A pathogen effector protein undergoes specific phosphorylation and associates with host 14-3-3 proteins, which affects its subcellular distribution and stability.
Abstract
HopQ1 (for Hrp outer protein Q), a type III effector secreted by Pseudomonas syringae pv phaseolicola, is widely conserved among diverse genera of plant bacteria. It promotes the development of halo blight in common bean (Phaseolus vulgaris). However, when this same effector is injected into Nicotiana benthamiana cells, it is recognized by the immune system and prevents infection. Although the ability to synthesize HopQ1 determines host specificity, the role it plays inside plant cells remains unexplored. Following transient expression in planta, HopQ1 was shown to copurify with host 14-3-3 proteins. The physical interaction between HopQ1 and 14-3-3a was confirmed in planta using the fluorescence resonance energy transfer-fluorescence lifetime imaging microscopy technique. Moreover, mass spectrometric analyses detected specific phosphorylation of the canonical 14-3-3 binding site (RSXpSXP, where pS denotes phosphoserine) located in the amino-terminal region of HopQ1. Amino acid substitution within this motif abrogated the association and led to altered subcellular localization of HopQ1. In addition, the mutated HopQ1 protein showed reduced stability in planta. These data suggest that the association between host 14-3-3 proteins and HopQ1 is important for modulating the properties of this bacterial effector.
A multicomponent defense response is initiated when plant pattern recognition receptors perceive microbially derived structural components (Nürnberger and Brunner, 2002), which are referred to as pathogen-associated molecular patterns. Many bacterial pathogens have developed type III secretion system (TTSS) effectors that can suppress or modulate pathogen-associated molecular pattern-triggered immunity (Jones and Dangl, 2006). Effector-triggered immunity represents a second layer of defense, whereby plants have evolved mechanisms that rely upon Resistance (R) proteins to sense and respond to cognate TTSS effectors. Thus, the expression of a specific bacterial effector can either sustain disease in susceptible plants or render the pathogen avirulent in resistant plants that express the corresponding R protein. Several lines of evidence suggest an involvement of scaffold proteins from the 14-3-3 family in mediating these defense responses at various levels (Yang et al., 2009; Oh et al., 2010). Some R proteins have been shown to bind 14-3-3 proteins directly. RPW2.8, which confers resistance to fungal pathogens of Golovinomyces spp., associates specifically with the 14-3-3 isoform λ (designated GF14λ) from Arabidopsis (Arabidopsis thaliana; Yang et al., 2009). Moreover, both types of resistance were compromised in Arabidopsis lacking the λ isoform. Consistently, ectopic overexpression of GF14λ in transgenic Arabidopsis results in enhanced resistance to powdery mildew (Golovinomyces cichoracearum; Yang et al., 2009). Tobacco (Nicotiana tabacum) N protein, which mediates resistance to Tobacco mosaic virus, also binds 14-3-3 protein (Ueda et al., 2006). The viral p50 replicase helicase domain is the cognate ligand for N protein. Since this domain also interacts with 14-3-3s, it is possible that 14-3-3s might function in the formation of the receptor-ligand recognition complex (Ueda et al., 2006). In addition, the tomato (Solanum lycopersicum) 14-3-3 protein TF7 has been shown to exhibit positive regulation on the mitogen-activated protein kinase cascade, which is activated rapidly by pathogen recognition (Oh et al., 2010; Oh and Martin, 2011).
There is increasing evidence that many intracellular pathways are regulated by the modulation of scaffold protein properties rather than the activities of integral components in the signaling cascades (Good et al., 2011). This strategy enables signal transduction to be turned on or off rapidly via the assembly or disassembly of complexes. This same mechanism also allows the intensity and kinetics of a response to be fine-tuned to the stimulus (Good et al., 2011). It was recently suggested that the manipulation of scaffolding may be one strategy employed by pathogens to interfere with the host defense response. The best-characterized example of scaffolding manipulation is the phytotoxin fusicoccin, which is secreted by the fungus Fusicoccum amygdali. Fusicoccin targets a 14-3-3 protein that regulates guard cell H+-ATPases, and its activity results in stomatal opening, facilitating pathogen entry (Oecking et al., 1994). Some bacterial virulence factors simply require scaffold proteins to reach their destination within host cells or to become enzymatically active, while others target the host scaffold proteins to suppress defenses. Yersinia species secrete the TTSS effector YopK (for Yersinia outer protein K), which binds to the Receptor for Activated C Kinase1 in mammals (Thorslund et al., 2011). It is hypothesized that this interaction blocks phagocytosis, allowing efficient extracellular proliferation of the bacteria. Yersinia spp. has also acquired the virulence factor YopM, which mimics eukaryotic scaffolds and forces bridging of host kinases (McDonald et al., 2003). Similarly, enterohemorrhagic Escherichia coli strains use EspG to form an artificial complex that effectively reprograms host signaling (Selyunin et al., 2011).
HopQ1 (for Hrp outer protein Q [also known as HopQ1-1]; AAZ37975.1) is a type III effector that has been acquired recently by Pseudomonas syringae strains (Rohmer et al., 2004), whereas its xenologs from Ralstonia solanacearum and Xanthomonas spp. appear to be ancient. HopQ1 contributes to host specificity, but its exact role in pathogenesis remains undefined. This study shows that HopQ1 must undergo a specific phosphorylation event in planta as a prerequisite for its binding to host 14-3-3 and that its properties depend upon the formation of the effector-host protein complex.
RESULTS
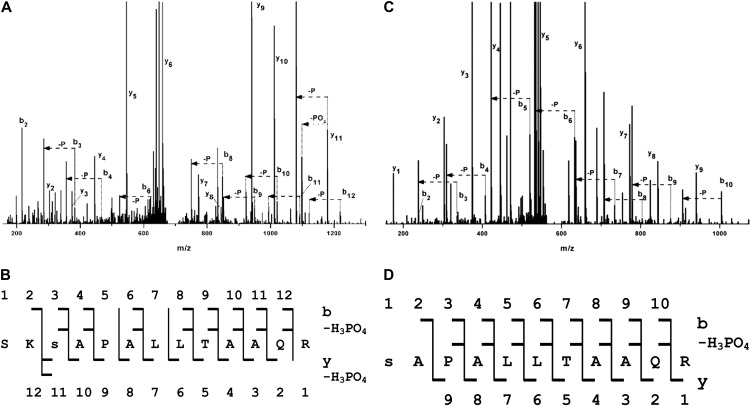
A number of bacterial effectors are activated once inside host cells, upon interaction with host proteins. Analysis in silico of the P. syringae pv phaseolicola 1448A type III effector inventory revealed that most of the effectors possess putative 14-3-3 binding motifs (Supplemental Table S1). This suggests that association with plant 14-3-3 proteins can be important for their function. HopQ1, which contains in the N-terminal domain (Fig. 1) a canonical mode 1 14-3-3 binding site (48–53 amino acids; high-stringency Scansite algorithm; http://scansite.mit.edu; Obenauer et al., 2003), was selected to test the significance of this motif and expressed transiently in planta. Leaf samples were collected 2 d after agroinfiltration of Nicotiana benthamiana, and HopQ1 was affinity purified via its Strep tag II. Liquid chromatography-tandem mass spectrometry (LC-MS/MS) analyses revealed that HopQ1 copurified with nine isoforms of 14-3-3 proteins. Two isoforms of this family, a and c, were identified in all four independent experiments (Table I). The mode 1 14-3-3 binding motif (RSXpSXP, where pS denotes phospho-Ser) contains a phospho-Ser; therefore, it is possible that HopQ1 is phosphorylated in host cells. Mass spectrometric analyses performed on HopQ1 expressed in planta showed that a central Ser residue in the predicted 14-3-3 binding site (Ser-51) had been phosphorylated in the peptide SKpSAPALLTAAQR (mass-to-charge ratio [m/z] = 687.362+; MASCOT score 85; Fig. 2, A and B). We detected up to four different putative phosphorylation sites in several independent experiments (data not shown), but only Ser-51 was rezpeatedly phosphorylated in all of them.
Figure 1.
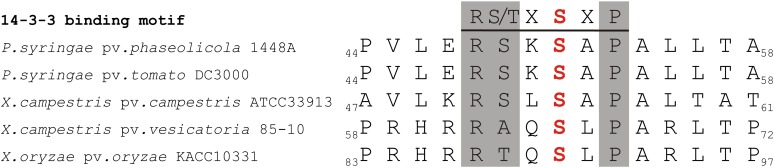
The 14-3-3 binding site is conserved in HopQ1, the TTSS effector from P. syringae, and XopQ, its xenolog from Xanthomonas spp. The canonical mode-1 14-3-3-interacting motif is shaded in gray, and the putative phospho-Ser is highlighted in red. [See online article for color version of this figure.]
Table I. LC-MS/MS data showing 14-3-3 family isoforms that copurify with HopQ1-Strep II transiently expressed in N. benthamiana leaves.
| 14-3-3 Isoform | Sequence Coverage in Separate Experimentsa |
|||
|---|---|---|---|---|
| a | 28 | 14 | 60 | 43 |
| b | 40 | 29 | 0b | 0 |
| c | 28 | 16 | 50 | 50 |
| d | 30 | 0 | 0 | 0 |
| e | 28 | 54 | 54 | 0 |
| f | 27 | 27 | 0 | 0 |
| g | 30 | 23 | 0 | 0 |
| h | 47 | 0 | 0 | 0 |
| i | 30 | 31 | 0 | 0 |
Protein coverage parameters calculated by Mascot software are shown for each experiment. Protein coverage parameters for HopQ1 in these experiments were 61, 36, 23, and 65, respectively. bProtein coverage 0 refers to the 14-3-3 isoform that has not been identified by LC-MS/MS analyses in this experiment.
Figure 2.
The predicted 14-3-3 binding motif of HopQ1 is phosphorylated by plant kinases. A and B, HopQ1-Strep tag II fusion protein expressed in N. benthamiana leaves was affinity purified and subjected to LC-MS/MS analyses. C and D, HopQ1-6xHis expressed in bacteria was incubated with total protein extracts from N. benthamiana and then affinity purified and analyzed by LC-MS/MS. A and C show the fragmentation spectra with peak assignment to b, y, b-H3PO4, and y-H3PO4, with “–P” denoting loss of an H3PO4 group. Major signals of the MS/MS spectra are identified by their corresponding fragment tags of the b, y series, but also of the y-H3PO4 and b-H3PO4 series, which were expected in the case of a phosphorylated peptide. B and D show the peptide sequences with daughter ions of the b, y, b-H3PO4, and y-H3PO4 series found in the spectra. In the peptide derived from HopQ1 phosphorylated in vitro (C and D), the presence of the full series of b2 to b10 fragments and the expected b-H3PO4 series allows for unequivocal localization of the phosphate at Ser-51. In the peptide derived from HopQ1 expressed in planta (A and B), the majority of b and b-H3PO4 pairs are also present. In addition, the presence of strong y, y-PO3, and y-H3PO4 signals indicates that the site of phosphorylation is Ser-51, not Ser-49.
As a complementary approach, in vitro kinase assays were performed. Recombinant HopQ1 was expressed in E. coli and then coincubated with crude plant extracts, which provided a source of kinase (Supplemental Fig. S1). While HopQ1 purified from E. coli was not modified (data not shown), mass spectrometry indicated that HopQ1 was phosphorylated when incubated with the crude extracts (Fig. 2, C and D), and an increase of 80 D was again detected at the Ser-51 residue within the peptide pSAPALLTAAQR (m/z = 589.792+; MASCOT score 107). To confirm these results, the relevant Ser residue was mutated to a phospho-null Ala (HopQ1-S51A) and the mutant construct was expressed in E. coli. In the in vitro assay, phosphorylation of purified HopQ1-S51A was almost completely abolished, which suggests that Ser-51 represents a major phosphorylation site (Fig. 3). In contrast, the modification of HopQ1 at position 49 (HopQ1-S49A) resulted in a partial reduction of the phosphorylation signal, not its elimination. The same modification pattern was observed with resistant (N. benthamiana) and susceptible (bean) plants (Supplemental Fig. S2). Interestingly, HopQ1 was phosphorylated by extracts from various plants, including nonhost species (Supplemental Fig. S1). The amino acid sequence surrounding Ser-51 suggests that HopQ1 could be a substrate for a kinase from the CDPK-SnRK superfamily (Hrabak et al., 2003). Consistent with this possibility, representatives of both CDPK and SnRK families phosphorylated HopQ1 in vitro (Supplemental Fig. S3). These findings support the suggestion that an S49A substitution might weaken the consensus site, and collectively, these data suggest that HopQ1 S51 might play a central role in interaction with 14-3-3. Consistent with this hypothesis, 14-3-3 proteins were not detected by mass spectrometry when HopQ1-S51A was transiently expressed in N. benthamiana, whereas HopQ1-S49A copurified with the same set of 14-3-3 isoforms as the wild-type HopQ1 (Table II).
Figure 3.
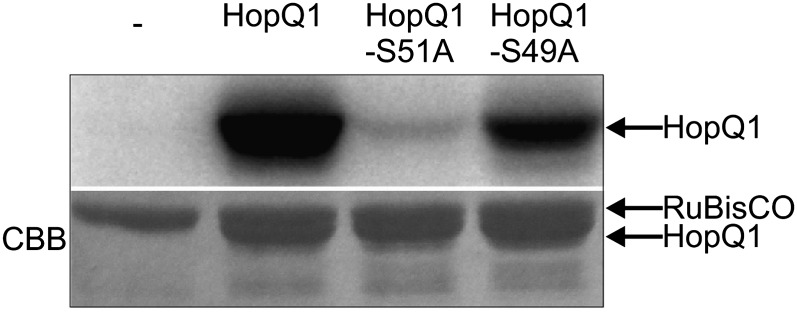
Ser-51 plays a critical role in the phosphorylation of HopQ1. Recombinant HopQ1 variants with a C-terminal 6xHis epitope were incubated with leaf protein extracts from N. benthamiana in buffer containing [γ-32P]ATP. Samples were resolved by SDS-PAGE and analyzed by autoradiography. Only residual phosphorylation was detected for HopQ1 mutated at position 51. As a loading control, the Coomassie Brilliant Blue (CBB)-stained gel is shown in the bottom panel. This experiment was performed three times with similar results.
Table II. LC-MS/MS data showing 14-3-3 family isoforms that copurify with variants of HopQ1-Strep II transiently expressed in N. benthamiana leaves.
| 14-3-3 Isoform | HopQ1-S51A (53, 43)a |
HopQ1-S49A (45, 63) |
||
|---|---|---|---|---|
| a | 0b | 0 | 43 | 61 |
| b | 0 | 0 | 29 | 34 |
| c | 0 | 0 | 50 | 47 |
| d | 0 | 0 | 13 | 0 |
| e | 0 | 0 | 54 | 38 |
| f | 0 | 0 | 27 | 34 |
| g | 0 | 0 | 23 | 14 |
| h | 0 | 0 | 46 | 46 |
| i | 0 | 0 | 31 | 26 |
Protein coverage parameters calculated by Mascot software are shown for each experiment. Protein coverage parameters for HopQ1-S51A and HopQ1-S49A in these experiments are shown in parentheses. bProtein coverage 0 refers to the 14-3-3 isoform that has not been identified by LC-MS/MS analyses in this experiment.
To corroborate these findings, fluorescence resonance energy transfer (FRET) efficiency was quantified for HopQ1 variants and 14-3-3a, an isoform identified in all copurification experiments. A fusion protein between 14-3-3a and enhanced cyan fluorescent protein (eCFP) was coexpressed with wild-type HopQ1-eYFP (for enhanced yellow fluorescent protein) or HopQ1-S51A-eYFP in the leaves of N. benthamiana plants. After 2 d, the average eCFP lifetime was assayed using fluorescence lifetime imaging microscopy (FLIM), and significant reductions were found when 14-3-3a-eCFP was coexpressed with HopQ1-eYFP (Table III; Supplemental Fig. S4). This finding confirms the presence of a physical interaction between 14-3-3a and HopQ1 in planta. In contrast, only a minor reduction in eCFP lifetime was observed in the presence of the HopQ1-S51A-eYFP fusion protein, which supports the idea that Ser-51 is involved in binding 14-3-3.
Table III. FRET-FLIM analysis showing that HopQ1 Ser-51 is critical for 14-3-3a binding.
Pull-down assays were used to determine whether HopQ1 phosphorylation is a prerequisite for 14-3-3 binding. Recombinant HopQ1-6xHis protein obtained from E. coli was coincubated with leaf protein extracts from N. benthamiana or bean (Table IV; Supplemental Table S2). Subsequently, HopQ1 was purified and eluates were subjected to mass spectrometric analysis. Under such conditions, no 14-3-3-derived peptides were found. However, when HopQ1-6xHis was first phosphorylated in vitro and then used in the pull-down assay, various 14-3-3 isoforms were detected. In contrast, no 14-3-3s were identified when in vitro phosphorylated HopQ1-S51A was used in the experiments.
Table IV. Various 14-3-3 isoforms from N. benthamiana interact with recombinant HopQ1-6xHis in a phosphorylation-dependent manner.
| 14-3-3 Isoform | Nonphosphorylated HopQ1-6xHisa | In Vitro Phosphorylated HopQ1-6xHisb | In Vitro Phosphorylated HopQ1-S51A-6xHisb |
|---|---|---|---|
| a | 0 | 47c | 0 |
| b | 0 | 0d | 0 |
| c | 0 | 44 | 0 |
| d | 0 | 0 | 0 |
| e | 0 | 28 | 0 |
| f | 0 | 0 | 0 |
| g | 0 | 16 | 0 |
| h | 0 | 36 | 0 |
| i | 0 | 23 | 0 |
Recombinant HopQ1-6xHis purified from E. coli was incubated with N. benthamiana crude protein extract. After consecutive purification steps, the sample was subjected to LC-MS/MS analyses. Protein coverage for HopQ1 was 87. bRecombinant HopQ1-6xHis and HopQ1-S51A-6xHis were phosphorylated in vitro prior to incubation with N. benthamiana crude protein extract. After consecutive purification steps, the samples were subjected to LC-MS/MS analyses. Protein coverage parameters for HopQ1 and HopQ1-S51A in these experiments were 83 and 86, respectively. cProtein coverage parameters of 14-3-3 isoforms that copurified with HopQ1-6xHis calculated by Mascot software following LC-MS/MS analyses. dProtein coverage 0 refers to the 14-3-3 isoform that has not been identified by LC-MS/MS analyses in this experiment. The experiment was performed twice.
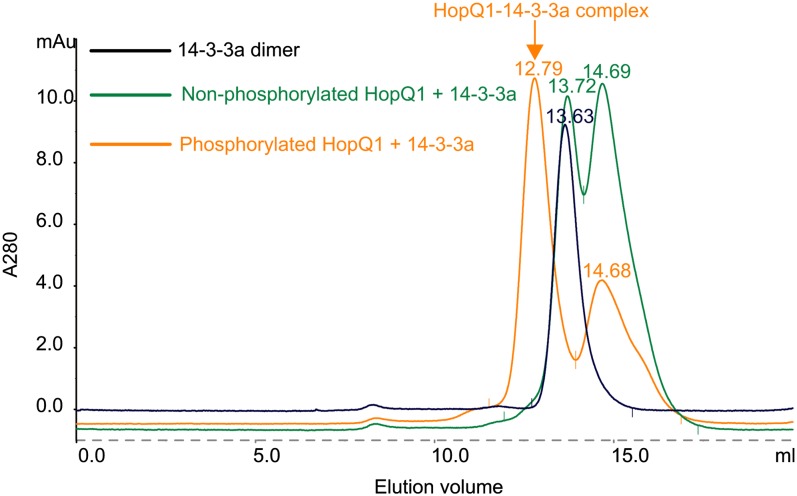
In an alternative experiment, we analyzed by gel filtration chromatography the formation of the HopQ1-14-3-3 complex in vitro (Fig. 4). Recombinant 14-3-3a protein was first incubated with HopQ1-6xHis, either nonphosphorylated or in vitro phosphorylated by CPK3. Next, the samples were separated using the Superdex 200 10/300GL column. A peak corresponding to the HopQ1-14-3-3 complex was detected only in the samples containing phosphorylated HopQ1-6xHIS (Fig. 4, orange trace). In contrast, the analyses revealed the presence of only 14-3-3 dimer and HopQ1 monomer in the samples containing nonphosphorylated HopQ1. The identity of the peaks was confirmed by SDS-PAGE analysis of the appropriate fractions (data not shown). Collectively, these results demonstrate that HopQ1 binds 14-3-3s in a phosphorylation-dependent manner and that this binding occurs both in plants where it triggers defense responses (N. benthamiana; Table IV) and in plants where it contributes to increased virulence of the bacteria (bean; Supplemental Table S2).
Figure 4.
HopQ1 binds to 14-3-3a in a phosphorylation-dependent manner. Representative gel filtration runs on a Superdex 200 column are shown. Recombinant HopQ1 with a C-terminal 6xHis epitope was incubated with recombinant CPK3 prior to binding to 14-3-3a (orange trace). As a control, nonphosphorylated HopQ1 was used (green trace). Under the conditions used (5 mm DTT), HopQ1 exists as a monomer (elution volumes of 14.68 and 14.69). 14-3-3a was eluted as dimers (peaks 13.63 and 13.72). Recombinant CPK3 was removed from the reaction by affinity capture on a GST column. The experiment was performed twice with similar results.
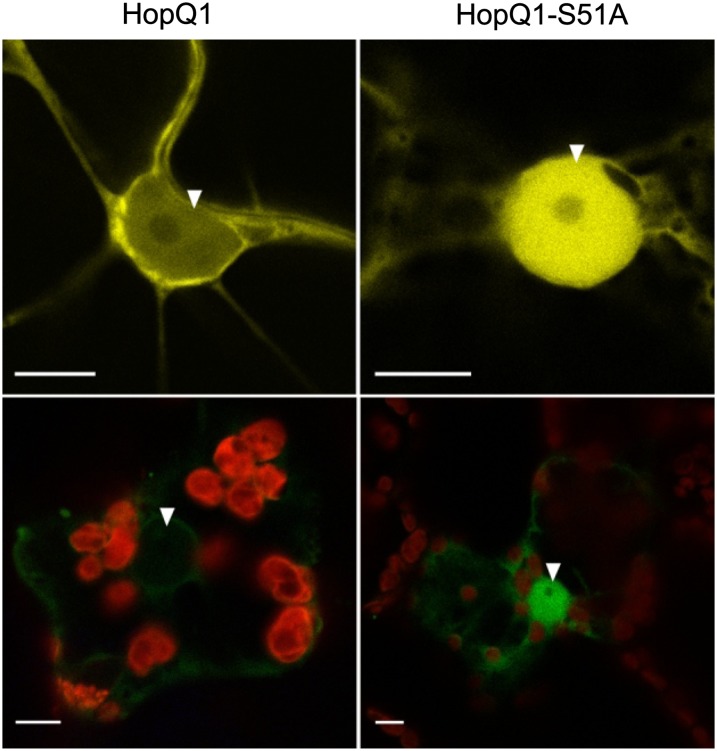
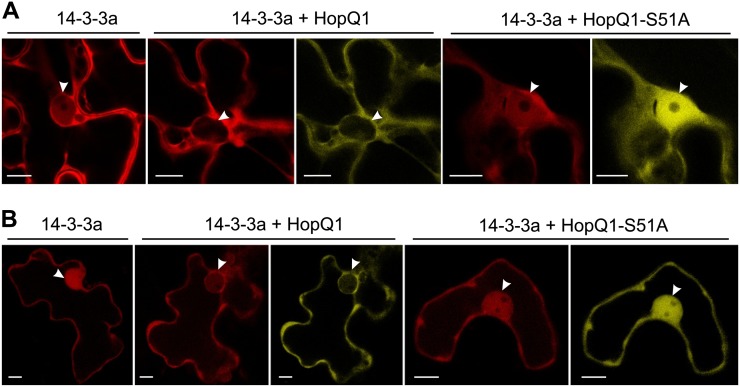
Since an association with 14-3-3 might modify the localization of a partner, the subcellular distribution patterns of HopQ1 and HopQ1-S51A were compared. As seen in Figure 5, in epidermal cells of N. benthamiana HopQ1-eYFP localized almost exclusively to the cytoplasm, whereas HopQ1-S51A-eYFP was directed to the nucleus. A similar pattern was observed in mesophyll cells of N. benthamiana. Thus, interaction with 14-3-3a might either facilitate nuclear export or hinder nuclear import of HopQ1. Interestingly, when 14-3-3a was expressed alone, it accumulated in both the cytosolic and nuclear compartments (Fig. 6), but in the presence of ligand, 14-3-3a was detected predominantly in the cytoplasm. In contrast, HopQ1-S51A expression did not affect 14-3-3a distribution. Susceptible and resistant plants showed similar localization patterns. These observations indicate that both interacting partners might be involved in the reciprocal determination of localization.
Figure 5.
Subcellular localization of HopQ1 variants. Confocal images show representative N. benthamiana leaf epidermal cells (top panels) or mesophyll cells (bottom panels) transiently expressing either wild-type HopQ1-eYFP (left panels) or HopQ1-S51A-eYFP (right panels). White arrowheads indicate the nuclei. The photographs were taken 72 h after agroinfiltration. For each variant, approximately 50 transformed cells were examined. Bars = 10 µm.
Figure 6.
Coexpression of 14-3-3a and HopQ1 affects the nuclear-cytoplasmic partitioning of the binding partners. A, N. benthamiana leaves were coinfiltrated with A. tumefaciens strains carrying constructs encoding 14-3-3a-mRFP and variants of HopQ1 fused to eYFP. For each variant, approximately 50 transformed cells were examined. B, The same constructs were transiently coexpressed in bean epidermal cells via particle bombardment. In both plant species, 14-3-3a-mRFP localized to the cytoplasm and nucleus. Coexpression with wild-type HopQ1-eYFP resulted in the relocation of 14-3-3a-mRFP from the nucleus to the cytoplasm. A noninteracting form of HopQ1 (HopQ1-S51A-eYFP) shows highly increased nuclear accumulation and does not alter the localization of 14-3-3a-mRFP. For each variant, approximately 20 transformed cells were examined. White arrowheads indicate the nuclei. Bars = 10 µm.
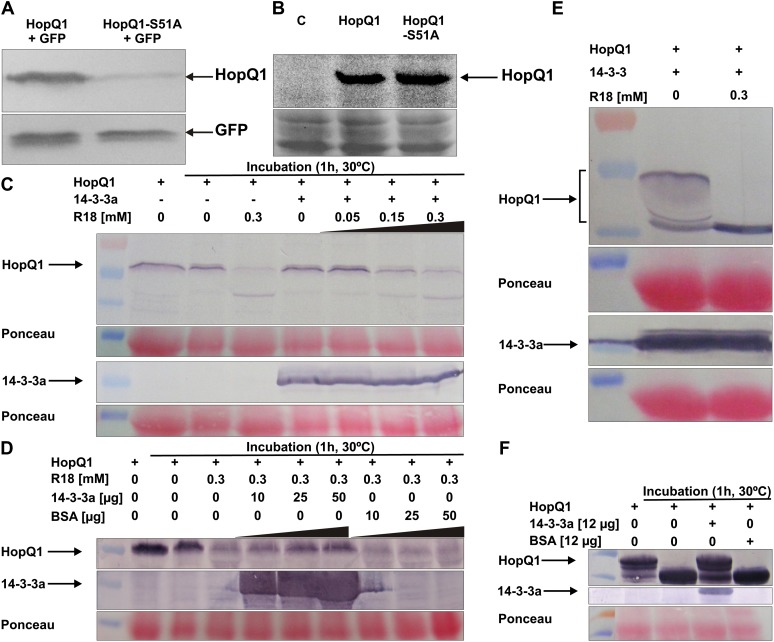
Another aspect of the association with 14-3-3 is revealed by the difference in the steady-state levels of C-terminally 3× hemagglutinin (HA) tagged HopQ1 variants when these were transiently expressed in N. benthamiana under the control of the cauliflower mosaic virus 35S promoter (Fig. 7A). In contrast, HopQ1 and HopQ1-S51A levels were similar when expressed in P. syringae pv tabaci in a broad-host-range plasmid under the control of the constitutive form of the tac promoter (Fig. 7B). This suggests that interaction with 14-3-3 may increase the stability of HopQ1 effector in host tissue. To test this hypothesis, we monitored levels of HopQ1 in protein extracts supplemented with R18, an inhibitor that disrupts 14-3-3-ligand associations (Wang et al., 1999). In the first experiment (Fig. 7C), crude protein extracts isolated from N. benthamiana leaves, which transiently expressed HopQ1-Flag protein, were incubated for 1 h at 30°C. Subsequently, HopQ1 was detected by immunoblot analysis using an antibody specific to Flag. While HopQ1 steady-state level remained unchanged under control conditions, coincubation with increasing amounts of the R18 proportionally reduced its level. The fact that application of R18 to the extract decreased the HopQ1 level indicates that R18 is able to displace HopQ1 from the complex with endogenous 14-3-3 proteins. We next tested whether application of exogenous 14-3-3a would outcompete the inhibitor. To this end, increasing amounts of the recombinant 14-3-3a from E. coli were added to the extract, from N. benthamiana leaves expressing HopQ1-Flag protein, which had been supplemented with the R18. As shown in Figure 7D, the effect of R18 on HopQ1 stability could be overcome by exogenous 14-3-3a but not bovine serum albumin (BSA). A similar set of experiments was performed with bean protein extracts (Fig. 7, E and F). After incubation of the in vitro reconstituted HopQ1-14-3-3a complex for 1 h at 30°C with the bean crude extract, a band corresponding to intact HopQ1 protein was visible on the immunoblot, while in samples containing R18 this band was not detected (Fig. 7E). Additionally, as shown in Figure 7F, application of the phosphorylated HopQ1 to the bean extract resulted in its degradation, while the presence of 14-3-3a suppressed this process. Collectively, these results suggest that binding to 14-3-3 might protect HopQ1 from degradation by plant proteases (Fig. 7, C–F). The fact that we have not found substantial differences in protein levels of HopQ1 and HopQ1-S51A C-terminally fused to eYFP suggests that the presence of longer tags might stabilize the effector in N. benthamiana cells (data not shown).
Figure 7.
Interaction with 14-3-3 proteins increases HopQ1 stability in plants. A, The presence of Ser-51 affects HopQ1 steady-state level in planta. Wild-type HopQ1 or HopQ1-S51A proteins carrying C-terminal 3xHA epitopes were transiently coexpressed with GFP in N. benthamiana leaves. Crude protein extracts (20 μg of proteins per lane) were isolated from leaves 48 h after agroinfiltration. HopQ1 variants were detected by immunoblot analysis using an antibody specific to HA. As an expression control, the level of GFP was checked using anti-GFP antibody. The experiment was repeated twice. B, Modification of the 14-3-3 binding motif does not change HopQ1 stability in P. syringae. C-terminally His-tagged HopQ1 variants were expressed in P. syringae pv tabaci DAPP-PG677. Crude protein extracts prepared from overnight bacterial cultures were fractionated on 12.5% SDS-PAGE and subjected to immunoblot analysis using a specific anti-His antibody. Equal protein loading is shown by Ponceau Red staining. The experiment was repeated twice. C, R18, the inhibitor of 14-3-3 binding, affects HopQ1 stability in plant extracts. Wild-type HopQ1 protein carrying a C-terminal Flag epitope was transiently expressed in N. benthamiana leaves. Crude protein extract was supplemented with recombinant 14-3-3a-Strep II protein isolated from E. coli or R18 peptide at various concentrations, as indicated. HopQ1 was detected by immunoblot analysis using specific primary antibodies. The level of 14-3-3a was monitored using Strep-Tactin AP conjugate. Equal protein loading is shown by Ponceau Red staining. D, Application of 14-3-3a reverts the effect of R18 on HopQ1 stability. Wild-type HopQ1 protein carrying a C-terminal Flag epitope was transiently expressed in N. benthamiana leaves. Crude protein extract was supplemented with R18 and increasing amounts of recombinant 14-3-3a-Strep II protein isolated from E. coli, as indicated. HopQ1 was detected by immunoblot analysis using specific primary antibodies. The level of 14-3-3a was monitored using Strep-Tactin AP conjugate. Equal protein loading is shown by Ponceau Red staining. E, Assembled in vitro complex of HopQ1-6xHis and 14-3-3a-Strep II was incubated with bean crude protein extract in the absence or presence of R18. HopQ1 and 14-3-3a were detected by immunoblot analyses using anti-His antibodies or Strep-Tactin AP conjugate, respectively. Equal protein loading is shown by Ponceau Red staining. F, In vitro phosphorylated HopQ1-6xHis was added to bean crude extract and incubated in the presence of 14-3-3a or BSA. HopQ1 and 14-3-3a were detected by immunoblot analysis using anti-His antibodies or Strep-Tactin AP conjugate, respectively. Equal protein loading is shown by Ponceau Red staining.
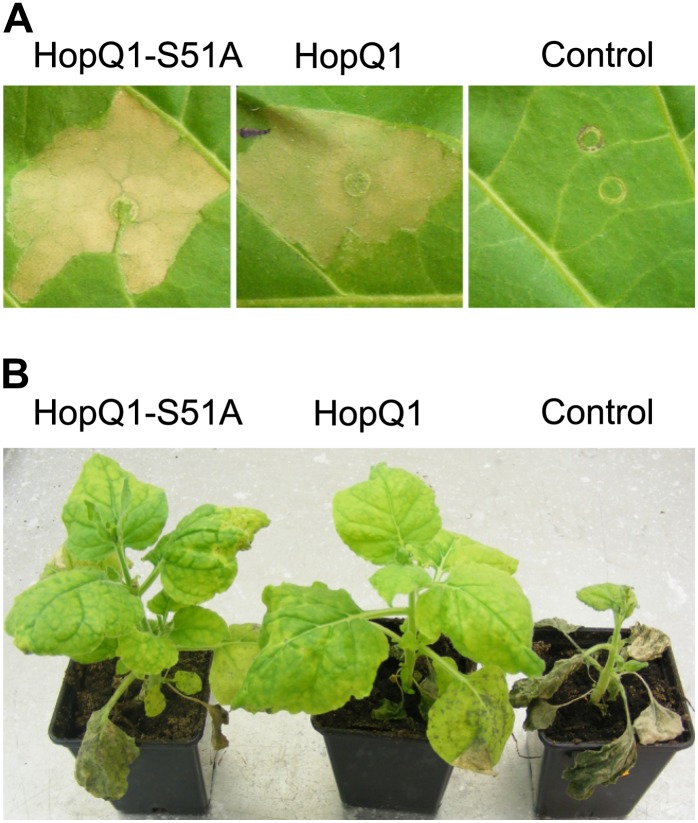
The efficient plant defense strategy, hypersensitive cell death, is induced when HopQ1 is transiently expressed in tobacco (Fig. 8A), in contrast to the severe chloroses triggered by the effector in N. benthamiana leaves (Wroblewski et al., 2009). To determine whether modification of the 14-3-3 binding site is critical for HopQ1 recognition by the plant immune system, tobacco leaves were infiltrated with Agrobacterium tumefaciens cultures carrying HopQ1 constructs. As shown in Figure 8A, the hypersensitive response is elicited by both HopQ1 and HopQ1-S51A, which suggests that this protein can function as an avirulence determinant, despite impaired 14-3-3 binding.
Figure 8.
HopQ1 interaction with 14-3-3s is not critical for its perception by host plants. A, Binary vectors encoding either wild-type HopQ1 or HopQ1-S51A were introduced via agroinfiltration into tobacco leaves. The hypersensitive response developed in the infiltrated area within 48 h in response to both A. tumefaciens strains. In contrast, no macroscopic signs of tissue necrotization developed in control leaves expressing GFP. B, N. benthamiana plants were inoculated with PsyB728a strains carrying pBBR1-MCS2 derivatives, which express HopQ1, HopQ1-S51A, or mCherry protein, as a control. Disease symptoms developed only in control plants treated with PsyB728a encoding mCherry protein. The photographs were taken 10 d post inoculation. The experiment was performed twice, with similar results.
HopQ1 is missing from P. syringae pv syringae B728a (PsyB728a), which is highly virulent in N. benthamiana (Vinatzer et al., 2006). To further test whether the association between HopQ1 and 14-3-3 is involved in the pathogen recognition process, PsyB728a was engineered to express HopQ1 variants. Next, whole N. benthamiana plants were inoculated by dipping in bacterial suspensions, and then plants were incubated for 10 d. As observed with the tobacco wildfire pathogen P. syringae pv tabaci 11528 (Wei et al., 2007), expression of HopQ1 or HopQ1-S51A rendered PsyB728a avirulent (Fig. 8B). In contrast, plants infected by the control strain of PsyB728a, which carried the mCherry protein construct, were severely affected and eventually died. In summary, these observations suggest that the mutation that inhibits 14-3-3 binding does not interfere with HopQ1 perception by tobacco plants. These findings are also consistent with a model in which the association between HopQ1 and 14-3-3 represents a part of the pathogenic strategy.
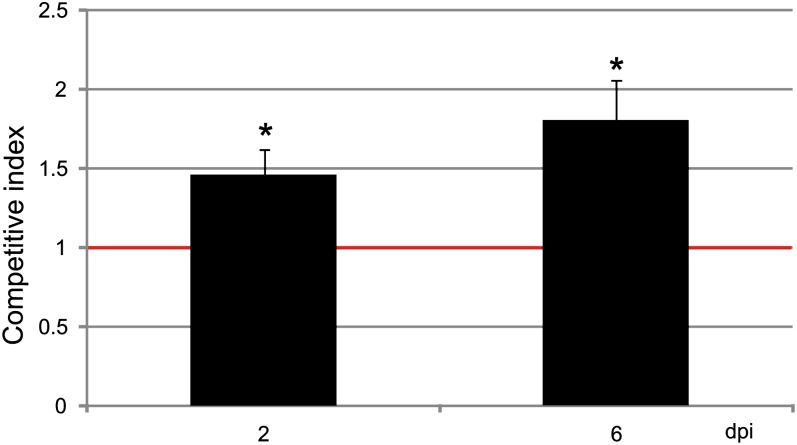
Previous studies demonstrated that HopQ1 expression positively affected the growth of P. syringae in common bean ‘Red Mexican’ (Ferrante et al., 2009). To assess the functional relevance of 14-3-3 binding in HopQ1 virulence, we employed a competitive index (CI) assay (Macho et al., 2007). Plasmids expressing HopQ1 variants were introduced to P. syringae pv tomato DC3000D28E, a strain deficient in 28 effectors. Bean leaves were infiltrated with a mixed inoculum of strains expressing HopQ1 or HopQ1-S51A. At selected time points, bacteria were isolated from leaf tissue and plated. To differentiate the strains, the resulting colonies were replicated onto new plates containing appropriate antibiotics. CI was calculated as the wild type-to-mutant ratio within an output sample normalized for bacterial load. As shown in Figure 9, the strain expressing wild-type HopQ1 was a superior competitor, since the CI was significantly different from 1. The differences between HopQ1 and HopQ1-S51A properties were subtle, yet reproducible. Although modification of the 14-3-3 binding motif did not attenuate virulence dramatically under growth chamber conditions, it may result in a significant reduction of pathogen fitness in the field (Wichmann and Bergelson, 2004).
Figure 9.
Assessment of virulence properties of the HopQ1 effector mutated to eliminate 14-3-3 binding. Bean leaves were inoculated with P. syringae pv tomato DC3000D28E (approximately 105 cfu mL−1) strains expressing HopQ1 or HopQ1-S51A. Immediately prior to infiltration, bacteria were mixed in a 1:1 ratio. Two and 6 d post inoculation (dpi), two leaf discs per plant were cut out from the infiltrated zones, ground in sterile 10 mm MgCl2, diluted, and plated on KB medium. Bacterial strains were distinguished by a selectable marker. The CI was calculated as the ratio of bacteria expressing the wild-type HopQ1 to bacteria expressing the mutated HopQ1 isolated from plant leaf and normalized to the input titers of the bacteria. Asterisks indicate that the index is significantly different from 1, as established using Student’s t test (P < 0.01). The experiment was performed three times with similar results. [See online article for color version of this figure.]
DISCUSSION
Previous studies have suggested that HopQ1 plays a role in determining the host range of P. syringae; however, its mode of action inside the plant cell has remained unclear. This study shows that HopQ1 associates with plant 14-3-3 proteins in a phosphorylation-dependent manner. The HopQ1 sequence contains one putative 14-3-3 binding site of high affinity encompassing the phosphorylated Ser-51. Strikingly, this motif resembles the most typical plant 14-3-3 binding site, with Leu at position −5 (with respect to the Ser-51 residue), Arg at position −3, and Ser at position −2 (Johnson et al., 2010). A single substitution within this motif (HopQ1-S51A) abrogates 14-3-3 binding (Tables II and III; Supplemental Fig. S4). Ser-51 represents possibly the major phosphorylation site of HopQ1, since a site-directed mutation that alters this residue almost completely eliminated the in vitro phosphorylation of HopQ1, in contrast to other substitutions tested (Supplemental Fig. S2). Mass spectrometric analysis of recombinant HopQ1-6xHis produced in P. syringae pv tabaci DAPP-PG677 did not reveal any phosphopeptides (F. Giska, unpublished data). This finding indicates that the phosphorylation of HopQ1 occurs upon delivery of the effector into plant cells. The context of amino acids surrounding Ser-51 indicates that the 14-3-3 binding site of HopQ1 can be recognized and phosphorylated by plant kinases from the CDPK-SnRK family. They are basophilic and have preference for Arg at position −3 (Vlad et al., 2008), and importantly, many of them are activated in response to various environmental cues, including pathogen attack (Romeis et al., 2001; Kulik et al., 2011). Consistently, HopQ1 is phosphorylated in vitro (Supplemental Fig. S3) by representatives of this family (CPK3 and SnRK2.4), but it deserves to be examined more closely in vivo.
It is assumed that client proteins with two low-affinity binding sites bind to dimeric 14-3-3 but not to monomeric 14-3-3 forms, while proteins with a high-affinity site bind also to the monomeric forms (Tzivion et al., 2001). Medium-stringency Scansite search and ELM prediction (for the Eukaryote Linear Motif resource for functional sites in proteins [http://elm.eu.org/links.html]; Gould et al., 2010) revealed in HopQ1 one (20–34 amino acids) or two (24–29 and 73–78 amino acids) additional putative 14-3-3 binding sites, respectively (Supplemental Table S1). This suggests that HopQ1 might interact with 14-3-3 monomerically (via the Ser-51-containing motif) or cooperatively with the dimeric form of 14-3-3. Our ongoing structural studies suggest that this latter possibility looks more likely, since the stoichiometry of the HopQ1-14-3-3 complex seems to be 1:2. However, the motif encompassing Ser-51 is a dominant binding site, as a single amino acid substitution that alters this motif completely abolished the interaction with 14-3-3s (Tables II–IV). Interestingly, one of the 14-3-3 binding sites overlaps one predicted by ELM phosphodegron sequence (22–29 amino acids). Phosphorylation within phosphodegrons often initiates the ubiquitination and proteolytic degradation cycle for proteasome substrates. As HopQ1 was more stable than HopQ1-S51A protein (Fig. 7) when expressed in N. benthamiana, this might suggest that protection of the phosphodegron by 14-3-3 protein could account for the differences in the protein steady-state levels. Consistently, disruption of the HopQ1-14-3-3 complex by the competitive inhibitor R18 significantly reduced HopQ1 stability in plant extracts (Fig. 7). Strikingly, some viruses use the same phosphodegron motif to perturb cellular regulation (Dinkel et al., 2012), suggesting that it can also play a similar role in bacterial pathogenesis.
While the HopQ1 mutated to eliminate 14-3-3 binding (HopQ1-S51A) is still recognized by the tobacco surveillance system (Fig. 8), its ability to enhance P. syringae pv tomato DC3000D28E growth in bean plants is slightly reduced (Fig. 9). This finding suggests that while the interaction with 14-3-3s might contribute to HopQ1 virulence, it is not related to avirulence. This suggestion is further supported by the presence of one or more predicted (Gould et al., 2010) 14-3-3 binding sites among HopQ1 xenologs from the genus Xanthomonas (XopQ; for Xanthomonas outer protein Q) and from R. solanacearum (RipB; for Ralstonia effector injected into plant cells B). In addition, RipB copurified with isoforms 14-3-3e and 14-3-3i when transiently expressed in N. benthamiana (M. Piechocki, unpublished data). We have not observed a drastic reduction in the virulence of hopQ1-S51A (Fig. 9). This is not unexpected, since wild-type HopQ1 caused only a slight increase in P. syringae pv tabaci DAPP-PG677 growth (5-fold), whereas P. syringae pv phaseolicola 1448AΔhopQ1 showed no growth attenuation in bean plants (Macho et al., 2012).
HopQ1 associated in vitro and in planta with several 14-3-3 isoforms from various phylogenetic groups and showed no apparent specificity for one isoform. Similarly, the TTSS effector XopN of Xanthomonas campestris pv vesicatoria has been shown to bind four tomato 14-3-3 isoforms in the yeast two-hybrid system (Kim et al., 2009). However, not necessarily all interactions of HopQ1 with 14-3-3s were direct, since 14-3-3s can form homodimers and heterodimers. It is often assumed that many 14-3-3 isoforms are functionally redundant, while the specificity of others can be achieved not only through protein structure but also due to distinct patterns of spatial and temporal expression, dimerization status, and phosphorylation (Boer et al., 2012; Paul et al., 2012).
Interaction with 14-3-3 can affect various features of partner proteins: their enzymatic activity, half-life, subcellular localization, and ability to bind other proteins. By interacting with 14-3-3, some bacterial pathogens have hijacked these properties. Exoenzyme S, an ADP-ribosyl-transferase toxin secreted by the TTSS of Pseudomonas aeruginosa, becomes active upon interaction with host 14-3-3 proteins (Ottmann et al., 2007). The nuclear accumulation of RolB, a bacterial oncoprotein involved in the pathogenesis of Agrobacterium rhizogenes, depends upon binding to a specific isoform of 14-3-3 (Moriuchi et al., 2004). Based on in silico predictions, HopQ1 is classified as a nucleoside hydrolase; however, this activity could not be detected in the recombinant protein produced in E. coli. The E. coli ribonucleoside hydrolase RihA (Petersen and Møller, 2001) is active under the same conditions (data not shown). Although it might be expected that posttranslational modification and/or 14-3-3 binding could result in catalytically active HopQ1 in planta, no increase in nucleoside hydrolase activity was found between protein extracts obtained from N. benthamiana control plants and plants transiently expressing HopQ1 (data not shown). It remains possible that HopQ1 nucleoside hydrolase activity requires low-Mr cofactors that were lost during extraction and/or that its activity is exerted on substrates that have not yet been tested. While wild-type HopQ1 localizes primarily to the cytoplasm, HopQ1-S51A accumulates in the nucleus (Fig. 5). Thus, the nuclear-cytoplasmic shuttling of HopQ1 might be regulated by its association with 14-3-3. Conversely, since HopQ1 copurified with 14-3-3 isoforms from various phylogenetic groups and showed no apparent specificity for one isoform, it is possible that 14-3-3s might serve as a virulence target for the bacterial effector. Although members of this protein family are quite abundant, it has been hypothesized that some intracellular processes are regulated by temporal redistribution of 14-3-3s, limiting its availability for other partner proteins (Tzivion et al., 2000). HopQ1 could exert its virulence activity by retaining 14-3-3s in the cytoplasm, thereby interfering with host defense signaling, and such a scenario might explain why 14-3-3a is redistributed when coexpressed with HopQ1. On the other hand, binding of 14-3-3s only contributes partially to HopQ1 virulence activity, so this interaction may play an accessory role, possibly in recruiting bona fide virulence targets. An association between HopQ1 and 14-3-3 could enable the binding of a putative virulence target via the second subunit of the 14-3-3 dimer. Questions pertaining to the catalytic activity of HopQ1 and whether multiprotein complexes are formed in plant cells remain to be explored.
MATERIALS AND METHODS
Plant Material
Nicotiana benthamiana, tobacco (Nicotiana tabacum ‘Xanthi-nc’), and common bean (Phaseolus vulgaris ‘Red Mexican’) plants were grown in soil under controlled environmental conditions (21°C, 16 h of light, 8 h of dark), as described previously (Talarczyk et al., 2002).
Recombinant Protein Preparation
A sequence encoding HopQ1 (AAZ37975.1) from Pseudomonas syringae pv phaseolicola 1448A was cloned into the pET30a vector. For site-directed mutagenesis, target plasmids carrying hopQ1 were PCR amplified with two phosphorylated primers (Supplemental Table S3). The primers were designed so that they annealed back to back with the plasmid, and one primer carried the desired mutations. Following amplification with Phusion Hot Start II DNA Polymerase (Thermo Fisher Scientific; www.thermoscientific.com), templates were removed by digestion with DpnI (Thermo Fisher Scientific), and mutated PCR products were circularized with T4 DNA ligase (Thermo Fisher Scientific). The resulting plasmids were transformed into Escherichia coli DH5α, and clones were screened by sequencing. Full-length complementary DNA (cDNA) encoding AtCPK3 (AEE84789.1) was cloned into the pGEX-6P-2 vector. Glutathione S-transferase (GST)-tagged AtSnRK2.4 (AEE28666.1) was a kind gift from Dr. G. Dobrowolska (Bucholc et al., 2011). Full-length cDNA encoding Nt14-3-3a-1 (BAD12168.1) was first cloned into the pGEM-T-easy Vector System and then recloned into pASK-IBA3 to express Nt14-3-3a-1 C-terminally fused with Strep tag II. Recombinant protein expression was performed in E. coli (BL21 Rosetta) cells, induced either with 0.25 mm isopropylthio-β-galactoside or 0.43 μm anhydrotetracycline hydrochloride (tet promoter), at 18°C for 4 h. His- and GST-tagged recombinant proteins were purified using HisTrap and GSTrap HP columns, respectively (GE Healthcare Bio-Sciences; www.gehealthcare.com). Strep-tagged proteins were purified using Strep-Tactin Macroprep columns (IBA; http://www.iba-go.com).
Protein Analysis
Four leaf discs (diameter of 1.2 cm) were collected and extracted in buffer containing 100 mm Tris-HCl, pH 8.0, 150 µm NaCl, 0.2% (v/v) Triton X-100, and protease inhibitor cocktail (BioShop Canada; http://www.bioshopcanada.com). Protein content was measured by the Bradford method using a commercially available reagent (Bio-Rad Laboratories; www.bio-rad.com). Protein samples were fractionated by 12.5% SDS-PAGE and subjected to immunoblot analysis using the specific primary antibodies monoclonal mouse anti-Flag (1:3,000; Sigma-Aldrich), anti-HA (1:3,000; Santa Cruz Biotechnology; www.scbt.com), anti-His (1:3,000 for colorimetric or 1:10,000 for chemiluminescent assay; Sigma-Aldrich), and anti-GFP (1:3,000; Covance; www.covance.com) and secondary anti-mouse antibodies conjugated either to alkaline phosphatase or horseradish peroxidase (both 1:10,000; Sigma-Aldrich). Strep-tagged proteins were detected using Strep-Tactin AP conjugate (1:2,000; IBA). Immunoblots were developed using the nitroblue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate colorimetric detection kit from Roche Applied Science (www.roche-applied-science.com) or ECL Plus Western Blotting Reagents (GE Healthcare).
In Vitro Phosphorylation Assay
Purified recombinant HopQ1-6xHis variants (approximately 3 µg) were incubated with the appropriate recombinant protein kinase (approximately 0.3 µg) or crude plant protein extract (approximately 2 µg) in reaction buffer (40 mm Tris-HCl [pH 8.0], 10 mm MgCl2, 1 mm dithiothreitol [DTT], and 0.1% [v/v] Triton X-100) containing 50 μm ATP supplemented with 1.5 μCi of [γ-32P]ATP. For CPK3 kinase assays, 0.25 mm CaCl2 was added to the buffer. Reactions were carried out at 30°C for 30 min and terminated by adding 4× Laemmli sample buffer. Samples were resolved by 12.5% SDS-PAGE and analyzed with a phosphor imager or by autoradiography. For mass spectrometry, samples were prepared as described above but using nonradioactive ATP. For pull-down assays, phosphorylated HopQ1 was obtained by incubating 2 mg of HopQ1 and 200 µg of CPK3 kinase under the conditions described above.
Strep Tag II Affinity Purification
DNA fragments encoding HopQ1 gene variants were cloned into the pROK2 vector containing a (6xHis)-(Ala-Ala)-Strep tag II-encoding sequence at the 3′ end. Next, the constructs were electroporated into Agrobacterium tumefaciens (GV3101) cells, which were then infiltrated into 4-week-old N. benthamiana leaves, as described previously (Romeis et al., 2001). Infiltrated leaves were collected after 48 h and ground in liquid nitrogen. Samples were then thawed in Ex-Strep buffer (100 mm Tris-HCl [pH 8.0], 150 mm NaCl, 10 mm DTT, 2 mm 4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride, 5 μg mL−1 leupeptin, 5 μg mL−1 bestatin, 50 mm NaF, 1% [v/v] Phosphatase Inhibitor Cocktail 1 [Sigma-Aldrich; www.sigmaaldrich.com], 0.5% [v/v] Triton X-100, and 200 μg mL−1 avidin) and purified using Strep-Tactin MacroPrep (IBA; http://www.iba-go.com).
Flag Affinity Purification
RipB (CAQ60397.1) was amplified using purified Ralstonia solanacearum DNA as a template (a gift from Dr. Stephane Genin), cloned into pENTR/D-TOPO vector, and then moved using Gateway LR clonase II enzyme mix (Invitrogen) into the appropriate pGWB vector. Following transient expression in N. benthamiana, RipB-Flag was purified using EZ View Red Anti-FLAG M2 Affinity Gel (Sigma-Aldrich), and the samples were subjected to mass spectrometry analysis.
Mass Spectrometry Analysis
Samples were reduced with 100 mm DTT for 30 min at 56°C and then alkylated with iodoacetamide in darkness for 45 min at room temperature. Following overnight digestion with sequencing-grade modified trypsin (Promega; www.promega.com) and gel separation, the resulting peptides were eluted from the gel with 0.1% trifluoroacetic acid and 2% acetonitrile. LC-MS analyses were carried out using the nano-Acquity (Waters; www.waters.com) liquid chromatography system coupled to an LTQ-FTICR (Thermo Fisher Scientific) mass spectrometer. Spectrometer parameters were as follows: capillary voltage, 2.5 kV; cone, 40 V; nitrogen gas flow, 0; and range, 300 to 2,000 m/z. The spectrometer was calibrated on a weekly basis using Calmix (caffeine; Met-Arg-Phe-Ala peptide; Ultramark 1621). Samples were loaded from the autosampler tray (cooled to 10°C) to the precolumn (Symmetry C18; 180 µm × 20 mm, 5 µm; Waters) using a mobile phase of 100% MilliQ water acidified by 0.1% formic acid. The peptides were then transferred to a nano-ultra-performance liquid chromatography column (BEH130 C18; 75 µm × 250 mm, 1.7 µm; Waters) with a gradient of 5% to 30% acetonitrile, 0.1% formic acid over 45 min. The column outlet was coupled directly to the electrospray ion source of the LTQ-FTICR (Thermo Fisher Scientific) mass spectrometer, working in the data-dependent mode to perform the switch automatically between mass spectrometry and MS/MS. A blank run preceded each analysis to ensure the absence of cross contamination between samples.
Database Search Procedures
After preprocessing of the raw data with Mascot Distiller software (version 2.1.1; Matrix Science), output lists of precursor and product ions were compared to the National Center for Biotechnology Information nonredundant database (www.ncbi.nlm.nih.gov) using the Mascot database search engine (version 2.1; Matrix Science). Search parameters included semitrypsin enzyme specificity, one missed cleavage site, Cys carbamidomethyl fixed modification, and variable modifications including Met oxidation and phosphorylation of Ser, Thr, or Tyr residues. The protein mass and taxonomy were unrestricted, peptide mass tolerance was 20 ppm, and MS/MS tolerance was 0.8 D. Proteins containing peptides with Mascot cutoff scores greater than 50, which indicated identity or extensive homology (P < 0.05) of peptides, were considered positive identifications.
HopQ1-14-3-3a Complex Formation and Size-Exclusion Chromatography
Recombinant HopQ1 tagged with 6xHis and CPK3 kinase fused to GST were expressed in E. coli and purified by affinity chromatography. Then, HopQ1-6xHis was in vitro phosphorylated by CPK3. To check the phosphorylation status of HopQ1, the reaction mixture was separated by SDS-PAGE and stained with Pro-Q Diamond. To remove the CPK3 kinase, the reaction was loaded onto a GST-binding column. HopQ1 was further purified by ion-exchange chromatography (Q-Sepharose column; GE Healthcare) and size-exclusion chromatography (Superdex 200 10/300GL; GE Healthcare). As a control, a nonphosphorylated form of HopQ1-6xHis was purified using affinity chromatography, ion-exchange chromatography, and size-exclusion chromatography. The recombinant 14-3-3a protein was produced in E. coli in fusion with Strep tag II and purified by affinity chromatography and ion-exchange chromatography. To reconstitute the complex, phosphorylated HopQ1 was mixed with 14-3-3a at a 2:1 mass ratio and incubated in buffer containing 20 mm Tris-HCl, 150 mm NaCl, and 5 mm DTT, pH 8.0, for 2 h at 4°C on a rotator. Nonphosphorylated HopQ1 was incubated with 14-3-3a under the same conditions. To check the formation of the complex, both reaction mixtures were analyzed by size-exclusion chromatography in buffer containing 20 mm Tris-HCl and 150 mm NaCl, pH 8.0. To determine the retention volume for the unbound 14-3-3a fraction, a sample containing only 14-3-3a was run.
Constructs for Subcellular Localization Experiments and FRET-FLIM Measurements
HopQ1 sequence variants or full-length cDNA encoding Nt14-3-3a were cloned into the pENTR/D-TOPO vector (Invitrogen; www.invitrogen.com). PCR amplification was performed using reverse primers (Supplemental Table S3) without the native stop codons to fuse open reading frames in frame with sequences encoding C-terminal tags. To generate expression clones, LR recombination was performed using the entry clones obtained and appropriate destination vectors from the pGWB series (Nakagawa et al., 2007a, 2007b): pGWB441, pGWB444, and pGWB454. To generate a construct expressing eCFP-eYFP chimeric fusion protein as a positive control, the eYFP cDNA was cloned into pSAT6-eCFP and subcloned into the destination vector PZP-RCS2. Subsequently, A. tumefaciens cultures containing the constructs were infiltrated into N. benthamiana leaves, and after 72 h, tissues were analyzed using confocal laser scanning microscopy.
Microbombardment
Leaves from 2- to 4-week-old plants were used to determine the subcellular locations of HopQ1-eYFP and 14-3-3a-mRFP (for monomeric red fluorescence protein) fusion proteins in bean. For transient gene expression in epidermal cells, plasmid DNA (2 µg) was adsorbed onto tungsten M17 particles (diameter, 1.1 µm; 350 µg), and then microbombardment was performed at a pressure of 1,100 pound-force per square inch using the Biolistic PDS-1000/He Particle Delivery System (Bio-Rad Laboratories; www.bio-rad.com). Tissues were analyzed 24 h after bombardment.
Confocal Laser Scanning Fluorescence Microscopy
Transient intracellular fluorescence was observed by confocal laser scanning microscopy using a Nikon TE2000E EZ-C1 inverted confocal microscope equipped with 60× oil-immersion objective lens (numerical aperture = 1.4). eYFP was excited with the 488-nm line from an argon ion laser, and images of mRFP were obtained using 543-nm helium-neon laser excitation. eYFP and mRFP fluorescence signals were detected using the 515/30 and 605/75 emission filters, respectively. Scanning was performed in sequential mode to prevent bleed through. Images were collected in z-stack series at a 0.5-μm focus interval and carefully processed using the freeware ImageJ. For publication, single optical sections with distinctly visible nucleoli were selected to ensure that similar focal planes were compared for all tested variants. FLIM was performed as described previously (Kwaaitaal et al., 2010).
Analysis of HopQ1 Steady-State Levels
To generate clones expressing HopQ1 variants C-terminally tagged with 3xHA or Flag epitopes, LR recombination was performed using the appropriate pENTR/D-TOPO constructs and destination vectors from the pGWB series.
To check protein levels of HopQ1 variants transiently expressed in N. benthamiana leaves, A. tumefaciens cultures containing the hopQ1-3xHA constructs were coinfiltrated with the strains expressing GFP, and after 48 h, tissues were analyzed by immunodetection using anti-HA and anti-GFP antibodies.
To analyze steady-state levels of HopQ1 variants in P. syringae, P. syringae pv tabaci DAPP-PG677 (a kind gift of Dr. B. Vinatzer) was transformed with pBBR1MCS-2 plasmids encoding hopQ1 or hopQ1-S51A in fusion with a 6xHis epitope. The strains were cultured overnight in King's B medium. Subsequently, 1.5-mL bacterial cultures were collected and centrifuged. The pellets were suspended in 300 µL of phosphate-buffered saline. The samples were subjected to immunoblot analysis using anti-His antibody. The strain carrying pBBR1MCS-2 encoding the mcherry gene was used as a control.
To demonstrate the role of 14-3-3 binding in HopQ1 stability, the competitive antagonist peptide R18 (Enzo Life Sciences; www.enzolifesciences.com) was used in the next two experiments. A. tumefaciens cultures containing the hopQ1-Flag constructs were infiltrated into N. benthamiana leaves, and after 48 h, tissues were collected, ground in liquid nitrogen, and thawed in buffer: 50 mm Tris-HCl, 75 mm NaCl, 50 mm NaF, 1× Phosphatase Inhibitor Cocktail (Sigma), 5 mm DTT, and 0.2% Triton, pH 7.0. Samples containing 50 µg of crude plant protein extract were supplemented with 5 µg of recombinant 14-3-3a-Strep tag II protein produced in E. coli and/or R18 peptide at concentrations 5, 150, and 300 mm or alternatively with 300 mm R18 and 10, 25, or 50 µg of 14-3-3a-Strep tag II, as indicated. Following incubation for 60 min at 30°C, the samples were fractionated by 12.5% SDS-PAGE and subjected to immunoblot analyses with anti-Flag antibody or Strep-Tactin AP conjugate.
For studies of HopQ1 stability in extracts from bean, 5 µg of in vitro-reconstituted HopQ1-14-3-3a complex was added to 50 µg of bean crude protein extract (obtained as described above). Then, R18 peptide at a concentration of 300 mm was added to the appropriate samples. In another experiment, 10 µg of in vitro-phosphorylated HopQ1 was added to 50 µg of bean crude protein extract and supplemented with 12 µg of 14-3-3a or BSA. In both experiments, the samples were incubated for 60 min at 30°C, fractionated by 12.5% SDS-PAGE, and subjected to immunoblot analyses with anti-His antibody or Strep-Tactin AP conjugate.
Assessment of the Hypersensitive Response in Tobacco
A. tumefaciens cultures (optical density at 600 nm [OD600] = 1) containing pGWB411-derived plasmids were infiltrated into fully expanded leaves of 6-week-old tobacco ‘Xanthi-nc’ plants. After 48 h, symptoms of the hypersensitive response were observed and photographed.
P. syringae Strains and Inoculation
The broad-host-range plasmids pBBR1MCS-2 and pBBR1MCS-5 (Kovach et al., 1994) were used to express hopq1 variants in P. syringae. The tac promoter was amplified from pGBT30 using a forward primer that added a SacI restriction site to the 5′ end and a reverse primer that added a ribosome-binding site in front of the BamHI restriction site at the 3′ end of the product. The promoter-containing fragment was cloned into pBBR1MCS-2. hopq1, hopq1-S51A, and mcherry sequences were amplified with primers (Supplemental Table S3) that introduced BamHI and XhoI sites to the opposite ends of the products, and then these fragments were cloned downstream of the tac promoter. The constructs were electroporated into PsyB728a and P. syringae pv tomato DC3000D28E (kind gifts of Drs. J. Greenberg and A. Collmer, respectively). The strains were maintained on nutrient agar and stored at 18°C. Bacteria were prepared for inoculation as described previously (Krzymowska et al., 2007), with the exception that following centrifugation at 3,500g for 10 min, the pellet was washed once and resuspended in sterile 10 mm MgCl2. The bacterial suspension was adjusted to OD600 = 0.2 and further diluted (assuming OD600 = 0.2 corresponds to approximately 108 colony-forming units [cfu] mL−1), as indicated. Bacterial titers were checked by plating.
For avirulence assays, bacterial suspensions were adjusted to 106 cfu mL−1 in MilliQ water and supplemented with Silwet L-77 (0.02%). Five-week-old N. benthamiana plants were dip inoculated by inverting whole plants into bacterial suspensions and gently agitating for 30 s. Following inoculation, plants were placed immediately under a plastic dome to maintain high humidity levels for 24 h. Development of symptoms was assessed within 10 d.
For virulence assays, P. syringae pv tomato DC3000D28E derivatives expressing HopQ1 variants were mixed at equal cfus prior to inoculation (105 cfu mL−1). Bacterial suspensions were infiltrated into leaves of 2-week-old bean ‘Red Mexican’ plants using a needleless hypodermic syringe. At selected time points, two 1-cm-diameter leaf discs were cut from infiltrated zones in each plant. Discs were superficially sterilized with 70% ethanol for 1 min, rinsed with sterile water for 1 min, and then ground in 300 µL of 10 mm MgCl2. Serial dilutions were plated onto KB agar plates. The bacteria were grown at 28°C and after 2 d replicated onto plates containing either kanamycin or gentamicin, which enabled strain differentiation and cfu counting. The CI was calculated as described previously (Macho et al., 2007). CI was defined as the ratio of the strain carrying wild-type hopQ1 to the strain expressing hopQ1S51A within the output samples, divided by the corresponding ratios in the input inocula. Data are reported as means ± sd. Mean results were calculated from five plants for each variant.
Statistical Analysis
Data are reported as means ± sd. The results were compared statistically by using a two-tailed Student’s t test, and differences were considered significant at P < 0.05 or as indicated in the figure legends.
Pull-Down Assays
Recombinant HopQ1 (0.5 mg) was phosphorylated in vitro and incubated with 30 mL of plant extracts (1 µg µL−1) for 2 h at 4°C in buffer containing 50 mm NaH2PO4, 150 mm NaCl, 2 mm 4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride, 5 μg mL−1 leupeptin, 5 μg mL−1 bestatin, 50 mm NaF, 1% (v/v) Phosphatase Inhibitor Cocktail 1, 0.5% Triton X-100, and 20 mm imidazole (pH 8.0). Samples were loaded onto a HisTrap HP column, washed with 20 column volumes of His tag wash buffer (50 mm NaH2PO4, 150 mm NaCl, and 20 mm imidazole [pH 8.0]), and then bound proteins were eluted by the addition of 500 mm imidazole. Mass spectrometry was used to identify proteins that copurified with HopQ1. Nonphosphorylated 6xHis-HopQ1 was used as a negative control.
Sequence data from this article can be found in the GenBank data libraries under accession numbers HopQ1, AAZ37975.1; Nt14-3-3a-1, BAD12168.1; AtCPK3, AEE84789.1; AtSnRK2.4, AEE28666.1; and RipB, CAQ60397.1.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Kinase activity capable of phosphorylating HopQ1 is ubiquitously conserved in plants.
Supplemental Figure S2. Ser-51 is the major phosphorylation site of HopQ1.
Supplemental Figure S3. CPK3 and SnRK2 phosphorylate HopQ1 in vitro.
Supplemental Figure S4. Interaction between HopQ1 variants and 14-3-3a in planta.
Supplemental Table S1. 14-3-3 binding motifs are ubiquitously present in TTSS effectors from P. syringae pv phaseolicola 1448A.
Supplemental Table S2. Various 14-3-3 isoforms from P. vulgaris interact with recombinant HopQ1-6xHis in a phosphorylation-dependent manner.
Supplemental Table S3. List of primers used for constructs.
Glossary
- TTSS
type III secretion system
- LC-MS
liquid chromatography-mass spectrometry
- MS/MS
tandem mass spectrometry
- m/z
mass-to-charge ratio
- FRET
fluorescence resonance energy transfer
- eCFP
enhanced cyan fluorescent protein
- eYFP
enhanced yellow fluorescent protein
- FLIM
fluorescence lifetime imaging microscopy
- BSA
bovine serum albumin
- PsyB728a
Pseudomonas syringae pv syringae B728a
- CI
competitive index
- GST
glutathione S-transferase
- HA
hemagglutinin
- DTT
dithiothreitol
- OD600
optical density at 600 nm
- CFU
colony-forming units
- cDNA
complementary DNA
- mRFP
monomeric red fluorescence protein
- cfu
colony-forming units
References
- Boer A, Kleeff PM, Gao J. (August 29, 2012) Plant 14-3-3 proteins as spiders in a web of phosphorylation. Protoplasma, http://dx.doi.org/10.1007/s00709-012-0437-z [DOI] [PubMed] [Google Scholar]
- Bucholc M, Ciesielski A, Goch G, Anielska-Mazur A, Kulik A, Krzywińska E, Dobrowolska G. (2011) SNF1-related protein kinases 2 are negatively regulated by a plant-specific calcium sensor. J Biol Chem 286: 3429–3441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinkel H, Michael S, Weatheritt RJ, Davey NE, Van Roey K, Altenberg B, Toedt G, Uyar B, Seiler M, Budd A, et al. (2012) ELM: the database of eukaryotic linear motifs. Nucleic Acids Res 40: D242–D251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrante P, Clarke CR, Cavanaugh KA, Michelmore RW, Buonaurio R, Vinatzer BA. (2009) Contributions of the effector gene hopQ1-1 to differences in host range between Pseudomonas syringae pv. phaseolicola and P. syringae pv. tabaci. Mol Plant Pathol 10: 837–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good MC, Zalatan JG, Lim WA. (2011) Scaffold proteins: hubs for controlling the flow of cellular information. Science 332: 680–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould CM, Diella F, Via A, Puntervoll P, Gemünd C, Chabanis-Davidson S, Michael S, Sayadi A, Bryne JC, Chica C, et al. (2010) ELM: the status of the 2010 eukaryotic linear motif resource. Nucleic Acids Res 38: D167–D180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrabak EM, Chan CW, Gribskov M, Harper JF, Choi JH, Halford N, Kudla J, Luan S, Nimmo HG, Sussman MR, et al. (2003) The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol 132: 666–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C, Crowther S, Stafford MJ, Campbell DG, Toth R, MacKintosh C. (2010) Bioinformatic and experimental survey of 14-3-3-binding sites. Biochem J 427: 69–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JD, Dangl JL. (2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Kim JG, Li X, Roden JA, Taylor KW, Aakre CD, Su B, Lalonde S, Kirik A, Chen Y, Baranage G, et al. (2009) Xanthomonas T3S effector XopN suppresses PAMP-triggered immunity and interacts with a tomato atypical receptor-like kinase and TFT1. Plant Cell 21: 1305–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovach ME, Phillips RW, Elzer PH, Roop RM, II, Peterson KM. (1994) pBBR1MCS: a broad-host-range cloning vector. Biotechniques 16: 800–802 [PubMed] [Google Scholar]
- Krzymowska M, Konopka-Postupolska D, Sobczak M, Macioszek V, Ellis BE, Hennig J. (2007) Infection of tobacco with different Pseudomonas syringae pathovars leads to distinct morphotypes of programmed cell death. Plant J 50: 253–264 [DOI] [PubMed] [Google Scholar]
- Kulik A, Wawer I, Krzywińska E, Bucholc M, Dobrowolska G. (2011) SnRK2 protein kinases: key regulators of plant response to abiotic stresses. OMICS 15: 859–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwaaitaal M, Keinath NF, Pajonk S, Biskup C, Panstruga R. (2010) Combined bimolecular fluorescence complementation and Forster resonance energy transfer reveals ternary SNARE complex formation in living plant cells. Plant Physiol 152: 1135–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macho AP, Zumaquero A, Gonzalez-Plaza JJ, Ortiz-Martín I, Rufián JS, Beuzón CR. (2012) Genetic analysis of the individual contribution to virulence of the type III effector inventory of Pseudomonas syringae pv. phaseolicola. PLoS ONE 7: e35871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macho AP, Zumaquero A, Ortiz-Martín I, Beuzón CR. (2007) Competitive index in mixed infections: a sensitive and accurate assay for the genetic analysis of Pseudomonas syringae-plant interactions. Mol Plant Pathol 8: 437–450 [DOI] [PubMed] [Google Scholar]
- McDonald C, Vacratsis PO, Bliska JB, Dixon JE. (2003) The Yersinia virulence factor YopM forms a novel protein complex with two cellular kinases. J Biol Chem 278: 18514–18523 [DOI] [PubMed] [Google Scholar]
- Moriuchi H, Okamoto C, Nishihama R, Yamashita I, Machida Y, Tanaka N. (2004) Nuclear localization and interaction of RolB with plant 14-3-3 proteins correlates with induction of adventitious roots by the oncogene rolB. Plant J 38: 260–275 [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T. (2007a) Development of series of Gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104: 34–41 [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Suzuki T, Murata S, Nakamura S, Hino T, Maeo K, Tabata R, Kawai T, Tanaka K, Niwa Y, et al. (2007b) Improved Gateway binary vectors: high-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci Biotechnol Biochem 71: 2095–2100 [DOI] [PubMed] [Google Scholar]
- Nürnberger T, Brunner F. (2002) Innate immunity in plants and animals: emerging parallels between the recognition of general elicitors and pathogen-associated molecular patterns. Curr Opin Plant Biol 5: 318–324 [DOI] [PubMed] [Google Scholar]
- Obenauer JC, Cantley LC, Yaffe MB. (2003) Scansite 2.0: proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res 31: 3635–3641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oecking C, Eckerskorn C, Weiler EW. (1994) The fusicoccin receptor of plants is a member of the 14-3-3 superfamily of eukaryotic regulatory proteins. FEBS Lett 352: 163–166 [DOI] [PubMed] [Google Scholar]
- Oh CS, Martin GB. (2011) Tomato 14-3-3 protein TFT7 interacts with a MAP kinase kinase to regulate immunity-associated programmed cell death mediated by diverse disease resistance proteins. J Biol Chem 286: 14129–14136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh CS, Pedley KF, Martin GB. (2010) Tomato 14-3-3 protein 7 positively regulates immunity-associated programmed cell death by enhancing protein abundance and signaling ability of MAPKKK α. Plant Cell 22: 260–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottmann C, Yasmin L, Weyand M, Veesenmeyer JL, Diaz MH, Palmer RH, Francis MS, Hauser AR, Wittinghofer A, Hallberg B. (2007) Phosphorylation-independent interaction between 14-3-3 and exoenzyme S: from structure to pathogenesis. EMBO J 26: 902–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul AL, Denison FC, Schultz ER, Zupanska AK, Ferl RJ. (2012) 14-3-3 phosphoprotein interaction networks: does isoform diversity present functional interaction specification? Front Plant Sci 3: 190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen C, Møller LB. (2001) The RihA, RihB, and RihC ribonucleoside hydrolases of Escherichia coli: substrate specificity, gene expression, and regulation. J Biol Chem 276: 884–894 [DOI] [PubMed] [Google Scholar]
- Rohmer L, Guttman DS, Dangl JL. (2004) Diverse evolutionary mechanisms shape the type III effector virulence factor repertoire in the plant pathogen Pseudomonas syringae. Genetics 167: 1341–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeis T, Ludwig AA, Martin R, Jones JD. (2001) Calcium-dependent protein kinases play an essential role in a plant defence response. EMBO J 20: 5556–5567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selyunin AS, Sutton SE, Weigele BA, Reddick LE, Orchard RC, Bresson SM, Tomchick DR, Alto NM. (2011) The assembly of a GTPase-kinase signalling complex by a bacterial catalytic scaffold. Nature 469: 107–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talarczyk A, Krzymowska M, Borucki W, Hennig J. (2002) Effect of yeast CTA1 gene expression on response of tobacco plants to tobacco mosaic virus infection. Plant Physiol 129: 1032–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorslund SE, Edgren T, Pettersson J, Nordfelth R, Sellin ME, Ivanova E, Francis MS, Isaksson EL, Wolf-Watz H, Fällman M. (2011) The RACK1 signaling scaffold protein selectively interacts with Yersinia pseudotuberculosis virulence function. PLoS ONE 6: e16784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzivion G, Luo ZJ, Avruch J. (2000) Calyculin A-induced vimentin phosphorylation sequesters 14-3-3 and displaces other 14-3-3 partners in vivo. J Biol Chem 275: 29772–29778 [DOI] [PubMed] [Google Scholar]
- Tzivion G, Shen YH, Zhu J. (2001) 14-3-3 proteins: bringing new definitions to scaffolding. Oncogene 20: 6331–6338 [DOI] [PubMed] [Google Scholar]
- Ueda H, Yamaguchi Y, Sano H. (2006) Direct interaction between the tobacco mosaic virus helicase domain and the ATP-bound resistance protein, N factor during the hypersensitive response in tobacco plants. Plant Mol Biol 61: 31–45 [DOI] [PubMed] [Google Scholar]
- Vinatzer BA, Teitzel GM, Lee MW, Jelenska J, Hotton S, Fairfax K, Jenrette J, Greenberg JT. (2006) The type III effector repertoire of Pseudomonas syringae pv. syringae B728a and its role in survival and disease on host and non-host plants. Mol Microbiol 62: 26–44 [DOI] [PubMed] [Google Scholar]
- Vlad F, Turk BE, Peynot P, Leung J, Merlot S. (2008) A versatile strategy to define the phosphorylation preferences of plant protein kinases and screen for putative substrates. Plant J 55: 104–117 [DOI] [PubMed] [Google Scholar]
- Wang B, Yang H, Liu Y-C, Jelinek T, Zhang L, Ruoslahti E, Fu H. (1999) Isolation of high-affinity peptide antagonists of 14-3-3 proteins by phage display. Biochemistry 38: 12499–12504 [DOI] [PubMed] [Google Scholar]
- Wei CF, Kvitko BH, Shimizu R, Crabill E, Alfano JR, Lin NC, Martin GB, Huang HC, Collmer A. (2007) A Pseudomonas syringae pv. tomato DC3000 mutant lacking the type III effector HopQ1-1 is able to cause disease in the model plant Nicotiana benthamiana. Plant J 51: 32–46 [DOI] [PubMed] [Google Scholar]
- Wichmann G, Bergelson J. (2004) Effector genes of Xanthomonas axonopodis pv. vesicatoria promote transmission and enhance other fitness traits in the field. Genetics 166: 693–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wroblewski T, Caldwell KS, Piskurewicz U, Cavanaugh KA, Xu H, Kozik A, Ochoa O, McHale LK, Lahre K, Jelenska J, et al. (2009) Comparative large-scale analysis of interactions between several crop species and the effector repertoires from multiple pathovars of Pseudomonas and Ralstonia. Plant Physiol 150: 1733–1749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Wang W, Coleman M, Orgil U, Feng J, Ma X, Ferl R, Turner JG, Xiao S. (2009) Arabidopsis 14-3-3 lambda is a positive regulator of RPW8-mediated disease resistance. Plant J 60: 539–550 [DOI] [PubMed] [Google Scholar]