A chromoplast protein may assist in the sequestration and stabilization of carotenoids.
Abstract
Tomato (Solanum lycopersicum) high-pigment mutants with lesions in diverse loci such as DNA Damage-Binding Protein1 (high pigment1 [hp1]), Deetiolated1 (hp2), Zeaxanthin Epoxidase (hp3), and Intense pigment (Ip; gene product unknown) exhibit increased accumulation of fruit carotenoids coupled with an increase in chloroplast number and size. However, little is known about the underlying mechanisms exaggerating the carotenoid accumulation and the chloroplast number in these mutants. A comparison of proteome profiles from the outer pericarp of hp1 mutant and wild-type (cv Ailsa Craig) fruits at different developmental stages revealed at least 72 differentially expressed proteins during ripening. Hierarchical clustering grouped these proteins into three clusters. We found an increased abundance of chromoplast-specific carotenoid-associated protein (CHRC) in hp1 fruits at red-ripe stage that is also reflected in its transcript level. Western blotting using CHRC polyclonal antibody from bell pepper (Capsicum annuum) revealed a 2-fold increase in the abundance of CHRC protein in the red-ripe stage of hp1 fruits compared with the wild type. CHRC levels in hp2 were found to be similar to that of hp1, whereas hp3 and Ip showed intermediate levels to those in hp1, hp2, and wild-type fruits. Both CHRC and carotenoids were present in the isolated plastoglobules. Overall, our results suggest that loss of function of DDB1, DET1, Zeaxanthin Epoxidase, and Ip up-regulates CHRC levels. Increase in CHRC levels may contribute to the enhanced carotenoid content in these high-pigment fruits by assisting in the sequestration and stabilization of carotenoids.
Tomato (Solanum lycopersicum) high-pigment mutants hp1 (hp1, hp1w), hp2 (hp2, hp2j, hp2dg), hp3, and Intense pigment (Ip) not only share characteristically high lycopene content but also exhibit an increased number and size of chloroplasts in leaves as well as in green fruits. This leads to a higher number of chromoplasts when the fruits ripen (Azari et al., 2010). HP1 codes for DNA Damage-Binding Protein1 (DDB1; Lieberman et al., 2004; Liu et al., 2004), while HP2 encodes the Arabidopsis (Arabidopsis thaliana) homolog of Deetiolated1 (DET1; Mustilli et al., 1999) and the hp3 mutant has a lesion in zeaxanthin epoxidase (Galpaz et al., 2008). However, the gene product for Ip is still unknown (Lavi et al., 2009). Similar to hp1 and hp2 mutants, Ip also shows exaggerated photomorphogenesis (Lavi et al., 2009). Due to their high lycopene, flavonoid, and vitamin contents, hp1 and hp2 mutations have been introgressed into various breeding populations. One such introgression from hp2dg resulted in a 3.5-fold increase in lycopene content in tomato fruits (Levin et al., 2003). Both DDB1 and DET1 seem to interact with Cullin4 (CUL4) and were found to be components of a CUL4-based E3 ubiquitin ligase complex (Wang et al., 2008). CUL4-DDB1 complexes have been shown to affect overall plant development (Bernhardt et al., 2006) and flowering (Chen et al., 2010), where they may affect the epigenetic control of flowering. Double mutants of ddb1a and ddb1b showed that in Arabidopsis, DDB1 is critical for embryo development (Bernhardt et al., 2010).
Manipulation of light signaling components appears to be a good strategy to improve tomato fruit quality, as shown by fruit-specific RNA interference (RNAi)-mediated suppression of DET1 and repression of LeCOP1LIKE genes, which resulted in increased carotenoid levels (Liu et al., 2004; Davuluri et al., 2005). Similarly, repression of CUL4 and fruit-specific repression of HP1/DDB1 by RNAi resulted in increased plastid compartment size and enhanced pigmentation of tomato fruits (Wang et al., 2008). Disrupting the function of all or any of these light signaling components seems to affect plastid biogenesis, leading to an increased number of plastids with greater storage capacity for the carotenoids and/or pigments (Liu et al., 2004; Kolotilin et al., 2007). Likewise, the deficiency of abscisic acid (ABA) also seems to result in a similar high-pigment phenotype (Galpaz et al., 2008). In all the above cases, the efficient conversion of chloroplasts to chromoplasts is necessary to accumulate the high amount of synthesized carotenoids.
A number of changes occur during the conversion of chloroplasts to chromoplasts. The first is the disintegration of thylakoid membranes, followed by loss of chlorophyll, increase in the number of plastoglobules, accumulation of lycopene, and increase in the number of stromules, etc. (Bian et al., 2011). Plastoglobules, besides accumulating lipids, also accumulate carotenoids either in the crystalline form, as seen in tomato (Klee and Giovannoni, 2011), or the fibrillar form, as observed in bell pepper (Capsicum annuum; Pozueta-Romero et al., 1997). In addition, these plastoglobules also appear to be involved in regulating carotenoid metabolism, as enzymes such as ζ-carotene desaturase, lycopene β-cyclase, and β-carotene hydroxylases are present in them (Ytterberg et al., 2006). Furthermore, proteins called plastoglobulins are associated with plastoglobules and help in the accumulation of carotenoids. These include fibrillin, also known as CHRB or plastid lipid-associated protein (PAP) from pepper chromoplast fibrils, and chromoplast-associated carotenoid-binding protein (CHRC) and CHRD from petals of cucumber (Cucumis sativus; Vishnevetsky et al., 1999). The expression of plastoglobulin genes appears to be under strict spatial and temporal regulation and well coordinated with the expression of carotenoid biosynthetic genes like PSY1 in tomato fruits (Giuliano et al., 1993; Vishnevetsky et al., 1999).
All the plastoglobulins share a hydrophobic domain of 17 to 19 amino acids, and this region seems to be important for carotenoid-protein interactions (Vishnevetsky et al., 1999). The significance of the above-mentioned plastoglobulins in the sequestration and storage of carotenoids was elegantly exemplified in two studies (Leitner-Dagan et al., 2006; Simkin et al., 2007). Overexpression of a pepper fibrillin gene in tomato resulted in a 2-fold increase in carotenoid levels as well as carotenoid-derived volatiles (Simkin et al., 2007), whereas down-regulation of CHRC in tomato led to a 30% reduction of carotenoids in the flowers (Leitner-Dagan et al., 2006). Moreover, a delayed loss of thylakoids was observed during chromoplastogenesis in fibrillin-overexpressing tomato (Simkin et al., 2007). In addition, the Or gene of cauliflower (Brassica oleracea), which encodes a DnaJ Cys-rich zinc finger domain-containing protein, is also targeted to plastids and seems to control carotenoid accumulation by stimulating the formation of chromoplasts (Lu et al., 2006; Zhou et al., 2008; Li et al., 2012). All these studies suggest that manipulation of carotenoid sequestration and storage by stimulating chromoplastogenesis to create a metabolic sink might be an alternative strategy to increase carotenoid levels in food crops.
Currently, using systems biology approaches, efforts are being made to understand the complex interplay of ripening-related pathways wherein transcriptome, proteome, and metabolome are examined and attempts are made to interlink them (Osorio et al., 2011). Such a study in rin, nor, and Nr mutants led to a comprehensive understanding of ethylene-regulated processes during ripening and also reinforced the need for the integration of transcript, proteome, and metabolite analyses (Osorio et al., 2011). In the hp1 mutant, transcription factor profiling was combined with microarray and metabolite analyses, and that study revealed that the secondary metabolism is controlled at the transcriptional level (Rohrmann et al., 2011). Using fruit-specifically down-regulated DET1 tomato lines, Enfissi et al. (2010) showed the significance of posttranscriptional regulation in modulating carotenoid and isoprenoid biosynthesis.
On the other hand, several proteomics studies have been carried out in tomato (Rocco et al., 2006; Faurobert et al., 2007; Manaa et al., 2011; Osorio et al., 2011) as well as other fruits like strawberry (Fragaria spp.; Bianco et al., 2009), grape (Vitis vinifera; Zhang et al., 2008), and Citrus spp. (Zeng et al., 2011) and also in isolated chromoplasts (Siddique et al., 2006; Barsan et al., 2010). This led to global understanding of the changes in protein profiles accompanying ripening. Several recent studies examining the linkage between gene expression and the metabolite levels during tomato fruit ripening also highlighted the need for more comprehensive network approaches (Carrari and Fernie, 2006; Mounet et al., 2009; Rohrmann et al., 2011). Considering this, and given the importance of the hp1 mutant for improving the phytonutrient quality, we undertook a proteomic approach to decipher the role of DDB1 in tomato fruit ripening, as information on the proteome complement of hp1 is lacking. We compared the proteome profiles of outer pericarp in both wild-type and hp1 fruits at different stages during ripening. Although we did not find significant differences between the wild type and the mutant, we found a development-specific fine-tuning in the abundance of several proteins controlling multiple metabolic pathways in both wild-type and hp1 fruits during ripening. We found higher levels of CHRC in hp1. Our results suggest a likely relationship between increased lycopene content and CHRC levels during fruit ripening.
RESULTS AND DISCUSSION
Influence of DDB1 on the Tomato Proteome
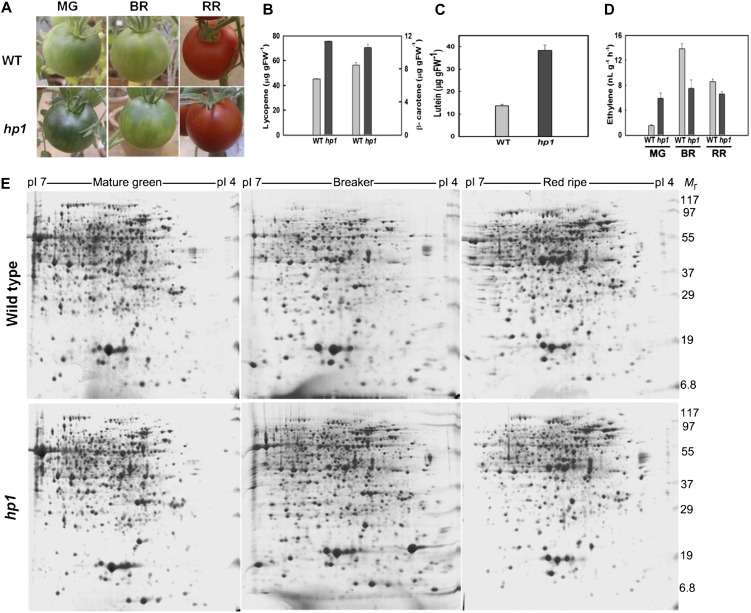
High pigment1 fruits are characteristically different from wild-type fruits in being more green in color at the mature green (MG) stage (Fig. 1A) and accumulating more lycopene at the red-ripe (RR) stage (Fig. 1, A and B). In view of this, we characterized the proteome profiles at both these stages. We also included the breaker (BR) stage, where the fruit makes transition, marking the onset of the ripening process. Consistent with earlier reports, loss of function of DDB1 led to an enhanced accumulation of lycopene in tomato fruits (Fig. 1B). In addition, the RR fruits of hp1 displayed 3-fold higher levels of lutein (Fig. 1C). In contrast, ethylene, an important regulator of fruit ripening, although elevated in hp1 at the MG stage, declined considerably at the BR stage of fruit development (Fig. 1D).
Figure 1.
Characterization of the phenotypic and metabolic differences between wild-type and hp1 fruits. A, Phenotypes of wild-type (WT) and hp1 fruits at MG, BR, and RR stages during ripening. B, Levels of lycopene (left) and β-carotene (right) in RR fruits of the wild type (WT) and hp1 (n > 3). C, Lutein levels in RR fruits of hp1 and the wild type (WT; n > 3). D, Ethylene emission from wild-type (WT) and hp1 fruits at different stages of ripening (n > 3). FW, Fresh weight. Error bars indicate se. E, Comparison of pericarp proteome profiles isolated from wild-type and hp1 fruits at MG, BR, and RR stages. Representative images from two-dimensional gel electrophoresis are shown. [See online article for color version of this figure.]
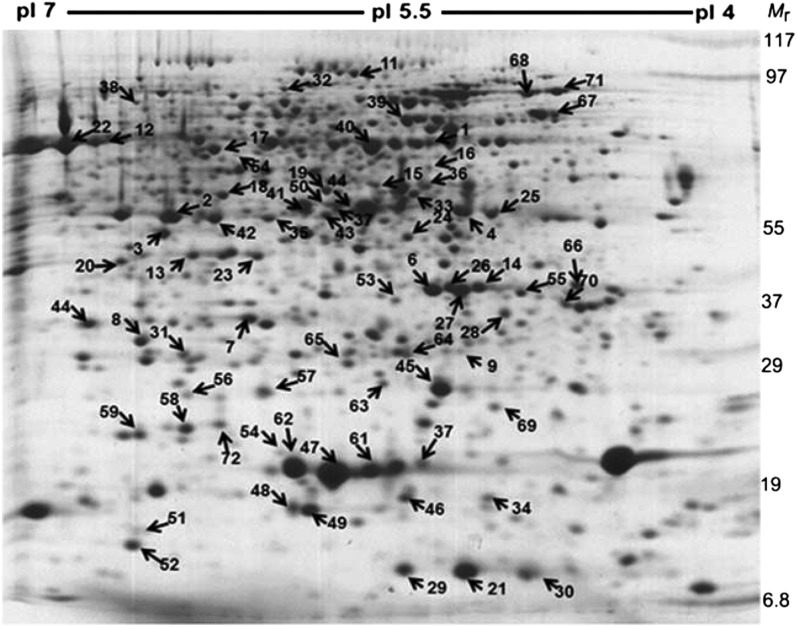
A randomized block design was used to grow the tomato plants. The fruit sampling for all three biological replicates was independently carried out according to the scheme shown in Supplemental Figure S1. For proteome profiling, we analyzed the proteins extracted from the outer pericarp of wild-type and hp1 fruits (Supplemental Fig. S2) at MG, BR, and RR stages. Representative two-dimensional gel electrophoresis images comparing the above three stages in wild-type and hp1 fruits are shown in Figure 1E. Image analysis of the gels (n = 3) with the Image Master Platinum software revealed about 800 proteins on each gel. Seventy-two proteins were found to be differentially expressed by at least 2-fold (overexpression or underexpression; statistically significant expression based on ANOVA with P ≤ 0.05) in both mutant and wild-type fruits in at least one stage during ripening, representing about 9% of spots on the gel. Notwithstanding the distinct phenotype of hp1 fruits, we did not find any proteins present specifically in the wild type or the mutant alone. This indicates that the subtle changes in proteome profile may be responsible for the specific phenotype of hp1 fruits. To ascertain this, we identified the molecular nature of differentially expressed proteins to reveal any development-specific fine-tuning in the expression of specific proteins in both the mutant and the wild type. The differentially regulated protein spots (Figs. 1E and 2) were excised from the gel and, after digestion with trypsin, subjected to matrix-assisted laser-desorption ionization (MALDI)-time of flight (TOF) analysis. Of the 72 spots, 51 could be successfully identified using Matrix Science’s Mascot search engine. For 17 spots, no firm identity could be assigned. For the remaining four spots, only a single peptide match was obtained that appeared to be statistically significant (Supplemental Table S1). On comparing these differentially expressed proteins with the proteins identified from cherry tomato (Faurobert et al., 2007), tomato ecotypes (Rocco et al., 2006), and the rin mutant (Qin et al., 2012), our study revealed 14 additional proteins (Supplemental Table S2) that were not detected in any of the above studies. Nevertheless, such variations between different studies are expected, considering the diversity in the genotypes of different cultivars and mutants, sampling tissue and stages, the methods used for protein extraction, etc.
Figure 2.
A representative two-dimensional gel electrophoresis colloidal Coomassie blue-stained gel showing the profile of proteins extracted from hp1 fruits at the BR stage. The numbers indicate differentially expressed proteins that were later identified by mass spectrometry, and the arrows indicate their relative positions on the gel.
Functional distribution of the above-identified proteins was performed according to the FunCat annotation and is shown in Supplemental Figure S3 (Ruepp at al, 2004). The differentially expressed proteins were distributed across 12 different categories, with nearly half of the proteins being involved in carbon metabolism (46%), in accordance with the massive up-regulation of metabolic processes such as respiration during fruit ripening. Among the remaining categories, stress response (14%) was maximally represented, followed by detoxification (7%), protein fate modification/degradation (6%), transport (6%), amino acid and hormone metabolism (4%), cell cycle regulation (3%), secondary metabolism (3%), replication (1%), protein synthesis and storage (1%), transcription (1%), and unclassified (8%).
Hierarchical Clustering of Identified Proteins
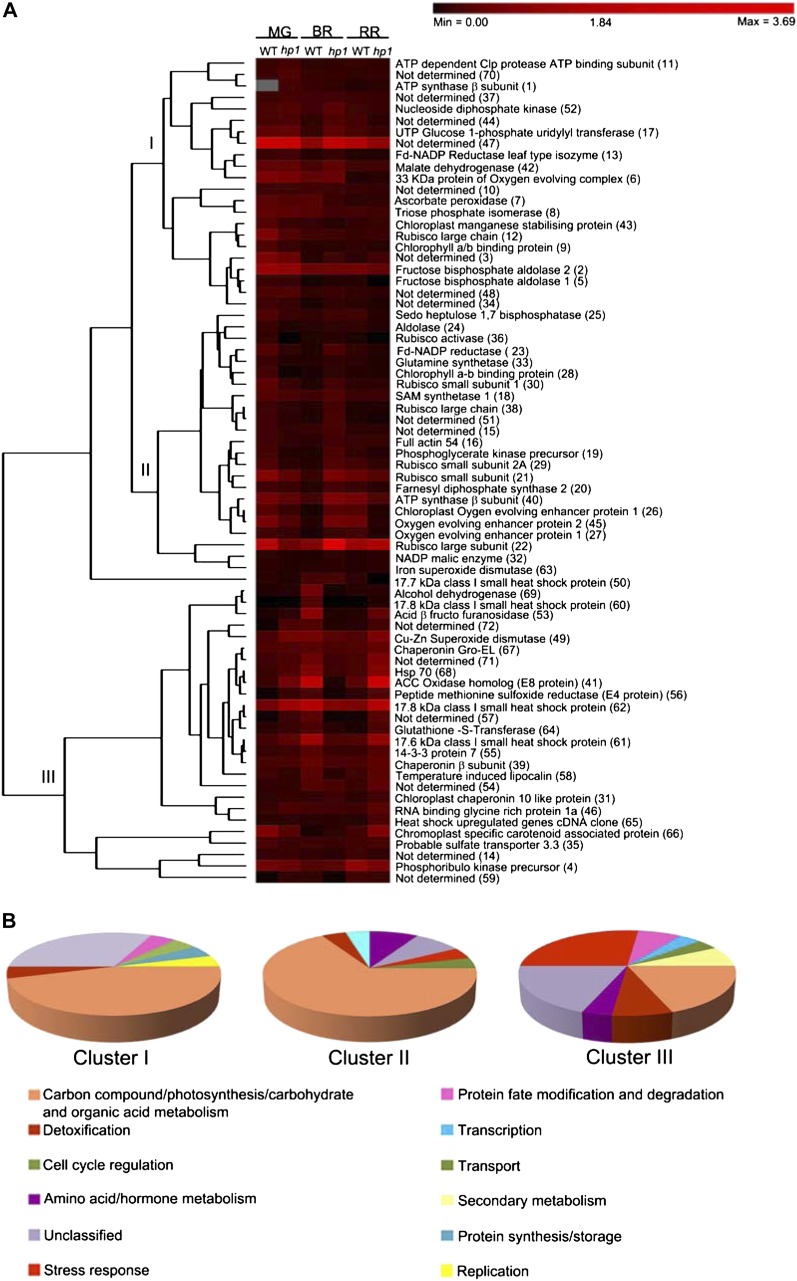
To reveal patterns of protein expression, the above differentially expressed proteins were grouped. These proteins were clustered based on the unweighted pair group method with arithmetic mean (Caraux and Pinloche, 2005) using the differences in protein spot intensity at different ripening stages of both wild-type and hp1 fruits. The above analysis distinguished three main clusters of proteins, hereafter named clusters I, II, and III (Fig. 3A). As expected, in all three clusters, proteins involved in carbohydrate metabolism were most abundant (Fig. 3B).
Figure 3.
Hierarchical clustering analysis of differentially expressed proteins in wild-type (WT) and hp1 fruits during ripening. A, Using PermutMatrix 1.9.3, proteins were clustered into three clusters based on their percentage spot volumes. A color bar corresponding to the relative levels of expression is given at the top of the heat map. The identity of each protein (spot no.) is indicated to the right of the rows. The columns represent levels of proteins at MG, BR, and RR stages of wild-type and hp1 fruits. B, Pie diagram showing the relative distribution of different functional classes of proteins in each cluster. [See online article for color version of this figure.]
Cluster I (30.55% of all protein spots) included three different subclusters reflecting protein expression patterns. In the first subcluster, the expression patterns of proteins remained nearly uniform during fruit development. This subcluster included 14 spots, of which the identity of six (3, 10, 37, 47, 48, and 70) was not known. The remaining eight spots (2, 8, 11, 12, 13, 42, 43, and 52) belonged to proteins involved in carbohydrate metabolism. The second subcluster consisted of proteins whose level was similar at the MG and RR stages but varied at the BR stage. This cluster consisted of five proteins, of which two were unidentified proteins (34 and 44) and the remaining three were involved in carbohydrate metabolism (5, 9, and 17). The third subcluster comprised three proteins that declined during fruit development (1, 6, and 7). While the majority of these proteins belonged to housekeeping functions, cluster I also included proteins regulating protein fate modification and degradation (11), detoxification (7), replication (52), and transport (1; Fig. 3B).
Similar to cluster I, cluster II (33.33% of proteins) also included three subclusters. Subcluster 1 included proteins most of which were down-regulated at the BR stage in the wild type (but with almost similar expression levels at MG and RR) but were up-regulated in hp1 and were specifically involved in carbohydrate metabolism (21, 23, 25, 29, 30, and 45). The proteins that showed higher expression in the BR stage of hp1, with reduced but similar expression levels at MG and RR, were classified in subcluster 2 (15, 19, 22, 28, 36, 38, 50, and 51). While most of these proteins were part of the photosynthesis machinery, this also included a small heat shock protein17.7 (50). Subcluster 3 consisted of proteins that showed almost constant expression levels in both the wild type and hp1 (16, 18, 20, 24, 32, 33, and 63) during ripening. These proteins belonged to carbon metabolism (24 and 32), amino acid/hormone metabolism (18 and 33), cell cycle regulation (16), secondary metabolism (20), transport (40), and detoxification (63; Fig. 3B).
Cluster III (36.12% of proteins) comprised proteins with diverse biological functions showing similar expression patterns in either the wild type or hp1 across different developmental stages during ripening. In wild-type fruits, this cluster included proteins modulating stress (60, 62, 65, and 68), detoxification (49), hormone metabolism (41), secondary metabolism (58), and unknown functions (54, 57, 71, and 72; Fig. 3B). The spots that were specifically modulated in hp1 fruit consisted of proteins involved in protein fate modification/degradation (46 and 56), detoxification (49), stress response (61), and a few of unknown function (39, 57, and 71; Fig. 3B). One of the spots (66) showed a diametrically different expression pattern in the wild type and hp1. It had higher expression in the wild type at the MG stage and declined at subsequent BR and RR stages, while in hp1, its expression progressively increased from MG to RR stages (Fig. 3A).
The metabolic assignment of variably expressed proteins during fruit ripening revealed three main pathways. The first pathway comprised proteins that are involved in the Calvin-Benson cycle, glycolysis, the tricarboxylic acid cycle, and the interconnecting pathways (Supplemental Fig. S4). Reactions related to photosynthetic electron transport in the plastids constituted the second pathway (Supplemental Fig. S5). The third pathway (Supplemental Fig. S6) is a model showing the probable mode of action of differentially expressed heat shock proteins detected in our study.
HP1 Shows an Accumulation of Proteins Related to Carotenoid Sequestration and Metabolism
High pigmentation of fruits in tomato appears to be regulated by a diverse set of genes encoded by distinct genetic loci: hp1, hp2, hp3, and Ip (Azari et al., 2010). At the RR stage, fruits of all these mutants possess elevated levels of lycopene and β-carotene, signifying commonality in the up-regulation of these carotenoids. In view of this, we specifically examined the 72 differentially abundant proteins for the presence of specific enzymes or proteins that may have assisted in carotenoid accumulation by regulating biosynthesis, sequestration, or degradation. We found three such proteins. Lipocalins are lipid-associated transporter molecules helping in the transport of certain lipids and small molecules (Frenette Charron et al., 2002). They also include enzymes like violaxanthin deepoxidase and zeaxanthin epoxidase (Hieber et al., 2000), which play roles in carotenoid degradation. During ripening, the abundance of temperature-induced lipocalins increased in both the wild type and hp1, although expression at the BR stage was less in the wild type and thereafter increased at the RR stage. Similar increases in levels of lipocalins were also observed in other tomato proteomics studies (Faurobert et al., 2007; Osorio et al., 2011). It is known that alcohol dehydrogenase is an important enzyme in the biosynthesis of volatiles in tomato (Speirs et al., 1998). We found alcohol dehydrogenase to be up-regulated in both the wild type and mutant at the RR stage, but the protein level was much higher in the wild type. The observed increase in alcohol dehydrogenase and lipocalins at the RR stage may be related to increases in the levels of compounds contributing to volatile/flavor formation in tomato fruits.
One protein that showed an interesting expression pattern is CHRC. This protein showed a gradual decline in abundance during ripening in the wild type. On the other hand, in hp1, the protein showed an opposite trend; it progressively increased, attaining a peak at RR (Fig. 3). This protein was also found in chromoplast proteome studies of tomato, bell pepper, and Citrus spp. (Siddique et al., 2006; Barsan et al., 2010; Zeng et al., 2011), and it is assumed that it plays a role in carotenoid storage.
Expression Profiling of Genes Involved in Carotenoid Precursor and Carotenoid Biosynthesis
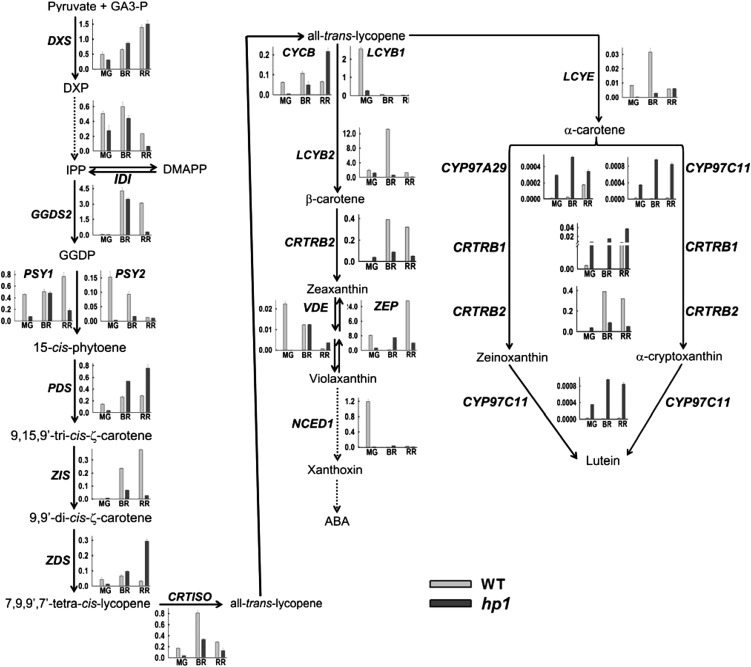
Although it is logically expected that high carotenoid levels in hp1 fruits should be reflected in up-regulation of enzymes of the pathway, our proteome analysis did not reveal any differentially expressed protein spots for any of the carotenoid biosynthetic enzymes. Apparently, the levels of these enzymes in both the wild type and hp1 were below the detection limits of colloidal Coomassie blue staining, and it is believed that the flux in the carotenoid biosynthetic pathway is mainly modulated by posttranscriptional, translational, or posttranslational regulation. To ascertain the influence of the hp1 mutation on transcript level, we examined the expression of genes encoding the entire carotenoid biosynthetic pathway. Figure 4 shows the expression profiles of 20 genes involved in carotenoid synthesis using quantitative real-time PCR analysis, including genes contributing to the formation of the first precursor phytoene. Despite the accumulation of higher levels of lycopene in hp1 fruits, the transcripts of genes belonging to the carotenoid pathway, regulating the formation of phytoene and its desaturation to lycopene, did not show any concerted up-regulation at the BR and RR stages when compared with the wild type. The absolute levels of different transcripts varied distinctly and could be classified into two groups. In wild-type fruit at the MG stage, LCYB1, LCYB2, ZEP, and NCED1 were the most abundant transcripts (greater than 1.0–8.0), whereas other genes were expressed at a lower level (0.0–1.0). A comparison of wild-type gene expression with hp1 fruit at the MG stage indicated that the expression of most genes was significantly down-regulated in the mutant, except CYP97A29, CYP97C11, CRTRB1, and CRTRB2, which were up-regulated.
Figure 4.
The biosynthetic pathway of carotenoids depicting the expression profiles of enzymes catalyzing different steps beginning from the precursors. Dotted arrows indicate multiple steps in the pathway. The transcripts were quantified in the wild type and hp1 at MG, BR, and RR stages of fruit development using real-time PCR. The transcripts were expressed after normalization with two internal controls, β-actin and ubiquitin. GA3-P, Glyceraldehyde-3-phosphate; DXP, deoxy-xylulose 5-phosphate; IPP, isopentenyl pyrophosphate; DMAPP, dimethylallyl pyrophosphate; GGDP, geranylgeranyl diphosphate; DXS, deoxy-xylulose 5-phosphate synthase; IDI, inositol diphosphate synthase; GGDS2, geranylgeranyl diphosphate synthase2; PSY1, phytoene synthase1; PSY2, phytoene synthase2; PDS, phytoene desaturase; ZIS, ζ-carotene isomerase; CRTISO, carotenoid isomerase; ZDS, ζ-carotene desaturase; LCYB1, lycopene β-cyclase1; LCYB2, lycopene β-cyclase2; CYCB, chromoplast-specific lycopene β-cyclase; LCYE, lycopene ε-cyclase; CRTRB2, β-carotene hydroxylase2; ZEP, zeaxanthin epoxidase; VDE, violoxanthin deepoxidase; NCED1, 9-cis-epoxycarotenoid dioxygenase1; CRTRB1, β-carotene hydroxylase1; CYP97A29, cytochrome P450 carotenoid β-hydroxylase A29; CYP97C11, cytochrome P450 carotenoid ε-hydroxylase C11; WT, wild type.
In the wild type, GGDS2 and LCYB2 were the most abundant transcripts (greater than 1.0–13.0) at the BR stage, whereas other genes were expressed at a lower level. While the expression levels of DXS, IDI, GGDS2, PSY1, ZDS, and VDE were nearly similar, other transcripts, such as PSY2, ZIS, CRTISO, LCYB2, CYCB, LCYE, and CRTRB2, were down-regulated in hp1 compared with the wild type. At the BR stage, the level of PDS, ZEP, CYP97A29, CYP97C11, and CRTRB1 transcripts were significantly higher in the hp1 mutant than in the wild type.
At the RR stage, DXS and ZEP were the most abundant transcripts (greater than 1.0–24.0) in the wild type, whereas other genes were expressed at a lower level. In RR fruits, transcript levels of DXS, PSY2, LCYB1, LCYE, and NCED1 were nearly similar in the wild type and hp1, whereas IDI, GGDS2, PSY1, ZIS, CRTISO, LCYB2, CRTRB2, and ZEP were down-regulated in hp1. In contrast, expression levels of PDS, ZDS, CYCB, VDE, CYP97A29, CYP97C11, and CRTRB1 were higher in the hp1 mutant. Among these seven transcripts, the gene products of only PDS and ZDS contribute to lycopene, whose levels are elevated in the hp1 mutant. The slight stimulation of β-carotene level in hp1 fruits may be related to an up-regulation of CYCB transcript at the RR stage. Similarly, the gene products of CYP97A29, CYP97C11, and CRTRB1 seem to contribute to the higher lutein levels found in RR fruits of hp1, as their transcripts are up-regulated. Our results are in agreement with the study of Stigliani et al. (2011) reporting higher expression of CYP97A29, CYP97C11, and CRTRB1 during tomato ripening.
Of interest was the opposite expression patterns of ZEP and VDE transcripts, with down-regulation of ZEP and up-regulation of VDE in the hp1 mutant. While the gene product of ZEP converts zeaxanthin to violaxanthin, the gene product of VDE reconverts violaxanthin to zeaxanthin. The reduced expression of ZEP and the enhanced expression of VDE in hp1 appear to be in agreement with the characteristic increase in chloroplast number and size, which could be correlated to reduced ABA levels in hp1 (Carvalho et al., 2011). A similar phenotype was also observed in the ABA-deficient hp3 mutant, which has a lesion in ZEP (Galpaz et al., 2008). A link between HP1 transcript levels and ABA level is also apparent from observations that the application of external ABA stimulates the expression of DDB1 mRNA in tomato seedlings (Wang et al., 2008).
Despite the distinctive increase in lycopene level in hp1 fruits, the transcript levels of most enzymes of the carotenoid biosynthesis pathway were lower than those of the wild type at all stages of fruit ripening. The predominant down-regulation of transcripts in hp1, barring a few such as PDS, ZDS, CYCB, and VDE that show up-regulation at the RR stage, indicates that the enhanced carotenoid formation in the hp1 mutant is most likely regulated at the posttranscriptional level. Similar to this study, Enfissi et al. (2010) also suggested the posttranscriptional regulation of the carotenoid biosynthesis pathway, as they did not find a correlation between transcript profiles of carotenoid biosynthetic genes and changes in the carotenoid levels in DET1 down-regulated tomato fruits. One of the enzymes that may regulate flux into the carotenoid biosynthesis pathway is phytoene synthase I. It is believed that an increase in phytoene level has a feed-forward effect on carotenoid biosynthesis (Fraser et al., 2007). It was also reported that the activity of the PSY1 enzyme in hp1 was up-regulated by 2-fold, although there was no corresponding increase in the transcripts of PSY1 and PDS (Cookson et al., 2003), suggesting a regulation of PSY1 activity at the posttranscriptional level.
A Systems Biology Approach Is Required to Uncover the Contribution of hp1 to Fruit Ripening
The emergence of “omics” has highlighted the need for a systems biology approach as a tool for deciphering links between coexpressed genes and pathways and identifying master regulatory points governing a biological response (Saito et al., 2008). The expression of a metabolome phenotype during the development of an organism results from a complex interaction between multiple responses at several levels, from the expression of genes regulating the pathway until the final biosynthesis of metabolites. In order to understand the complexities of the regulation of metabolic networks during fruit development in tomato, it is essential to correlate data from protein, transcript, and metabolite experiments. In view of this, we attempted to correlate the proteome data obtained in this study with the transcript data available at www.ted.bti.cornell.edu and metabolite data from Rohrmann et al. (2011). Out of 72 differentially expressed proteins that varied during fruit development, transcripts for only 18 proteins were available on the TOM1 microarray. Furthermore, a detailed examination of the gene expression data revealed that in the TOM1 microarray, the transcript profiles obtained at 7 DPA of fruit development were used for normalizing all other data at the different stages (7–57 DPA). However, we obtained proteome data only at three different ripening stages, MG, BR, and RR. Since we did not include a time point corresponding to 7 DPA in our study, a direct correlation to this data set was ruled out.
As an alternative, we compared the transcript profiles for five carotenoid pathway genes obtained in this study with the corresponding metabolite data obtained earlier in the wild type and hp1 at the RR stage (Cookson et al., 2003). These transcript-metabolite pairs include PSY1-phytoene, ZIS-ζ-carotene, ZDS-lycopene, CRTISO-lycopene, and CYCB-β-carotene. The above pairs were correlated using the Pearson correlation algorithm with XLSTAT 2012 software (Supplemental Fig. S7A). The correlation coefficient values indicated a negative correlation between transcripts and metabolites in the wild type. Despite the high expression of PSY1 and CRTISO in the wild type, low lycopene levels were observed at the RR stage. On the contrary, a poor correlation was observed in the mutant, where ZDS was up-regulated but CRTISO was down-regulated, yet resulted in higher lycopene levels in hp1 fruits. Similar to our study, a lack of correlation between transcript and metabolite profiles leading to lycopene formation was also observed in the case of DET1 down-regulated tomato lines (Enfissi et al., 2010) and also in PSY1-overexpressing tomato lines (Fraser et al., 2007). At the same time, in DET1 down-regulated tomato lines (Enfissi et al., 2010), a strong correlation between ripening-associated transcripts and specific metabolites, such as organic acids, sugars, and cell wall-related metabolites, was observed, underlining the diversity in the regulation of metabolic networks in fruit ripening. The negative correlation between carotenoid pathway transcripts and carotenoids indicates the importance of posttranscriptional regulation in elevating carotenoid levels in hp1 fruits. This view is supported by the observation that although there was no corresponding increase in the transcripts of PSY1 and PDS, the activity of the PSY1 enzyme in hp1 was up-regulated by 2-fold (Cookson et al., 2003). A similar lack of correlation was also observed between the transcript profiles of CYP97A29, CYP97C11, CRTRB1, and CRTRB2 and the lutein levels in wild-type and hp1 fruits (Supplemental Fig. S7B). Although the correlation coefficient values obtained were higher in hp1 compared with the wild type, they do not indicate a strong correlation between the transcripts and the lutein levels. The absence of correlation between transcript levels and their respective metabolites (lutein and lycopene) indicate that alternative mechanisms like posttranscriptional regulation might be controlling the carotenoid pathway.
Similarly, we correlated the protein (for five differentially expressed proteins) and the corresponding metabolite data across different developmental stages in both the wild type and hp1 (Rohrmann et al., 2011). The protein-metabolite pairs acid invertase (spot 53)-Glc, UDP-Glc pyrophosphorylase (spot 17)-Suc, SAM synthetase (spot 18)-ethylene, E8 protein (spot 41)-ethylene, and malate dehydrogenase (spot 42)-malate were analyzed by XLSTAT (Supplemental Fig. S7, C and D). The resulting correlation coefficient value suggested a negative correlation between proteins and metabolites at MG and BR stages for both the wild type and the mutant. On the contrary, at the RR stage, the correlation coefficient values were very low, indicating weak or no correlation between proteins and metabolites in both the wild type and hp1. On the whole, our results highlight the need for a comprehensive systems approach, including posttranslational regulation, feedback regulation, metabolite turnover, and partitioning between different organelles and/or locations in the cell, to uncover the linkage between metabolic networks regulating fruit ripening (Fernie and Stitt, 2012).
Loss of Function of DDB1 Enhances the Abundance of CHRC
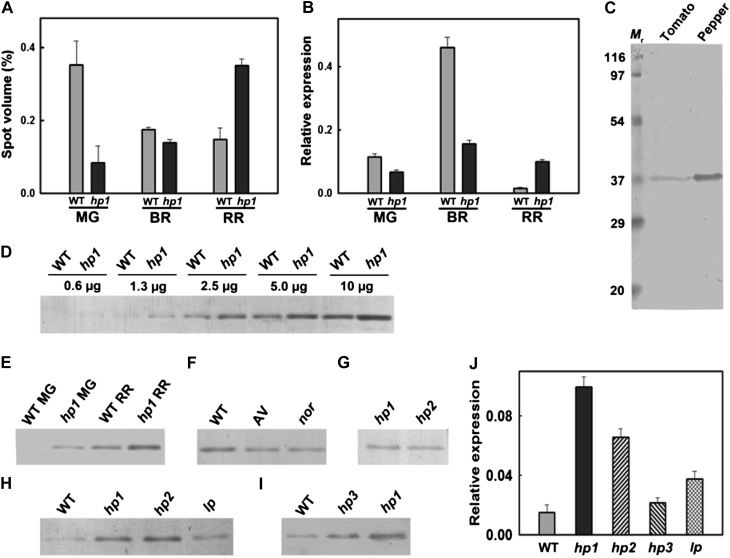
A comparison of the protein profiles in wild-type and hp1 fruits revealed an increase in the abundance of CHRC (spot 66) at the RR stage of hp1, whereas its level declined in the wild type at the same stage. A detailed chronological analysis revealed an interesting pattern of abundance of CHRC, with the wild type and hp1 following dramatically opposite patterns. Based on the intensity of spot volumes, in the wild type, CHRC level was highest at MG, followed by a decline to nearly similar levels in both BR and RR. In contrast, in hp1, CHRC level increased from MG to RR, with maximal level at the RR stage (Fig. 5A). Analysis of CHRC transcript levels (Fig. 5B) corroborated the above pattern of CHRC protein levels, albeit with a few differences. In the MG stage, the wild type exhibited slightly higher CHRC expression (1.7-fold) than hp1. At the BR stage, the wild type showed a 3-fold higher CHRC expression than hp1, but at the RR stage, hp1 showed around 6.35-fold higher expression than the wild type. hp1 showed highest CHRC expression in BR, followed by RR (Fig. 5B). But the protein profile revealed the opposite, with high abundance at the RR stage, suggesting posttranscriptional regulation of CHRC.
Figure 5.
Comparison of CHRC levels in wild-type (WT) and hp1 fruits. Unless specifically indicated, equal amounts of protein were loaded on all the immunoblots. A, Relative spot volume of CHRC protein in wild-type and hp1 fruits at different stages of ripening. B, Real-time PCR quantification of CHRC transcript in wild-type and hp1 fruits at different stages of ripening. C, Immunoblotting of proteins from bell pepper and tomato fruits. The polyclonal antibody raised against bell pepper PAP recognized a protein of 35 ± 2 kD in tomato. Since PAP levels were very high in bell pepper, a reduced amount of protein (0.3 μg) was loaded compared with wild-type tomato fruits (10 μg). D, Comparison of CHRC levels in wild-type and hp1 fruits at the RR stage. Progressively increasing concentrations of proteins were loaded on the gel. The equivalence in the band intensity of hp1 (2.5 μg) with that of the wild type (5.0 μg) indicates that the CHRC level in hp1 is approximately 2-fold higher than in the wild type. E, Comparison of CHRC levels in wild-type and hp1 fruits at MG and RR stages. Ripening stimulated an increase in the level of CHRC protein from MG to RR stages, with higher CHRC levels in hp1 fruits. F, Comparison of CHRC levels in RR fruits of AC, cv Arka vikas (AV), and nor. Note that the nor mutant, although it accumulates very little lycopene, exhibits a normal level of CHRC equal to the other two cultivars. G, Relative abundance of CHRC protein in hp1 and hp2 fruits at the MG stage. H, Relative levels of CHRC in hp1, hp2, and Ip fruits at the RR stage. Note that hp1 and hp2 exhibit similar levels of CHRC, while Ip shows an intermediate level of CHRC between the wild type and hp1 and hp2 fruits. I, Analysis of CHRC levels in RR fruits of the wild type, hp3, and hp1. The order of abundance of CHRC is hp1 > hp3 > wild type. J, Comparison of CHRC transcript levels in RR fruits of hp1, hp2, hp3, and Ip in relation to the wild type. Note that all the mutants exhibit high CHRC transcript level compared with the wild type.
CHRC was first discovered in cucumber corollas (Smirra et al., 1993) and was found to be a lipid-binding protein. A survey of the literature revealed that this protein was extensively studied in bell pepper (Deruère et al., 1994) and was named PAP, and antibodies against this protein were also raised. Comparison of protein sequences of bell pepper PAP and tomato CHRC revealed 89% homology (Supplemental Fig. S8, A and C). Also, tomato CHRC protein showed 100% sequence similarity to the tomato PAP protein sequence, suggesting that these two proteins are similar (Supplemental Fig. S8A). On the contrary, alignment of tomato CHRC/PAP with tomato fibrillin family members (10 members based on BLAST search in the Solanaceae Genome Network [SGN] database; Supplemental Fig. S8B) revealed a high degree of mismatches except for two sequences (Solyc08g005220.2.1 and Solyc08g076480.2.1), where homology of 41% and 50%, respectively, was found (Supplemental Fig. S8B). The divergence in CHRC sequence with fibrillin indicated that while these two proteins are different, the occurrence of PAP domains in fibrillin protein sequences led to their classification as PAP family proteins. Nevertheless, both CHRC and fibrillin appear to play major roles in the storage of carotenoids in chromoplasts (Vainstein et al., 1994; Pozueta-Romero et al., 1997).
Given the strong homology between bell pepper PAP and tomato CHRC, bell pepper PAP antibody recognized a tomato protein of 35 ± 2 kD at a location identical to bell pepper PAP, although the level of this protein was considerably higher in bell pepper than in tomato (Fig. 5C). We considered the possibility of whether bell pepper antibody recognized any protein other than CHRC. A comparison of Mr values of fibrillin proteins showed that among all fibrillins, only one (hit 7; accession no. Solyc08g076480.2.1) had a Mr of 37.48, which was very similar to that of tomato CHRC (Supplemental Fig. S8C). However, this accession showed only 50% homology to tomato CHRC (Supplemental Fig. S8B). Therefore, the probability of this protein being recognized by bell pepper antibody is minimal, as an alignment of peptides obtained after MALDI analysis of tryptic peptides from spot 66 showed 100% alignment with CHRC (Supplemental Fig. S8D) but little homology with fibrillin hit 7 (Supplemental Fig. S8E). The proteome analysis also did not show any other differentially expressed spot belonging to fibrillin hit 7. Therefore, in all probability, the bell pepper antibody recognized tomato CHRC protein on immunoblots. Moreover, the observed increase in the level of CHRC during ripening is also supported by an increase in its transcript level (Fig. 5, A and B).
A comparison of CHRC protein levels in both wild-type and hp1 fruits at the RR stage revealed at least 2-fold higher CHRC level in hp1 than in the wild type (Fig. 5D). At the MG stage, while the CHRC band was discernible in hp1, in the wild type it was below the detection limit. Immunoblotting indicated that hp1 fruits show a higher level of CHRC protein, and its level increased during ripening (Fig. 5E). We considered the possibility that an increase in lycopene level in hp1 may have stimulated an increase in the CHRC level in hp1 fruits. To ascertain this, we examined the CHRC levels in the nor mutant, which accumulates little lycopene, and in cv Arka vikas, an Indian cultivar that has a lycopene level similar to cv Ailsa Craig (AC; the progenitor of the hp1 mutant). Despite the low level of lycopene in nor, it accumulated CHRC protein at a level almost equal to the other two cultivars, Arka vikas and AC (Fig. 5F), thus ruling out the possibility that an increase in lycopene level may have stimulated the CHRC increase.
In another study, Lenucci et al. (2012) observed the enhanced level of a 41.6-kD protein (unidentified) in chromoplasts purified from mesocarp of a high-lycopene-accumulating cultivar of tomato, HLY-18, in comparison with a traditional cultivar. However, the protein profiles of hp1 fruits did not reveal any specific increase in the 41.6-kD protein. The observed variation between our study and that of Lenucci et al. (2012) could also be due to cultivar differences or differences in the tissue used, as we performed protein profiling from the outer pericarp only.
Since mutation in DET1 (hp2; Mustilli et al., 1999) also leads to a high-pigment phenotype, it was of interest to examine whether this mutant too exhibits high levels of CHRC protein. Analysis of CHRC levels from hp2 mutant fruits at both the MG (Fig. 5G) and RR (Fig. 5H) stages revealed an abundance level similar to hp1, suggesting an overlap in the action of hp1 and hp2 loci in regulating CHRC levels. Although DDB1 (hp1) and DET1 (hp2) are diverse loci, the observed overlap may have emanated from their coaction as part of the CUL4-DDB1-DET1 complex (Wang et al., 2008), where loss of function of either of these proteins may contribute to higher CHRC and also pigment levels. Consistent with the enhanced expression of CHRC protein in hp2, Enfissi et al. (2010) showed increases in the number and size of plastoglobules in RR fruits of DET1 down-regulated lines. Similarly, it was reported earlier that the chloroplast-to-chromoplast transition in tomato is accompanied by an increase in the number and size of plastoglobules (Harris and Spurr, 1969). Taken together with the above observations, the increased levels of CHRC protein in both hp1 and hp2 fruits may be linked to an increase in the abundance of plastoglobules, where higher CHRC levels in chromoplasts assist in the sequestration and stabilization of carotenoids, thus boosting their levels.
The specific role of CHRC in chromoplasts is also supported by the observation that a down-regulation of tomato CHRC using an RNAi strategy led to a 30% reduction of total carotenoids in tomato flowers (Leitner-Dagan et al., 2006). Since RNAi suppression of CHRC reduced the carotenoid level, overexpression should increase its level. Although there are no reports of CHRC overexpression in the literature, overexpression of a related protein, fibrillin from pepper, which shares the PAP domain with CHRC in tomato, resulted in almost a doubling of carotenoid levels and carotenoid-derived aroma compounds in tomato fruits (Simkin et al., 2007). Evidence is emerging for additional roles for plastoglobulin proteins like CHRC, which not only participate in the sequestration of carotenoids but also appear to influence chromoplastogenesis as well as carotenoid metabolism (Bréhélin and Kessler, 2008; Li et al., 2012). Taken together, the up-regulation of CHRC in chromoplasts supports the role of this protein in chromoplast-specific carotenoid storage (Leitner-Dagan et al., 2006).
Similar to hp1 and hp2 mutants, the Ip mutant exhibits high lycopene content in fruits and also has similar increases in the number and size of chloroplasts, but the gene responsible for this phenotype is not yet identified (Jones, 2000; Lavi et al., 2009). Kendrick et al. (1997) reported that Ip displayed photomorphogenic responses intermediate between hp1 and normal genotypes. In view of its high-pigment phenotype, we examined whether Ip also exhibits a high level of CHRC protein in ripened fruits. Western analysis revealed that, similar to hp1 and hp2, Ip also up-regulates CHRC protein but has levels intermediate between those in the wild type and hp1 and hp2 fruits (Fig. 5H). Consistent with the intermediate CHRC level, the RR fruits of the Ip mutant also showed intermediate levels of lycopene and β-carotene between those of the wild type and hp1 (Jones, 2000). This suggests that increases in chloroplast number, size, and lycopene level in Ip are likely regulated by a mechanism similar to that in high-pigment mutants and thus shows an intermediate increase in CHRC level.
Since a deficiency in ABA could also lead to a high-pigment phenotype (hp3), it was of interest to examine the CHRC levels in this mutant. Monitoring of CHRC levels by western blotting in the RR fruits of the hp3 mutant revealed a lower abundance of CHRC compared with that in hp1; nonetheless, it was higher than that of the wild type (Fig. 5I). This result is corroborated by the observation that the deficiency of ABA in hp3 is accompanied by an increase in plastid compartment size due to increased plastid division (Galpaz et al., 2008). This could result in a higher storage capacity for the carotenoids similar to hp1 and hp2 mutants. Although hp3 is not a member of the CUL4-DDB1-DET1 complex (Wang et al., 2008), this mutation may affect the carotenoid levels through feedback regulation (Galpaz et al., 2008).
In view of the observed differences in the CHRC abundance in hp1, hp2, hp3, and Ip, it was of interest to examine the transcript levels of CHRC in the RR fruits of these four mutants (Fig. 5J). In comparison with the wild type, hp1 showed the highest expression of CHRC (6.35-fold), followed by hp2 (4.12-fold), Ip (2.37-fold), and hp3 (1.37-fold; Fig. 5J). These transcript profiles appear to be consistent with the protein profiles determined by western blotting, except in the case of hp2. Nevertheless, all four mutants exhibited high transcript as well as protein levels for CHRC compared with the wild type, which is consistent with their high-pigment phenotype, where this protein might have a major role in sequestering the elevated carotenoids.
Both Carotenoids and CHRC Are Present in Plastoglobules
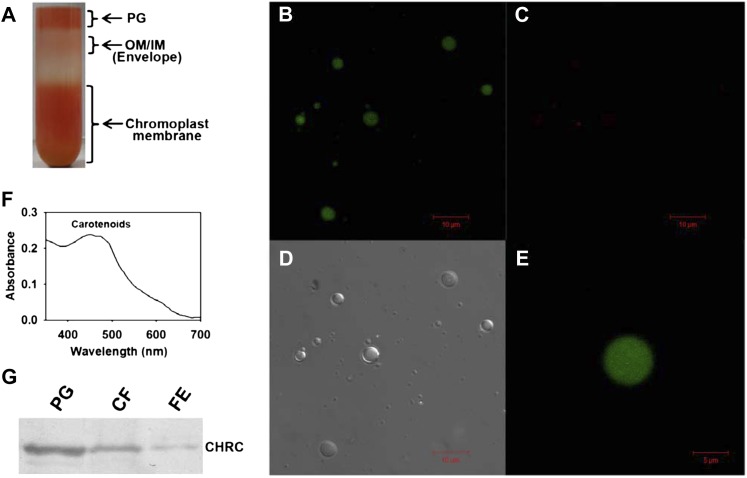
The transition from chloroplasts to chromoplasts during ripening is accompanied by an increase in osmiophilic bodies associated with the thylakoids named plastoglobules. The chromoplasts of the hp1 mutant show a 1.3-fold increase in size compared with the wild type (Cookson et al., 2003). Likewise, down-regulation of DET1 leads to an increase in plastoglobule number in the chromoplasts (Enfissi et al., 2010). Biochemical analysis of plastoglobules revealed that these are enriched in lipids and also contain several proteins, including enzymes participating in carotenoid metabolism (Austin et al., 2006; Bréhélin and Kessler, 2008). Based on the enrichment of lipids in the plastoglobules, it is assumed that carotenoids are also present in them. Since CHRC has been implicated as a protein that assists in the sequestration and storage of carotenoids, and it may be localized in plastoglobules, we examined the presence of carotenoids and CHRC in the isolated plastoglobules. We purified the plastoglobules from RR fruits of tomato by taking advantage of their lower density compared with other membranous fractions on Suc density gradients (Fig. 6A) by adapting the procedures of Besagni et al. (2011) and Lundquist et al. (2012). The purity of the plastoglobule fraction and fluorescence emission from globuli were examined by confocal microscopy (Fig. 6, B–D), which revealed the presence of intact globules (Fig. 6D). Moreover, the purified plastoglobules exhibited a bright green autofluorescence on exposure to a 488-nm argon laser, indicating the enrichment of carotenoids (emission in the range of 500–510 nm; Fig. 6, B and E), whereas they exhibited only a trace level of red autofluorescence (740–750 nm; Fig. 6C), indicating a near absence of chlorophylls. Since the plastoglobules were isolated from RR fruits, the absence of chlorophyll was expected, akin to an earlier observation for the isolated chromoplasts (Egea et al., 2011). In addition, carotenoids were extracted from the plastoglobule fraction, and the absorbance spectrum of the extract showed a distinct peak in the 440- to 475-nm range, which is characteristic of carotenoids (Fig. 6F). From the foregoing observations, it is evident that the plastoglobules isolated from RR fruits are enriched in carotenoids.
Figure 6.
Confocal microscopy imaging and western blotting of isolated plastoglobules. A, Suc density gradient purification of plastoglobules (PG). Note that the distinct upper layer of purified plastoglobules is well separated from the underlying chromoplast membranous fractions. OM/IM indicate outer and inner membranes of chromoplasts. B to D, Purified plastoglobules were visualized using a confocal microscope. On excitation with a 488-nm laser, the plastoglobules exhibit strong green fluorescence (emission between 500 and 510 nm; B) but show negligible red fluorescence (emission between 740 and 750 nm; C). D shows a bright-field image of the plastoglobules. E, Enlarged view of a single plastoglobule. Note that the green fluorescence emitted from plastoglobules is uniformly distributed. F, Absorption spectrum of the extract obtained from purified plastoglobules showing characteristic carotenoid peaks at 440 to 475 nm. G, Western analysis of CHRC levels in the purified plastoglobule fraction (PG; 0.33 μg) in comparison with the chromoplast membrane fraction (CF [the chromoplast fraction prior to sonication]; 2.5 μg) and total protein from the outer pericarp of RR fruits of hp1 (FE, fruit extract; 2.5 μg). Note that the plastoglobule fraction shows enrichment of CHRC protein. [See online article for color version of this figure.]
To ascertain the presence of CHRC in the purified plastoglobules, the proteins were extracted from the plastoglobule fraction and resolved by SDS-PAGE, followed by western blotting. Figure 6G shows the presence of CHRC protein in purified plastoglobules. The comparison of CHRC levels in plastoglobules with other fractions indicated a substantial enrichment of the CHRC protein in the plastoglobules over the chromoplast membrane fraction or the initial fruit extract. The presence of CHRC in the chromoplast membrane fraction (prior to its sonication to release the plastoglobules) is also in conformity with an earlier transmission electron microscopy observation that the plastoglobules are associated with chromoplast membranes in the ripe fruits (Harris and Spurr, 1969).
The up-regulation of CHRC in the fruits of four diverse mutants sharing a high-carotenoid phenotype suggests an interrelationship between carotenoid accumulation and CHRC levels. Several studies have indicated that the CHRC and related proteins, fibrillins, enhance the sequestration of carotenoids into plastoglobules (Bréhélin and Kessler, 2008). The importance of carotenoid sequestration and storage is highlighted in studies where attempts to enhance the carotenoid levels were mitigated by the absence of a proper sink. In potato (Solanum tuberosum) tubers, a massive loss of carotenoids occurs during long-term storage due to enzymatic or nonenzymatic oxidation of carotenoids. However, such loss can be prevented by expression of the cauliflower Or gene, which stimulates the formation of plastoglobule-like structures storing carotenoids in potato tubers. The sequestration of the carotenoids in the above structures prevents the oxidation of carotenoids and ensures the continued formation of carotenoids during the storage of tubers (Li et al., 2012). It is likely that higher CHRC levels function to abet the storage of carotenoids and protect them from degradation in high-pigment mutants. However, it remains to be investigated whether the up-regulation of CHRC by genetic or transgenic means would improve the storage capacity of carotenoids in tomato fruits.
In summary, our results suggest that the loss of function of DDB1, DET1, Zeaxanthin Epoxidase, and Ip in tomato fruits appears to up-regulate CHRC protein levels in a manner that is currently unknown. More studies are needed to determine how these diverse alleles regulate CHRC levels in tomato fruits.
CONCLUSION
In this study, using a proteomic approach, we examined the influence of DDB1 on tomato fruit ripening. At least 72 proteins were differentially expressed in both the wild type and hp1 whose expression was fine-tuned in response to developmental cues during fruit ripening. We found an additional role for DDB1 and DET1 in up-regulating the level of CHRC by 2-fold by an unidentified mechanism. The hp3 mutant, which is deficient in zeaxanthin epoxidase, also exhibits a higher CHRC level, albeit at a lower level than in hp1 and hp2 mutants. This is consistent with its enlarged plastid compartment size and elevated carotenoid content. The intermediate level of CHRC in Ip appears to be related to its intermediate photomorphogenic phenotype and carotenoid levels between the wild type and hp1 and hp2 mutants. Interestingly, transcript profiling of carotenoid pathway genes revealed an overall up-regulation of the pathway in the wild type rather than in hp1, except the desaturation steps catalyzed by PDS and ZDS, suggesting posttranscriptional control of the pathway. Moreover, none of the carotenoid biosynthetic enzymes were detected in our proteomics studies, either because their levels were below the detection limits or they were regulated at the translation level. However, the transcript levels of cytochrome P450 carotenoid β- and ε-hydroxylases responsible for the conversion of α-carotene to lutein were up-regulated in the hp1 mutant but not in the wild type, consistent with the elevated lutein levels found in the mutant. Compared with the wild type, transcript levels for ABA biosynthetic genes were down-regulated in hp1, suggesting a relation between low ABA levels and increases in chloroplast number and size, as reported previously in the hp3 mutant. A poor correlation was observed between transcript, protein, and metabolite levels, reinforcing the need for systems biology approaches for complete understanding of the metabolite networks. In summary, our study revealed that CHRC likely regulates carotenoid sequestration and storage, which seems to be important for the enhanced accumulation of carotenoids in high-pigment tomato fruits.
MATERIALS AND METHODS
Plant Material and Experimental Design
Fruits of tomato (Solanum lycopersicum) photomorphogenic mutant hp1 and its isogenic wild type AC (Srinivas et al., 2004) were used for proteomic analysis in this study. The tomato accessions AC (LA2838), hp1 (LA3538), hp2dg (LA2451), Ip (LA1500), hp3 (3-343), and nor (LA3770) were obtained from the Tomato Genetics Resource Center (University of California, Davis). Tomato plants were grown under a natural photoperiod in the greenhouse at 28°C ± 1°C during the day and ambient temperature at night. Plants of both the wild type and hp1 were grown in three randomized blocks (each block constitutes one biological replicate), with each block consisting of three rows, with five plants each for the wild type and hp1 per row (Supplemental Fig. S1). To minimize variation, fruits developed only on the first and second truss were used for experiments. The flowers were tagged at the time of anthesis for chronological monitoring of fruit developmental stages, and different ripening stages were identified: MG (35 DPA), BR (40 DPA), and RR (49 DPA; Fig. 1A). From each block (biological replicate), 30 to 35 fruits per ripening stage per genotype were harvested, and the outer pericarp was immediately isolated as described below, pooled separately at every ripening stage for both the wild type and hp1 (Supplemental Fig. S1), flash frozen in liquid nitrogen, and stored at −80°C until use.
Plant Material for Protein, Transcript, and Carotenoid Analyses
Fruits of the wild type and hp1 were collected at different ripening stages as described above. Since a temporal difference in the synthesis of carotenoids in the outer pericarp and inner pericarp was observed in the hp1 mutant (Y. Sreelakshmi and R. Sharma, unpublished data), we initiated an independent study on both outer and inner pericarp. The outer pericarp is the outer layer of the fruit including the skin, as indicated in Supplemental Figure S2; its thickness is measured with a Vernier calipers and varies from 0.5 ± 0.1 mm in MG/BR to 0.3 ± 0.1 mm in RR (Supplemental Fig. S2). From fruits at different ripening stages and from all biological replicates, the outer pericarp was carefully excised using a scalpel, pooled, flash frozen in liquid nitrogen, and stored at −80°C until further use. Unless specifically indicated, this tissue was used for proteome, transcript, and carotenoid analyses.
Protein Extraction
Proteins were extracted from the outer pericarp of fruit tissue using a previously described protocol (Isaacson et al., 2006) with slight modifications. About 1 g of tissue was homogenized in liquid nitrogen and suspended in 7 mL of extraction buffer containing 0.7 m Suc, 0.1 m KCl, 0.5 m Tris, pH 7.5, 50 mm EDTA, 50 mm dithiothreitol (DTT), 1 mm phenylmethylsulfonyl fluoride (PMSF), and 25 µL of protease inhibitor cocktail (Sigma-Aldrich). To this, an equal volume of Tris-saturated phenol was added, and the sample was mixed thoroughly by shaking at 4°C for 30 min. The mixture was centrifuged at 20,000g for 30 min at 4°C. The upper phenolic phase was collected and reextracted two more times as described above. The protein in the phenolic phase was precipitated at −80°C by adding 5 volumes of 0.1 m ammonium acetate containing 50 mm DTT, and the protein was pelleted by centrifugation at 26,200g for 30 min at 4°C. The protein pellet was washed twice with methanol containing 10 mm DTT and once with acetone with 10 mm DTT and finally dissolved in lysis buffer (7 m urea, 2 m thiourea, and 4% [w/v] CHAPS). The protein was quantified using the Amido Black method (Goldring and Ravaioli, 1996). For each developmental stage, at least three biological replicates were used in three independent extractions.
Two-Dimensional Gel Electrophoresis
Proteins (500 µg) were dissolved in 250 μL of rehydration buffer (7 m urea, 2 m thiourea, 4% [w/v] CHAPS, 50 mm DTT, and 1% [v/v]) ampholytes [pH 4–7]) and applied to 13-cm Immobiline dry strips, pH 4 to 7 (GE Healthcare) by passive rehydration overnight. Isoelectric focusing was carried out at 20°C in Ettan IPGPhor 3 (GE Healthcare), and the settings used were as follows: 150 V for 1 h, 250 V for 1 h, 500 V for 1 h, 1,000 V for 30 min, 8,000 V for a total of 40,000 V h, with a maximum current of 50 μA per strip. After isoelectric focusing, the strips were equilibrated first in buffer containing 6 m urea, 0.075 m Tris, 2% (w/v) SDS, 30% (v/v) glycerol, and 2% (w/v) DTT for 20 min. In the second step, 2.5% (w/v) iodoacetamide was used instead of DTT in the same buffer for another 20 min. Then the strips were layered on 13% (w/v) acrylamide gels and sealed with agarose (0.65% [w/v] in 1× electrode buffer) for separation in the second dimension using the Ruby SE600 electrophoresis setup (GE Healthcare). SDS-PAGE was carried out at 20 mA for an initial 1 h and at 30 mA for the next 3.5 h or until bromphenol blue dye reached the end of the gel. Following SDS-PAGE, the gels were rinsed with MilliQ water two or three times, fixed for 1 h in methanol:water:acetic acid (5:4:1, v/v/v), and stained for 16 to 18 h in colloidal Coomassie stain (0.08% [w/v] Coomassie Brilliant Blue G-250, 20% [v/v] ethanol, 8% [w/v] ammonium sulfate, and 0.35 m phosphoric acid). Gels were destained for 24 h in deionized water with several changes on a shaker, and after completion of destaining, gels were stored in 20% (v/v) methanol.
Image Visualization and Data Analysis
A total of 18 gels (two genotypes, three ripening stages, and three biological replicates at every ripening stage) were scanned using Image Scanner (GE Healthcare), and images were analyzed with Image Master 2D Platinum Software 6.0 (GE Healthcare) according to the manufacturer’s instructions. About 800 protein spots were detected and matched. Spots that matched automatically on all the gels in both the wild type and hp1 were also manually verified. The percentage spot volumes were statistically analyzed using one-way ANOVA (Sigmaplot version 11.0), and the spots showing 2-fold differences (up- and down-regulation in at least one of the stages during ripening) in all three biological replicates with P ≤ 0.05 were selected for identification by mass spectrometry. The power of the test was determined as 0.9 to 0.99 using one-way ANOVA from the percentage spot volumes.
Trypsin Digestion
The spots selected for further identification by MALDI-TOF/TOF analysis were manually excised and digested with trypsin (sequencing grade; Promega) as described previously (Shevchenko et al., 2006). Eluted peptide fragments were concentrated in a speed vacuum concentrator (Thermo Scientific) and were reconstituted in 5 μL of 1:1 (v/v) acetonitrile and 1% (v/v) trifluoroaetic acid.
Mass Spectrometry
Peptide fragments (2 μL) obtained after trypsin digestion was mixed with 2 μL of α-cyano-4-hydroxycinnamic acid matrix (freshly made in 50% [v/v] acetonitrile and 1% [v/v] trifluoroacetic acid [1:1, v/v]), spotted onto a MALDI target plate, and dried. Tandem mass spectrometry (MS/MS) analyses were carried out using the AutoFlex III smartbeam MALDI-TOF mass spectrometer (Bruker Daltonics). TOF spectra within the range of 500 to 4,000 mass-to-charge ratio (m/z) were obtained using Flex Control software 3.0 (Bruker Daltonics) in the reflector mode with a laser intensity of 6,000 V and an average of 2,000 shots per spectrum. The mass spectra were calibrated with external standards with masses ranging from 757.399 to 3,147.4710 m/z. Trypsin autolysis peaks were used for internal calibration, and they are as follows: trypsin (108–115; m/z 842.509400), trypsin (98–107; m/z 1,045.563700), trypsin (58–77; m/z 2,211.104000), trypsin (78–97; m/z 2,283.180200), trypsin (58–72; m/z 1,713.808400), trypsin (62–77; m/z1,774.897500), and trypsin (58–76; m/z 2,083.009600). A range of two to 10 precursor ions were selected for MS/MS analysis. Monoisotopic mass was attributed using Bruker’s Sophisticated Numerical Annotation Procedure by collecting data from an average of 5,000 laser shots at a threshold intensity of 2,500 over the mass range. Peak lists from mass spectrometry and MS/MS analyses were generated using Flex Analysis software 3.0 (Bruker Daltonics). Proteins were identified using MASCOT version 2.4.01 (http://www.matrixscience.com; Matrix Science) with the help of Biotools software (Bruker Daltonics). The following parameters were set for searches against target databases such as National Center for Biotechnology Information nonredundant (downloaded on October 1, 2012; 20,543,454 sequences, 7,050,788,919 residues), SwissProt (downloaded on October 1, 2012; 537,505 sequences, 190,795,142 residues), and Plant EST (downloaded on October 1, 2012; 157,998,786 sequences, 27,903,646,510 residues): (1) tryptic cleavage with a maximum of two missed cleavages; (2) mass tolerance of 30 to 100 ppm; (3) peptide mass tolerance of 0.1 to 1.0 D; (4) a minimum of two peptides matching the protein; (5) fixed and variable modifications like carbamidomethylation of Cys and Met oxidation, respectively. In addition, we also searched in SGN (offline from Mascot) at the link ftp://ftp.solgenomics.net/genomes/Solanum_lycopersicum/annotation/ITAG2.3_release/ITAG2.3_proteins.fasta. The false discovery rate for the statistically significant proteins was found to be 5% using the Benjamini and Hochberg method (Benjamini and Hochberg, 1995; Eravci et al., 2009).
Hierarchical Clustering
PermutMatrix software version 1.9.3 was used to generate hierarchical clustering (Pearson’s algorithm; Caraux and Pinloche, 2005). The data file with all identified proteins together with their expression profiles were given as input, and a cluster map along with the protein expression profile was generated.
RNA Isolation and Quantitative Real-Time PCR
RNA was isolated from the outer pericarp of wild-type and hp1 fruits at three ripening stages, MG, BR, and RR (each with three biological replicates), using TRI reagent (Sigma-Aldrich; T9424) according to the manufacturer’s protocol. The isolated RNA was treated with RQ1 RNase Free DNase (Promega) according to the manufacturer’s recommendations to remove DNA. Reverse transcription was performed with 2 µg of RNA using the SuperScript III RT First Strand Synthesis Kit (Invitrogen) according to the manufacturer’s protocol. Quantitative real-time PCR was performed with about 4 ng of total RNA in a 10-µL reaction volume using SYBR Green PCR Master Mix (Takara) on a 7300 Fast Real Time PCR System (Applied Biosystems; http://www.appliedbiosystems.com/). Gene-specific primers were designed from the sequences obtained from the SGN database using Primer 3 software and are represented in Supplemental Table S3. The relative fold differences for each sample were determined by normalizing cycle threshold (Ct) values of each gene to the mean expression of both β-Actin and Ubiquitin genes and were calibrated using the equation 2−ΔCt, which gives the relative expression of each gene. Mean ± se (Sigmaplot version 11.0) cycle threshold values obtained from three biological replicates were used for the calculation of expression levels.
Western Blotting
The outer pericarp of tomato fruit (from each genotype, three biological replicates were used) was homogenized in extraction buffer (200 mm Tris, pH 8.0, 150 mm NaCl, 2.5 μL of protease inhibitor cocktail per 1 mL of extraction buffer [Sigma], 1 mm PMSF, and 0.125% [v/v] Triton X-100) and centrifuged at 13,000g at 4°C for 15 min, and the supernatant was collected. The protein content was quantified by the method of Bradford (1976). After 12% SDS-PAGE (Laemmli, 1970), proteins were electroblotted onto a polyvinylidene difluoride membrane (Millipore; Towbin et al., 1979). The blots were incubated with a 1:2,500 dilution of antibodies raised against bell pepper (Capsicum annuum) PAP protein followed by anti-rabbit IgG alkaline phosphatase conjugate (1:80,000 dilution; Sigma-Aldrich). The blots were finally developed using chromogenic substrates for alkaline phosphatase, 66 µL of nitroblue tetrazolium (50 mg mL−1) and 33 µL of 5-bromo-4-chloro-3-indolyl phosphate (50 mg mL−1), in 10 mL of buffer (100 mm Tris-HCl, 100 mm NaCl, and 5 mm MgCl2, pH 9.5; Sreelakshmi, 2000).
Determination of Ethylene Evolution
To measure ethylene evolution after harvesting, fruits were enclosed in air-tight boxes (250 mL). After 4 h, 1 mL of head-space gas was removed from the boxes and injected into a Porapak T column connected to a gas chromatograph (GC-17A; Shimadzu) as described previously (Santisree et al., 2011). For every sample, ethylene in the head-space gas was measured at least three times using a minimum of three biological replicates at each ripening stage.
Estimation of Carotenoids
Carotenoids were extracted from fruits and estimated using a previously described procedure (Barba et al., 2006). Carotenoids were separated and analyzed by HPLC (Shimadzu) on a C-18 column (250 × 4.6 mm, 5 μm; Phenomenex) using a 60-min isocratic gradient of methanol:acetonitrile (90:10, v/v) plus triethylamine (9 µm) as mobile phase. Carotenoids were identified and quantified based on the retention times and peak areas of the standards (lycopene [L-9879] from tomato, β-carotene [C-4582], and lutein [X-6250]; Sigma-Aldrich), and β-carotene, lutein, and lycopene contents were determined by monitoring A475 (Barba et al., 2006) using a UV-visible detector (SPD 20 A; Shimadzu).
Isolation of Plastoglobules and Western Blotting
Plastoglobules were isolated from tomato fruits at the RR stage using a method adapted from two protocols: the procedure of Besagni et al. (2011) and that of Lundquist et al. (2012). About 400 g of fruit tissue was ground in 400 mL of cold homogenization buffer (450 mm sorbitol, 20 mm Tricine-KOH, pH 8.4, 10 mm EDTA, 10 mm NaHCO3, 1 mm MnCl2, 5 mm sodium-ascorbate, 0.05% [w/v] bovine serum albumin fraction V, and 1 mm PMSF). The homogenate was filtered immediately through four layers of 20-μm Miracloth, and the filtrate was distributed equally into eight 50-mL tubes and then centrifuged for 10 min at 1,200g. Each chromoplast membrane pellet was gently resuspended in 4 to 5 mL of 0.6 m Suc/1× TrE (10× stock solution has 50 mm Tricine- KOH, pH 7.5, 2 mm EDTA, and 2 mm DTT), and these were pooled into a 50-mL Falcon tube. The membrane fraction was then sonicated five times for 40 s each on ice at an amplitude of 23% with a 3-min interval between sonications (Sonics and Materials; Vibracell VCX 130PB). The samples were disbursed in 5- to 6-mL aliquots into the required number of UltraClear SW28 tubes and then carefully overlaid with the Suc/1× TrE solutions in the following order: 6 mL of 38% (w/v) Suc, 6 mL of 20% (w/v) Suc, 4 mL of 15% (w/v) Suc, and finally, 8 mL of 5% (w/v) Suc to the top of the tube. These gradients were centrifuged for 16 h in a Beckman Coulter Optima L-90K centrifuge at 100,000g. The plastoglobule-rich fraction was then collected from the top layer.
The plastoglobule fraction thus obtained was dialyzed against 1× TrE buffer using a 3500 MWCO membrane (Thermo Scientific), and the protein was precipitated from the plastoglobule fraction using methanol-chloroform as essentially described by Besagni et al. (2011). After precipitation, the protein was resuspended in sample buffer, boiled, and then separated by 12% SDS-PAGE followed by western blotting with CHRC antibody as described above.
Confocal Microscopy of Isolated Plastoglobules
Confocal images of purified plastoglobules were obtained with a laser scanning confocal microscope (Carl Zeiss; LSM 710 NLO) coupled to an inverted microscope (with non-descanned detector multiphoton laser). Freshly isolated plastoglobules were placed on a glass slide and covered with a coverslip. The carotenoid autofluorescence from the isolated plastoglobules was examined by measuring the emission between 500 and 510 nm (green), and chlorophyll emission was measured between 740 and 750 nm (red) after excitation with a laser at 488 nm.
In addition, the absorption spectrum for carotenoids from the plastoglobule fraction was obtained as follows. To about 100 μL of freshly isolated plastoglobule fraction, an equal volume of n-hexane was added and mixed thoroughly. The mixture was centrifuged at 12,000g for 5 min, and the supernatant was dried in a SpeedVac. After drying, the sample was dissolved in 6 μL of n-hexane, and the absorption spectra for 1.5 μL were recorded three times in a Nanodrop spectrophotometer (ND1000; Thermo Scientific) from 350 to 700 nm.
The following genes (with accession numbers) were examined in this study: DXS, Solyc01g067890.2.1; IDI, Solyc05g055760.2.1; GGDS2, Solyc04g079960.1.1; PSY1, Solyc03g031860.2.1; PSY2, Solyc02g081330.2.1; PDS, Solyc03g123760.2.1; ZIS, AK326152.1; CRTISO, Solyc10g081650.1.1; ZDS, Solyc01g097810.2.1; LCYB1, Solyc04g040190.1.1; LCYB2, Solyc10g079480.1.1; CYCB, Solyc06g074240.1.1; LCYE, Solyc12g008980.1.1; CRTRB2, Solyc03g007960.2.1; ZEP, Solyc02g090890.2.1; VDE, Solyc04g050930.2.1; NCED1, Solyc07g056570.1.1; CYP97A29, Solyc04g051190.2.1; CYP97C11, Solyc10g083790.1.1; CRTRB1, Solyc06g036260.2.1; β-ACTIN, FJ532351.1; UBIQUITIN3, X58253.1; and CHRC, Solyc02g081170.2.1.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Experimental design for proteomic study.
Supplemental Figure S2. Visualization of the outer pericarp in tomato fruits.
Supplemental Figure S3. Functional classification of differentially expressed proteins isolated from tomato fruits.
Supplemental Figure S4. Schematic representation of metabolic pathways and abundance of proteins identified in wild-type and mutant fruits.
Supplemental Figure S5. Schematic representation of the electron transport chain operating in the thylakoid membranes of chloroplasts.
Supplemental Figure S6. Hypothetical model representing the induction of heat-shock proteins during fruit ripening.
Supplemental Figure S7. Comparison of protein, transcript, and metabolite levels in wild-type and hp1 fruits during ripening.
Supplemental Figure S8. Homology between sequences of different chromoplast-associated proteins.
Supplemental Table S1. Identification of differentially expressed proteins in both wild-type and hp1 fruits by MALDI-TOF/TOF analysis.
Supplemental Table S2. The differentially expressed proteins (Supplemental Table S1) were compared with other studies to identify similarities and differences in the proteome complement.
Supplemental Table S3. Primers used in this study for quantitative real-time PCR analysis.
Acknowledgments
We thank Dr. Mireille Faurobert and Dr. Mark Lohse for helpful discussions. We also thank Dr. Guy Houlne for his generous gift of bell pepper PAP antibody. We thank Ms. Monica Kannan for her help with MALDI. We thank Ms. Nalini Manthapuram for her help with confocal microscopy. We thank the Tomato Genetics Resource Centre for providing tomato AC, hp1, hp2, hp3, Ip, and nor seeds and the Indian Institute of Horticultural Research for tomato ‘Arka vikas’ seeds. We are also thankful to the Department of Biotechnology, Government of India Centre for Teaching and Research in Biology and Biotechnology, the Department of Science and Technology, Government of India Fund for Improvement of Science and Technology Infrastructure, the University Grants Commission, and the New Delhi Centre for Advanced Studies for providing necessary equipment support for this work.
Glossary
- RNAi
RNA interference
- MG
mature green
- RR
red-ripe
- BR
breaker
- MALDI
matrix-assisted laser-desorption ionization
- TOF
time of flight
- ABA
abscisic acid
- SGN
Solanaceae Genome Network
- AC
cv Ailsa Craig
- DTT
dithiothreitol
- PMSF
phenylmethylsulfonyl fluoride
- MS/MS
tandem mass spectrometry
- m/z
mass-to-charge ratio
References
- Austin JR, II, Frost E, Vidi PA, Kessler F, Staehelin LA. (2006) Plastoglobules are lipoprotein subcompartments of the chloroplast that are permanently coupled to thylakoid membranes and contain biosynthetic enzymes. Plant Cell 18: 1693–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azari R, Tadmor Y, Meir A, Reuveni M, Evenor D, Nahon S, Shlomo H, Chen L, Levin I. (2010) Light signaling genes and their manipulation towards modulation of phytonutrient content in tomato fruits. Biotechnol Adv 28: 108–118 [DOI] [PubMed] [Google Scholar]
- Barba AIO, Hurtado MC, Mata MCS, Ruiz VF, de Tejada MLS. (2006) Application of a UV-vis detection-HPLC method for a rapid determination of lycopene and β-carotene in vegetables. Food Chem 95: 328–336 [Google Scholar]
- Barsan C, Sanchez-Bel P, Rombaldi C, Egea I, Rossignol M, Kuntz M, Zouine M, Latché A, Bouzayen M, Pech JC. (2010) Characteristics of the tomato chromoplast revealed by proteomic analysis. J Exp Bot 61: 2413–2431 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 57: 289–300 [Google Scholar]
- Bernhardt A, Lechner E, Hano P, Schade V, Dieterle M, Anders M, Dubin MJ, Benvenuto G, Bowler C, Genschik P, et al. (2006) CUL4 associates with DDB1 and DET1 and its downregulation affects diverse aspects of development in Arabidopsis thaliana. Plant J 47: 591–603 [DOI] [PubMed] [Google Scholar]
- Bernhardt A, Mooney S, Hellmann H. (2010) Arabidopsis DDB1a and DDB1b are critical for embryo development. Planta 232: 555–566 [DOI] [PubMed] [Google Scholar]
- Besagni C, Piller LE, Brehelin C (2011) Preparation of plastoglobules from Arabidopsis plastids for proteomic analysis and other studies. Methods Mol Biol 775: 223–239 [DOI] [PubMed] [Google Scholar]
- Bian W, Barsan C, Egea I, Purgatto E, Chervin C, Zouine M, Latché A, Bouzayen M, Pech JC. (2011) Metabolic and molecular events occurring during chromoplast biogenesis. J Bot 2011: 1–13 [Google Scholar]
- Bianco L, Lopez L, Scalone AG, Di Carli M, Desiderio A, Benvenuto E, Perrotta G. (2009) Strawberry proteome characterization and its regulation during fruit ripening and in different genotypes. J Proteomics 72: 586–607 [DOI] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Bréhélin C, Kessler F. (2008) The plastoglobule: a bag full of lipid biochemistry tricks. Photochem Photobiol 84: 1388–1394 [DOI] [PubMed] [Google Scholar]
- Caraux G, Pinloche S. (2005) PermutMatrix: a graphical environment to arrange gene expression profiles in optimal linear order. Bioinformatics 21: 1280–1281 [DOI] [PubMed] [Google Scholar]
- Carrari F, Fernie AR. (2006) Metabolic regulation underlying tomato fruit development. J Exp Bot 57: 1883–1897 [DOI] [PubMed] [Google Scholar]
- Carvalho RF, Aidar ST, Azevedo RA, Dodd IC, Peres LE. (2011) Enhanced transpiration rate in the high pigment 1 tomato mutant and its physiological significance. Plant Biol (Stuttg) 13: 546–550 [DOI] [PubMed] [Google Scholar]
- Chen H, Huang X, Gusmaroli G, Terzaghi W, Lau OS, Yanagawa Y, Zhang Y, Li J, Lee JH, Zhu D, et al. (2010) Arabidopsis CULLIN4-damaged DNA binding protein 1 interacts with CONSTITUTIVELY PHOTOMORPHOGENIC1-SUPPRESSOR OF PHYA complexes to regulate photomorphogenesis and flowering time. Plant Cell 22: 108–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cookson PJ, Kiano JW, Shipton CA, Fraser PD, Romer S, Schuch W, Bramley PM, Pyke KA. (2003) Increases in cell elongation, plastid compartment size and phytoene synthase activity underlie the phenotype of the high pigment-1 mutant of tomato. Planta 217: 896–903 [DOI] [PubMed] [Google Scholar]
- Davuluri GR, van Tuinen A, Fraser PD, Manfredonia A, Newman R, Burgess D, Brummell DA, King SR, Palys J, Uhlig J, et al. (2005) Fruit-specific RNAi-mediated suppression of DET1 enhances carotenoid and flavonoid content in tomatoes. Nat Biotechnol 23: 890–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deruère J, Bouvier F, Steppuhn J, Klein A, Camara B, Kuntz M. (1994) Structure and expression of two plant genes encoding chromoplast-specific proteins: occurrence of partially spliced transcripts. Biochem Biophys Res Commun 199: 1144–1150 [DOI] [PubMed] [Google Scholar]
- Egea I, Bian W, Barsan C, Jauneau A, Pech JC, Latché A, Li Z, Chervin C. (2011) Chloroplast to chromoplast transition in tomato fruit: spectral confocal microscopy analyses of carotenoids and chlorophylls in isolated plastids and time-lapse recording on intact live tissue. Ann Bot (Lond) 108: 291–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enfissi EM, Barneche F, Ahmed I, Lichtlé C, Gerrish C, McQuinn RP, Giovannoni JJ, Lopez-Juez E, Bowler C, Bramley PM, et al. (2010) Integrative transcript and metabolite analysis of nutritionally enhanced DE-ETIOLATED1 downregulated tomato fruit. Plant Cell 22: 1190–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eravci M, Mansmann U, Broedel O, Weist S, Buetow S, Wittke J, Brunkau C, Hummel M, Eravci S, Baumgartner A. (2009) Strategies for a reliable biostatistical analysis of differentially expressed spots from two-dimensional electrophoresis gels. J Proteome Res 8: 2601–2607 [DOI] [PubMed] [Google Scholar]
- Faurobert M, Mihr C, Bertin N, Pawlowski T, Negroni L, Sommerer N, Causse M. (2007) Major proteome variations associated with cherry tomato pericarp development and ripening. Plant Physiol 143: 1327–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernie AR, Stitt M. (2012) On the discordance of metabolomics with proteomics and transcriptomics: coping with increasing complexity in logic, chemistry, and network interactions scientific correspondence. Plant Physiol 158: 1139–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser PD, Enfissi EM, Halket JM, Truesdale MR, Yu D, Gerrish C, Bramley PM. (2007) Manipulation of phytoene levels in tomato fruit: effects on isoprenoids, plastids, and intermediary metabolism. Plant Cell 19: 3194–3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenette Charron JB, Breton G, Badawi M, Sarhan F. (2002) Molecular and structural analyses of a novel temperature stress-induced lipocalin from wheat and Arabidopsis. FEBS Lett 517: 129–132 [DOI] [PubMed] [Google Scholar]
- Galpaz N, Wang Q, Menda N, Zamir D, Hirschberg J. (2008) Abscisic acid deficiency in the tomato mutant high-pigment 3 leading to increased plastid number and higher fruit lycopene content. Plant J 53: 717–730 [DOI] [PubMed] [Google Scholar]
- Giuliano G, Bartley GE, Scolnik PA. (1993) Regulation of carotenoid biosynthesis during tomato development. Plant Cell 5: 379–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldring JP, Ravaioli L. (1996) Solubilization of protein-dye complexes on nitrocellulose to quantify proteins spectrophotometrically. Anal Biochem 242: 197–201 [DOI] [PubMed] [Google Scholar]
- Harris WM, Spurr AR. (1969) Chromoplasts of tomato fruits. II. The red tomato. Am J Bot 56: 380–389 [Google Scholar]
- Hieber AD, Bugos RC, Yamamoto HY. (2000) Plant lipocalins: violaxanthin de-epoxidase and zeaxanthin epoxidase. Biochim Biophys Acta 1482: 84–91 [DOI] [PubMed] [Google Scholar]
- Isaacson T, Damasceno CM, Saravanan RS, He Y, Catalá C, Saladié M, Rose JK. (2006) Sample extraction techniques for enhanced proteomic analysis of plant tissues. Nat Protoc 1: 769–774 [DOI] [PubMed] [Google Scholar]
- Jones CM (2000) Evaluation of carotenoids and anthocyanins in high pigment, processing, heirloom and anthocyanin fruit tomatoes. MS thesis. Oregon State University, Corvallis [Google Scholar]
- Kendrick RE, Kerckhoffs LH, van Tuinen A, Koornneef M. (1997) Photomorphogenic mutants of tomato. Plant Cell Environ 20: 746–751 [PubMed] [Google Scholar]
- Klee HJ, Giovannoni JJ. (2011) Genetics and control of tomato fruit ripening and quality attributes. Annu Rev Genet 45: 41–59 [DOI] [PubMed] [Google Scholar]
- Kolotilin I, Koltai H, Tadmor Y, Bar-Or C, Reuveni M, Meir A, Nahon S, Shlomo H, Chen L, Levin I. (2007) Transcriptional profiling of high pigment-2dg tomato mutant links early fruit plastid biogenesis with its overproduction of phytonutrients. Plant Physiol 145: 389–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Lavi N, Tadmor Y, Meir A, Bechar A, Oren-Shamir M, Ovadia R, Reuveni M, Nahon S, Shlomo H, Chen L, et al. (2009) Characterization of the intense pigment tomato genotype emphasizing targeted fruit metabolites and chloroplast biogenesis. J Agric Food Chem 57: 4818–4826 [DOI] [PubMed] [Google Scholar]
- Leitner-Dagan Y, Ovadis M, Shklarman E, Elad Y, Rav David D, Vainstein A. (2006) Expression and functional analyses of the plastid lipid-associated protein CHRC suggest its role in chromoplastogenesis and stress. Plant Physiol 142: 233–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenucci MS, Serrone L, De Caroli M, Fraser PD, Bramley PM, Piro G, Dalessandro G. (2012) Isoprenoid, lipid, and protein contents in intact plastids isolated from mesocarp cells of traditional and high-pigment tomato cultivars at different ripening stages. J Agric Food Chem 60: 1764–1775 [DOI] [PubMed] [Google Scholar]
- Levin I, Frankel P, Gilboa N, Tanny S, Lalazar A. (2003) The tomato dark green mutation is a novel allele of the tomato homolog of the DEETIOLATED1 gene. Theor Appl Genet 106: 454–460 [DOI] [PubMed] [Google Scholar]
- Li L, Yang Y, Xu Q, Owsiany K, Welsch R, Chitchumroonchokchai C, Lu S, Van Eck J, Deng XX, Failla M, et al. (2012) The Or gene enhances carotenoid accumulation and stability during post-harvest storage of potato tubers. Mol Plant 5: 339–352 [DOI] [PubMed] [Google Scholar]
- Lieberman M, Segev O, Gilboa N, Lalazar A, Levin I. (2004) The tomato homolog of the gene encoding UV-damaged DNA binding protein 1 (DDB1) underlined as the gene that causes the high pigment-1 mutant phenotype. Theor Appl Genet 108: 1574–1581 [DOI] [PubMed] [Google Scholar]
- Liu Y, Roof S, Ye Z, Barry C, van Tuinen A, Vrebalov J, Bowler C, Giovannoni J. (2004) Manipulation of light signal transduction as a means of modifying fruit nutritional quality in tomato. Proc Natl Acad Sci USA 101: 9897–9902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S, Van Eck J, Zhou X, Lopez AB, O’Halloran DM, Cosman KM, Conlin BJ, Paolillo DJ, Garvin DF, Vrebalov J, et al. (2006) The cauliflower Or gene encodes a DnaJ cysteine-rich domain-containing protein that mediates high levels of β-carotene accumulation. Plant Cell 18: 3594–3605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundquist PK, Poliakov A, Bhuiyan NH, Zybailov B, Sun Q, van Wijk KJ. (2012) The functional network of the Arabidopsis plastoglobule proteome based on quantitative proteomics and genome-wide coexpression analysis. Plant Physiol 158: 1172–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manaa A, Ben Ahmed H, Valot B, Bouchet JP, Aschi-Smiti S, Causse M, Faurobert M. (2011) Salt and genotype impact on plant physiology and root proteome variations in tomato. J Exp Bot 62: 2797–2813 [DOI] [PubMed] [Google Scholar]
- Mounet F, Moing A, Garcia V, Petit J, Maucourt M, Deborde C, Bernillon S, Le Gall G, Colquhoun I, Defernez M, et al. (2009) Gene and metabolite regulatory network analysis of early developing fruit tissues highlights new candidate genes for the control of tomato fruit composition and development. Plant Physiol 149: 1505–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustilli AC, Fenzi F, Ciliento R, Alfano F, Bowler C. (1999) Phenotype of the tomato high pigment-2 mutant is caused by a mutation in the tomato homolog of DEETIOLATED1. Plant Cell 11: 145–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osorio S, Alba R, Damasceno CM, Lopez-Casado G, Lohse M, Zanor MI, Tohge T, Usadel B, Rose JK, Fei Z, et al. (2011) Systems biology of tomato fruit development: combined transcript, protein, and metabolite analysis of tomato transcription factor (nor, rin) and ethylene receptor (Nr) mutants reveals novel regulatory interactions. Plant Physiol 157: 405–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozueta-Romero J, Rafia F, Houlné G, Cheniclet C, Carde JP, Schantz ML, Schantz R. (1997) A ubiquitous plant housekeeping gene, PAP, encodes a major protein component of bell pepper chromoplasts. Plant Physiol 115: 1185–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin G, Wang Y, Cao B, Wang W, Tian S. (2012) Unraveling the regulatory network of the MADS box transcription factor RIN in fruit ripening. Plant J 70: 243–255 [DOI] [PubMed] [Google Scholar]
- Rocco M, D’Ambrosio C, Arena S, Faurobert M, Scaloni A, Marra M. (2006) Proteomic analysis of tomato fruits from two ecotypes during ripening. Proteomics 6: 3781–3791 [DOI] [PubMed] [Google Scholar]
- Rohrmann J, Tohge T, Alba R, Osorio S, Caldana C, McQuinn R, Arvidsson S, van der Merwe MJ, Riaño-Pachón DM, Mueller-Roeber B, et al. (2011) Combined transcription factor profiling, microarray analysis and metabolite profiling reveals the transcriptional control of metabolic shifts occurring during tomato fruit development. Plant J 68: 999–1013 [DOI] [PubMed] [Google Scholar]
- Ruepp A, Zollner A, Maier D, Albermann K, Hani J, Mokrejs M, Tetko I, Güldener U, Mannhaupt G, Münsterkötter M, et al. (2004) The FunCat, a functional annotation scheme for systematic classification of proteins from whole genomes. Nucleic Acids Res 32: 5539–5545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Hirai MY, Yonekura-Sakakibara K. (2008) Decoding genes with coexpression networks and metabolomics: ‘majority report by precogs.’ Trends Plant Sci 13: 36–43 [DOI] [PubMed] [Google Scholar]
- Santisree P, Nongmaithem S, Vasuki H, Sreelakshmi Y, Ivanchenko MG, Sharma R. (2011) Tomato root penetration in soil requires a coaction between ethylene and auxin signaling. Plant Physiol 156: 1424–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. (2006) In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc 1: 2856–2860 [DOI] [PubMed] [Google Scholar]
- Siddique MA, Grossmann J, Gruissem W, Baginsky S. (2006) Proteome analysis of bell pepper (Capsicum annuum L.) chromoplasts. Plant Cell Physiol 47: 1663–1673 [DOI] [PubMed] [Google Scholar]
- Simkin AJ, Gaffé J, Alcaraz JP, Carde JP, Bramley PM, Fraser PD, Kuntz M. (2007) Fibrillin influence on plastid ultrastructure and pigment content in tomato fruit. Phytochemistry 68: 1545–1556 [DOI] [PubMed] [Google Scholar]
- Smirra I, Halevy AH, Vainstein A. (1993) Isolation and characterization of a chromoplast-specific carotenoid-associated protein from Cucumis sativus corollas. Plant Physiol 102: 491–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speirs J, Lee E, Holt K, Yong-Duk K, Scott NS, Loveys B, Schuch W. (1998) Genetic manipulation of alcohol dehydrogenase levels in ripening tomato fruit affects the balance of some flavor aldehydes and alcohols. Plant Physiol 117: 1047–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreelakshmi Y (2000) Regulation and characterization of tomato phenylalanine ammonia-lyase(s). PhD thesis. University of Hyderabad, Hyderabad, India [Google Scholar]
- Srinivas A, Behera RK, Kagawa T, Wada M, Sharma R. (2004) High pigment1 mutation negatively regulates phototropic signal transduction in tomato seedlings. Plant Physiol 134: 790–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stigliani AL, Giorio G, D’Ambrosio C. (2011) Characterization of P450 carotenoid beta- and epsilon-hydroxylases of tomato and transcriptional regulation of xanthophyll biosynthesis in root, leaf, petal and fruit. Plant Cell Physiol 52: 851–865 [DOI] [PubMed] [Google Scholar]
- Towbin H, Staehelin T, Gordon J. (1979) Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA 76: 4350–4354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vainstein A, Halevy AH, Smirra I, Vishnevetsky M. (1994) Chromoplast biogenesis in Cucumis sativus corollas (rapid effect of gibberellin A3 on the accumulation of a chromoplast-specific carotenoid-associated protein). Plant Physiol 104: 321–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishnevetsky M, Ovadis M, Vainstein A. (1999) Carotenoid sequestration in plants: the role of carotenoid-associated proteins. Trends Plant Sci 4: 232–235 [DOI] [PubMed] [Google Scholar]
- Wang S, Liu J, Feng Y, Niu X, Giovannoni J, Liu Y. (2008) Altered plastid levels and potential for improved fruit nutrient content by downregulation of the tomato DDB1-interacting protein CUL4. Plant J 55: 89–103 [DOI] [PubMed] [Google Scholar]
- Ytterberg AJ, Peltier JB, van Wijk KJ. (2006) Protein profiling of plastoglobules in chloroplasts and chromoplasts: a surprising site for differential accumulation of metabolic enzymes. Plant Physiol 140: 984–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Pan Z, Ding Y, Zhu A, Cao H, Xu Q, Deng X. (2011) A proteomic analysis of the chromoplasts isolated from sweet orange fruits [Citrus sinensis (L.) Osbeck]. J Exp Bot 62: 5297–5309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Ma H, Feng J, Zeng L, Wang Z, Chen S. (2008) Grape berry plasma membrane proteome analysis and its differential expression during ripening. J Exp Bot 59: 2979–2990 [DOI] [PubMed] [Google Scholar]
- Zhou X, Van Eck J, Li L. (2008) Use of the cauliflower Or gene for improving crop nutritional quality. Biotechnol Annu Rev 14: 171–190 [DOI] [PubMed] [Google Scholar]