Chloroplast DNA replication is regulated by the redox state in the cell, which is sensed by the chloroplast nucleoids in Chlamydomonas reinhardtii.
Abstract
Chloroplasts arose from a cyanobacterial endosymbiont and multiply by division. In algal cells, chloroplast division is regulated by the cell cycle so as to occur only once, in the S phase. Chloroplasts possess multiple copies of their own genome that must be replicated during chloroplast proliferation. In order to examine how chloroplast DNA replication is regulated in the green alga Chlamydomonas reinhardtii, we first asked whether it is regulated by the cell cycle, as is the case for chloroplast division. Chloroplast DNA is replicated in the light and not the dark phase, independent of the cell cycle or the timing of chloroplast division in photoautotrophic culture. Inhibition of photosynthetic electron transfer blocked chloroplast DNA replication. However, chloroplast DNA was replicated when the cells were grown heterotrophically in the dark, raising the possibility that chloroplast DNA replication is coupled with the reducing power supplied by photosynthesis or the uptake of acetate. When dimethylthiourea, a reactive oxygen species scavenger, was added to the photoautotrophic culture, chloroplast DNA was replicated even in the dark. In contrast, when methylviologen, a reactive oxygen species inducer, was added, chloroplast DNA was not replicated in the light. Moreover, the chloroplast DNA replication activity in both the isolated chloroplasts and nucleoids was increased by dithiothreitol, while it was repressed by diamide, a specific thiol-oxidizing reagent. These results suggest that chloroplast DNA replication is regulated by the redox state that is sensed by the nucleoids and that the disulfide bonds in nucleoid-associated proteins are involved in this regulatory activity.
Chloroplasts are semiautonomous organelles that possess their own genome, which is complexed with proteins to form nucleoids and also certain machinery needed for protein synthesis, as is the case in prokaryotes. It is generally accepted that chloroplasts arose from a bacterial endosymbiont closely related to the currently extant cyanobacteria (Archibald, 2009; Keeling, 2010). In a manner reminiscent of their free-living ancestor, chloroplasts proliferate by the division of preexisting organelles that are coupled to the duplication and segregation of the nucleoids (Kuroiwa, 1991) and have retained the bulk of their bacterial biochemistry. However, chloroplasts have subsequently been substantially remodeled by the host cell so as to function as complementary organelles within the eukaryotic host cell (Rodríguez-Ezpeleta and Philippe, 2006; Archibald, 2009; Keeling, 2010). For example, most of the genes that were once in the original endosymbiont genome have been either lost or transferred into the host nuclear genome. As a result, the size of the chloroplast genome has been reduced to less than one-tenth that of the free-living cyanobacterial genome. Thus, the bulk of the chloroplast proteome consists of nucleus-encoded proteins that are translated on cytoplasmic ribosomes and translocated into chloroplasts. In addition, chloroplast division ultimately came to be a process tightly regulated by the host cell, which ensured permanent inheritance of the chloroplasts during the course of cell division and from generation to generation (Rodríguez-Ezpeleta and Philippe, 2006; Archibald, 2009; Keeling, 2010).
Chloroplast division is performed by constriction of the ring structures at the division site, encompassing both the inside and the outside of the two envelopes (Yang et al., 2008; Maple and Møller, 2010; Miyagishima, 2011; Pyke, 2013). One part of the division machinery is derived from the cyanobacterial cytokinetic machinery that is based on the FtsZ protein. In contrast, other parts of the division machinery involve proteins specific to eukaryotes, including one member of the dynamin family. The majority of algae (both unicellular and multicellular), which diverged early within the Plantae, have just one or at most only a few chloroplasts per cell. In algae, the chloroplast divides once per cell cycle before the host cell completes cytokinesis (Suzuki et al., 1994; Miyagishima et al., 2012). In contrast, land plants and certain algal species contain dozens of chloroplasts per cell that divide nonsynchronously, even within the same cell (Boffey and Lloyd, 1988). Because land plants evolved from algae, there is likely to have been a linkage between the cell cycle and chloroplast division in their algal ancestor that was subsequently lost during land plant evolution. Our recent study showed that the timing of chloroplast division in algae is restricted to the S phase by S phase-specific formation of the chloroplast division machinery, which is based on the cell cycle-regulated expression of the components of the chloroplast division machinery (Miyagishima et al., 2012).
Because chloroplasts possess their own genome, chloroplast DNA must be duplicated so that each daughter chloroplast inherits the required DNA after division. However, it is still unclear how the replication of chloroplast DNA is regulated and whether the replication is coupled with the timing of chloroplast division, even though certain studies have addressed this issue, as described below.
Bacteria such as Escherichia coli and Bacillus subtilis possess a single circular chromosome. In these bacteria, the process of DNA replication is tightly coupled with cell division (Boye et al., 2000; Zakrzewska-Czerwińska et al., 2007), in which the initiation of replication is regulated such that it occurs only once per cell division cycle (Boye et al., 2000). In contrast, cyanobacteria contain multiple copies of their DNA (e.g. three to five copies in Synechococcus elongatus PCC 7942; Mann and Carr, 1974; Griese et al., 2011). In some obligate photoautotrophic cyanobacterial species, replication is initiated only when light is available (Binder and Chisholm, 1990; Mori et al., 1996; Watanabe et al., 2012). Replication is initiated asynchronously among the multiple copies of the DNA. Although the regulation of the initiation of DNA replication is less stringent than that in E. coli and B. subtilis, as described above, a recent study using S. elongatus PCC 7942 showed that this replication peaks prior to cell division, as in other bacteria.
Chloroplasts also contain multiple copies of DNA (approximately 1,000 copies; Boffey and Leech, 1982; Miyamura et al., 1986; Baumgartner et al., 1989; Oldenburg and Bendich, 2004; Oldenburg et al., 2006; Shaver et al., 2008). In algae, chloroplast DNA is replicated in a manner that keeps pace with chloroplast and cell division in order to maintain the proper DNA content per chloroplast (i.e. per cell). In contrast, in land plants, the copy number of DNA in each chloroplast (plastid) changes during the course of development and differentiation, although contradictory results were reported about leaf development (Lamppa and Bendich, 1979; Boffey and Leech, 1982; Hashimoto and Possingham, 1989; Kuroiwa, 1991; Rowan and Bendich, 2009; Matsushima et al., 2011). Previous studies that synchronized the algal cell cycle by means of a 24-h light/dark cycle showed that chloroplast DNA is replicated only during the G1 phase, after which it is separated into daughter chloroplasts during the S phase by chloroplast division, implying that chloroplast DNA replication and division are temporally separated (Chiang and Sueoka, 1967; Grant et al., 1978; Suzuki et al., 1994). However, under these experimental conditions, G1 cells grow and the chloroplast DNA level increases during the light period. Cells enter into the S phase, chloroplast DNA replication ceases, and the chloroplasts divide at the beginning of the dark period. Thus, it is still unclear whether chloroplast DNA replication is directly controlled by the cell cycle, as is the case in chloroplast division, or chloroplast DNA replication occurs merely when light energy is available.
We addressed this issue using a synchronous culture as well as a heterotrophic culture of the mixotrophic green alga Chlamydomonas reinhardtii. The results show that chloroplast DNA replication occurs independently of either the cell cycle or the timing of chloroplast division. Instead, it is shown that chloroplast DNA replication occurs when light is available in photoautotrophic culture and even under darkness in heterotrophic culture. Further experimental results suggest that chloroplast DNA replication is regulated by the redox state in the cell, which is sensed by the chloroplast nucleoids.
RESULTS
The Relationship between Chloroplast DNA Replication and the Timing of Chloroplast Division
In order to determine the changes in the chloroplast DNA level that take place during the cell cycle by means of quantitative real-time PCR (qPCR), the green alga C. reinhardtii was used in this study for the following reasons. The cell cycle in this alga is synchronized by a light/dark cycle (Surzycki, 1971). Because the cell cycle is linked to circadian rhythms, cell cycle synchrony is maintained even under continuous light after initial entrainment by a light/dark cycle (Goto and Johnson, 1995). C. reinhardtii cells contain a single chloroplast, and this chloroplast divides during the S/M phase. These features allow an examination of the relationship between chloroplast DNA replication and the cell/chloroplast division cycle. In addition, cells grow in size by many fold during the G1 phase and then enter into one to four rounds of the S/M phases to produce two to 16 daughter cells during a single 24-h light/dark cycle. Thus, the levels of the nuclear and chloroplast DNA increase many fold in a single cycle, which makes it easier to detect any change in the DNA level by qPCR.
Previous studies using a 24-h light/dark synchronous culture of the red alga Cyanidioschyzon merolae (Suzuki et al., 1994) and the green alga Scenedesmus quadricauda (Zachleder et al., 1995) showed that the level of chloroplast DNA increases only during the light period (the G1 phase) and that chloroplasts divide early in the dark period (the S phase). Thus, it has remained unclear whether the timing of chloroplast DNA replication is defined by the cell/chloroplast division cycle or is coupled with the availability of light.
To address this issue, we synchronized two populations of C. reinhardtii using a light/dark cycle, and then one population was cultured in the same light/dark cycle while the other was cultured under continuous light. The nuclear and chloroplast DNA levels were examined by qPCR of the Rubisco small subunit (RBCS) gene (the nuclear genome) and the Rubisco large subunit (rbcL) gene (the chloroplast genome), respectively.
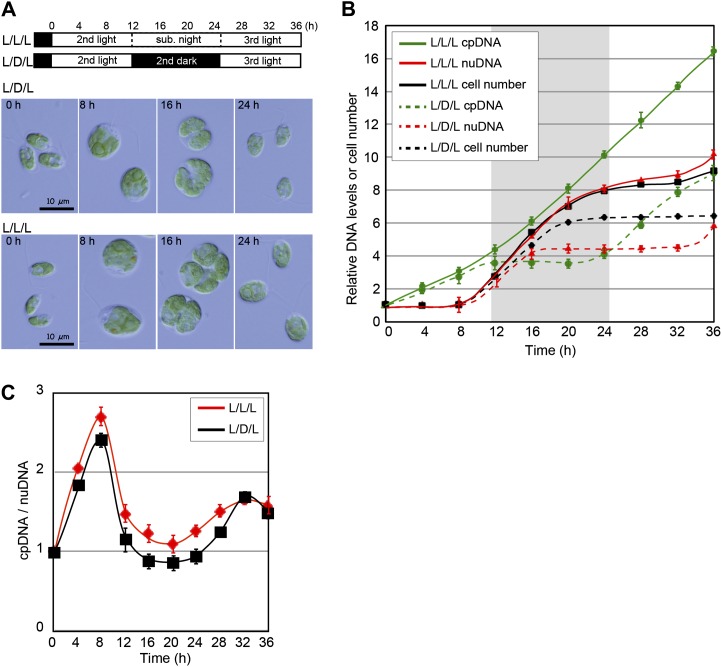
Under the light/dark cycle (L/D/L in Fig. 1B), the chloroplast DNA level increased only during the light period (corresponding to the G1 phase: 0–12 h and 24–36 h) prior to the increase of the nuclear DNA level and cell division (corresponding to the S/M phase when the chloroplast divides: 8–20 h; Fig. 1, A and B), as observed in previous studies on other algae (Chiang and Sueoka, 1967; Suzuki et al., 1994; Zachleder et al., 1995). In contrast, when cells were cultured under continuous light (L/L/L in Fig. 1B) after the entrainment by a light/dark cycle, the level of chloroplast DNA kept increasing, even though nuclear DNA replication (8–24 h) and cell division (8–24 h) were restricted to subjective night. These results indicate that chloroplast DNA replication is linked to the availability of light and not the cell/chloroplast division cycle.
Figure 1.
Changes in the nuclear and chloroplast DNA levels in the synchronous culture. C. reinhardtii 137c mt+ cells were cultured in the photoautotrophic medium (HSM). A, Progression of the cell cycle in the synchronous culture. Cells were entrained by a 12-h-light/12-h-dark cycle and then grown under a light/dark cycle (L/D/L) or continuous light condition (L/L/L). Bars = 10 µm. B, Changes in cell number and nuclear and chloroplast DNA levels in the synchronous culture were determined by qPCR analyses using the primer sets for rbcL (chloroplast DNA [cpDNA]) and RBCS1 (nuclear DNA [nuDNA]). The gray and white regions in the graph indicate the subjective night and light periods, respectively. Error bars represent the se of three technical repeats of qPCR. C, Ratio of the chloroplast DNA relative to the nuclear DNA in cells grown under the L/D/L or L/L/L condition. The ratio was calculated from the DNA levels in B. Error bars represent the se. Two independent experiments showed similar results. [See online article for color version of this figure.]
Although the chloroplast DNA level kept increasing under continuous light, there was little difference in the ratio of chloroplast DNA to nuclear DNA between the light/dark culture and the continuous light culture (Fig. 1C). This is because both the nuclear DNA level and the cell number increased more under continuous light than under the light/dark cycle (Fig. 1A). In addition, it is known that both the cell and chloroplast size increase, even in the S/M phase, when light is available (Fig. 1A; Rollins et al., 1983). Thus, it appears that chloroplast DNA replication is linked to increases of the cell and chloroplast size.
The Relationship between Chloroplast DNA Replication and Photosynthesis
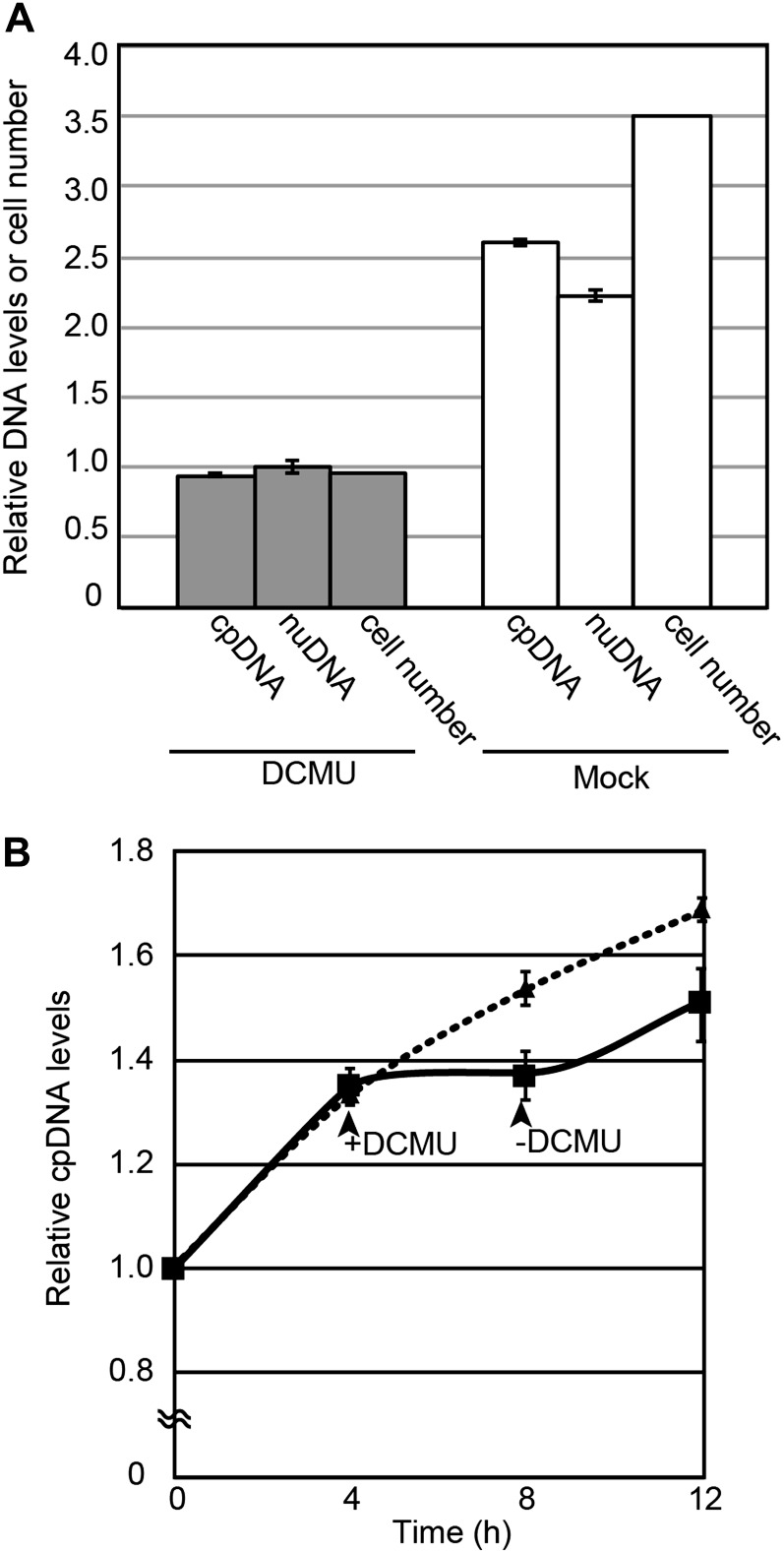
The obtained results indicate that the chloroplast DNA is replicated when light is available during photoautotrophic growth. In order to investigate the relationship between photosynthesis and chloroplast DNA replication, 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU), which blocks electron transfer between PSII and plastoquinone (Taiz and Zeiger, 2010), was added to a nonsynchronous log-phase culture that was grown under continuous light. After the addition of DCMU, cells were cultured for 12 h under continuous light and the changes in the chloroplast and nuclear DNA levels over 12 h were examined by qPCR (Fig. 2A). After the addition of DCMU, the increases in the chloroplast and nuclear DNA levels were blocked (Fig. 2A). When DCMU was removed by replacing the medium with a fresh one lacking DCMU, the increase in the chloroplast DNA level was resumed (Fig. 2B). These results indicate that chloroplast DNA replication requires photosynthesis during photoautotrophic growth.
Figure 2.
Effect of an inhibition of photosynthetic electron transfer on chloroplast DNA replication. C. reinhardtii 137c mt+ cells were cultured in the photoautotrophic medium. To inhibit photosynthetic electron transfer, DCMU was added to the nonsynchronous log-phase culture that was grown under continuous light. The nuclear (nuDNA) and chloroplast (cpDNA) DNA levels were determined by qPCR analyses. A, Levels of chloroplast and nuclear DNA and cell number 12 h after treatment with DCMU (DCMU dissolved in ethanol was added) or without DCMU (Mock; ethanol was added). The DNA levels and the cell number just before the addition of DCMU are defined as 1.0. Error bars represent the se of three technical repeats of qPCR. Two independent experiments showed similar results. B, Change in the level of chloroplast DNA after the removal of DCMU. Four hours after the addition of DCMU, medium was replaced by fresh medium without DCMU and further cultured for 4 h. The solid line indicates the chloroplast DNA level in the culture treated with DCMU, and the broken line represents the culture without DCMU treatment. Error bars represent the se of three technical repeats of qPCR. Two independent experiments showed similar results.
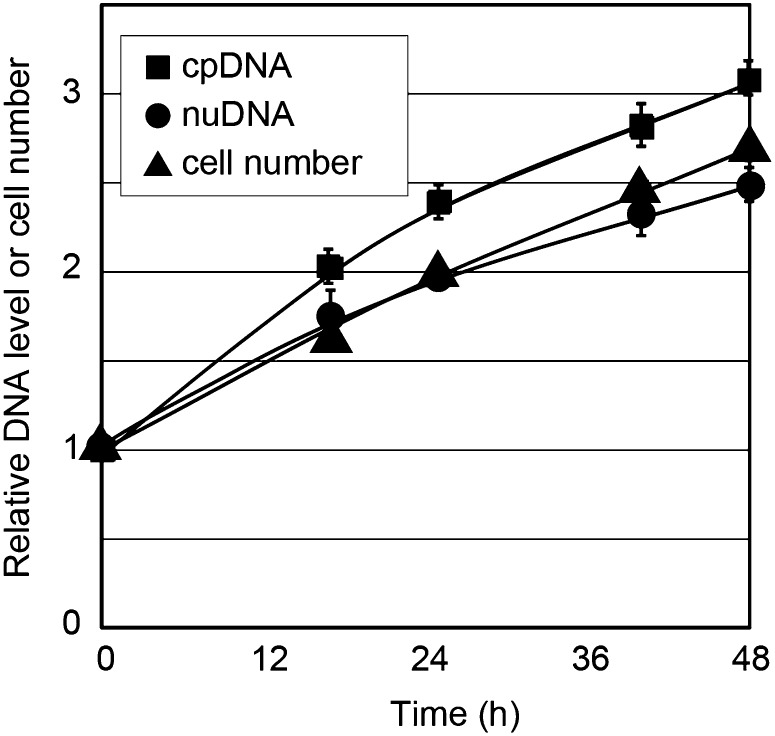
In order to examine whether there are factors specific to photosynthetic electron transfer that are required for chloroplast DNA replication, we cultured the cells heterotrophically under darkness. C. reinhardtii cells are capable of efficient heterotrophic growth in the presence of acetate (Harris, 1989). qPCR analyses showed that the chloroplast DNA level keeps increasing in accordance with the replication of the nuclear DNA and cell division (Fig. 3). Given these results, it is suggested that chloroplast DNA replication is coupled with cell growth by photosynthesis or the uptake of acetate, which supply a carbon source and reducing power to the cell.
Figure 3.
Changes in the chloroplast DNA level in the heterotrophic culture under the dark condition. C. reinhardtii 137c mt+ cells were cultured in the medium containing acetate (Tris-acetate-phosphate medium) under complete darkness for 2 d. The nuclear (nuDNA) and chloroplast (cpDNA) DNA levels were determined by qPCR analyses. Error bars represent the se of three technical repeats of qPCR. Two independent experiments showed similar results.
Relationship between Chloroplast DNA Replication and Redox State
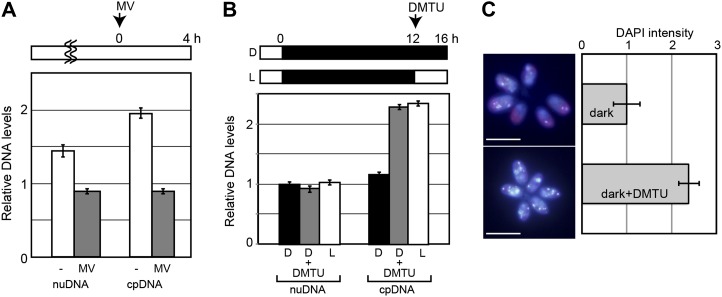
One possibility raised by the above results is that chloroplast DNA replication is linked to the cellular redox state, because photosynthesis and acetate uptake metabolism supply reducing power to the cell. Therefore, we examined the effects of the oxidative and reductive states on chloroplast DNA replication using membrane-permeable redox reagents. Methylviologen accepts electrons from PSI and transfers them to oxygen molecules, resulting in the production of reactive oxygen species in chloroplasts (Taiz and Zeiger, 2010). Dimethylthiourea (DMTU) quenches reactive oxygen species and reduces oxidative stress (Levine et al., 1994). Under the culturing conditions we used, treatment with DMTU for more than 4 h perturbed the cell shape and decreased both the chloroplast and nuclear DNA levels, probably as the result of cell death. Therefore, we examined the effects of methylviologen and DMTU on DNA replication 4 h after the addition of these reagents (Fig. 4; Table I).
Figure 4.
Effects of redox reagents on chloroplast DNA replication in vivo. C. reinhardtii 137c mt+ cells were cultured in the photoautotrophic medium. A, Effect of methylviologen (MV; a hydrogen peroxide producer dependent on PSII) on chloroplast DNA (cpDNA) and nuclear DNA (nuDNA) replication. Methylviologen was added to a nonsynchronous log-phase culture that was grown under continuous light. DNA levels in the culture 4 h after the treatment with (MV) or without (−) methylviologen were examined by qPCR analyses. Error bars represent the se of three technical repeats of qPCR. Two independent experiments showed similar results. B, Effect of DMTU (a hydrogen peroxide scavenger) on chloroplast DNA and nuclear DNA replication. DMTU was added to synchronous culture at the end of a 12-h dark period. Nuclear and chloroplast DNA levels in the cultures that were treated with or without DMTU for 4 h under dark (D) or light (L) were examined by qPCR analyses. Error bars represent the se of three technical repeats of qPCR. Three independent experiments showed similar results. C, DAPI-stained images of cells treated with or without DMTU for 4 h under darkness. The intensity of the fluorescent DAPI staining that overlapped with the chloroplast red autofluorescence was measured using ImageJ. Error bars represent the sd (n = 50 cells). The levels just before the addition of the reagents (at 0 h for MV, at 12 h for DMTU) are defined as 1.0. Bars = 10 µm. [See online article for color version of this figure.]
Table I. Change in levels of GSH and GSSG.
Cells were cultured in conditions as described in Figures 1, 4, and 6. GSH and GSSG were extracted from cells at the end of a 12-h subjective night (Light), a dark period (Dark), after 4 h of treatment with (MV+, DMTU+) or without (MV−, DMTU−) the redox reagents methylviologen and DMTU, or from isolated chloroplasts. The values are means ± se of three technical repeats. Two independent experiments showed similar results. Asterisks indicate significant differences by Student’s t test (P < 0.005).
| Culture | GSSG | GSH | GSH/GSSG |
|---|---|---|---|
| nmol mg–1 protein | |||
| Light | 0.114 ± 0.015 | 0.377 ± 0.006 | 3.3 ± 0.5 |
| Dark | 0.110 ± 0.016 | 0.218 ± 0.021 | 2.0 ± 1.0 |
| MV+ (light) | 0.094 ± 0.038 | 0.057 ± 0.004 | 0.7 ± 0.2 |
| MV– (light) | 0.111 ± 0.017 | 0.324 ± 0.008 | 3.0 ± 0.2 |
| DMTU+ (dark) | 0.120 ± 0.046 | 0.425 ± 0.009 | 7.8 ± 3.2 |
| DMTU– (dark) | 0.104 ± 0.029 | 0.229 ± 0.011 | 2.2 ± 0.7 |
| Isolated chloroplasts (light) | 0.250 ± 0.067 | 0.778 ± 0.058 | 3.2 ± 0.7 |
| Isolated chloroplasts (dark) | 0.267 ± 0.054 | 0.550 ± 0.060 | 2.1 ± 0.2 |
First, we examined the effect of redox reagents on cellular redox state by quantifying cellular glutathione level (Table I). When methylviologen was added to a nonsynchronous log-phase culture that was grown photoautotrophically under continuous light, the level of reduced glutathione (GSH) decreased by one-fifth of that of untreated cells. In addition, the ratio of GSH to oxidized glutathione (GSSG) also decreased from 3.0 ± 0.5 to 0.7 ± 0.2, which was lower than the ratio in cells cultured in the dark without methylviologen (2.0 ± 0.1). In contrast, when DMTU was added to photoautotrophic synchronous culture at the end of a 12-h dark period, the amount of GSH increased 2-fold and the GSH/GSSG ratio also increased from 2.2 ± 0.7 to 7.8 ± 3.2, which was higher than the ratio in cells cultured under light without DMTU (ratio of 3.3 ± 0.5). These results indicate that methylviologen and DMTU treatment changes the cellular redox state as expected. Furthermore, we confirmed that the light and dark treatment also changes redox state in chloroplasts.
When methylviologen was added to culture as described above, the replication activities of the chloroplast and nuclear DNA were blocked (Fig. 4A). In contrast, when DMTU was added to dark culture as described above, the chloroplast DNA level, but not the nuclear DNA level, increased under the dark condition to the same level as that in cells grown under light without DMTU (Fig. 4B). We also examined the effect of DMTU on chloroplast DNA replication by 4′,6-diamidino-phenylindole (DAPI) staining (Fig. 4C). The DAPI fluorescence intensity in the chloroplast (the foci overlapping the red chloroplast autofluorescence) did not change under darkness without DMTU. In contrast, DMTU treatment increased the fluorescent intensity up to 2.5-fold over 4 h. These results suggest that the chloroplast DNA replication is regulated by the cellular redox state and that replication occurs when the cells are in a reduced state.
The Connection between the Cellular Redox State and Chloroplast DNA Synthesis
In order to understand how chloroplast DNA replication is linked to the cellular redox state, we tested the following three hypotheses: (1) the chloroplast DNA polymerase level increases under a reducing condition; (2) the chloroplast DNA polymerase activity is enhanced under a reducing condition by a modification of the polymerase or other regulatory proteins; and (3) the intrachloroplast deoxynucleotide level (deoxyribonucleotide triphosphate [dNTP], the substrate of DNA replication) increases in the reduced state.
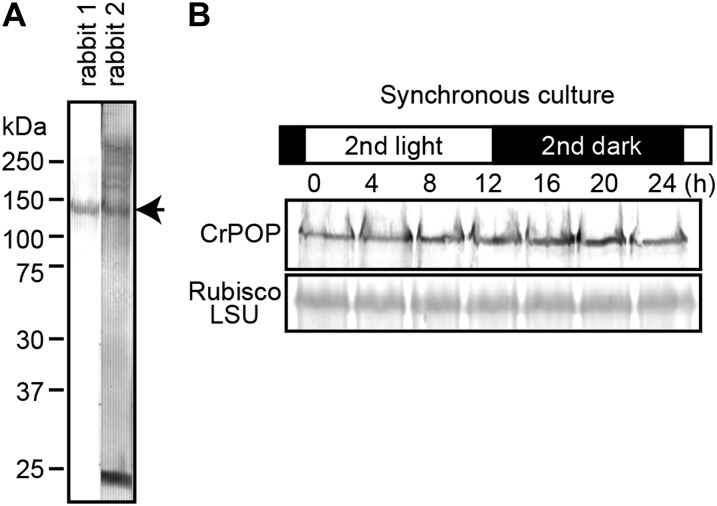
To test the first hypothesis, we examined the level of chloroplast DNA polymerase in synchronous culture. Plants and algae do not possess any DNA polymerase of cyanobacterial origin. Recent studies showed that the replication of chloroplast DNA is performed by the plant organellar DNA polymerase (POP; Ono et al., 2007; Moriyama et al., 2008; Parent et al., 2011; Udy et al., 2012), which is similar to bacterial DNA polymerase I and is targeted to both chloroplasts and mitochondria. We performed BLAST searches in the C. reinhardtii database in an effort to identify POP orthologs. We observed that the C. reinhardtii POP gene was misannotated as two adjacent but separate genes (accession nos. XM_001695976 and XM_001695977). XM_001695976 and XM_001695977 encode the 3′ to 5′ exonuclease domain, which is in the N terminus, and the PolA domain, which is in the C terminus of the POP protein in other species. By means of reverse transcription-PCR, we verified that XM_001695976 and XM_001695977 are indeed transcribed as a single gene. We prepared polyclonal antibodies against CrPOP that detect a band of 130 kD (Fig. 5), which is similar to the molecular mass of the POP proteins in other species (Ono et al., 2007; Moriyama et al., 2008). Immunoblot analysis showed that the CrPOP level was constant throughout the cell cycle when synchronized by a light/dark cycle (Fig. 5). This result indicates that chloroplast DNA is not replicated in phototrophic culture grown in the dark, even though the POP protein is present at the same level in culture under light.
Figure 5.
Chloroplast DNA polymerase levels in the light and dark cycles. Immunoblot analyses were performed using anti-chloroplast DNA polymerase (CrPOP) antibodies. Twenty micrograms of protein was separated in each lane. A, Total proteins from asynchronous log-phase cells were detected by the CrPOP antibodies that were raised in two rabbits. B, Aliquots of the synchronous culture were collected at the indicated time points, and the total proteins were separated. The level of CrPOP protein was analyzed with the anti-CrPOP antibodies (rabbit 1), and the Rubisco large subunit (Rubisco LSU) was detected by Coomassie Brilliant Blue staining as the quantitative control.
To test the second and third possibilities, we performed an in vitro chloroplast DNA synthesis assay using isolated chloroplasts. Intact chloroplasts were isolated from photoautotrophic synchronous culture either from the end of the dark period or the end of the subjective night phase of continuous light culture. After chloroplasts were osmotically permeabilized in a hypertonic buffer, an excess amount of substrates for DNA replication (dNTPs and digoxigenin [DIG]-labeled dUTP) was added, and then the DNA synthesis activity was determined as the incorporation of DIG into the chloroplast DNA. The assay showed that the level of DIG-labeled dUTP incorporation into the chloroplast DNA was larger in the chloroplasts isolated from culture under light than under dark (Fig. 6). The incorporation of DIG-labeled dUTP was blocked when ethidium bromide, which blocks DNA replication, was added to the reaction mixture, indicating that this DIG incorporation is specific to DNA replication.
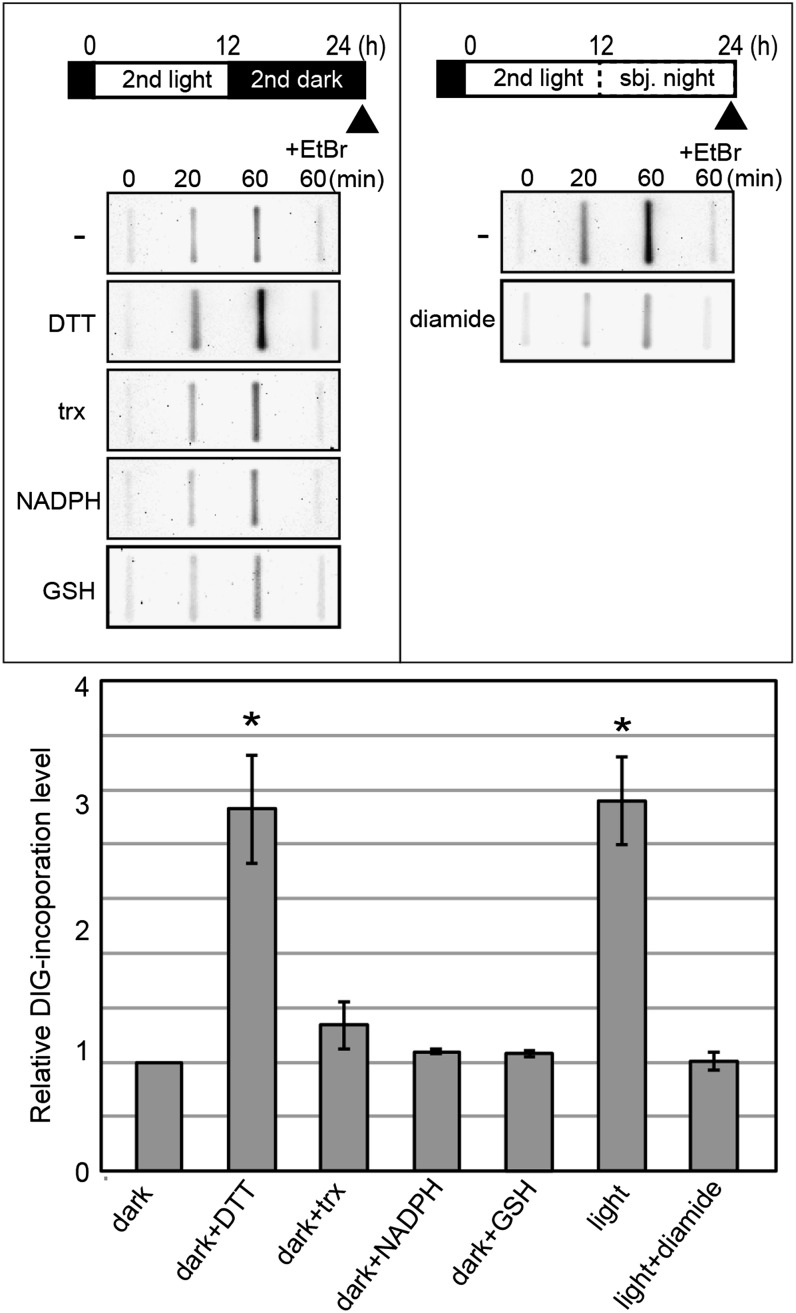
Figure 6.
Effects of redox reagents on chloroplast DNA replication in isolated chloroplasts. C. reinhardtii cw-15 cells were cultured in the photoautotrophic medium. Chloroplasts were isolated from synchronous culture either at the end of the second dark period or the end of the second subjective night under light (indicated by the arrowheads) and then osmotically permeabilized in hypertonic buffer. To investigate the effects of reducing reagents on chloroplast DNA replication activity, DTT, reduced thioredoxin (trx), NADPH, or GSH was added to chloroplasts isolated from the dark culture. To investigate the involvement of disulfide bonds on the regulation of the chloroplast DNA replication activity, diamide was added to chloroplasts isolated from the light culture. Ethidium bromide (EtBr), which inhibits DNA replication, was added as a negative control. Each reagent was added to the permeabilized chloroplasts. After preincubation for 15 min, chloroplasts were further incubated for 60 min with certain substrates (dNTP and DIG-labeled dUTP). DNA was extracted at the indicated time points and blotted onto a Hybond N+ membrane, and DIG, which was incorporated into the chloroplast DNA, was detected using anti-DIG antibodies. The relative levels of the incorporated DIG were plotted. The level of the culture grown under darkness at 60 min is defined as 1.0. Error bars represent the sd of three biological replicates. Asterisks indicate significant differences from the dark sample by Student’s t test (P < 0.0001).
To examine whether the redox state has an effect on the activity of chloroplast DNA replication as observed in vivo (Fig. 4), redox reagents were added to the reaction mixture (Fig. 6). The addition of reduced thioredoxin, NADPH, and GSH, which are reducing agents in chloroplasts (Scheibe, 1991), had no significant effect on DNA synthesis. However, the addition of dithiothreitol (DTT) elevated the DNA synthesis activity of chloroplasts prepared from dark-grown culture so as to be as high as that in the light-grown culture. In accordance with this result, the addition of diamide, a sulfhydryl group-specific oxidizing agent, reduced the DNA synthesis in the chloroplasts prepared from the light-grown culture to as low as that from dark-grown culture. Even though excess substrate levels were included in the reaction mixture, DNA synthesis activity was still affected by either the culture condition (light or dark) or redox reagents. Although it cannot completely be ruled out that a change in the intrachloroplast dNTP level might also be involved in the regulation of DNA replication, the above results suggest that replication is regulated by the redox state of a sulfhydryl group in chloroplast proteins.
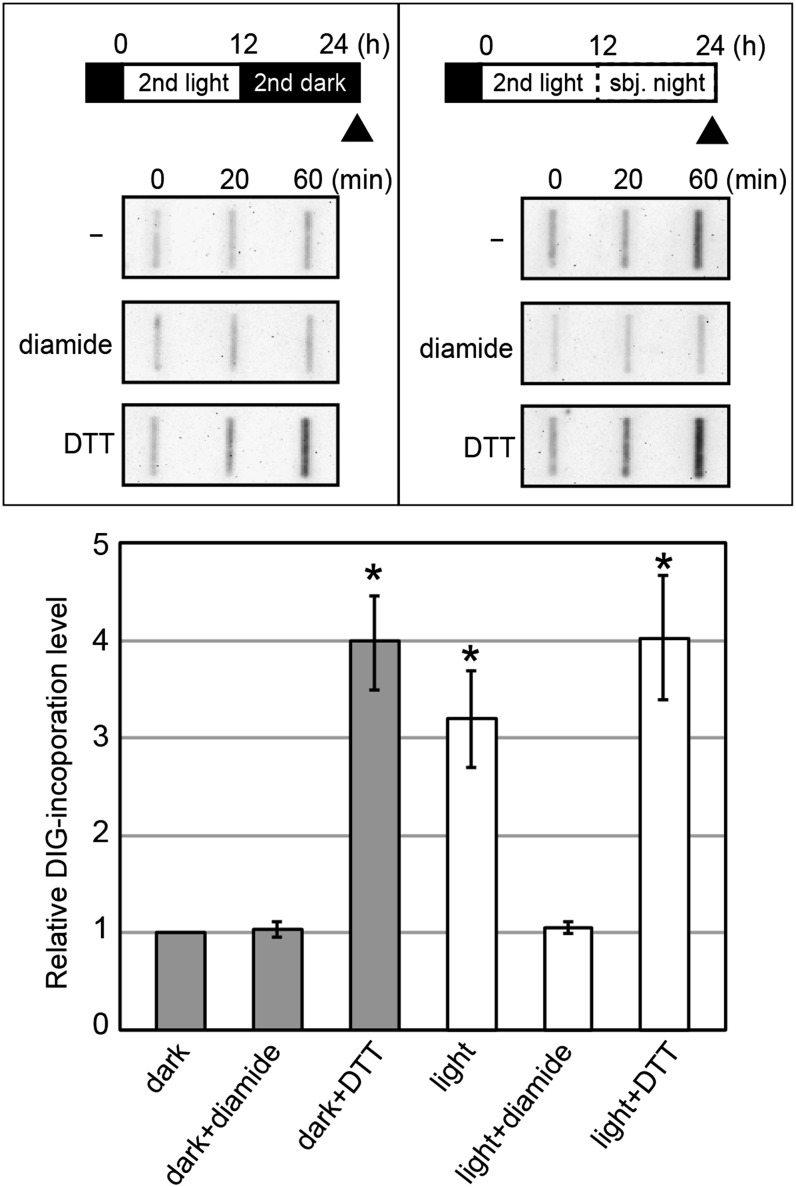
Next, we asked whether chloroplast nucleoids are sufficient to sense the redox state and thus affect DNA replication activity. To this end, we performed an in vitro chloroplast DNA synthesis assay using isolated chloroplast nucleoids (Fig. 7). The isolated chloroplasts were solubilized by the nonionic detergent Nonidet P-40, and the nucleoids were isolated. As in the case of isolated chloroplasts, DNA synthesis activity was found to be higher in the nucleoids isolated from the light-exposed culture than that from the culture under darkness. DTT increased while diamide reduced the DNA synthesis activity. These results suggest that the chloroplast nucleoid is sufficient to respond to changes in the redox state in order to regulate DNA replication.
Figure 7.
Effects of redox reagents on chloroplast DNA replication in isolated chloroplast nucleoids. Chloroplast nucleoids were isolated from synchronous culture (the isolated time points were the same as in Fig. 6). Diamide or DTT was added to chloroplast nucleoids. After preincubation for 15 min, DNA synthesis was examined as the incorporation of DIG-labeled dUTP, as described in Figure 6. The relative levels of the incorporated DIG are plotted. The level of the culture grown under darkness at 60 min is defined as 1.0. Error bars represent the sd of three biological replicates. Asterisks indicate significant differences from the dark by Student’s t test (P < 0.001).
DISCUSSION
Previous studies using a 24-h light/dark synchronous culture of algae showed that the chloroplast DNA is replicated only during the light period (G1 phase), prior to nuclear DNA replication (S phase), chloroplast division, and cytokinesis, which occur during the dark period (Chiang and Sueoka, 1967; Suzuki et al., 1994; Zachleder et al., 1995). Based on these results, it was concluded that the DNA replication and division phases are temporally separated in chloroplasts. However, here we have shown that the chloroplast DNA is replicated independently of the timing of chloroplast division and the cell cycle and that chloroplast DNA continues to be replicated as long as light energy is available to drive photosynthetic electron transfer during photoautotrophic growth (Figs. 1 and 2).
Under the continuous light condition after entrainment with a light/dark cycle in the photoautotrophic medium, chloroplast DNA continued to be replicated and the chloroplast and cell sizes also continued to increase, even in the S/M phase (Fig. 1, A and B). Because cells grown under continuous light undergo more rounds of the S/M phase than cells grown in the light/dark condition, the chloroplast DNA level per cell/chloroplast does not change in daughter cells in the two conditions (Fig. 1C). Given these results, it appears that chloroplast DNA replication is correlated with cell/chloroplast growth, thereby maintaining the proper DNA content per cell/chloroplast volume. The significance of chloroplast polyploidy has been much debated. Mutation in the organellar DNA polymerase in maize (Zea mays) reduces the chloroplast DNA copy number as well as the chloroplast-encoded transcripts and proteins, suggesting that the DNA copy number is a limiting factor for the expression of chloroplast-encoded genes (Udy et al., 2012). Other recent studies revealed a reduction in the chloroplast DNA copy number under a phosphate-limited condition in C. reinhardtii (Yehudai-Resheff et al., 2007) and an increase in the copy number under a phosphate-rich condition in the green alga Nannochloris bacillaris (Sumiya et al., 2008). These studies raise the possibility that polyploidal chloroplast DNA may at least serve as a repository of phosphorus. Taken together, one possibility is that the chloroplast DNA replication is correlated with cell/chloroplast growth by phosphorus that is assimilated by reducing power supplied by photosynthesis or the uptake of acetate. On this point, further studies will be required.
When cells were cultured in a photoautotrophic medium, photosynthetic electron flow was required for the chloroplast DNA replication (Fig. 2). However, even under darkness, chloroplast DNA replicated in a heterotrophic medium that contained acetate as sources of carbon and reducing power (Fig. 3). These results raised the possibility that the chloroplast DNA replication is linked to the cellular redox state. Consistent with this assumption, Lau et al. (2000) showed that cells treated with cadmium and cells growing in a nitrogen-replete medium, in which levels of chloroplast DNA decreased, contained low GSH levels. In this study, we showed that artificial alteration of the cellular redox state by redox reagents changed the activity of chloroplast DNA replication (Fig. 4). In our study, chloroplast DNA replication was blocked by the addition of methylviologen under light (Fig. 4A). In contrast, replication of the chloroplast DNA, but not that of nuclear DNA, was activated by the addition of DMTU under dark (Fig. 4, B and C). In addition, we confirmed that cellular redox state was changed when cells were treated with the above redox reagents (Table I). Thus, our results suggest that the cellular redox state affects the level of chloroplast DNA replication.
For factors that link chloroplast DNA replication and the cellular redox state, three candidates were considered: (1) the level or (2) the replication activity of the POP protein (chloroplast DNA polymerase) and (3) the level of dNTPs (substrates for DNA replication). The results show that the POP level is constant under the light and dark conditions during photoautotrophic growth (Fig. 5). The results obtained showed that DNA replication activity is higher under light than under dark even when excessive levels of dNTPs were supplied in vitro (Figs. 6 and 7). Thus, our results suggest that the activity of the DNA polymerase (probably POP) is a rate-limiting factor in chloroplast DNA replication. However, in vitro DNA synthesis was still observed in chloroplasts isolated from culture grown under darkness (Figs. 6 and 7). Therefore, at this point, we cannot completely rule out the possibility that change in the intrachloroplast dNTP level might also be involved in the regulation of the chloroplast DNA replication. However, replication of the nuclear DNA occurs during the dark period in photoautotrophic synchronous culture, indicating that there is at least a sufficient dNTP level for DNA synthesis in the whole cell.
Chloroplast DNA replication in vitro was activated by DTT but was inactivated by diamide, which is a sulfhydryl group-specific oxidative agent (Figs. 6 and 7). These results suggest that the activity of the chloroplast DNA polymerase is regulated by the redox state of the sulfhydryl group, at least in some of the chloroplast proteins. Besides, given that chloroplast nucleoids are sufficient to sense the redox state to change the DNA replication activity (Fig. 7), the sulfhydryl group in certain chloroplast nucleoid-associated proteins, including POP itself, may link the redox state to DNA replication activity. In this regard, FrxB protein is likely a candidate. FrxB binds to the chloroplast DNA replicative origin in C. reinhardtii. In addition, FrxB is an iron-sulfur redox protein subunit of chloroplast NADH dehydrogenase, raising the possibility that FrxB links the redox state to chloroplast DNA replication (Wu et al., 1989, 1993).
For a better understanding of the regulation of chloroplast DNA replication, identification of the redox-sensing, replication-related proteins will be required. Plant and algal genomes do not encode any DNA replication-related proteins of cyanobacterial origin, such as the DNA polymerase and DnaA protein, which are involved in the initiation of replication in bacteria. Therefore, biochemical approaches, including disulfide proteomics of the chloroplast nucleoids, will be needed to determine the eukaryote-specific mechanism of the redox state-based regulation of chloroplast DNA replication.
MATERIALS AND METHODS
Synchronous Culture
Chlamydomonas reinhardtii 137c mt+ and cw-15 cells were cultured in Sueoka’s high-salt medium (HSM; Sueoka, 1967) photoautotrophically. For synchronization, the cells in log phase were subcultured to 1 × 106 cells mL−1 and were subjected to a 12-h-light/12-h-dark cycle (100 µmol photons m−2 s−1) at 24°C under aeration with ambient air (Surzycki, 1971). Cells in the second and third cycles were used for further analyses.
Nonsynchronous Heterotrophic Culture
For heterotrophic culture, the 137c mt+ cells were cultured in Tris-acetate-phosphate (Gorman and Levine, 1965) medium under complete darkness at 24°C and ambient aeration with ambient air (Chen and Johnson, 1996).
Drug Treatments
For the drug treatments, 137c mt+ cells were used. For DCMU treatment, a 1:1,000 volume of 50 mm DCMU stock solution in ethanol was added to nonsynchronous log-phase culture that was grown under continuous light. For methylviologen treatment, a 1:1,000 volume of 1 mm methylviologen stock solution in water was added to nonsynchronous log-phase cultures that were grown under continuous light. For DMTU treatment, a 1:100 volume of 2 m DMTU stock solution dissolved in HSM was added to the synchronous culture at the end of the second dark period.
Quantification of Nuclear and Chloroplast DNA by qPCR
The DNA level was analyzed by qPCR. Cells were harvested from 0.5 mL of culture by centrifugation at 1,000g for 5 min and stored at −80°C until DNA extraction. Cells were resuspended in 50 mm Tris-HCl, pH 8.0, 20 mm EDTA, 200 mm NaCl, and 1% SDS buffer, and then DNA was extracted by the phenol/chloroform method. The extracted DNA was dissolved in 100 µL of distilled water. qPCR amplification was performed using Power SYBR Green PCR Master Mix (Applied Biosystems) in a real-time PCR system (StepOne Plus; Applied Biosystems). qPCR was performed using the primers 5′-GCGCCCGCAGCTGTACT-3′ and 5′-AACATGGGCAGCTTCCACAT-3′ for the nuclear DNA (RBCS1 gene) and 5′-GTCGTGACCTTGCTCGTGAA-3′ and 5′-TCTGGAGACCATTTACAAGCTGAA-3′ for the chloroplast DNA (rbcL gene).
Quantification of GSH and GSSG
Cells were harvested by centrifugation at 1,000g for 5 min at 4°C and then washed with phosphate-buffered saline. Cells were resuspended in 5% sulfosalicylic acid solution and disrupted by sonication. Supernatants were obtained by centrifugation at 15,000g for 5 min at 4°C. Chloroplasts isolated as described below were harvested by centrifugation at 700g for 5 min at 4°C and then were washed with 50 mm HEPES-KOH, pH 7.5, containing 300 mm sorbitol. The isolated chloroplasts were resuspended in 5% sulfosalicylic acid solution and centrifuged at 15,000g for 5 min at 4°C to remove debris. GSH and GSSG extracted into the supernatants were quantified by using the GSSG/GSH quantification kit (Dojindo) according to the manufacturer’s instructions.
Antibody Preparation
The complementary DNA sequence encoding a partial fragment of CrPOP was amplified by PCR using the primers 5′-CACCAGCCGCAGCGGCAAGAAGACCTACGG-3′ and 5′-CTACACATTCATGTAGCGGCTCACCTTGAT-3′. The PCR product was cloned into a pET100 expression vector (Invitrogen), and the 6xHis fusion polypeptide was expressed in Rosetta (DE3) Escherichia coli cells and purified using a HisTrap HP column (GE Healthcare). The polypeptide was further separated electrophoretically on a preparative acrylamide gel. The antibodies against CrPOP were raised in rabbits using the gel band containing the recombinant polypeptide, and then antibodies were affinity purified from the antisera using an NHS-activated HiTrap HP column (GE Healthcare), to which recombinant polypeptide (eluate of the HisTrap HP column) was covalently bound.
Immunoblot Analyses
Cells were harvested from 5 mL of culture by centrifugation and stored at −80°C. Cells were suspended in the protein extraction buffer (50 mm Tris, pH 7.5, 8 m urea, and 0.1% Triton X-100) and sonicated. Protein content was determined by Bradford assay, and equal amounts of the total proteins were subjected to immunoblot analyses. Immunoblotting assays were performed as described previously (Kabeya et al., 2010). Anti-CrPOP was used at a dilution of 1:1,000.
Isolation of Intact Chloroplasts
Chloroplasts were isolated using a modified version of the method of Mason et al. (2006). All manipulations were done at 4°C or on ice. The cw-15 cells were synchronously cultured to less than 1.0 × 107 cells mL−1. Cells were harvested by centrifugation at 3,000g for 10 min and resuspended in chloroplast (CP) isolation buffer (50 mm HEPES-KOH, pH 7.5, 2 mm EDTA, 1 mm MgCl2, 1% bovine serum albumin, and 300 mm sorbitol). The cells were then broken by a single passage through an airbrush (0.2-mm-aperture airbrush, HP-60; Olympos) at a pressure of 0.7 kg cm−2 according to the reported airbrush method (Nishimura et al., 2002). The lysate was centrifuged at 750g for 2 min, and the pellet was gently resuspended in 3 mL of CP isolation buffer using a paintbrush. The sample was layered on top of a discontinuous Percoll gradient (20%/45%/65% Percoll in CP isolation buffer). After centrifugation at 3,220g for 30 min, the green band that appeared at the interface between the 45% and 65% layers was collected. It was confirmed by DAPI staining that the collected fraction contained scant nuclear or mitochondrial contamination. The chloroplast-rich fraction was diluted with CP isolation buffer and then centrifuged at 670g for 1 min to remove the Percoll. The purified chloroplasts were used for in vitro DNA synthesis or isolation of the nucleoids.
Isolation of the Chloroplast Nucleoids
Chloroplast nucleoids were isolated using a modified version of the method of Sato et al. (1998). All manipulations were performed at 4°C or on ice. Isolated chloroplasts were lysed with 2% Nonidet P-40 dissolved in TAN buffer (20 mm Tris-HCl, pH 7.5, 0.5 mm EDTA, 7 mm 2-mercaptoethanol, 1.2 mm spermidine, and 0.5 m Suc) for 1 h. Then the sample was centrifuged at 20,000g for 30 min. The pellet with the enriched chloroplast nucleoids was resuspended with TAN buffer containing 2% Nonidet P-40 and centrifuged again. The isolated nucleoids were used for in vitro DNA synthesis.
Detection of Newly Synthesized Chloroplast DNA Using the Isolated Chloroplasts and Chloroplast Nucleoids
Chloroplasts (1 × 107) were suspended in 300 µL of hypertonic replication buffer (50 mm Tris-HCl, pH 8.0, 50 mm KCl, and 5 mm MgCl2). Nucleoids isolated from 1 × 109 chloroplasts were suspended in 300 µL of replication buffer. After incubation with or without 10 mm DTT, 0.5 mm NADPH, 0.1 mm thioredoxin (203-13041; Wako) reduced by DTT, 10 mm GSH, 1 mm diamide, or 10 µg mL−1 ethidium bromide for 15 min at 24°C, a one-tenth-volume of DIG labeling mix (Roche) was added and then incubated at 24°C up to 1 h. After incubation, DNA was extracted at 0, 20, and 60 min and slot blotted onto a Hybond-N+ membrane. The signals representing newly synthesized chloroplast DNA were detected by anti-DIG-alkaline phosphatase conjugates (11585550910; Roche) and the CDP-Star system (Applied Biosystems).
Acknowledgments
We thank Y. Ono and A. Yamashita for technical support. We also thank Dr. N. Sumiya, Dr. S. Hirooka, and Ms. M. Nakamura for useful discussions.
Glossary
- qPCR
quantitative real-time PCR
- DCMU
3-(3′,4′-dichlorophenyl)-1,1-dimethylurea
- DMTU
dimethylthiourea
- GSH
reduced glutathione
- GSSG
oxidized glutathione
- DAPI
4′6′-diamidino-phenylindole
- dNTP
deoxyribonucleotide triphosphate
- POP
plant organellar DNA polymerase
- DIG
digoxigenin
- DTT
dithiothreitol
- HSM
high-salt medium
- CP
chloroplast
- DCMU
3-(3,4-dichlorophenyl)-1,1-dimethylurea
References
- Archibald JM. (2009) The puzzle of plastid evolution. Curr Biol 19: R81–R88 [DOI] [PubMed] [Google Scholar]
- Baumgartner BJ, Rapp JC, Mullet JE. (1989) Plastid transcription activity and DNA copy number increase early in barley chloroplast development. Plant Physiol 89: 1011–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder BJ, Chisholm SW. (1990) Relationship between DNA cycle and growth rate in Synechococcus sp. strain PCC 6301. J Bacteriol 172: 2313–2319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boffey SA, Leech RM. (1982) Chloroplast DNA levels and the control of chloroplast division in light-grown wheat leaves. Plant Physiol 69: 1387–1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boffey SA, Lloyd D, editors (1988) The Division and Segregation of Organelles. Cambridge University Press, Cambridge, UK [Google Scholar]
- Boye E, Løbner-Olesen A, Skarstad K. (2000) Limiting DNA replication to once and only once. EMBO Rep 1: 479–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Johnson MR. (1996) Heterotrophic growth of Chlamydomonas reinhardtii on acetate in chemostat culture. Process Biochem 31: 601–604 [Google Scholar]
- Chiang KS, Sueoka N. (1967) Replication of chloroplast DNA in Chlamydomonas reinhardtii during vegetative cell cycle: its mode and regulation. Proc Natl Acad Sci USA 57: 1506–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman DS, Levine RP. (1965) Cytochrome f and plastocyanin: their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 54: 1665–1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto K, Johnson CH. (1995) Is the cell division cycle gated by a circadian clock? The case of Chlamydomonas reinhardtii. J Cell Biol 129: 1061–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant D, Swinton D, Chiang K. (1978) Differential patterns of mitochondrial, chloroplastic and nuclear DNA synthesis in the synchronous cell cycle of Chlamydomonas reinhardtii. Planta 141: 259–267 [DOI] [PubMed] [Google Scholar]
- Griese M, Lange C, Soppa J. (2011) Ploidy in cyanobacteria. FEMS Microbiol Lett 323: 124–131 [DOI] [PubMed] [Google Scholar]
- Harris EH, editor (1989) The Chlamydomonas Sourcebook. Academic Press, San Diego [Google Scholar]
- Hashimoto H, Possingham JV. (1989) Effect of light on the chloroplast division cycle and DNA synthesis in cultured leaf discs of spinach. Plant Physiol 89: 1178–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y, Nakanishi H, Suzuki K, Ichikawa T, Kondou Y, Matsui M, Miyagishima SY. (2010) The YlmG protein has a conserved function related to the distribution of nucleoids in chloroplasts and cyanobacteria. BMC Plant Biol 10: 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling PJ. (2010) The endosymbiotic origin, diversification and fate of plastids. Philos Trans R Soc Lond B Biol Sci 365: 729–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroiwa T. (1991) The replication, differentiation, and inheritance of plastids with emphasis on the concept of organelle nuclei. Int Rev Cytol 128: 1–62 [Google Scholar]
- Lamppa GK, Bendich AJ. (1979) Changes in chloroplast DNA levels during development of pea (Pisum sativum). Plant Physiol 64: 126–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau KW, Ren J, Wu M. (2000) Redox modulation of chloroplast DNA replication in Chlamydomonas reinhardtii. Antioxid Redox Signal 2: 529–535 [DOI] [PubMed] [Google Scholar]
- Levine A, Tenhaken R, Dixon R, Lamb C. (1994) H2O2 from the oxidative burst orchestrates the plant hypersensitive disease resistance response. Cell 79: 583–593 [DOI] [PubMed] [Google Scholar]
- Mann N, Carr NG. (1974) Control of macromolecular composition and cell division in the blue-green algae Anacystis nidulans. J Gen Microbiol 83: 399–405 [DOI] [PubMed] [Google Scholar]
- Maple J, Møller SG. (2010) The complexity and evolution of the plastid-division machinery. Biochem Soc Trans 38: 783–788 [DOI] [PubMed] [Google Scholar]
- Mason CB, Bricker TM, Moroney JV. (2006) A rapid method for chloroplast isolation from the green alga Chlamydomonas reinhardtii. Nat Protoc 1: 2227–2230 [DOI] [PubMed] [Google Scholar]
- Matsushima R, Tang LY, Zhang L, Yamada H, Twell D, Sakamoto W. (2011) A conserved, Mg²+-dependent exonuclease degrades organelle DNA during Arabidopsis pollen development. Plant Cell 23: 1608–1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagishima SY. (2011) Mechanism of plastid division: from a bacterium to an organelle. Plant Physiol 155: 1533–1544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagishima SY, Suzuki K, Okazaki K, Kabeya Y. (2012) Expression of the nucleus-encoded chloroplast division genes and proteins regulated by the algal cell cycle. Mol Biol Evol 29: 2957–2970 [DOI] [PubMed] [Google Scholar]
- Miyamura S, Nagata T, Kuroiwa T. (1986) Quantitative fluorescence microscopy on dynamic changes of plastid nucleoids during wheat development. Protoplasma 133: 66–72 [Google Scholar]
- Mori T, Binder B, Johnson CH. (1996) Circadian gating of cell division in cyanobacteria growing with average doubling times of less than 24 hours. Proc Natl Acad Sci USA 93: 10183–10188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama T, Terasawa K, Fujiwara M, Sato N. (2008) Purification and characterization of organellar DNA polymerases in the red alga Cyanidioschyzon merolae. FEBS J 275: 2899–2918 [DOI] [PubMed] [Google Scholar]
- Nishimura Y, Misumi O, Kato K, Inada N, Higashiyama T, Momoyama Y, Kuroiwa T. (2002) An mt(+) gamete-specific nuclease that targets mt(−) chloroplasts during sexual reproduction in C. reinhardtii. Genes Dev 16: 1116–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldenburg DJ, Bendich AJ. (2004) Most chloroplast DNA of maize seedlings in linear molecules with defined ends and branched forms. J Mol Biol 335: 953–970 [DOI] [PubMed] [Google Scholar]
- Oldenburg DJ, Rowan BA, Zhao L, Walcher CL, Schleh M, Bendich AJ. (2006) Loss or retention of chloroplast DNA in maize seedlings is affected by both light and genotype. Planta 225: 41–55 [DOI] [PubMed] [Google Scholar]
- Ono Y, Sakai A, Takechi K, Takio S, Takusagawa M, Takano H. (2007) NtPolI-like1 and NtPolI-like2, bacterial DNA polymerase I homologs isolated from BY-2 cultured tobacco cells, encode DNA polymerases engaged in DNA replication in both plastids and mitochondria. Plant Cell Physiol 48: 1679–1692 [DOI] [PubMed] [Google Scholar]
- Parent JS, Lepage E, Brisson N. (2011) Divergent roles for the two PolI-like organelle DNA polymerases of Arabidopsis. Plant Physiol 156: 254–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyke KA. (2013) Divide and shape: an endosymbiont in action. Planta 237: 381–387 [DOI] [PubMed] [Google Scholar]
- Rodríguez-Ezpeleta N, Philippe H. (2006) Plastid origin: replaying the tape. Curr Biol 16: R53–R56 [DOI] [PubMed] [Google Scholar]
- Rollins MJ, Harper JDI, John PCL. (1983) Synthesis of individual proteins, including tubulins and chloroplast membrane proteins, in synchronous cultures of the eukaryote Chlamydomonas reinhardtii: elimination of periodic changes in protein synthesis and enzyme activity under constant environmental conditions. J Gen Microbiol 129: 1899–1919 [Google Scholar]
- Rowan BA, Bendich AJ. (2009) The loss of DNA from chloroplasts as leaves mature: fact or artefact? J Exp Bot 60: 3005–3010 [DOI] [PubMed] [Google Scholar]
- Sato N, Ohshima K, Watanabe A, Ohta N, Nishiyama Y, Joyard J, Douce R. (1998) Molecular characterization of the PEND protein, a novel bZIP protein present in the envelope membrane that is the site of nucleoid replication in developing plastids. Plant Cell 10: 859–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheibe R. (1991) Redox-modulation of chloroplast enzymes: a common principle for individual control. Plant Physiol 96: 1–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaver JM, Oldenburg DJ, Bendich AJ. (2008) The structure of chloroplast DNA molecules and the effects of light on the amount of chloroplast DNA during development in Medicago truncatula. Plant Physiol 146: 1064–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sueoka N. (1967) Mitotic replication of deoxyribonucleic acid in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 46: 83–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumiya N, Hirata A, Kawano S. (2008) Multiple FtsZ ring formation and reduplicated chloroplast DNA in Nannochloris bacillaris (Chlorophyta, Trebouxiophyceae) under phosphate-enriched culture. J Phycol 44: 1476–1489 [DOI] [PubMed] [Google Scholar]
- Surzycki S. (1971) Synchronously grown cultures of Chlamydomonas reinhardtii. Methods Enzymol 23: 67–73 [Google Scholar]
- Suzuki K, Ehara T, Osafune T, Kuroiwa H, Kawano S, Kuroiwa T. (1994) Behavior of mitochondria, chloroplasts and their nuclei during the mitotic cycle in the ultramicroalga Cyanidioschyzon merolae. Eur J Cell Biol 63: 280–288 [PubMed] [Google Scholar]
- Taiz L, Zeiger E, editors (2010) Plant Physiology, Ed 5. Sinauer Associates, Sunderland, MA [Google Scholar]
- Udy DB, Belcher S, Williams-Carrier R, Gualberto JM, Barkan A. (2012) Effects of reduced chloroplast gene copy number on chloroplast gene expression in maize. Plant Physiol 160: 1420–1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S, Ohbayashi R, Shiwa Y, Noda A, Kanesaki Y, Chibazakura T, Yoshikawa H. (2012) Light-dependent and asynchronous replication of cyanobacterial multi-copy chromosomes. Mol Microbiol 83: 856–865 [DOI] [PubMed] [Google Scholar]
- Wu M, Chang C, Yang J, Zhang Y, Nie Z, Hsieh C. (1993) Regulation of chloroplast DNA replication in Chlamydomonas reinhardtii. Bot Bull Acad Sin 34: 115–131 [Google Scholar]
- Wu M, Nie ZQ, Yang J. (1989) The 18-kD protein that binds to the chloroplast DNA replicative origin is an iron-sulfur protein related to a subunit of NADH dehydrogenase. Plant Cell 1: 551–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Glynn JM, Olson BJ, Schmitz AJ, Osteryoung KW. (2008) Plastid division: across time and space. Curr Opin Plant Biol 11: 577–584 [DOI] [PubMed] [Google Scholar]
- Yehudai-Resheff S, Zimmer SL, Komine Y, Stern DB. (2007) Integration of chloroplast nucleic acid metabolism into the phosphate deprivation response in Chlamydomonas reinhardtii. Plant Cell 19: 1023–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachleder V, Kawano S, Kuroiwa T. (1995) The course of chloroplast DNA replication and its relationship to other reproductive processes in the chloroplast and nucleocytoplasmic compartment during the cell cycle of the alga Scenedesmus quadricauda. Protoplasma 188: 245–251 [Google Scholar]
- Zakrzewska-Czerwińska J, Jakimowicz D, Zawilak-Pawlik A, Messer W. (2007) Regulation of the initiation of chromosomal replication in bacteria. FEMS Microbiol Rev 31: 378–387 [DOI] [PubMed] [Google Scholar]