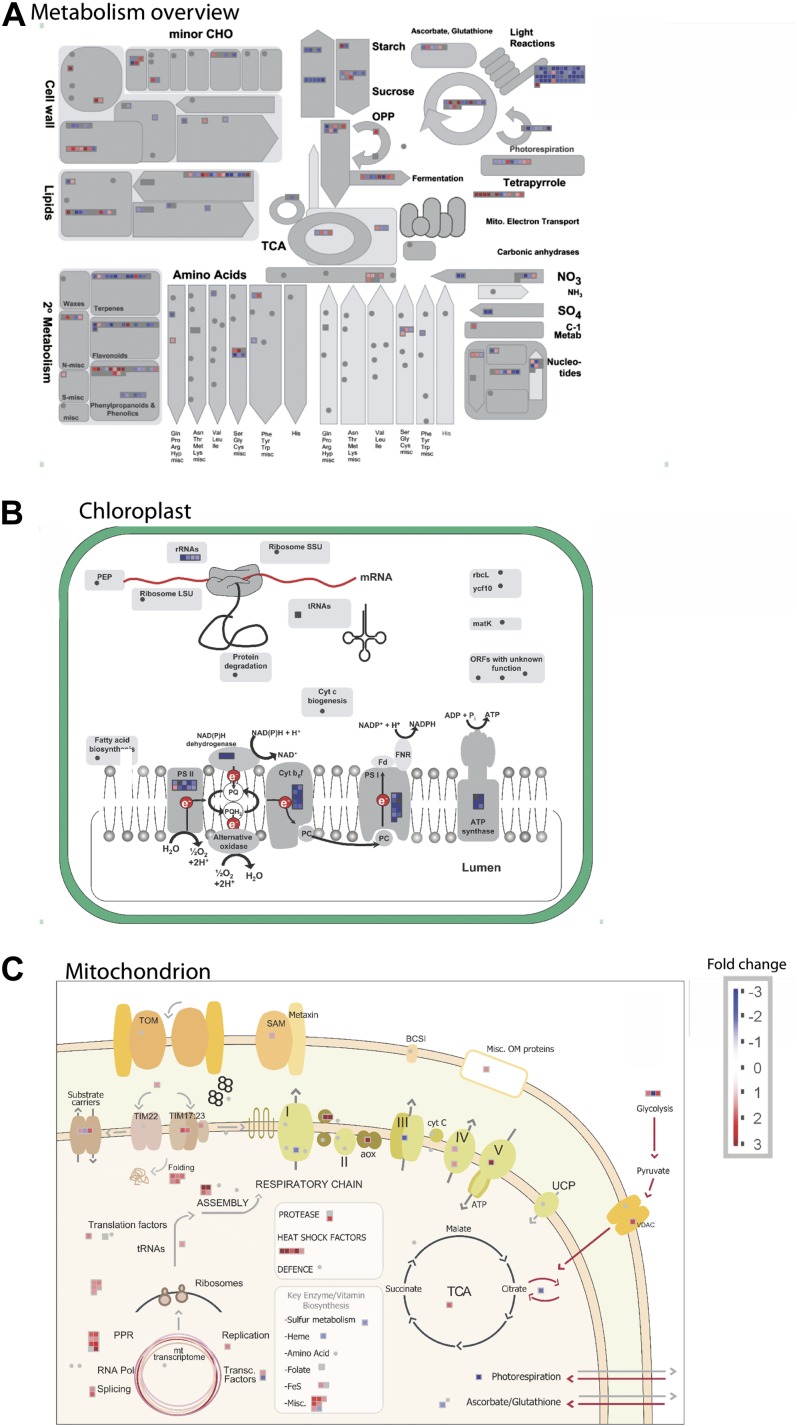
As PAPST1 is a dual-targeted protein, the defect in early chloroplast development could be due to a role in chloroplasts, mitochondria, or both. Thus, whole-genome transcriptomic analysis was carried out to determine processes that were disrupted in the mutant at the molecular level. An analysis of processes that were affected in the mutant revealed that both photosynthesis and major carbohydrate metabolism were overrepresented in down-regulated processes in shoots (Supplemental Fig. S5; Supplemental Table S1), consistent with the observed phenotype. A more detailed analysis of metabolism (Fig. 8A) revealed that genes encoding proteins involved in photosynthesis (light reactions) and carbohydrate synthesis were significantly down-regulated, and an analysis of this with MapMan of chloroplast functions indicated that this involved the four multisubunit protein complexes associated with photosynthetic electron transport, PSI, PSII, cytochrome b6f, and ATP synthase (Fig. 8B). In contrast to the few mitochondrial processes that were affected, they were largely up-regulated, although subunit 8 of the cytochrome bc1 complex was down-regulated (Fig. 8C). Also notable was that many other processes in chloroplasts were not affected in such a dramatic manner (Fig. 8B).
Figure 8.
Overview of differences in the transcriptome between wild-type and papst1 plants. A, MapMan overview of changes in metabolism between wild-type and papst1 plants. The most notable changes are a decrease in transcript abundance for the light reaction of photosynthesis, carbohydrate synthesis. Additionally, changes in SO4 and secondary metabolism were noticeable. B and C, MapMan overviews of changes in transcript abundance for genes encoding chloroplastic (B) and mitochondrial (C) proteins.
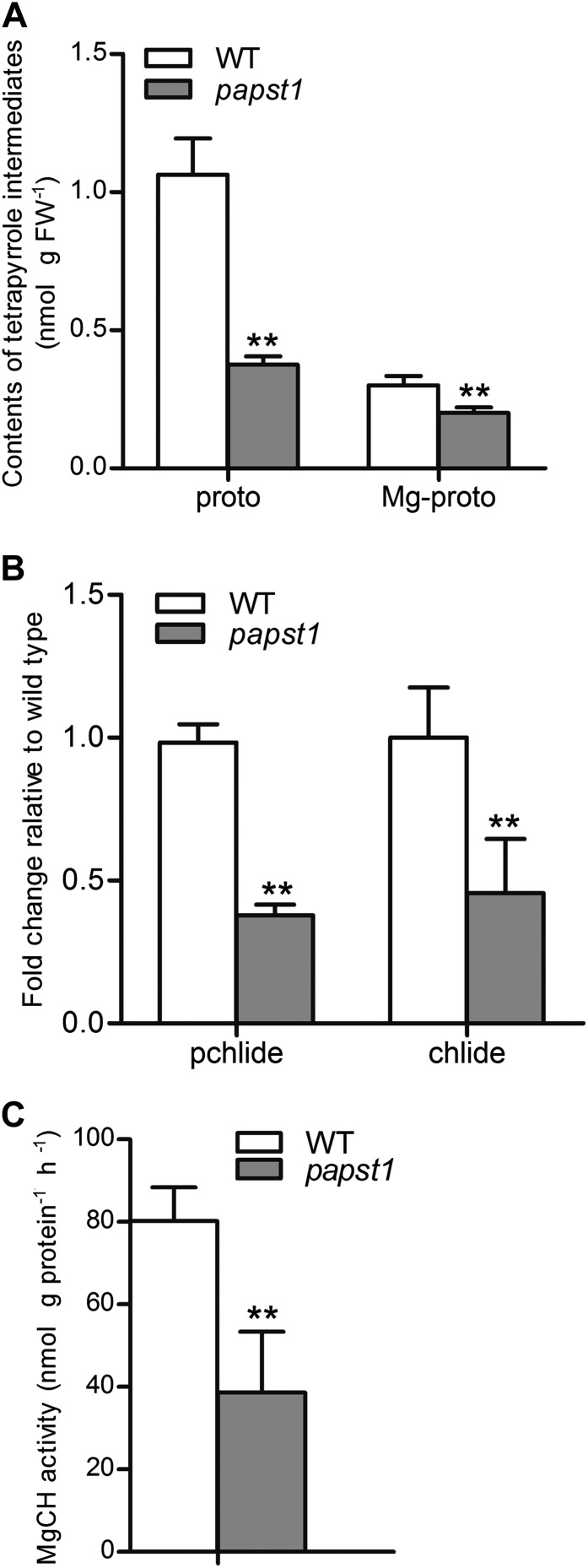
We also investigated the transcript abundance of genes encoding proteins involved in magnesium protoporphyrin IX (Mg-proto IX) synthesis, the concentrations of protoporphyrin and magnesium protoporphyrin, and the relative change of magnesium chelatase activity in newly emerged leaves of papst1 mutant seedlings compared with wild-type seedlings. The results showed that the concentrations of protoporphyrin IX and Mg-proto IX in the newly emerged leaves of papst1 mutants were significantly reduced compared with the wild type (Fig. 9A). The relative levels of protochlorophyllide (Pchlide) and chlorophyllide (Chlide) were also significantly decreased in the newly emerged leaves of papst1 (Fig. 9B). The measurement of magnesium chelatase activity also showed that in papst1 mutants, the activity of magnesium chelatase is significantly repressed (Fig. 9C). The expression pattern of all three of these genes encoding three magnesium chelatase subunits, CHLH, CHLD, and CHLI, was analyzed in the newly emerged leaves of papst1 mutants and the wild type. The expression of CHLD and CHLI was similar in wild-type and papst1 leaves (Supplemental Fig. S6), while the CHLH gene exhibited a significant increase in papst1 mutants (Supplemental Fig. S6).
Figure 9.
Contents of tetrapyrrole intermediates and magnesium chelatase activities in the third leaf 1 d after emergence. A, Protoporphyrin IX and Mg-proto IX levels. B, Pchlide and Chlide levels. C, Magnesium chelatase activity. All data represent means of three independent experiments ± sd. papst1 values that are significantly different from the corresponding wild-type (WT) controls are indicated by asterisks (**P < 0.01; Student’s t test). FW, Fresh weight.
Temporal and Spatial Expression Patterns of the OsPAPST1 Gene
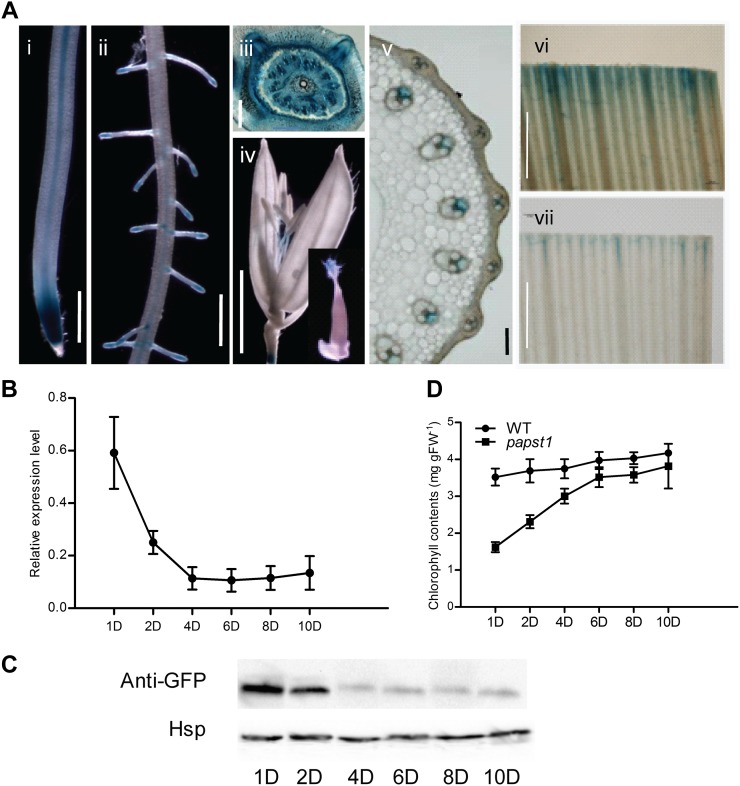
To investigate the expression patterns of OsPAPST1, seven independent transgenic lines with the GUS reporter gene driven by a promoter of OsPAPST1 were analyzed. Six of these lines showed very similar expression patterns, varying only slightly in the intensity of GUS staining. OsPAPST1 expression is mainly visible in root tip and the central cylinder (Fig. 10A, i and ii). The expression of OsPAPST1 could also be detected in the stem base (Fig. 10A, iii), the stigma and filament (Fig. 10A, iv), and the vascular bundle of the stem (Fig. 10A, v). OsPAPST1 was also observed to be expressed in leaves (Fig. 10A, vi and vi). As the altered phenotype of papst1 was apparent only during early leaf development, this suggested that either the function was only required at this stage or some other protein can compensate for the function at later developmental stages. Quantitative reverse transcription (qRT)-PCR analysis for the expression of OsPAPST1 during leaf development indicates that the OsPAPST1 gene is highly expressed in the third leaf 1 d after emergence, while 4 d after emergence the transcript abundance of OsPAPST1 is substantially decreased (Fig. 10B). Western-blot analysis using a GFP antibody with OsPAPST1:eGFP-complemented papst1 transgenic plants confirmed that the protein abundance followed transcript abundance, in that while it was high in the third leaf at days 1 and 2 after emergence, it decreased substantially afterward (Fig. 10C). The chlorophyll contents of the leaves tested follow an inverse relationship in the papst1 mutant (Fig. 10D), in that they are substantially lower than in the wild type at days 1 and 2 after emergence, but by day 10 it has almost reached wild-type levels.
Figure 10.
Expression pattern of OsPAPST1. A, The 3.44-kb region of the promoter of the OsPAPST1 gene was linked to GUS, and GUS staining was examined in six individual transformants. i, Root showing expression at the tip and central cylinder. Bar = 20 µm. ii, Root elongation zone where the lateral roots emerge. Bar = 20 µm. iii, Stem base. Bar = 200 µm. iv, Flower. Bar = 20 µm. v, Stem. Bar = 100 µm. vi, Leaf. Bar = 1 mm. vii, Leaf. Bar = 1 mm. B, qRT-PCR analysis of OsPAPST1 in the third leaf from days 1 to 10 after emergence. C, Western-blot analysis of OsPAPST1:eGFP-complemented papst1 in the third leaf from days 1 to 10 after emergence. Rice heat shock protein (Hsp; LOC_Os09g30418) was used as the reference protein (Li et al., 2011). D, Chlorophyll contents in the third leaf from days 1 to 10 after emergence in the wild type (WT) and the papst1 mutant. Values represent means ± sd of 10 seedlings. FW, Fresh weight.
DISCUSSION
A protein belonging to the mitochondrial carrier family was identified as being required for early leaf development in rice. While the immediate reaction may be to speculate that this protein is an ATP carrier, several lines of evidence are not consistent with this assumption. First, as this protein is located on the chloroplast envelope, there are specific plastid ATP carrier proteins that should fulfill this function, specifically LOC_Os02g11740 and LOC_Os01g45910 that encode the NTTs in rice and appear to be constitutively expressed (Supplemental Fig. S7). Furthermore, the expression pattern of PAPST1 suggests that it is expressed in a variety of tissues; thus, if it had a primary role as an ATP transporter, altered phenotypes would be expected, especially in lateral roots and flowers, where altered ATP transport would be expected to have phenotypic effects. A number of lines of evidence suggest that PAPST1 may encode a PAPS transporter: phylogenetic analysis reveals that PAPST1 branches most closely with proteins encoded by At3g51870 and At5g01500 (Fig. 4). While the initial report indicated that At5g01500 encoded an ATP/ADP transported in the thylakoid membrane (Thuswaldner et al., 2007), it was shown recently to have a primary role in transporting plastidic PAPS to the cytosol (Gigolashvili et al., 2012). Our results showing that PAPST1 is located on the chloroplast envelope is consistent with this role. Additionally, with a limited phylogenetic analysis, it branches with human and Arabidopsis adenosine-3′,5′-bisphosphate (PAP) transporters rather than ATP/ADP transporters (Fig. 6E).
The decreased transcript abundance of genes associated with the light reaction of photosynthesis suggests that normal chloroplast-to-nuclear signaling has been disrupted in papst1. A close analysis of the transcriptomic data reveals that in addition to those involved in the light reaction of photosynthesis, genes associated with sulfur metabolism, amino acid metabolism involving sulfur, are also changed (Fig. 8A; Supplemental Table S2). Most notably, in shoots, only a gene encoding Adenosine 5′-Phosphosulfate Reductase (APR) has transcript abundance reduced by more than 4- to 5-fold (LOC_Os7g32570.1). In the primary pathway to assimilate sulfate in plants, sulfate is activated by ATP sulfurylase to form adenosine 5′-phosphosulfate, which is reduced by APR to sulfite that is reduced to sulfide reductase, the latter used in the biosynthesis of Cys (Takahashi et al., 2011; Chen et al., 2012). Notably, two genes encoding this step of Cys synthesis (LOC_Os01g59920 and LOC_Os12g42980) are also changed in transcript abundance, one induced and one decreased (Supplemental Table S2). Additionally, transcripts of genes encoding a sulfide oxidase, glutathione S-transferase, and glutathione peroxidase are also changed in abundance (Supplemental Table S2). Tyr aminotransferase was reduced in abundance that contributed 4-hydroxyphenylpyruvate for tocopherol synthesis, and is an important antioxidant linked with sulfur metabolism (Chan et al., 2013). There were also changes in transcript abundance for several genes encoding proteins involved in secondary metabolism, including terpenes, flavonoids, and phenylpropanoids, and S-adenosyl-Met-dependent proteins, all consistent with alteration in sulfur assimilation (Supplemental Table S1).
It should be noted that while papst1 displayed an early leaf developmental phenotype, this is likely due to the fact that additional transporters may also exist that act as PAP/PAPS transporters on the envelope membrane; thus, the altered phenotype is relatively mild. The phenotype is most severe when OsPAPST1 displays the highest level of expression at the transcript and protein levels, but the phenotype reverts to normal when expression decreases. This suggests that additional PAP/PAPS transporters exist, as has been suggested in Arabidopsis (Palmieri et al., 2011; Gigolashvili et al., 2012), that likely display developmental and possibly tissue-specific expression patterns. Thus, the altered phenotype is confined to young developing leaves in rice with papst1, which suggests that additional carriers are present in other tissues. Thus, while PAPS has previously been classified as an operational signal, the delay in chloroplast development suggests that it may also operate as a biogenic signal (Pogson et al., 2008).
The papst1 mutant can be used to identify seed purity and authenticity in hybrid rice. In two-line system hybrid rice, the purity of hybrid seeds is a prerequisite for attaining potential yield. For the effective application of two-line system hybrid rice, an applicable strategy is to breed new thermo/photoperiod-sensitive genic male sterile (T/PGMS) lines with a phenotypic marker, which can facilitate the elimination of contaminated T/PGMS seeds (Shu et al., 1996). Through consecutive crosses or backcrosses with the mutants possessing leaf color markers, several new T/PGMS rice lines with pale or purple leaves have been released (Dong et al., 1995; Cao et al., 1999). As the phenotypes of papst1 mutants are leaf virescent and recessive, it can be used to identify seed purity and authenticity in hybrid rice.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
The papst1 mutant was isolated from an ethylmethane sulfonate-generated rice (Oryza sativa japonica ‘Nipponbare’) mutant library grown in nutrient solution. Hydroponic experiments were performed using normal rice culture solution (Yoshida et al., 1976). Rice plants were grown in growth chambers at 30°C/22°C (12-h day/12-h night) after germination, with approximately 60% humidity.
Chlorophyll and Chlorophyll Intermediate Measurements and TEM Analysis
Leaves at different development stages were used for chlorophyll analysis and TEM analysis. Total chlorophyll (a + b) content was calculated as described previously (Lichtenthaler, 1987). The chlorophyll intermediates, including protoporphyrin IX, Mg-proto IX, Pchlide, and Chlide, were assayed as described by Reinhold et al. (2007) and Wu et al. (2007). TEM analysis was carried out as described previously (Zhao et al., 2012).
Map-Based Cloning
For map-based cloning of the OsPAPST1 gene, 385 F2 mutants were selected from an F2 population derived from a cross between the papst1 mutant and indica var Kasalath. SSR markers on chromosome 1 were used for fine-mapping. The OsPAPST1 gene was selected out of eight putative genes on an approximately 49-kb region as the candidate gene. Genomic DNA and the coding sequence of OsPAPST1 were amplified by PCR and reverse transcription-PCR from the papst1 mutant and wild-type plants for sequence analysis. A CAPS marker was also developed to confirm the mutated site of Ospapst1. These PCR products were digested by NotI. Primers used in the map-based cloning are listed in Supplemental Table S3.
Southern-Blot Analysis of Transgenic Plants
Genomic DNA was isolated as described previously (Murray and Thompson, 1980) and was digested with the restriction enzyme HindIII. Southern-blot analysis was carried out as described previously (Zhao et al., 2012).
Phylogenetic Analysis of the Rice Mitochondrial Carrier Protein Family
Sequences for putative mitochondrial carrier proteins were downloaded from The Arabidopsis Information Resource Web site (http://www.arabidopsis.org/) and from the rice genome annotation project Web site (http://rice.plantbiology.msu.edu/index.shtml). A multiple sequence alignment was first produced using the Web-based program Multiple Alignment using Fast Fourier Transform (http://mafft.cbrc.jp/alignment/server/; Katoh and Toh, 2007). The phylogenetic tree was calculated with MEGA5 using the maximum likelihood tree method and the Jones-Thornton-Taylor model of amino acid substitution (Tamura et al., 2011). The bootstrapping values indicate the maximum likelihood and maximum parsimony after 1,000 replications. Carrier protein families and functional groups were annotated based on Haferkamp and Schmitz-Esser (2012).
Gene Expression and Microarray Analysis
For all samples, RNA was isolated using the Qiagen RNeasy Plant RNA isolation kit with on-column DNase treatment. Total RNA of leaves at different development stages of the wild type and the papst1 mutant was extracted. The first-strand complementary DNA was synthesized from 5 mg of DNaseI-treated total RNA using SuperScript II reverse transcriptase (Invitrogen). The primers for qRT-PCR are listed in Supplemental Table S3.
The Affymetrix IVT express and hybridization, wash, and stain kits were also used following the manufacturer’s instructions, as carried out previously using the Affymetrix Rice Genome Arrays (Zheng et al., 2009). The 12 samples analyzed by microarrays included root and shoot samples of wild-type plants (cv Nipponbare) and papst1 mutant plants that were 5 d old (three independent biological replicates per sample).
The microarray preprocessing and normalization were carried out as done previously by Zheng et al. (2009), where CEL files were normalized by MAS5 and GC-robust multiarray analysis normalization, which showed that 28,557 probe sets were present in one or more of the samples. Differential expression analysis was carried out using Cyber-T (Baldi et al., 2001), where genes were defined as differentially expressed at P < 0.05 and posterior probability of differential expression > 0.96. The Pageman tool (overrepresentation analysis) was used to find overrepresented functional categories in differentially expressed gene sets (Usadel et al., 2006).
Functional Analysis of PAPST1
To construct Escherichia coli plasmids expressing OsPAPST1 and Ospapst1 with an N-terminal His tag, the coding region of OsPAPST1 and Ospapst1 was amplified by PCR on first-strand complementary DNA from wild-type and papst1 leaf tissues and introduced into the isopropylthio-β-galactoside (IPTG)-inducible expression vector pTrCHisB (Invitrogen). The primers used for PCR amplification are listed in Supplemental Table S3. The E. coli strain Top10 F′ (Invitrogen) was exploited for heterologous synthesis. Uptake experiments with E. coli cells after the synthesis of OsPAPST1 and Ospapst1 were performed as described earlier (Thuswaldner et al., 2007). In vitro import studies into rice mitochondria isolated from 7-d-old seedlings were carried out as described previously (Howell et al., 2007) using the Tim23 protein as a mitochondrial control (Wang et al., 2012) and the small subunit of 1,5-ribulose bisphosphate as a plastidial control (Carrie et al., 2009). In vivo localization studies were carried out as described previously (Carrie et al., 2009) using GFP linked to the small subunit of 1,5-ribulose bisphosphate as a plastidial marker and GFP linked to the mitochondrial alternative oxidase protein as a mitochondrial marker. Western-blot analysis was carried out as outlined previously (Wang et al., 2012) using antibodies to mitochondrial porin (Wang et al., 2012), the small subunit of 1,5-ribulose bisphosphate (Yue et al., 2010), and GFP (Sigma).
Measurement of Magnesium Chelatase Activity
Magnesium chelatase activity was examined as described previously (Zhang et al., 2006).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Phenotypic characterization of roots of papst1 mutant plants.
Supplemental Figure S2. TEM images of mitochondrial structure in wild-type and papst1 mutant plants.
Supplemental Figure S3. Multiple sequence alignment of OsPAPST1 with various carrier proteins.
Supplemental Figure S4. Phenotype of OsPAPST1:eGFP-complemented papst1 transgenic plants.
Supplemental Figure S5. Pageman classification of overrepresentation or underrepresentation of the functional categories of genes whose transcript abundance increased or decreased in papst1 compared with wild-type plants.
Supplemental Figure S6. Relative transcript levels of CHLI (Os03g36540), CHLD (Os03g59640), and CHLH (Os03g20700).
Supplemental Figure S7. Relative transcript abundance of plastid NTT genes and genes encoding proteins of the mitochondrial carrier family in rice from various tissues.
Supplemental Table S1. Genes that are significantly changed in papst1 plants in shoots and roots.
Supplemental Table S2. Genes associated with sulfur assimilation that are significantly changed in papst1 mutant plants in shoots and roots.
Supplemental Table S3. Primers used in this research.
Acknowledgments
We thank Sophia Ng for her help with manuscript formatting.
Glossary
- NTT
nucleotide transporter
- TAAC
thylakoid ATP/ADP carrier
- PAPS
3′-phosphoadenosine 5′-phosphosulfate
- SSR
simple sequence repeat
- CAPS
cleaved-amplified polymorphic sequence
- RNAi
RNA interference
- eGFP
enhanced GFP
- Mg-proto IX
magnesium protoporphyrin IX
- qRT
quantitative reverse transcription
- T/PGMS
thermo/photoperiod-sensitive genic male sterile
- TEM
transmission electron microscopy
- IPTG
isopropylthio-β-galactoside
- CAPS
cleaved amplified polymorphic sequence
- Pchlide
protochlorophyllide
- Chlide
chlorophyllide
- PAP
adenosine-3',5'-bisphosphate
References
- Arai Y, Hayashi M, Nishimura M. (2008) Proteomic identification and characterization of a novel peroxisomal adenine nucleotide transporter supplying ATP for fatty acid β-oxidation in soybean and Arabidopsis. Plant Cell 20: 3227–3240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahaji A, Li J, Ovecka M, Ezquer I, Muñoz FJ, Baroja-Fernández E, Romero JM, Almagro G, Montero M, Hidalgo M, et al. (2011a) Arabidopsis thaliana mutants lacking ADP-glucose pyrophosphorylase accumulate starch and wild-type ADP-glucose content: further evidence for the occurrence of important sources, other than ADP-glucose pyrophosphorylase, of ADP-glucose linked to leaf starch biosynthesis. Plant Cell Physiol 52: 1162–1176 [DOI] [PubMed] [Google Scholar]
- Bahaji A, Muñoz FJ, Ovecka M, Baroja-Fernández E, Montero M, Li J, Hidalgo M, Almagro G, Sesma MT, Ezquer I, et al. (2011b) Specific delivery of AtBT1 to mitochondria complements the aberrant growth and sterility phenotype of homozygous Atbt1 Arabidopsis mutants. Plant J 68: 1115–1121 [DOI] [PubMed] [Google Scholar]
- Baldi P, Long AD. (2001) A Bayesian framework for the analysis of microarray expression data: regularized t-test and statistical inferences of gene changes. Bioinformatics 17: 509–519 [DOI] [PubMed] [Google Scholar]
- Breuers FK, Bräutigam A, Geimer S, Welzel UY, Stefano G, Renna L, Brandizzi F, Weber AP. (2012) Dynamic remodeling of the plastid envelope membranes: a tool for chloroplast envelope in vivo localizations. Front Plant Sci 3: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L, Qian Q, Zhu X, Zheng D, Min S, Xiong Z. (1999) Breeding of a photoperiod-sensitive genic male sterile indica rice Zhongsi S with a purple leaf marker and the heterosis of its hybrid rice produced with it. Acta Agron Sin 25: 44–49 [Google Scholar]
- Carrie C, Kühn K, Murcha MW, Duncan O, Small ID, O’Toole N, Whelan J. (2009) Approaches to defining dual-targeted proteins in Arabidopsis. Plant J 57: 1128–1139 [DOI] [PubMed] [Google Scholar]
- Chan KX, Wirtz M, Phua SY, Estavillo GM, Pogson BJ. (2013) Balancing metabolites in drought: the sulfur assimilation conundrum. Trends Plant Sci 18: 18–29 [DOI] [PubMed] [Google Scholar]
- Chen H, Zhang B, Hicks LM, Xiong L. (2011) A nucleotide metabolite controls stress-responsive gene expression and plant development. PLoS ONE 6: e26661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Liu R, Sun C, Zhang P, Feng C, Shen Z. (2012) Spatial and temporal variations in nitrogen and phosphorous nutrients in the Yangtze River estuary. Mar Pollut Bull 64: 2083–2089 [DOI] [PubMed] [Google Scholar]
- Chiou TJ, Lin SI. (2011) Signaling network in sensing phosphate availability in plants. Annu Rev Plant Biol 62: 185–206 [DOI] [PubMed] [Google Scholar]
- Dong F, Zhu X, Xiong Z. (1995) Release of temperature-sensitive genic male sterile M2s with greenish leaf color marker. Chin Rice Sci 9: 65–70 [Google Scholar]
- Estavillo GM, Crisp PA, Pornsiriwong W, Wirtz M, Collinge D, Carrie C, Giraud E, Whelan J, David P, Javot H, et al. (2011) Evidence for a SAL1-PAP chloroplast retrograde pathway that functions in drought and high light signaling in Arabidopsis. Plant Cell 23: 3992–4012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferro M, Brugière S, Salvi D, Seigneurin-Berny D, Court M, Moyet L, Ramus C, Miras S, Mellal M, Le Gall S, et al. (2010) AT_CHLORO, a comprehensive chloroplast proteome database with subplastidial localization and curated information on envelope proteins. Mol Cell Proteomics 9: 1063–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferro M, Salvi D, Brugière S, Miras S, Kowalski S, Louwagie M, Garin J, Joyard J, Rolland N. (2003) Proteomics of the chloroplast envelope membranes from Arabidopsis thaliana. Mol Cell Proteomics 2: 325–345 [DOI] [PubMed] [Google Scholar]
- Fiermonte G, Paradies E, Todisco S, Marobbio CM, Palmieri F. (2009) A novel member of solute carrier family 25 (SLC25A42) is a transporter of coenzyme A and adenosine 3′,5′-diphosphate in human mitochondria. J Biol Chem 284: 18152–18159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gigolashvili T, Geier M, Ashykhmina N, Frerigmann H, Wulfert S, Krueger S, Mugford SG, Kopriva S, Haferkamp I, Flügge UI. (2012) The Arabidopsis thylakoid ADP/ATP carrier TAAC has an additional role in supplying plastidic phosphoadenosine 5′-phosphosulfate to the cytosol. Plant Cell 24: 4187–4204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haferkamp I, Fernie AR, Neuhaus HE. (2011) Adenine nucleotide transport in plants: much more than a mitochondrial issue. Trends Plant Sci 16: 507–515 [DOI] [PubMed] [Google Scholar]
- Haferkamp I, Hackstein JH, Voncken FG, Schmit G, Tjaden J. (2002) Functional integration of mitochondrial and hydrogenosomal ADP/ATP carriers in the Escherichia coli membrane reveals different biochemical characteristics for plants, mammals and anaerobic chytrids. Eur J Biochem 269: 3172–3181 [DOI] [PubMed] [Google Scholar]
- Haferkamp I, Schmitz-Esser S. (2012) The plant mitochondrial carrier family: functional and evolutionary aspects. Front Plant Sci 3: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell KA, Cheng K, Murcha MW, Jenkin LE, Millar AH, Whelan J. (2007) Oxygen initiation of respiration and mitochondrial biogenesis in rice. J Biol Chem 282: 15619–15631 [DOI] [PubMed] [Google Scholar]
- Katoh K, Toh H. (2007) PartTree: an algorithm to build an approximate tree from a large number of unaligned sequences. Bioinformatics 23: 372–374 [DOI] [PubMed] [Google Scholar]
- Kirchberger S, Tjaden J, Neuhaus HE. (2008) Characterization of the Arabidopsis Brittle1 transport protein and impact of reduced activity on plant metabolism. Plant J 56: 51–63 [DOI] [PubMed] [Google Scholar]
- Leroch M, Neuhaus HE, Kirchberger S, Zimmermann S, Melzer M, Gerhold J, Tjaden J. (2008) Identification of a novel adenine nucleotide transporter in the endoplasmic reticulum of Arabidopsis. Plant Cell 20: 438–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Bai H, Wang X, Li L, Cao Y, Wei J, Liu Y, Liu L, Gong X, Wu L, et al. (2011) Identification and validation of rice reference proteins for western blotting. J Exp Bot 62: 4763–4772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler FW. (1987) Karl Freudenberg, Burckhardt Helferich, Hermann O. L. Fischer: a centennial tribute. Carbohydr Res 164: 1–22 [DOI] [PubMed] [Google Scholar]
- Linka N, Theodoulou FL, Haslam RP, Linka M, Napier JA, Neuhaus HE, Weber AP. (2008) Peroxisomal ATP import is essential for seedling development in Arabidopsis thaliana. Plant Cell 20: 3241–3257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray MG, Thompson WF. (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8: 4321–4325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenführ A. (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Palmieri F, Pierri CL, De Grassi A, Nunes-Nesi A, Fernie AR. (2011) Evolution, structure and function of mitochondrial carriers: a review with new insights. Plant J 66: 161–181 [DOI] [PubMed] [Google Scholar]
- Pogson BJ, Woo NS, Förster B, Small ID. (2008) Plastid signalling to the nucleus and beyond. Trends Plant Sci 13: 602–609 [DOI] [PubMed] [Google Scholar]
- Reinhold T, Alawady A, Grimm B, Beran KC, Jahns P, Conrath U, Bauer J, Reiser J, Melzer M, Jeblick W, et al. (2007) Limitation of nocturnal import of ATP into Arabidopsis chloroplasts leads to photooxidative damage. Plant J 50: 293–304 [DOI] [PubMed] [Google Scholar]
- Rieder B, Neuhaus HE. (2011) Identification of an Arabidopsis plasma membrane-located ATP transporter important for anther development. Plant Cell 23: 1932–1944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu Q, Wu D, Xia Y. (1996) Marker-assisted elimination of contaminations in two-line hybrid rice seed production and multiplication. Acta Agriculturae Universitatis Zhejiangenesis 22: 56–60 [Google Scholar]
- Sun Q, Zybailov B, Majeran W, Friso G, Olinares PD, van Wijk KJ. (2009) PPDB, the Plant Proteomics Database at Cornell. Nucleic Acids Res 37: D969–D974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Kopriva S, Giordano M, Saito K, Hell R. (2011) Sulfur assimilation in photosynthetic organisms: molecular functions and regulations of transporters and assimilatory enzymes. Annu Rev Plant Biol 62: 157–184 [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuswaldner S, Lagerstedt JO, Rojas-Stütz M, Bouhidel K, Der C, Leborgne-Castel N, Mishra A, Marty F, Schoefs B, Adamska I, et al. (2007) Identification, expression, and functional analyses of a thylakoid ATP/ADP carrier from Arabidopsis. J Biol Chem 282: 8848–8859 [DOI] [PubMed] [Google Scholar]
- Usadel B, Nagel A, Steinhauser D, Gibon Y, Bläsing OE, Redestig H, Sreenivasulu N, Krall L, Hannah MA, Poree F, et al. (2006) PageMan: an interactive ontology tool to generate, display, and annotate overview graphs for profiling experiments. BMC Bioinformatics 7: 535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Carrie C, Giraud E, Elhafez D, Narsai R, Duncan O, Whelan J, Murcha MW. (2012) Dual location of the mitochondrial preprotein transporters B14.7 and Tim23-2 in complex I and the TIM17:23 complex in Arabidopsis links mitochondrial activity and biogenesis. Plant Cell 24: 2675–2695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber AP, Linka N. (2011) Connecting the plastid: transporters of the plastid envelope and their role in linking plastidial with cytosolic metabolism. Annu Rev Plant Biol 62: 53–77 [DOI] [PubMed] [Google Scholar]
- Wu Z, Zhang X, He B, Diao L, Sheng S, Wang J, Guo X, Su N, Wang L, Jiang L, et al. (2007) A chlorophyll-deficient rice mutant with impaired chlorophyllide esterification in chlorophyll biosynthesis. Plant Physiol 145: 29–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin L, Lundin B, Bertrand M, Nurmi M, Solymosi K, Kangasjärvi S, Aro EM, Schoefs B, Spetea C. (2010) Role of thylakoid ATP/ADP carrier in photoinhibition and photoprotection of photosystem II in Arabidopsis. Plant Physiol 153: 666–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S, Forno DA, Cock JH, Gomez KA (1976) Routine Procedures for Growing Rice Plants in Culture Solution, Ed 3. International Rice Research Institute, Los Banos, Philippines [Google Scholar]
- Yue R, Wang X, Chen J, Ma X, Zhang H, Mao C, Wu P. (2010) A rice stromal processing peptidase regulates chloroplast and root development. Plant Cell Physiol 51: 475–485 [DOI] [PubMed] [Google Scholar]
- Zhang H, Li J, Yoo JH, Yoo SC, Cho SH, Koh HJ, Seo HS, Paek NC. (2006) Rice Chlorina-1 and Chlorina-9 encode ChlD and ChlI subunits of Mg-chelatase, a key enzyme for chlorophyll synthesis and chloroplast development. Plant Mol Biol 62: 325–337 [DOI] [PubMed] [Google Scholar]
- Zhao C, Xu J, Chen Y, Mao C, Zhang S, Bai Y, Jiang D, Wu P. (2012) Molecular cloning and characterization of OsCHR4, a rice chromatin-remodeling factor required for early chloroplast development in adaxial mesophyll. Planta 236: 1165–1176 [DOI] [PubMed] [Google Scholar]
- Zheng L, Huang F, Narsai R, Wu J, Giraud E, He F, Cheng L, Wang F, Wu P, Whelan J, et al. (2009) Physiological and transcriptome analysis of iron and phosphorus interaction in rice seedlings. Plant Physiol 151: 262–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zrenner R, Stitt M, Sonnewald U, Boldt R. (2006) Pyrimidine and purine biosynthesis and degradation in plants. Annu Rev Plant Biol 57: 805–836 [DOI] [PubMed] [Google Scholar]