Abstract
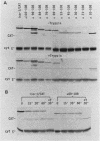
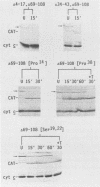
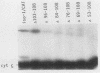
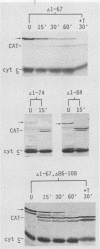
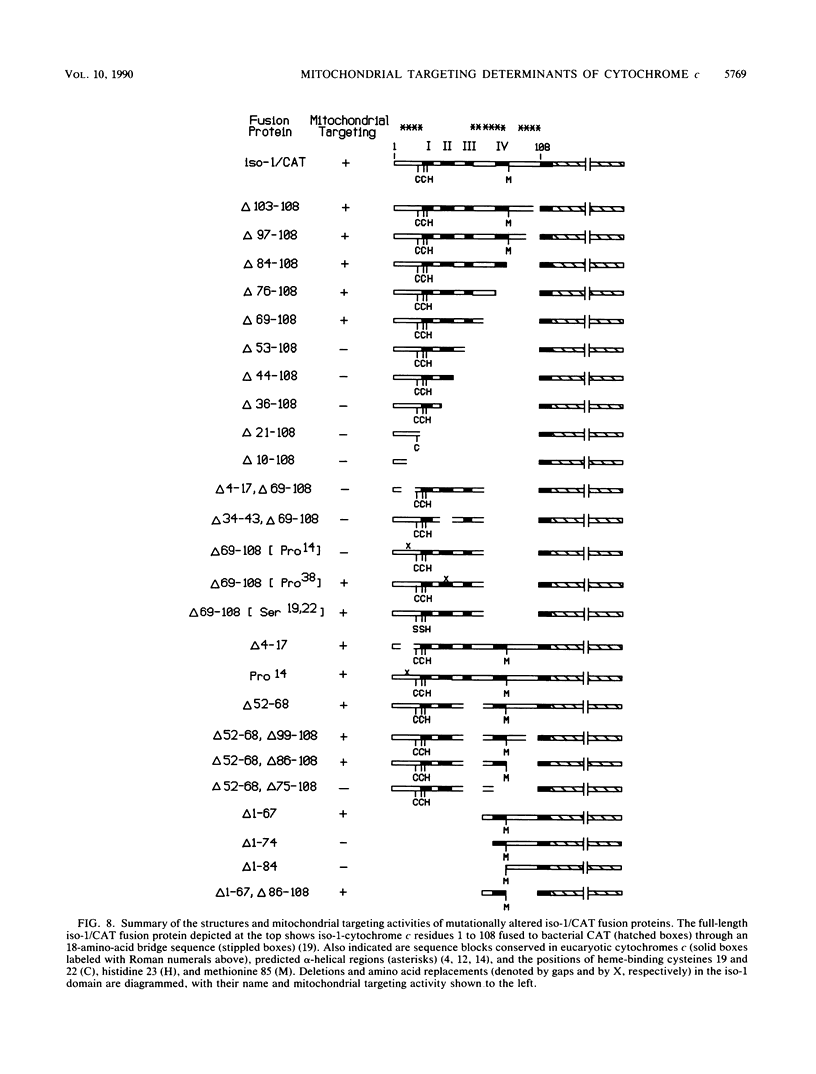
An iso-1-cytochrome c-chloramphenicol acetyltransferase fusion protein (iso-1/CAT) was expressed in Saccharomyces cerevisiae and used to delineate two stages in the cytochrome c import pathway in vivo (S. H. Nye and R. C. Scarpulla, Mol. Cell. Biol. 10:5753-5762, 1990 [this issue]). Fusion proteins with the CAT reporter domain in its native conformation were arrested at the initial stage of mitochondrial membrane recognition and insertion. In contrast, those with a deletional disruption of the CAT moiety were relieved of this block and allowed to translocate to the intermembrane space, where they functioned in respiratory electron transfer. In the present study, iso-1/CAT was used to map structural determinants in apoiso-1-cytochrome c involved in the initial step of targeting to the mitochondrial membrane. Carboxy-terminal deletions revealed that one of these determinants consisted of the amino-terminal 68 residues. Deletion mutations either within or at the ends of this determinant destroyed mitochondrial targeting activity, suggesting that functionally important information spans the length of this fragment. Disruption of an alpha-helix near the amino terminus by a helix-breaking proline substitution for leucine 14 also eliminated the targeting activity of the 1 to 68 determinant, suggesting a contribution from this structure. A second, functionally independent targeting determinant was found in the carboxy half of the apoprotein between residues 68 and 85. This determinant coincided with a stretch of 11 residues that are invariant in nearly 100 eucaryotic cytochromes c. Therefore, in lieu of an amino-terminal presequence, apocytochrome c has redundant structural information located in both the amino and carboxy halves of the molecule that can function independently to specify mitochondrial targeting and membrane insertion in vivo.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bedwell D. M., Klionsky D. J., Emr S. D. The yeast F1-ATPase beta subunit precursor contains functionally redundant mitochondrial protein import information. Mol Cell Biol. 1987 Nov;7(11):4038–4047. doi: 10.1128/mcb.7.11.4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W. J., Douglas M. G. The role of protein structure in the mitochondrial import pathway. Unfolding of mitochondrially bound precursors is required for membrane translocation. J Biol Chem. 1987 Nov 15;262(32):15605–15609. [PubMed] [Google Scholar]
- Cohen J. D., Eccleshall T. R., Needleman R. B., Federoff H., Buchferer B. A., Marmur J. Functional expression in yeast of the Escherichia coli plasmid gene coding for chloramphenicol acetyltransferase. Proc Natl Acad Sci U S A. 1980 Feb;77(2):1078–1082. doi: 10.1073/pnas.77.2.1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickerson R. E. The structure and history of an ancient protein. Sci Am. 1972 Apr;226(4):58–passim. doi: 10.1038/scientificamerican0472-58. [DOI] [PubMed] [Google Scholar]
- Douglas M. G., McCammon M. T., Vassarotti A. Targeting proteins into mitochondria. Microbiol Rev. 1986 Jun;50(2):166–178. doi: 10.1128/mr.50.2.166-178.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont M. D., Mathews A. J., Nall B. T., Baim S. B., Eustice D. C., Sherman F. Differential stability of two apo-isocytochromes c in the yeast Saccharomyces cerevisiae. J Biol Chem. 1990 Feb 15;265(5):2733–2739. [PubMed] [Google Scholar]
- Dumont M. E., Richards F. M. Insertion of apocytochrome c into lipid vesicles. J Biol Chem. 1984 Apr 10;259(7):4147–4156. [PubMed] [Google Scholar]
- Hampsey D. M., Das G., Sherman F. Amino acid replacements in yeast iso-1-cytochrome c. Comparison with the phylogenetic series and the tertiary structure of related cytochromes c. J Biol Chem. 1986 Mar 5;261(7):3259–3271. [PubMed] [Google Scholar]
- Hartl F. U., Ostermann J., Guiard B., Neupert W. Successive translocation into and out of the mitochondrial matrix: targeting of proteins to the intermembrane space by a bipartite signal peptide. Cell. 1987 Dec 24;51(6):1027–1037. doi: 10.1016/0092-8674(87)90589-7. [DOI] [PubMed] [Google Scholar]
- Hennig B., Koehler H., Neupert W. Receptor sites involved in posttranslational transport of apocytochrome c into mitochondria: specificity, affinity, and number of sites. Proc Natl Acad Sci U S A. 1983 Aug;80(16):4963–4967. doi: 10.1073/pnas.80.16.4963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig B., Neupert W. Assembly of cytochrome c. Apocytochrome c is bound to specific sites on mitochondria before its conversion to holocytochrome c. Eur J Biochem. 1981 Dec;121(1):203–212. doi: 10.1111/j.1432-1033.1981.tb06450.x. [DOI] [PubMed] [Google Scholar]
- Jordi W., Zhou L. X., Pilon M., Demel R. A., de Kruijff B. The importance of the amino terminus of the mitochondrial precursor protein apocytochrome c for translocation across model membranes. J Biol Chem. 1989 Feb 5;264(4):2292–2301. [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Louie G. V., Hutcheon W. L., Brayer G. D. Yeast iso-1-cytochrome c. A 2.8 A resolution three-dimensional structure determination. J Mol Biol. 1988 Jan 20;199(2):295–314. doi: 10.1016/0022-2836(88)90315-4. [DOI] [PubMed] [Google Scholar]
- Maccecchini M. L., Rudin Y., Blobel G., Schatz G. Import of proteins into mitochondria: precursor forms of the extramitochondrially made F1-ATPase subunits in yeast. Proc Natl Acad Sci U S A. 1979 Jan;76(1):343–347. doi: 10.1073/pnas.76.1.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura S., Arpin M., Hannum C., Margoliash E., Sabatini D. D., Morimoto T. In vitro synthesis and posttranslational uptake of cytochrome c into isolated mitochondria: role of a specific addressing signal in the apocytochrome. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4368–4372. doi: 10.1073/pnas.78.7.4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson D. W., Hergersberg C., Neupert W. Role of cytochrome c heme lyase in the import of cytochrome c into mitochondria. J Biol Chem. 1988 Dec 15;263(35):19034–19042. [PubMed] [Google Scholar]
- Nye S. H., Scarpulla R. C. In vivo expression and mitochondrial targeting of yeast apoiso-1-cytochrome c fusion proteins. Mol Cell Biol. 1990 Nov;10(11):5753–5762. doi: 10.1128/mcb.10.11.5753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfanner N., Hoeben P., Tropschug M., Neupert W. The carboxyl-terminal two-thirds of the ADP/ATP carrier polypeptide contains sufficient information to direct translocation into mitochondria. J Biol Chem. 1987 Nov 5;262(31):14851–14854. [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpulla R. C., Agne K. M., Wu R. Isolation and structure of a rat cytochrome c gene. J Biol Chem. 1981 Jun 25;256(12):6480–6486. [PubMed] [Google Scholar]
- Scarpulla R. C., Nye S. H. Functional expression of rat cytochrome c in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1986 Sep;83(17):6352–6356. doi: 10.1073/pnas.83.17.6352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F., Stewart J. W., Schweingruber A. M. Mutants of yeast initiating translation of iso-1-cytochrome c within a region spanning 37 nucleotides. Cell. 1980 May;20(1):215–222. doi: 10.1016/0092-8674(80)90249-4. [DOI] [PubMed] [Google Scholar]
- Smith M., Leung D. W., Gillam S., Astell C. R., Montgomery D. L., Hall B. D. Sequence of the gene for iso-1-cytochrome c in Saccharomyces cerevisiae. Cell. 1979 Apr;16(4):753–761. doi: 10.1016/0092-8674(79)90091-6. [DOI] [PubMed] [Google Scholar]
- Sprinkle J. R., Hakvoort T. B., Koshy T. I., Miller D. D., Margoliash E. Amino acid sequence requirements for the association of apocytochrome c with mitochondria. Proc Natl Acad Sci U S A. 1990 Aug;87(15):5729–5733. doi: 10.1073/pnas.87.15.5729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart R. A., Nicholson D. W., Neupert W. Early steps in mitochondrial protein import: receptor functions can be substituted by the membrane insertion activity of apocytochrome c. Cell. 1990 Jan 12;60(1):31–43. doi: 10.1016/0092-8674(90)90713-o. [DOI] [PubMed] [Google Scholar]
- Woods R. A., Sanders H. K., Briquet M., Foury F., Drysdale B. E., Mattoon J. R. Regulation of mitochondrial biogenesis: enzymatic changes in cytochrome-deficient yeast mutants requiring delta-aminolevulinic acid. J Biol Chem. 1975 Dec 10;250(23):9090–9098. [PubMed] [Google Scholar]
- Zimmermann R., Hennig B., Neupert W. Different transport pathways of individual precursor proteins in mitochondria. Eur J Biochem. 1981 Jun 1;116(3):455–460. doi: 10.1111/j.1432-1033.1981.tb05357.x. [DOI] [PubMed] [Google Scholar]