Protein-coding overlapping genes in Arabidopsis have unexpectedly high levels and breadths of expression.
Abstract
Pairs of genes within eukaryotic genomes are often located on opposite DNA strands such that transcription generates cis-natural sense antisense transcripts (cis-NATs). This orientation of genes has been associated with the biogenesis of splice variants and natural antisense small RNAs. Here, in an analysis of currently available data, we report that within Arabidopsis (Arabidopsis thaliana), protein-coding cis-NATs are also characterized by high abundance, high coexpression, and broad expression. Our results suggest that a permissive chromatin environment may have led to the proximity of these genes. Compared with other genes, cis-NAT-encoding genes have enriched low-nucleosome-density regions, high levels of histone H3 lysine-9 acetylation, and low levels of H3 lysine-27 trimethylation. Promoters associated with broadly expressed genes are preferentially found in the 5′ regulatory sequences of cis-NAT-encoding genes. Our results further suggest that natural antisense small RNA production from cis-NATs is limited. Small RNAs sequenced from natural antisense small RNA biogenesis mutants including dcl1, dcl2, dcl3, and rdr6 map to cis-NATs as frequently as small RNAs sequenced from wild-type plants. Future work will investigate if the positive transcriptional regulation of overlapping protein-coding genes contributes to the prevalence of these genes within other eukaryotic genomes.
Among eukaryotic genomes, a widespread arrangement of protein-coding (PC) genes is one in which the genes are encoded by complementary strands of DNA at the same locus, thereby generating cis-natural sense antisense transcripts (cis-NATs). In higher eukaryotic organisms, 4% to 26% of PC genes are predicted to generate cis-NATs, ranging from 4% of the PC genes in zebrafish to 26% in Drosophila spp. (Soldà et al., 2008). Overall, about 8% of PC genes in Arabidopsis (Arabidopsis thaliana) are transcribed to cis-NATs, about the same frequency as in human and mouse (Chen et al., 2004; Jen et al., 2005; Wang et al., 2005; Zhang et al., 2006; Soldà et al., 2008).
Within Arabidopsis, cis-NATs have been associated with two regulatory functions. First, one cis-NAT may regulate the transcript abundance of its complement by triggering the biogenesis of natural antisense small RNAs (nat-siRNAs) that subsequently guide transcript cleavage (Borsani et al., 2005; Katiyar-Agarwal et al., 2006; Ron et al., 2010). Abiotic or biotic stresses have been proposed to induce nat-siRNA production from cis-NATs (Borsani et al., 2005; Katiyar-Agarwal et al., 2006; Jin et al., 2008). More than 10,000 small RNAs (sRNAs) from plant stress sRNA libraries map to overlapping regions of cis-NAT transcripts (Zhang et al., 2012). Second, Jen et al., (2005) noted that cis-NAT-encoding genes had far more alternative splice variants and alternative polyadenylation sites than other genes, suggesting a function in antisense transcript-induced RNA splicing, alternative splicing, and polyadenylation.
Transcription occurs within physically distinct regions of the nucleus enriched in active RNA polymerase II as well as other transcriptional regulatory and accessory factors (Edelman and Fraser, 2012). The concentration of transcriptional machinery within a small part of the nuclear space has been proposed to enhance the efficiency of transcription. We postulated that the physical proximity of cis-NAT-encoding genes favors their high and broad expression. Here, we test this theory by analyzing RNA-Seq data from Arabidopsis plants grown in six distinct conditions. We detect cis-NAT transcripts at high frequency and find that they have remarkably broad and high transcript levels. Promoter sequences of broadly expressed genes are highly represented in cis-NAT-encoding genes’ upstream regulatory regions, and the chromatin of cis-NAT-encoding genes is enriched for euchromatic marks. sRNAs appear to play a limited role in cis-NAT regulation, as sRNAs sequenced from sRNA biogenesis mutants map to PC cis-NATs at a similar frequency as in wild-type plants.
RESULTS
PC cis-NATs Have High Abundance
The Arabidopsis genome contains 33,239 genes and 33,234 adjacent gene pairs (Table I), as annotated by The Arabidopsis Information Resource (release 9). A total of 5.1% (1,710 of 33,234) of the adjacent genes, evenly distributed across chromosomes, may generate cis-NATs. We classified the pairs into three types. In type I, sense and antisense transcripts are complementary in their 3′ ends. In type II, they are complementary in their 5′ ends. In type III, one entire transcript has homology to a subsequent second transcript. PC gene cis-NAT pairs are overrepresented among cis-NATs, and type I pairs are highly likely to be PC. Among all cis-NAT-encoding gene pairs, 82% (1,402 of 1,710) encode for two PC transcripts (Table II), a significantly larger proportion than expected given the frequency of PC genes within the genome (P < 1e-10). Of the 1,363 type I cis-NAT pairs, 93.3% are PC cis-NATs (1,272 of 1,363; Table II), again significantly more than expected (P < 1e-10).
Table I. Chromosomal distribution of cis-NAT pairs.
| Chromosome | Type of cis-NAT Pairsa |
All Genes |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Type I |
Type II |
Type III |
Total |
|||||||
| Pairs | %b | Pairs | % | Pairs | % | Pairs | % | Pairs | Genes | |
| 1 | 368 | 4.4 | 35 | 0.4 | 51 | 0.6 | 454 | 5.4 | 8,397 | 8,398 |
| 2 | 223 | 4.1 | 32 | 0.6 | 44 | 0.8 | 299 | 5.4 | 5,496 | 5,497 |
| 3 | 236 | 3.5 | 29 | 0.4 | 39 | 0.6 | 304 | 4.5 | 6,727 | 6,728 |
| 4 | 203 | 4.0 | 27 | 0.5 | 25 | 0.5 | 255 | 5.0 | 5,121 | 5,122 |
| 5 | 333 | 4.4 | 29 | 0.4 | 36 | 0.5 | 398 | 5.3 | 7,493 | 7,493 |
| Total | 1,363 | 4.1 | 152 | 0.5 | 195 | 0.6 | 1,710 | 5.1 | 33,234 | 33,239 |
Type I, tail to tail (3′–3′); type II, head to head (5′–5′); type III, one transcript lies within another transcript. bProportion of cis-NAT pairs in the transcript pairs of its corresponding chromosome.
Table II. Chromosomal locations and orientations of putative PC cis-NAT gene pairs.
| Chromosome | Type of PC
cis-NAT Pairsa |
|||||||
|---|---|---|---|---|---|---|---|---|
| Type I |
Type II |
Type III |
Total |
|||||
| Pairs | %b | Pairs | % | Pairs | % | Pairs | % | |
| 1 | 341 | 92.7 | 16 | 45.7 | 11 | 21.6 | 368 | 81.1 |
| 2 | 199 | 89.2 | 8 | 25.0 | 23 | 52.3 | 230 | 76.9 |
| 3 | 227 | 96.2 | 4 | 13.8 | 14 | 35.9 | 245 | 80.6 |
| 4 | 191 | 94.1 | 15 | 55.6 | 8 | 32.0 | 214 | 83.9 |
| 5 | 314 | 94.3 | 19 | 65.5 | 12 | 33.3 | 345 | 86.7 |
| Total | 1,272 | 93.3 | 62 | 40.8 | 68 | 34.9 | 1,402 | 82.0 |
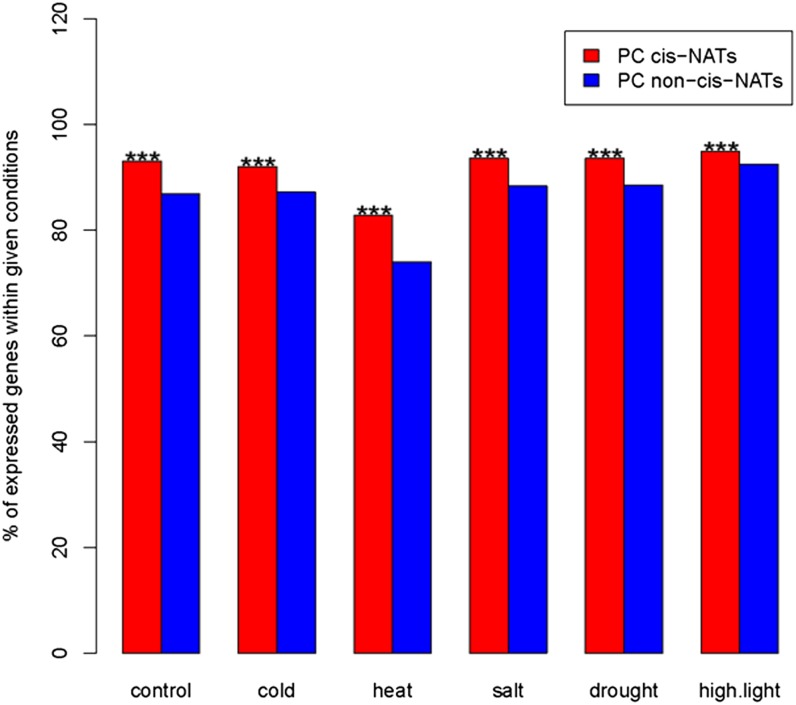
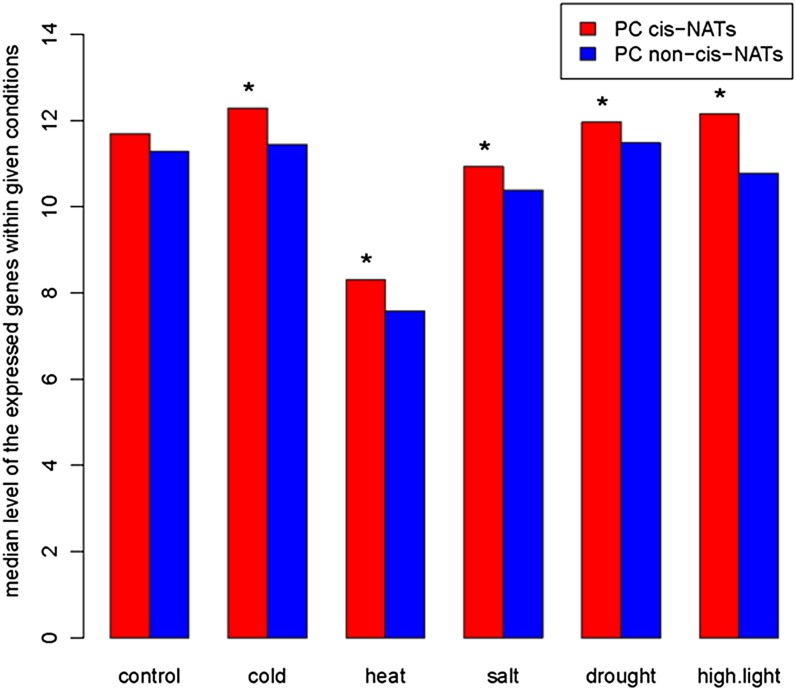
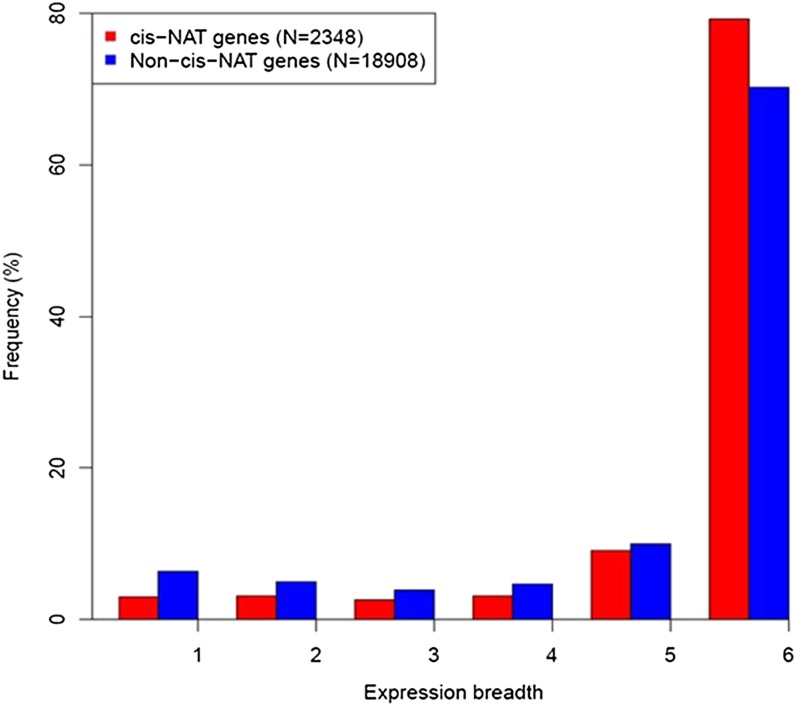
To assay transcript patterns of PC cis-NATs, we mapped RNA-Seq microreads from plants grown in standard, cold, heat, salt, drought, and high-light conditions to PC genes (Filichkin et al., 2010). Of the 2,804 cis-NAT-encoding genes within PC gene pairs, 2,338 (83%) had evidence of transcription in at least one condition. In contrast, among the 24,365 PC genes that do not encode cis-NATs, 18,908 (77%) had evidence for transcription, a significant difference (P < 0.01). Examining only genes with evidence for transcription, we first compared the frequency with which microreads mapped to cis-NATs compared with other transcripts in each growth condition. A higher proportion of cis-NAT-encoding genes were expressed relative to non-cis-NAT-encoding genes in each condition (P < 0.001; Fig. 1). The proportion of expressed cis-NAT-encoding genes in each condition also exceeded the proportion of non-cis-NATs when we used stricter criteria for defining evidence for transcription (Supplemental Fig. S1). Second, we investigated if cis-NAT-encoding genes had higher transcript abundances than non-cis-NAT-encoding genes. In all conditions except standard growth conditions, median cis-NAT abundances were significantly higher than non-cis-NAT abundances (P < 0.05; Fig. 2). Summing across conditions, the median expression level of the 2,338 genes within the 1,174 protein-encoding cis-NAT pairs was 62.8 reads per kilobase of exon model per million mapped reads (RPKM), significantly higher (Wilcoxon rank sum test, P = 1.19e-11) than the median expression level of the 18,908 expressed PC non-cis-NAT genes (52.5 RPKM). Third, PC cis-NAT genes were also more broadly expressed than were PC non-cis-NAT genes. The distribution of expression breadths of the PC cis-NAT genes was shifted to the right compared with the distribution of expression breadths of the PC non-cis-NAT genes (Wilcoxon rank sum test, P < 2.2e-16; Fig. 3). Most notably, the proportion of PC cis-NAT genes expressed in all six conditions was 9% higher than the proportion of PC non-cis-NAT genes (79.2% versus 70.2%; Fig. 3).
Figure 1.
PC genes that generate cis-NATs are more frequently expressed than are PC genes that do not. The y axis denotes the percentage of expressed PC genes out of 2,338 cis-NATs and 18,908 non-cis-NATs. The x axis denotes the six different growth conditions. Asterisks indicate that the proportion of PC cis-NAT-encoding genes expressed within a given condition is significantly greater than the proportion of PC non-cis-NAT-encoding genes at P < 1e-03. [See online article for color version of this figure.]
Figure 2.
Expressed PC cis-NATs accumulate to higher abundances than do expressed PC non-cis-NATs. The median expression levels of expressed PC cis-NATs and PC non-cis-NATs are noted on the y axis in RPKM for six growth conditions. Asterisks indicate that the median RPKM of expressed PC cis-NATs within a given condition is significantly greater than the median RPKM of expressed PC non-cis-NATs at P < 0.05. [See online article for color version of this figure.]
Figure 3.
Distributions of the expression breadths of PC cis-NAT genes and PC non-cis-NAT genes. Expression breadth is the number of growth conditions in which each PC cis-NAT gene and each PC non-cis-NAT gene was expressed. The six conditions are control, high light, heat, cold, salt, and drought. Frequencies of PC cis-NAT genes and PC non-cis-NAT genes with different expression breadths are plotted. [See online article for color version of this figure.]
A standard method for estimating the transcript levels of a gene from RNA-Seq data is to distribute reads that map to multiple transcripts across the target transcripts. In our case, because each cis-NAT overlaps another cis-NAT and our RNA-Seq reads were not strand specific, it is possible that the number of expressed cis-NAT genes and the breadth of expressed cis-NAT genes were inflated. Thus, we also assayed patterns of transcript abundance using only RNA-Seq reads that mapped to a single transcript. The results were similar to the findings when using weighted RNA-Seq microreads. In each condition, unique RNA-Seq microreads mapped to a higher proportion of cis-NAT-encoding genes than to non-cis-NAT-encoding genes (P < 0.001; Supplemental Fig. S2). Across all conditions except for the standard growth condition, unique reads mapped more frequently to cis-NATs than to other, non-cis-NAT genes (Wilcoxon rank sum test, P < 0.05; Supplemental Fig. S3). cis-NATs were also detected in more conditions than were non-cis-NATs (Wilcoxon rank sum test, P < 2.2e-16; Supplemental Fig. S4).
cis-NATs are present in more conditions than are non-cis-NATs, but transcripts within a cis-NAT pair may occur in different subsets of conditions. We used the Index of Co-Expression (ICE) to determine if the 1,174 Arabidopsis PC cis-NAT pairs tended to cooccur in the same conditions. A high ICE value indicates that two transcripts are frequently found in the same conditions (Chen et al., 2005). For all tested ICE levels, cis-NATs from the same gene pair cooccurred significantly more often than did transcripts from nonoverlapping genes (P < 0.0001; Table III). For example, 94% of PC cis-NAT pairs had ICE values greater than 0.5 and 78% had ICE values greater than 0.9 (Table III). In contrast, 87% of PC non-cis-NAT pairs had ICE values greater than 0.5 and 64% had ICE values greater than 0.9 (Table III). We considered a pair of cis-NATs to cooccur across a set of conditions if their ICE value was greater than 0.6, as suggested by Chen et al. (2005). The percentage of PC cis-NAT pairs with ICE values greater than 0.6 was 89% versus 80% for PC non-cis-NAT pairs (P < 0.0001; Table III). The ICE metric changes when different criteria are used to make transcript presence/absence calls. However, the proportion of cis-NATs that cooccurred also greatly exceeded the expected proportion using more stringent criteria for determining transcript presence (Supplemental Table S1).
Table III. Percentages of cooccurring PC cis-NAT pairs at different ICE criteria.
| ICE | PC cis-NATsa | PC non-cis-NATsb | Pc |
|---|---|---|---|
| % | |||
| >0.5 | 93.78 | 87.49 | <0.0001 |
| >0.6 | 88.84 | 79.81 | <0.0001 |
| >0.7 | 88.25 | 78.58 | <0.0001 |
| >0.8 | 83.13 | 71.52 | <0.0001 |
| >0.9 | 78.02 | 64.16 | <0.0001 |
The percentage of gene pair transcripts that cooccur. bThe average proportion of cooccurring gene pairs of 100,000 randomized sets of expressed PC non-cis-NAT gene pairs. cThe probability that a randomly selected group of PC non-cis-NATs would have a higher percentage of cooccurrence than the PC cis-NATs by chance.
Evidence for the Transcriptional Regulation of cis-NATs
To investigate if transcriptional regulation may have contributed to high cis-NAT abundances, we mapped cis-regulatory elements to gene 5′ upstream regions. We ranked each of 200 motifs by the frequency with which it was found upstream of a cis-NAT-encoding gene and the frequency with which it was found upstream of a nonoverlapping gene. Eight motifs were 10 or more ranks higher within the cis-NAT-encoding genes than within the non-cis-NAT-encoding genes (Supplemental Table S2). The eight motifs included GGGCC and GGCCCAWWW, which are enriched in regulatory regions of genes represented by Gene Ontology categories “structural constituent of ribosome” (GO:0003735), “ribosome” (GO:0005840), and “ribosome biogenesis and assembly” (GO:0042254). Nine motifs were more than 10 ranks higher in PC non-cis-NAT-encoding genes relative to PC cis-NAT-encoding genes (Supplemental Table S2). Three motifs (TGCAAAG, CATGCA, and CATGCAY) were associated with seed storage protein expression (Supplemental Table S2). Other highly represented motifs included GA and auxin response elements.
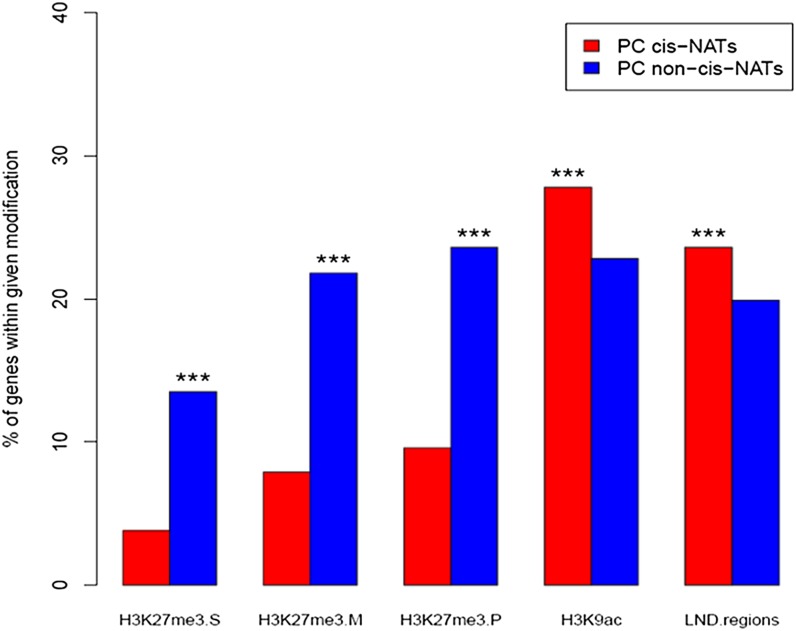
Gene transcriptional activities are reflected in chromatin modifications. Among the 2,338 overlapping PC genes with detected transcripts, 89 (3.8%) were among the 4,979 genes reported by Zhang et al. (2007) as targeted by histone H3 lysine-27 (H3K27) trimethylation, and 185 (7.9%) were among the 7,463 genes reported by Lafos et al. (2011) as H3K27 trimethylated (Fig. 4). In contrast, 13.5% (2,550 of 18,908) of nonoverlapping genes were among the genes Zhang et al. (2007) identified as targeted by H3K27 trimethylation, and 21.8% (4,126 of 18,908) were among the genes identified by Lafos et al. (2011; Fig. 4). Thus, significantly more nonoverlapping genes than cis-NAT-encoding genes were targeted by H3K27 trimethylation in both studies (P < 2.2e-16). The proportion of cis-NAT-encoding genes marked with histone H3 lysine-9 (H3K9) acetylation (Zhou et al., 2010) was significantly higher (27.8%, 650 of 2,338) than the proportion of PC non-cis-NAT-encoding genes (22.8%, 4,313 of 18,908; P < 1.0e-03; Fig. 4). Twenty-four percent (553 of 2,338) of PC cis-NAT-encoding genes were among a set of genes reported to have low-nucleosome-density promoters (Zhang et al., 2007), significantly more than the proportion of PC non-cis-NATs with low-nucleosome-density promoters (19.9%, 3,757 of 18,908; P < 1.0e-03; Fig. 4). As genes with low H3K27 trimethylation, more low-nucleosome-density regions, and high H3K9 acetylation have high transcript levels and are broadly expressed across conditions (Zhang et al., 2007), we suggest that the high transcript abundance of cis-NAT genes is due to transcriptional control.
Figure 4.
Euchromatic marks are overrepresented in PC cis-NAT-encoding genes relative to PC non-cis-NAT-encoding genes. The proportions of PC cis-NAT-encoding genes and PC non-cis-NAT-encoding genes with distinct chromatin modifications are noted on the y axis. The abbreviations on the x axis are as follows: H3K27me3.S, trimethylation of histone H3K27 (Zhang et al., 2007); H3K27me3.M, trimethylation of histone H3K27 (Lafos et al., 2011); H3K27me3.P, the union of H3K27me3.S and H3K27me3.M; H3K9ac, acetylation of histone H3K9 (Zhou et al., 2010); LND regions, low-nucleosome-density regions (Zhang et al., 2007). Asterisks indicate that the proportion of PC cis-NAT (PC non-cis-NAT) genes with a given chromatin modification is significantly greater than the proportion of PC non-cis-NAT (PC cis-NAT) genes at P < 1e-03. [See online article for color version of this figure.]
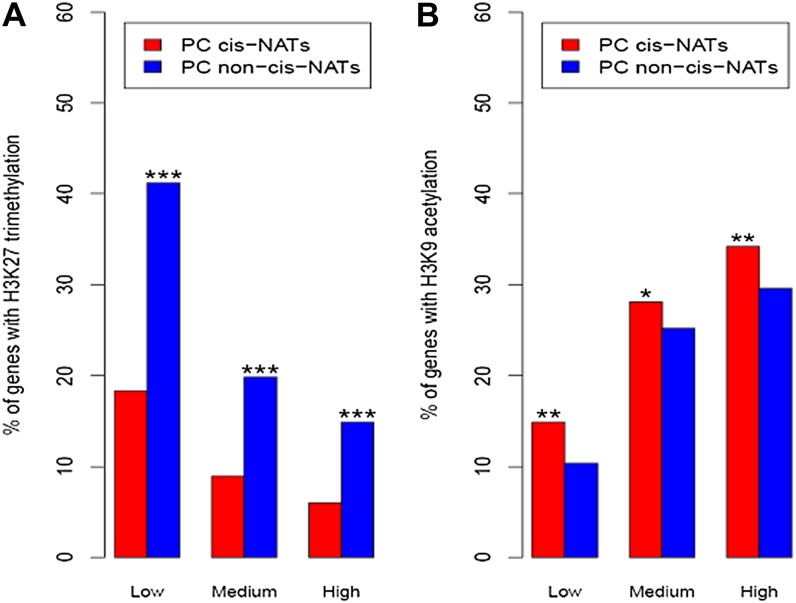
We further investigated if the chromatin states of PC cis-NAT-encoding genes differed from other PC genes with similar transcript abundances. Among genes expressed at similar levels, H3K27 trimethylation among cis-NAT-encoding genes was significantly less frequent than among non-cis-NAT-encoding genes (Fig. 5A). PC cis-NAT-encoding genes also had high levels of H3K9 acetylation relative to PC non-cis-NAT-encoding genes expressed at similar levels (Fig. 5B). Thus, the chromatin state of cis-NAT-encoding genes is more open than that of non-cis-NAT-encoding genes when transcripts from both accumulate to the same level. We also investigated if chromatin attributes differed for genes in different cis-NAT orientations. The chromatin status of genes encoding type III cis-NATs did not significantly differ from that of non-cis-NAT-encoding genes (Supplemental Table S3). Genes in tail-to-tail orientation (type I) and genes in head-to-head orientation (type II) had similar, high frequencies of H3K9 acetylation and low frequencies of H3K27 trimethylation (Supplemental Table S3).
Figure 5.
H3K9 acetylation (H3K9ac) is overrepresented and H3K27 trimethylation (H3K27me3) is underrepresented in PC cis-NAT-encoding genes relative to PC non-cis-NAT-encoding genes with similar expression levels. PC cis-NAT genes and PC non-cis-NAT genes are partitioned to three bins based on expression levels. Genes were binned into low, medium, and high groups based on their transcript abundances (2–16 RPKM, 16–128 RPKM, and more than 128 RPKM, respectively). Asterisks indicate that the proportion of PC cis-NAT (PC non-cis-NAT) genes with a given chromatin modification is significantly greater than the proportion of PC non-cis-NAT (PC cis-NAT) genes as follows: ***P < 1e-03, **P < 1e-02, *P < 5e-02. A, The proportions of PC cis-NAT-encoding genes and PC non-cis-NAT-encoding genes with H3K27me3 are noted on the y axis. A significantly lower proportion of PC cis-NAT genes are marked with H3K27me3 than are PC non-cis-NAT genes across all expression levels. B, The proportions of PC cis-NAT-encoding genes and PC non-cis-NAT-encoding genes marked with H3K9ac are noted on the y axis. The frequency of H3K9ac among PC cis-NAT genes is significantly higher than the frequency among PC non-cis-NAT genes. [See online article for color version of this figure.]
sRNAs Match PC cis-NATs Less Frequently than PC non-cis-NATs
As described above, one explanation for the frequency of overlapping genes is their capacity to generate small regulatory RNAs. However, genes to which sRNAs map have low transcript abundances relative to other genes (Groszmann et al., 2011). Henz et al. (2007) reported that sRNA target sites are underrepresented in cis-NATs compared with non-cis-NATs (Henz et al., 2007). However, Zhang et al. (2012), mapping a much larger number of unique sRNAs (17,141) to cis-NATs, reported that small interfering RNA (siRNA) target sites were overrepresented (P < 0.04) among the 3′ untranslated regions (UTRs) of cis-NAT-encoding genes compared with 3′ UTRs of non-cis-NAT-encoding genes (Zhang et al., 2012). We mapped sRNAs sequenced from wild-type and seven sRNA biogenesis mutant plants (Rajagopalan et al., 2006; Kasschau et al., 2007) and found that PC non-cis-NATs matched sRNAs more frequently than did PC cis-NATs across the eight genotypes (Supplemental Table S4). Across all genotypes, sRNAs matched PC non-cis-NATs from 1.4- to 5.2-fold more frequently than PC cis-NATs (Table IV). The frequency with which an sRNA sequenced from a putative nat-siRNA biogenesis mutant (dcl1, dcl2, dcl3, rdr2, rdr6; Borsani et al., 2005; Zhang et al., 2012) matched a PC cis-NAT was similar to the frequency with which an sRNA sequenced from wild-type plants and other sRNA mutants (dcl4, rdr1; Xie et al., 2004, 2005) matched PC cis-NATs (Table IV).
Table IV. Number and proportion of sRNAs in RNA silencing mutants matching PC cis-NAT transcripts and PC non-cis-NAT transcripts.
| Genotype | Total Unique sRNAsa |
sRNA Matches |
||
|---|---|---|---|---|
| PC cis-NATs (No. per Million)b | PC non-cis-NATs (No. per Million)b | Fold Differencec | ||
| dcl1-7 | 14,692 | 14 (0.408) | 467 (1.681) | 4.1 |
| dcl2-1 | 10,405 | 10 (0.411) | 340 (1.728) | 4.2 |
| dcl3-1 | 22,951 | 35 (0.652) | 714 (1.645) | 2.5 |
| dcl4-2 | 22,448 | 14 (0.267) | 587 (1.383) | 5.2 |
| rdr1-1 | 14,755 | 32 (0.928) | 529 (1.896) | 2.0 |
| rdr2-1d | 6,249 | 74 (5.065) | 841 (7.118) | 1.4 |
| rdr6-15 | 37,774 | 30 (0.340) | 934 (1.308) | 3.8 |
| Wild type | 57,966 | 75 (0.553) | 1,697 (1.548) | 2.8 |
sRNAs from RNA silencing mutants and wild-type plants were processed from sequences available from the National Center for Biotechnology Information Gene Expression Omnibus (accession no. GSE6682; Kasschau et al., 2007). bsRNAs that matched to 2,338 PC cis-NATs or 18,908 PC non-cis-NATs. Number per million, in parentheses, is the expected number of sRNAs out of 106 sRNAs that match a single cis-NAT or a single non-cis-NAT transcript. cThe frequency of sRNAs matching PC non-cis-NATs divided by the frequency of sRNAs matching PC cis-NATs. dThe number of sRNAs matching a PC transcript was higher in rdr2-1 than in other genotypes because rdr2-1 fails to produce many repeat-associated-siRNAs, leading to a high representation of sRNAs with homology to PC sequences (Kasschau et al., 2007).
DISCUSSION
Here, we propose that a central function of overlapping PC gene pairs is to promote high and broad gene transcription. We found that PC cis-NATs are detected more frequently and at a higher abundance than transcripts from nonoverlapping PC genes (Figs. 1 and 2). We also report that PC cis-NATs are broadly expressed and cooccur within harvested tissues more often than are PC non-cis-NAT genes (Fig. 3; Table III). PC genes are significantly overrepresented among cis-NAT-encoding genes, suggesting that genes that give rise to noncoding RNAs may be more sensitive to antisense transcripts than are PC genes (Table I). Among the PC cis-NAT-encoding genes, gene pairs with 3′ overlaps (type I) were significantly overrepresented (Table II). Recombination and/or mutation likely gives rise to all three orientations of cis-NAT-encoding genes, but most type II and III gene pairs are eliminated because the creation of one gene has deleterious effects on the regulatory or coding sequences of the complementary gene.
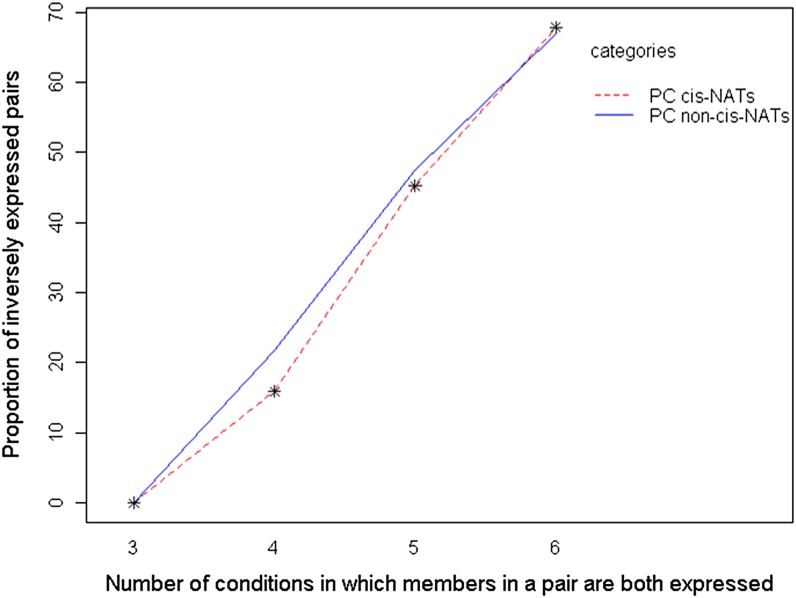
Our results seem to be at odds with a previous finding that cis-NATs exhibit inverse expression (e.g. in some conditions, one cis-NAT is highly abundant while its antisense transcript has low abundance, while in other conditions, the cis-NAT has low abundance while its antisense transcript has high abundance; Jin et al., 2008). However, the frequency of inverse expression is strongly correlated with expression breadth. Because gene pairs are declared inversely expressed if their abundances differ across conditions, a gene pair expressed across a large number of conditions is more likely to be inversely expressed than a gene pair expressed across a small number of conditions. Thus, we also found that PC cis-NATs were frequently inversely expressed across treatments (51% of PC cis-NATs compared with 42% of other PC transcripts; P = 3.5e-10). However, we compared the frequency of inverse expression among the PC cis-NAT pairs and random, PC non-cis-NAT gene pairs in which both members are expressed in three, four, five, and six conditions. Inverse expression increased with expression breadth, and the proportion of PC cis-NAT gene pairs that were inversely expressed did not significantly differ from nonoverlapping genes at any given breadth (Fig. 6). High levels of cis-NAT inverse expression have also been reported for human cis-NAT-encoding genes (Chen et al., 2005), and this result too may be a function of expression breadth.
Figure 6.
Proportions of inversely expressed PC cis-NATs and PC non-cis-NATs at different expression breadths. The proportion of inversely expressed gene pairs is plotted against the number of conditions in which both members of a pair are present. The dotted line represents the observed proportion of inversely expressed PC gene pairs. The solid line represents the mean proportions of inversely expressed gene pairs from 10,000 data sets of randomly picked, expressed PC non-cis-NAT pairs. Asterisks indicate no significant difference between PC cis-NAT and PC non-cis-NAT values at P < 0.05. [See online article for color version of this figure.]
Patterns of cis-NAT Abundances Are Likely Due to Transcriptional Control
Although antisense transcripts may posttranscriptionally stabilize sense transcripts and be generated by sense transcripts (Faghihi et al., 2008; Matsui et al., 2008a, 2008b), the high frequency of certain promoter motifs, low nucleosome density, low H3K27 trimethylation, and high H3K9 acetylation among the cis-NAT-encoding genes suggest that the transcript abundance patterns of cis-NATs are due to transcriptional regulation. Previous studies have reported that overlapping transcripts tend to be coregulated. Although nonsense-mediated mRNA decay widely suppresses non-PC, antisense transcripts (Kurihara et al., 2009), non-PC transcripts complementary to highly expressed, sense transcripts are expressed at relatively high levels (Luo et al., 2012). COOLAIR, the regulatory, noncoding antisense transcript of the floral repressor FLOWERING LOCUS C (FLC), has transcript levels that positively correlate with FLC transcript levels in most conditions (Ietswaart et al., 2012). Antisense genes may be under the regulatory control of shared enhancer elements that recruit RNA polymerase to both promoters (Tagoh et al., 2004; Ebralidze et al., 2008), and locus control regions can also establish extensive activated chromatin domains encompassing target promoters (Cajiao et al., 2004).
The transcription of overlapping PC genes itself may be a positive regulatory mechanism. As in our study, Katayama et al. (2005) found that PC cis-NATs were highly coexpressed in mouse cell lines. Furthermore, depleting transcripts derived from one of two overlapping PC genes with RNA interference reduced the transcript abundance of the second gene (Katayama et al., 2005). The expression of enhancer RNAs also positively correlates with the expression of nearby genes (Kim et al., 2010; Ørom et al., 2010). Investigating the transcript abundance and chromatin status of one gene within an overlapping cis-NAT pair when the other gene has been silenced would test if the transcription of a cis-NAT positively regulates its antisense transcript. As enhancers act independently of orientation and position, both overlapping PC genes and nonoverlapping, nearby PC genes may have high levels of coexpression.
Antisense transcription through, or terminating in, a sense promoter can promote sense gene transcription, likely by chromatin remodeling (Uhler et al., 2007). For example, the abundance of COOLAIR antisense transcripts with poly(A) sites within the sense FLC promoter region is correlated with FLC sense transcript abundance (Hornyik et al., 2010). Nonetheless, antisense transcription likely affects the kinetics but not the final level of gene transcription (Uhler et al., 2007). Furthermore, the 80% of PC cis-NAT gene pairs that are type I (tail-to-tail) do not have overlapping promoters.
As mentioned above, some cis-NATs are processed into siRNAs that target transcripts for cleavage. Thus, one may expect that cellular sRNAs map to cis-NATs at high frequencies. Consistent with this idea, Zhang et al. (2012) recently reported that siRNAs are overrepresented (P < 0.04) among the 3′ UTRs of cis-NATs compared with 3′ UTRs of non-cis-NATs (Zhang et al., 2012). In contrast, Henz et al. (2007) found that sRNAs are more enriched in non-cis-NATs compared with cis-NATs (Henz et al., 2007). Our results are consistent with Henz et al. (2007). We found that among wild-type plants, sRNAs matched PC non-cis-NATs 2.8 times more frequently than they matched PC cis-NATs (Table IV). In addition, we compared the number of sRNAs that map to cis-NATs in putative nat-siRNA biogenesis mutant plants (dcl1-7, dcl2-1, dcl3-1, and rdr6-15) with the number that match in other genotypes (dcl4-2, rdr1-1, and the wild type) and found them to be similar (Table IV). We suggest that nat-siRNA regulation of PC gene expression is infrequent. Our results and those of Henz et al. (2007) may disagree with the findings of Zhang et al. (2012), because this latter work included noncoding RNAs. Fifty-four of the 84 cis-NAT-encoding gene pairs associated with more than 10 siRNAs per million reads reported by Zhang et al. (2012) had one transcript annotated as “other RNAs.” A number of the nat-siRNAs reported by Zhang et al. (2012) were DCL1 or DCL3/RDR2 dependent and may be microRNAs or siRNAs derived from the noncoding transcript alone. RNA structure is important for guiding molecules into sRNA biogenesis pathways (Pouch-Pélissier et al., 2008), and a high frequency of noncoding RNAs form stable RNA structures and act as precursors for repeat associated siRNA and microRNA biogenesis (Hirsch et al., 2006; Ben Amor et al., 2009).
We can speculate on how highly and broadly expressed complementary transcripts avoid nat-siRNA production and recruitment into long noncoding RNA-related ribonucleoprotein-silencing complexes (Swiezewski et al., 2009). Perhaps these genes are highly and broadly transcribed together in the same tissue, but they are not simultaneously transcribed. Indeed, Osborne et al. (2004) noted that active genes undergo discontinuous transcription (Osborne et al., 2004). Computational analyses support this model, as PC cis-NAT levels are rarely correlated. Matsui et al. (2008a) reported that strong linear correlations between PC cis-NAT levels were rare. We previously reported 29 highly correlated transcript pairs from convergently oriented genes, of which type I genes are a subset (Zhan et al., 2006). Among the 29 gene pairs, not one is a type I cis-NAT pair, significantly fewer than expected (Table V; binomial test, P = 6.6e-4), and not one type II cis-NAT gene pair is among the 53 highly correlated, divergently oriented gene pairs (Table V). Investigating if positive transcriptional regulation with low cotranscription characterizes cis-NAT genes in other species will determine the generality of these findings.
Table V. Highly correlated neighboring genes and highly correlated cis-NAT genes.
| Orientation of Transcription | All Pairs | Highly Correlated Pairsa,b | cis-NAT Pairsb | Highly Correlated cis-NAT Pairsc |
|
|---|---|---|---|---|---|
| Observed | Expected | ||||
| Convergent | 5,412 | 29 (0.54%) | 1,363 (25.18%) | 0 | 7 |
| Divergent | 5,413 | 53 (0.98%) | 152 (2.81%) | 0 | 1 |
Highly correlated neighboring gene pairs were identified from microarray data sets across 128 experimental conditions with r > 0.7 (Zhan et al., 2006). bParentheses give the percentage of cis-NAT pairs in the defined orientation of transcription. cValues are given as counts.
MATERIALS AND METHODS
Identification of cis-NATs and RNA-Seq Analyses
To identify candidate cis-NAT-encoding genes, we used PERL scripts to retrieve gene model orientation and transcript start and stop positions from the GenBank Refseqs NC_003070.9, NC_003071.7, NC_003074.8, NC_003075.7, and NC_003076.8. We defined cis-NAT pairs as overlapping transcript pairs that arise from a pair of genes adjacently located on opposite strands of the same genomic locus. Although non-PC antisense transcripts outnumber PC antisense transcripts (Matsui et al., 2008a), we focused on the cis-NATs of overlapping PC genes. Using a χ2 test, we calculated the probability of the observed number of PC genes that encoded cis-NATs, given the null expectation that the same proportion of PC genes and non-PC genes encode cis-NATs. Similarly, we calculated the probability of the observed number of PC cis-NAT-encoding gene pairs that were type I, type II, and type III, given the null expectation that PC gene pairs would be type I, type II, and type III at the same frequency as all gene pairs.
To assay transcript abundance, we obtained Arabidopsis (Arabidopsis thaliana) RNA-Seq data from 3-week-old Columbia-0 plants grown in normal, high-light, heat, cold, salt, and drought conditions from Dr. Todd Mockler. Filichkin et al. (2010) described plant growth conditions, RNA isolation, and the preparation of complementary DNA for the Illumina 1G Genome Analyzer. A total of 53.44 million 36-base RNA-Seq microreads were truncated to the first 30 bases (Jiang and Wong, 2008; Filichkin et al., 2010), and we used SeqMap (Jiang and Wong, 2008) to map these reads to the annotated mRNAs. We used rSeq (Jiang and Wong, 2009) to compute expression levels in RPKM (Mortazavi et al., 2008). rSeq weights reads by the number of sites to which they map. For overlapping transcripts, the transcript that gave rise to the RNA-Seq read is unknown, as reads are not strand specific. Thus, we also investigated reads that mapped only to a single transcript. To calculate RPKM, we used the length of a gene’s longest transcript to compute the exon model length.
We only compared PC cis-NATs and non-cis-NATs with evidence of transcription, defined as having at least 1 RPKM in one condition and at least a sum of 2 RPKM across the six conditions. We used the two-sample proportion test to evaluate the significance of differences in the proportion of expressed genes within each condition (Fig. 1). To determine if a gene is expressed within a condition, we considered a gene with transcript abundance of 1 RPKM or greater to be present. The distribution of cis-NAT and non-cis-NAT RPKM values was nonnormal, and we used the Wilcoxon rank sum test to evaluate the significance of the median expression abundance differences between cis-NATs and non-cis-NATs (Fig. 2). Similarly, we used the Wilcoxon rank sum test to compare the expression breadths of cis-NATs and non-cis-NATs (Fig. 3). To evaluate if transcripts from both members of a gene pair tended to be detected in the same treatment, we calculated the ICE as described by Lercher et al. (2002). In this paper, we call this metric the “index of cooccurrence,” as the term coexpression is often used to signify correlation. The ICE metric ranges from 0 to 1, corresponding to no cooccurrence and perfect cooccurrence of two genes’ transcripts across samples. To determine if cooccurrence of cis-NAT pairs was more than expected by chance, we first calculated the cooccurrence rate for all cis-NAT pairs. We then generated a control data set by replacing each gene in the cis-NAT set with a randomly picked gene from expressed, nonoverlapping PC genes. We compared the observed frequency of cis-NAT cooccurrence with the frequencies calculated from 100,000 permuted, control data sets. The significance of the cis-NAT cooccurrence rates was calculated using this null distribution (Table III). The ICE metric depends in part on the criteria used to make transcript presence/absence calls. We also used more stringent criteria to assay ICE. Under stringent criteria, a gene was expressed if its RPKM was 4 or greater across six conditions and 3 or greater in one condition. Finally, we adopted a method proposed by Chen et al. (2005) to measure inverse expression (Fig. 6). We first Studentized the expression of each gene, allowing the expression differences of genes with high or low transcript abundances to be compared. We then labeled two genes (A and B) as inversely expressed if they had three or more contrasting expression patterns among their 15 pairwise comparisons across the six RNA-Seq experiments. A gene pair was termed contrasting between two experiments if the expression of gene A in one experiment was greater than the expression of gene B by greater than 0.5 and if the expression of gene B in the second experiment was greater than the expression of gene A by greater than 0.5. We again used permutations of expressed PC non-cis-NAT genes to calculate statistical significance. All analyses were performed with the R statistical software (R Development Core Team, 2010).
Epigenetic and Promoter Analyses of cis-NAT-Encoding Genes
To investigate epigenetic marks on cis-NAT-encoding genes, we obtained from online, supplemental data lists of genes that contain the chromatin attributes: H3K27 trimethylation (Zhang et al., 2007; Lafos et al., 2011), H3K9 acetylation (Zhou et al., 2010), and low nucleosome density (Zhang et al., 2007). We obtained epigenetic information for all genes with evidence of expression. We then calculated the proportions of cis-NAT-encoding PC genes that have each epigenetic attribute and the proportions of non-cis-NAT PC genes that have each epigenetic attribute. The significance of differences was calculated with the two-sample proportions test (Fig. 4). We took a similar approach to compare the frequency of chromatin attributes in PC cis-NAT-encoding genes and other PC genes expressed at a similar level (Fig. 5).
To evaluate the differences in cis-regulatory elements, we obtained a list of motifs and motif positions from ATCOECIS (Vandepoele et al., 2009). We recorded if a motif was identified within each gene’s upstream region. The upstream region was restricted to the first 1,000 bp upstream from the translation start site or to a shorter region if genes were closer than 1,000 bp (Vandepoele et al., 2009). The gene list was divided into cis-NAT-encoding genes and non-cis-NAT-encoding genes. For each group, we ranked the motifs by the frequency with which they were found among the cis-NATs and the non-cis-NATs. We report motifs that differed by more than 10 ranks between the two groups (Supplemental Table S2). Associations of the identified motifs with Gene Ontology categories were retrieved from ATCOECIS (Vandepoele et al., 2009).
Mapping sRNAs to cis-NATs
The nat-siRNA biogenesis is dependent on DCL1, DCL2, and RDR6 (Borsani et al., 2005; Ron et al., 2010). Zhang et al. (2012) also found that some nat-siRNAs were dependent on DCL1, DCL3, and RDR2. To investigate the frequency with which sRNAs mapped to cis-NATs in the wild type, nat-siRNA silencing mutants, and other sRNA silencing mutants, we obtained sRNA sequences from accessions GSE5228 (dcl1-7, dcl2-1, dcl3-1, dcl4-2, rdr1-1, rdr2-1, rdr6-15, and the wild type; Kasschau et al., 2007) and GSE6682 (the wild type; Rajagopalan et al., 2006) from the National Center for Biotechnology Information Gene Expression Omnibus. We tallied perfect matches between the sRNA sequences and PC transcripts using PERL scripts. An sRNA matching the target transcript was counted once if the sRNA matched multiple transcripts from the same gene or the same transcript multiple times. We calculated the number of sRNAs that mapped to cis-NATs and the average frequency with which an sRNA will match each cis-NAT per million sRNAs (Table IV).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Percentage of expressed PC cis-NAT-encoding genes and PC non-cis-NAT-encoding genes with high transcript abundances.
Supplemental Figure S2. Percentage of cis-NAT-encoding genes and non-cis-NAT-encoding genes with uniquely mapped RNA-Seq reads.
Supplemental Figure S3. RNA-Seq reads that map to unique positions are more abundant within PC cis-NATs than PC non-cis-NATs.
Supplemental Figure S4. Percentage of expressed PC cis-NAT and PC non-cis-NAT genes that had uniquely mapped RNA-Seq reads.
Supplemental Table S1. Percentage of cooccurring PC cis-NAT pairs using stringent expression criteria at different ICE cutoff values.
Supplemental Table S2. Motifs overrepresented within PC cis-NATs and PC non-cis-NATs.
Supplemental Table S3. Proportions of H3K9 acetylation and H3K27 trimethylation among types of PC cis-NAT-encoding genes.
Supplemental Table S4. Number and frequency of PC cis-NATs and PC non-cis-NATs matching sRNAs in RNA silencing mutants.
Acknowledgments
We thank all the laboratories whose data were used in our analyses. This work was made possible by the facilities of the Shared Hierarchical Academic Research Computing Network.
Glossary
- PC
protein-coding
- cis-NAT
cis-natural sense antisense transcript
- nat-siRNA
natural antisense small RNA
- sRNA
small RNA
- RPKM
reads per kilobase of exon model per million mapped reads
- ICE
Index of Co-Expression
- H3K27
histone H3 lysine-27
- H3K9
histone H3 lysine-9
- UTRs
untranslated regions
- siRNA
small interfering RNA
References
- Ben Amor B, Wirth S, Merchan F, Laporte P, d’Aubenton-Carafa Y, Hirsch J, Maizel A, Mallory A, Lucas A, Deragon JM, et al. (2009) Novel long non-protein coding RNAs involved in Arabidopsis differentiation and stress responses. Genome Res 19: 57–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsani O, Zhu J, Verslues PE, Sunkar R, Zhu JK. (2005) Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell 123: 1279–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cajiao I, Zhang A, Yoo EJ, Cooke NE, Liebhaber SA. (2004) Bystander gene activation by a locus control region. EMBO J 23: 3854–3863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Sun M, Hurst LD, Carmichael GG, Rowley JD. (2005) Genome-wide analysis of coordinate expression and evolution of human cis-encoded sense-antisense transcripts. Trends Genet 21: 326–329 [DOI] [PubMed] [Google Scholar]
- Chen J, Sun M, Kent WJ, Huang X, Xie H, Wang W, Zhou G, Shi RZ, Rowley JD. (2004) Over 20% of human transcripts might form sense-antisense pairs. Nucleic Acids Res 32: 4812–4820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebralidze AK, Guibal FC, Steidl U, Zhang P, Lee S, Bartholdy B, Jorda MA, Petkova V, Rosenbauer F, Huang G, et al. (2008) PU.1 expression is modulated by the balance of functional sense and antisense RNAs regulated by a shared cis-regulatory element. Genes Dev 22: 2085–2092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelman LB, Fraser P. (2012) Transcription factories: genetic programming in three dimensions. Curr Opin Genet Dev 22: 110–114 [DOI] [PubMed] [Google Scholar]
- Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, Finch CE, St Laurent G, III, Kenny PJ, Wahlestedt C. (2008) Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of beta-secretase. Nat Med 14: 723–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filichkin SA, Priest HD, Givan SA, Shen R, Bryant DW, Fox SE, Wong WK, Mockler TC. (2010) Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res 20: 45–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groszmann M, Greaves IK, Albertyn ZI, Scofield GN, Peacock WJ, Dennis ES. (2011) Changes in 24-nt siRNA levels in Arabidopsis hybrids suggest an epigenetic contribution to hybrid vigor. Proc Natl Acad Sci USA 108: 2617–2622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henz SR, Cumbie JS, Kasschau KD, Lohmann JU, Carrington JC, Weigel D, Schmid M. (2007) Distinct expression patterns of natural antisense transcripts in Arabidopsis. Plant Physiol 144: 1247–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch J, Lefort V, Vankersschaver M, Boualem A, Lucas A, Thermes C, d’Aubenton-Carafa Y, Crespi M. (2006) Characterization of 43 non-protein-coding mRNA genes in Arabidopsis, including the MIR162a-derived transcripts. Plant Physiol 140: 1192–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornyik C, Terzi LC, Simpson GG. (2010) The spen family protein FPA controls alternative cleavage and polyadenylation of RNA. Dev Cell 18: 203–213 [DOI] [PubMed] [Google Scholar]
- Ietswaart R, Wu Z, Dean C. (2012) Flowering time control: another window to the connection between antisense RNA and chromatin. Trends Genet 28: 445–453 [DOI] [PubMed] [Google Scholar]
- Jen CH, Michalopoulos I, Westhead DR, Meyer P. (2005) Natural antisense transcripts with coding capacity in Arabidopsis may have a regulatory role that is not linked to double-stranded RNA degradation. Genome Biol 6: R51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Wong WH. (2008) SeqMap: mapping massive amount of oligonucleotides to the genome. Bioinformatics 24: 2395–2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Wong WH. (2009) Statistical inferences for isoform expression in RNA-Seq. Bioinformatics 25: 1026–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Vacic V, Girke T, Lonardi S, Zhu JK. (2008) Small RNAs and the regulation of cis-natural antisense transcripts in Arabidopsis. BMC Mol Biol 9: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasschau KD, Fahlgren N, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Carrington JC. (2007) Genome-wide profiling and analysis of Arabidopsis siRNAs. PLoS Biol 5: e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, et al. (2005) Antisense transcription in the mammalian transcriptome. Science 309: 1564–1566 [DOI] [PubMed] [Google Scholar]
- Katiyar-Agarwal S, Morgan R, Dahlbeck D, Borsani O, Villegas A, Jr, Zhu JK, Staskawicz BJ, Jin H. (2006) A pathogen-inducible endogenous siRNA in plant immunity. Proc Natl Acad Sci USA 103: 18002–18007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et al. (2010) Widespread transcription at neuronal activity-regulated enhancers. Nature 465: 182–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara Y, Matsui A, Hanada K, Kawashima M, Ishida J, Morosawa T, Tanaka M, Kaminuma E, Mochizuki Y, Matsushima A, et al. (2009) Genome-wide suppression of aberrant mRNA-like noncoding RNAs by NMD in Arabidopsis. Proc Natl Acad Sci USA 106: 2453–2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafos M, Kroll P, Hohenstatt ML, Thorpe FL, Clarenz O, Schubert D. (2011) Dynamic regulation of H3K27 trimethylation during Arabidopsis differentiation. PLoS Genet 7: e1002040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lercher MJ, Urrutia AO, Hurst LD. (2002) Clustering of housekeeping genes provides a unified model of gene order in the human genome. Nat Genet 31: 180–183 [DOI] [PubMed] [Google Scholar]
- Luo C, Sidote DJ, Zhang Y, Kerstetter RA, Michael TP, Lam E. (2013) Integrative analysis of chromatin states in Arabidopsis identified potential regulatory mechanisms for natural antisense transcript production. Plant J 73: 77–90 [DOI] [PubMed] [Google Scholar]
- Matsui A, Ishida J, Morosawa T, Mochizuki Y, Kaminuma E, Endo TA, Okamoto M, Nambara E, Nakajima M, Kawashima M, et al. (2008a) Arabidopsis transcriptome analysis under drought, cold, high-salinity and ABA treatment conditions using a tiling array. Plant Cell Physiol 49: 1135–1149 [DOI] [PubMed] [Google Scholar]
- Matsui K, Nishizawa M, Ozaki T, Kimura T, Hashimoto I, Yamada M, Kaibori M, Kamiyama Y, Ito S, Okumura T. (2008b) Natural antisense transcript stabilizes inducible nitric oxide synthase messenger RNA in rat hepatocytes. Hepatology 47: 686–697 [DOI] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5: 621–628 [DOI] [PubMed] [Google Scholar]
- Ørom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, et al (2010) Long noncoding RNAs with enhancer-like function in human cells. Cell 143: 46–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne CS, Chakalova L, Brown KE, Carter D, Horton A, Debrand E, Goyenechea B, Mitchell JA, Lopes S, Reik W, et al (2004) Active genes dynamically colocalize to shared sites of ongoing transcription. Nat Genet 36: 1065–1071 [DOI] [PubMed] [Google Scholar]
- Pouch-Pélissier MN, Pélissier T, Elmayan T, Vaucheret H, Boko D, Jantsch MF, Deragon JM. (2008) SINE RNA induces severe developmental defects in Arabidopsis thaliana and interacts with HYL1 (DRB1), a key member of the DCL1 complex. PLoS Genet 4: e1000096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team (2011) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria
- Rajagopalan R, Vaucheret H, Trejo J, Bartel DP. (2006) A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev 20: 3407–3425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron M, Alandete Saez M, Eshed Williams L, Fletcher JC, McCormick S. (2010) Proper regulation of a sperm-specific cis-nat-siRNA is essential for double fertilization in Arabidopsis. Genes Dev 24: 1010–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldà G, Suyama M, Pelucchi P, Boi S, Guffanti A, Rizzi E, Bork P, Tenchini ML, Ciccarelli FD. (2008) Non-random retention of protein-coding overlapping genes in Metazoa. BMC Genomics 9: 174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiezewski S, Liu F, Magusin A, Dean C. (2009) Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature 462: 799–802 [DOI] [PubMed] [Google Scholar]
- Tagoh H, Schebesta A, Lefevre P, Wilson N, Hume D, Busslinger M, Bonifer C. (2004) Epigenetic silencing of the c-fms locus during B-lymphopoiesis occurs in discrete steps and is reversible. EMBO J 23: 4275–4285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhler JP, Hertel C, Svejstrup JQ. (2007) A role for noncoding transcription in activation of the yeast PHO5 gene. Proc Natl Acad Sci USA 104: 8011–8016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandepoele K, Quimbaya M, Casneuf T, De Veylder L, Van de Peer Y. (2009) Unraveling transcriptional control in Arabidopsis using cis-regulatory elements and coexpression networks. Plant Physiol 150: 535–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XJ, Gaasterland T, Chua NH. (2005) Genome-wide prediction and identification of cis-natural antisense transcripts in Arabidopsis thaliana. Genome Biol 6: R30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Allen E, Wilken A, Carrington JC. (2005) DICER-LIKE 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana. Proc Natl Acad Sci USA 102: 12984–12989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, Zilberman D, Jacobsen SE, Carrington JC. (2004) Genetic and functional diversification of small RNA pathways in plants. PLoS Biol 2: E104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan S, Horrocks J, Lukens LN. (2006) Islands of co-expressed neighbouring genes in Arabidopsis thaliana suggest higher-order chromosome domains. Plant J 45: 347–357 [DOI] [PubMed] [Google Scholar]
- Zhang X, Clarenz O, Cokus S, Bernatavichute YV, Pellegrini M, Goodrich J, Jacobsen SE. (2007) Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis. PLoS Biol 5: e129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Xia J, Lii YE, Barrera-Figueroa BE, Zhou X, Gao S, Lu L, Niu D, Chen Z, Leung C, et al. (2012) Genome-wide analysis of plant nat-siRNAs reveals insights into their distribution, biogenesis and function. Genome Biol 13: R20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu XS, Liu QR, Wei L. (2006) Genome-wide in silico identification and analysis of cis natural antisense transcripts (cis-NATs) in ten species. Nucleic Acids Res 34: 3465–3475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Wang X, He K, Charron JB, Elling AA, Deng XW. (2010) Genome-wide profiling of histone H3 lysine 9 acetylation and dimethylation in Arabidopsis reveals correlation between multiple histone marks and gene expression. Plant Mol Biol 72: 585–595 [DOI] [PubMed] [Google Scholar]