Abstract
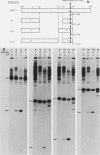
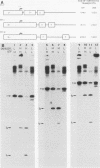
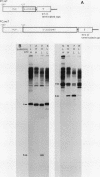
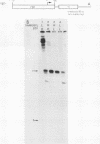
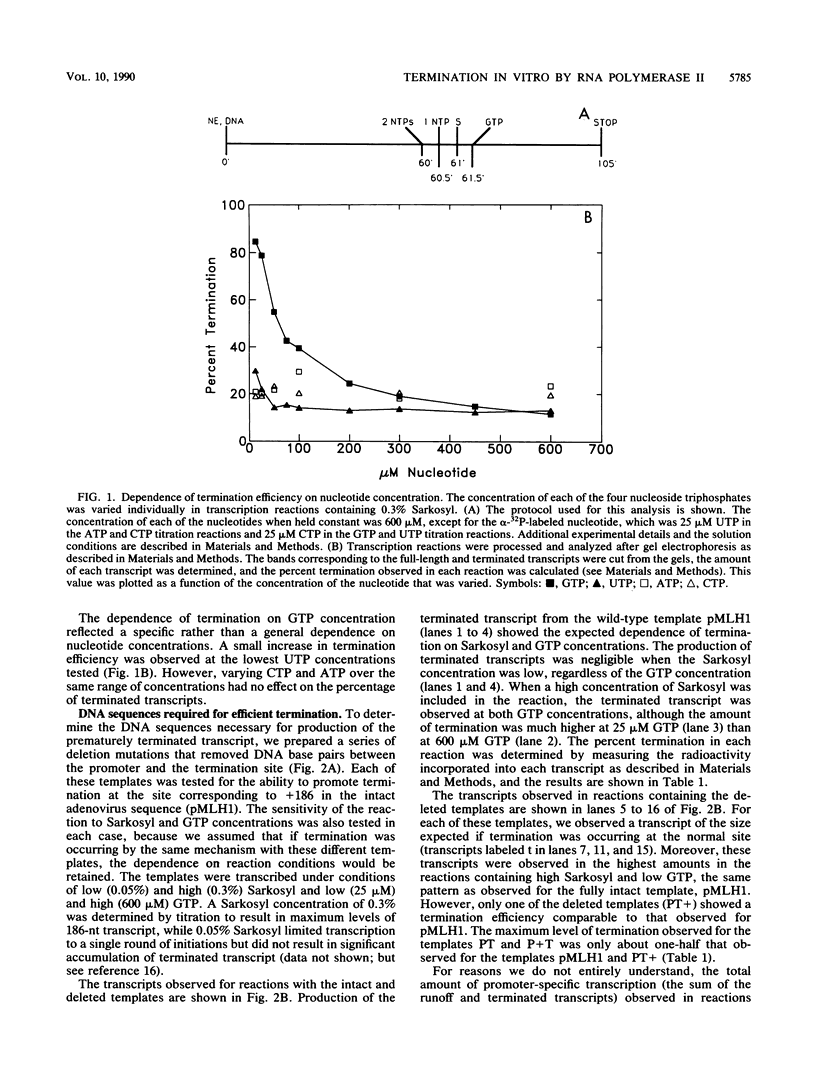
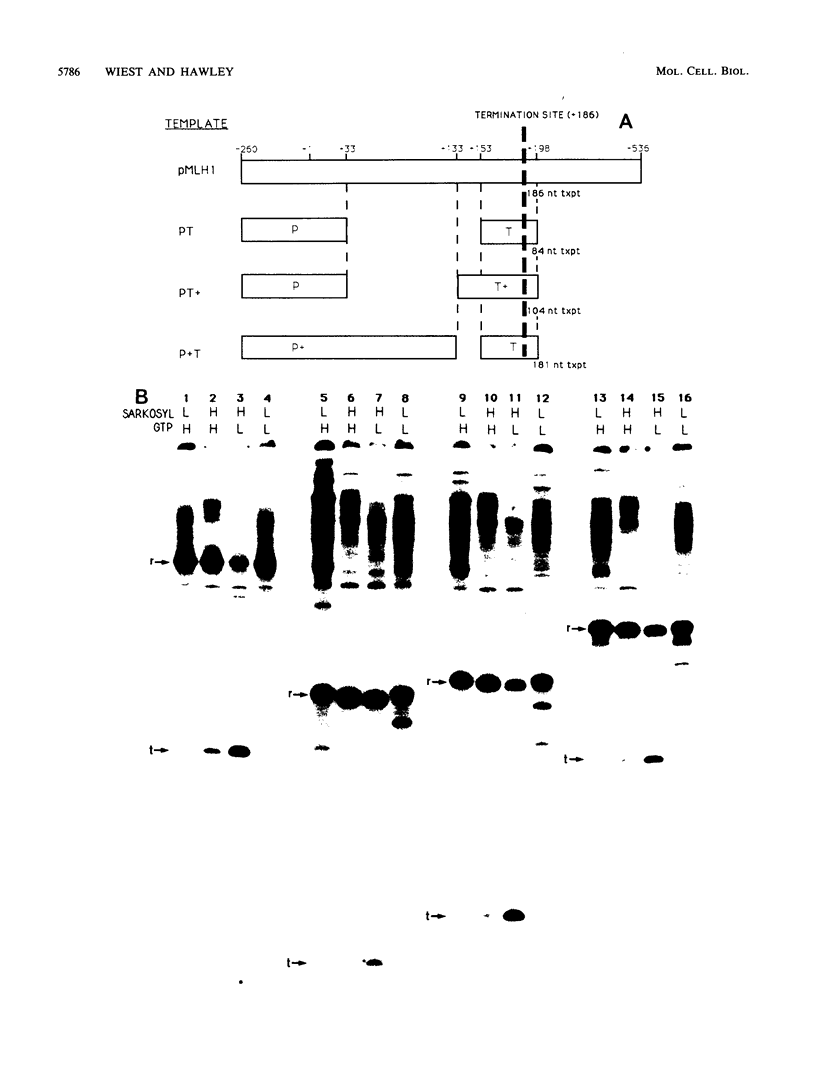
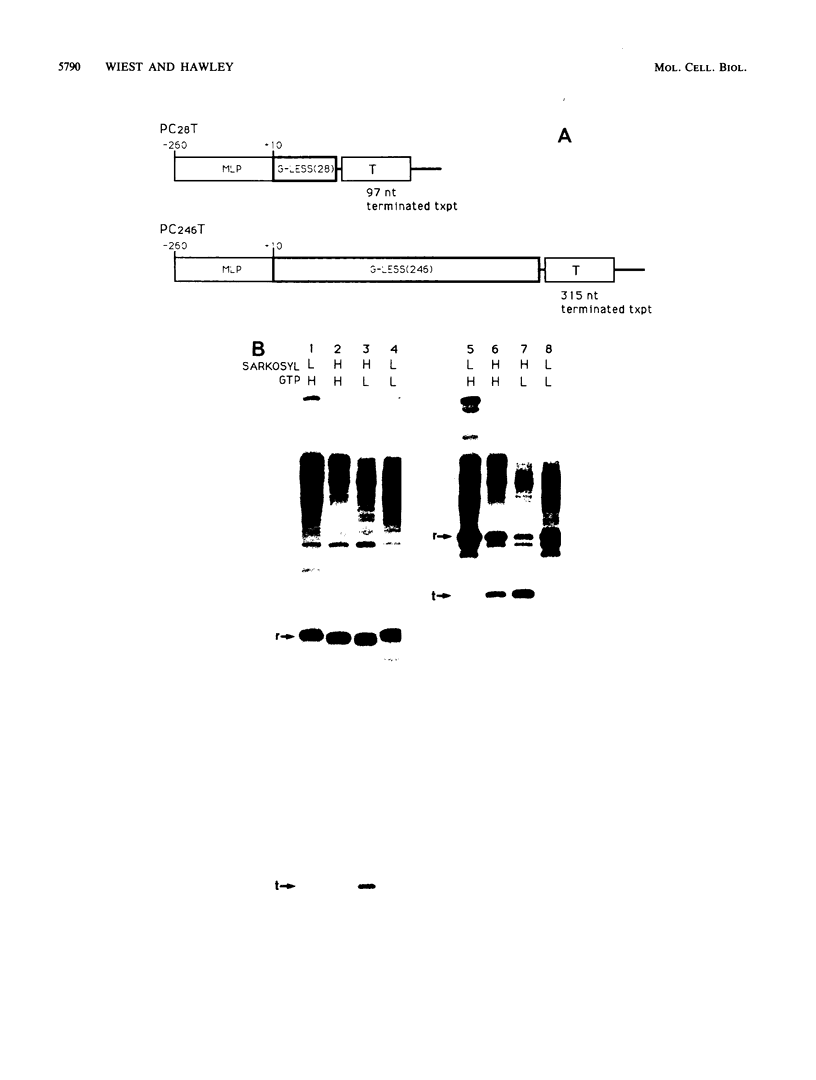
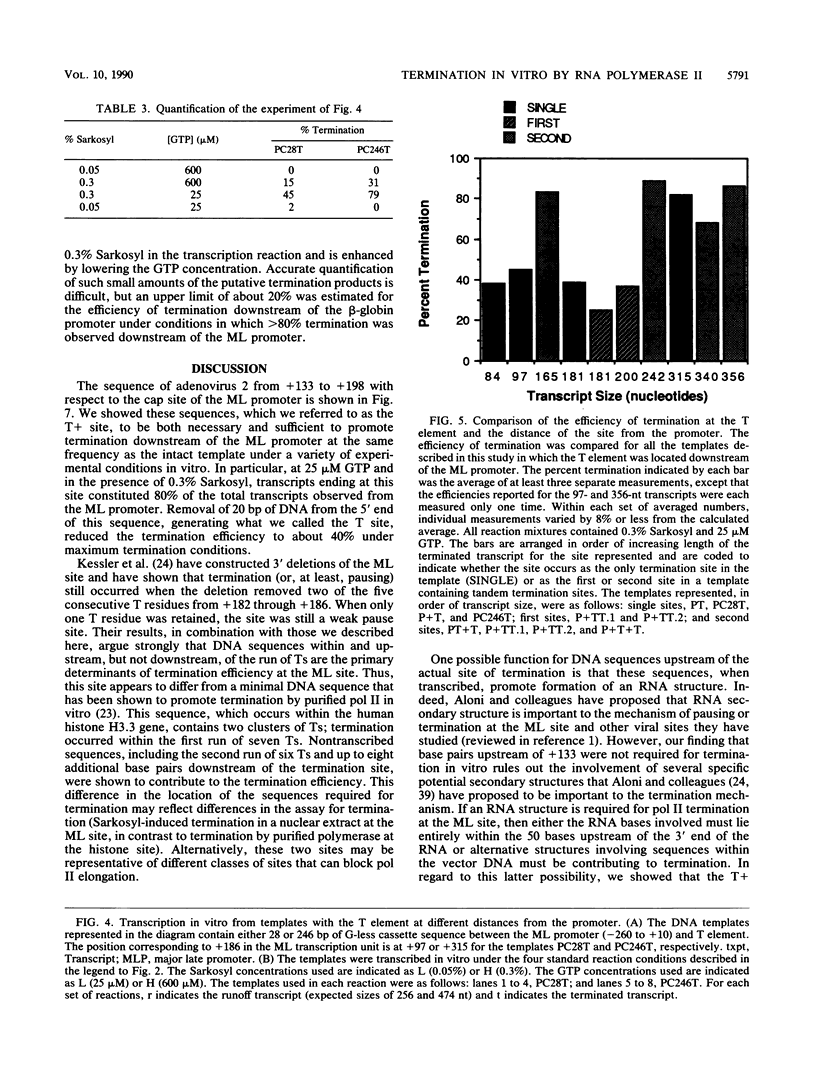
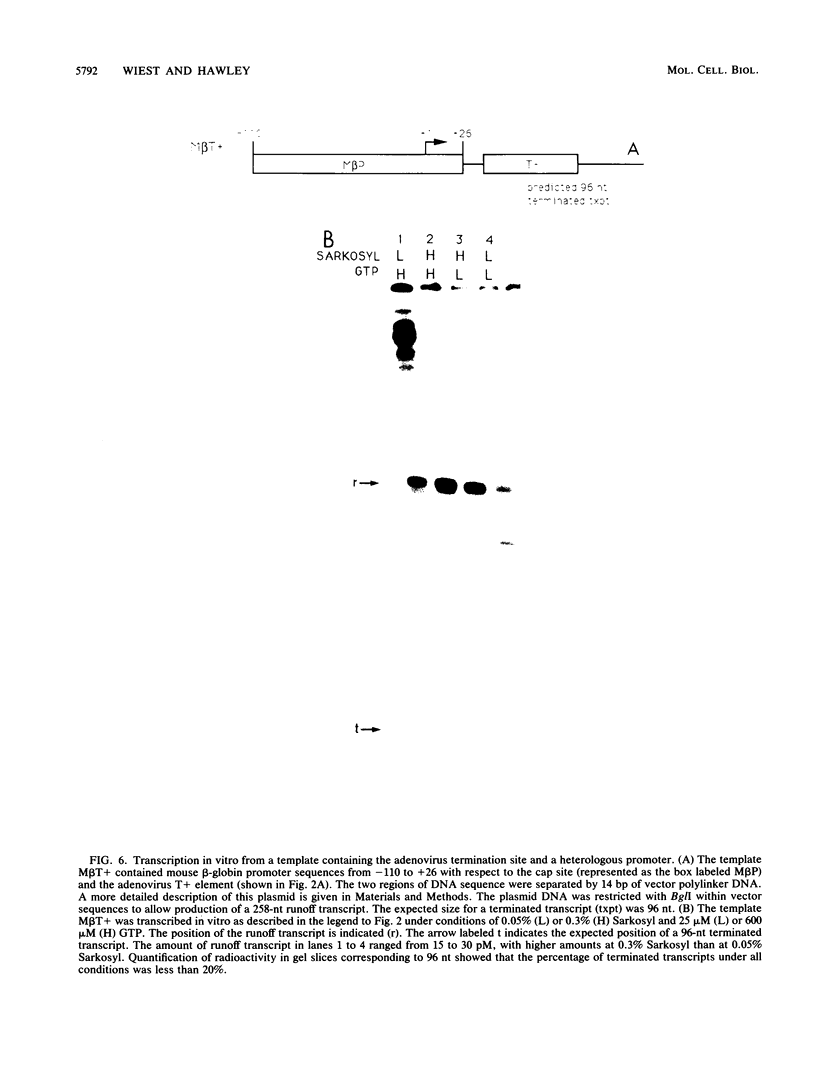
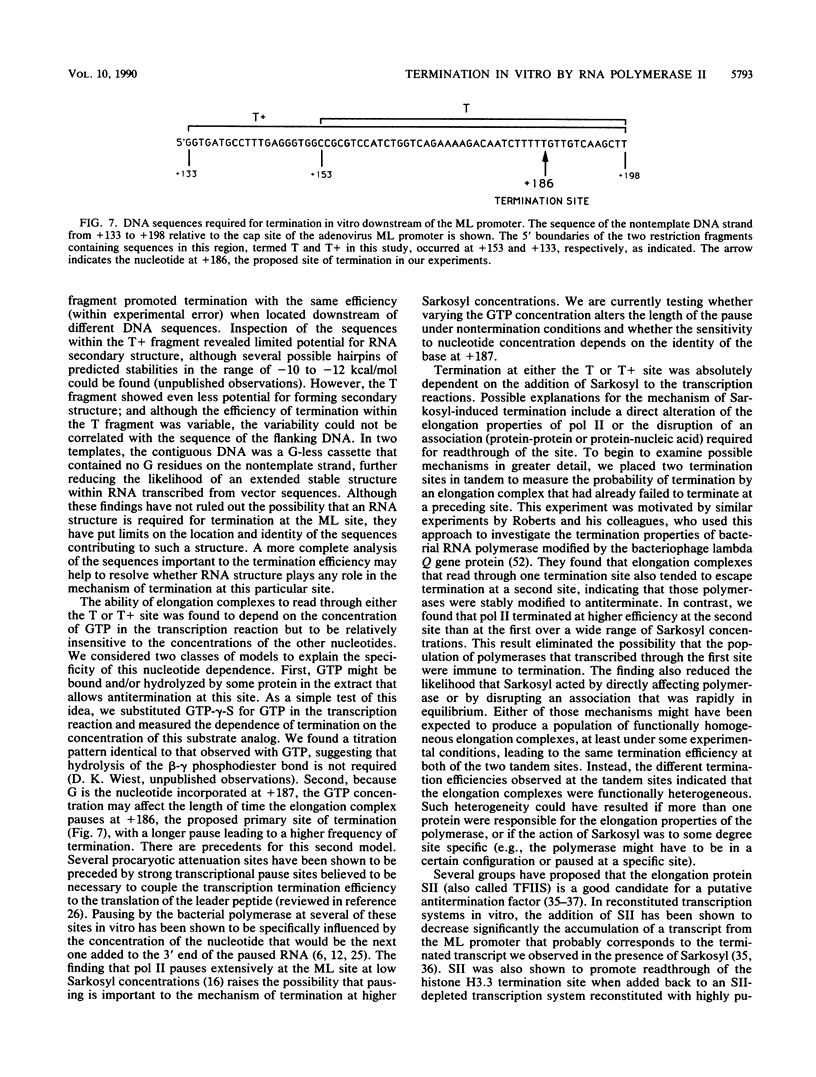
Transcription from the adenovirus major late (ML) promoter has previously been shown to pause or terminate prematurely in vivo and in vitro at a site within the first intron of the major late transcription unit. We are studying the mechanism of elongation arrest at this site in vitro to define the DNA sequences and proteins that determine the elongation behavior of RNA polymerase II. Our assay system consists of a nuclear extract prepared from cultured human cells. With standard reaction conditions, termination is not observed downstream of the ML promoter. However, in the presence of Sarkosyl, up to 80% of the transcripts terminate 186 nucleotides downstream of the start site. Using this assay, we showed that the DNA sequences required to promote maximal levels of termination downstream of the ML promoter reside within a 65-base-pair region and function in an orientation-dependent manner. To test whether elongation complexes from the ML promoter were functionally homogeneous, we determined the termination efficiency at each of two termination sites placed in tandem. We found that the behavior of the elongation complexes was different at these sites, with termination being greater at the downstream site over a wide range of Sarkosyl concentrations. This result ruled out a model in which the polymerases that read through the first site were stably modified to antiterminate. We also demonstrated that the ability of the elongation complexes to respond to the ML termination site was promoter specific, as the site did not function efficiently downstream of a heterologous promoter. Taken together, the results presented here are not consistent with the simplest class of models that have been proposed previously for the mechanism of Sarkosyl-induced termination.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aloni Y., Hay N. Attenuation may regulate gene expression in animal viruses and cells. CRC Crit Rev Biochem. 1985;18(4):327–383. doi: 10.3109/10409238509086785. [DOI] [PubMed] [Google Scholar]
- Bender T. P., Thompson C. B., Kuehl W. M. Differential expression of c-myb mRNA in murine B lymphomas by a block to transcription elongation. Science. 1987 Sep 18;237(4821):1473–1476. doi: 10.1126/science.3498214. [DOI] [PubMed] [Google Scholar]
- Bentley D. L., Groudine M. Sequence requirements for premature termination of transcription in the human c-myc gene. Cell. 1988 Apr 22;53(2):245–256. doi: 10.1016/0092-8674(88)90386-8. [DOI] [PubMed] [Google Scholar]
- Burton Z. F., Killeen M., Sopta M., Ortolan L. G., Greenblatt J. RAP30/74: a general initiation factor that binds to RNA polymerase II. Mol Cell Biol. 1988 Apr;8(4):1602–1613. doi: 10.1128/mcb.8.4.1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai H., Luse D. S. Transcription initiation by RNA polymerase II in vitro. Properties of preinitiation, initiation, and elongation complexes. J Biol Chem. 1987 Jan 5;262(1):298–304. [PubMed] [Google Scholar]
- Chan C. L., Landick R. The Salmonella typhimurium his operon leader region contains an RNA hairpin-dependent transcription pause site. Mechanistic implications of the effect on pausing of altered RNA hairpins. J Biol Chem. 1989 Dec 5;264(34):20796–20804. [PubMed] [Google Scholar]
- Chinsky J. M., Maa M. C., Ramamurthy V., Kellems R. E. Adenosine deaminase gene expression. Tissue-dependent regulation of transcriptional elongation. J Biol Chem. 1989 Aug 25;264(24):14561–14565. [PubMed] [Google Scholar]
- Conaway R. C., Conaway J. W. ATP activates transcription initiation from promoters by RNA polymerase II in a reversible step prior to RNA synthesis. J Biol Chem. 1988 Feb 25;263(6):2962–2968. [PubMed] [Google Scholar]
- Dignam J. D., Lebovitz R. M., Roeder R. G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983 Mar 11;11(5):1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam J. D., Martin P. L., Shastry B. S., Roeder R. G. Eukaryotic gene transcription with purified components. Methods Enzymol. 1983;101:582–598. doi: 10.1016/0076-6879(83)01039-3. [DOI] [PubMed] [Google Scholar]
- Evans R., Weber J., Ziff E., Darnell J. E. Premature termination during adenovirus transcription. Nature. 1979 Mar 22;278(5702):367–370. doi: 10.1038/278367a0. [DOI] [PubMed] [Google Scholar]
- Fisher R. F., Das A., Kolter R., Winkler M. E., Yanofsky C. Analysis of the requirements for transcription pausing in the tryptophan operon. J Mol Biol. 1985 Apr 5;182(3):397–409. doi: 10.1016/0022-2836(85)90199-8. [DOI] [PubMed] [Google Scholar]
- Fort P., Rech J., Vie A., Piechaczyk M., Bonnieu A., Jeanteur P., Blanchard J. M. Regulation of c-fos gene expression in hamster fibroblasts: initiation and elongation of transcription and mRNA degradation. Nucleic Acids Res. 1987 Jul 24;15(14):5657–5667. doi: 10.1093/nar/15.14.5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gariglio P., Buss J., Green M. H. Sarkosyl activation of RNA polymerase activity in mitotic mouse cells. FEBS Lett. 1974 Aug 30;44(3):330–333. doi: 10.1016/0014-5793(74)81170-1. [DOI] [PubMed] [Google Scholar]
- Hai T. W., Horikoshi M., Roeder R. G., Green M. R. Analysis of the role of the transcription factor ATF in the assembly of a functional preinitiation complex. Cell. 1988 Sep 23;54(7):1043–1051. doi: 10.1016/0092-8674(88)90119-5. [DOI] [PubMed] [Google Scholar]
- Hawley D. K., Roeder R. G. Functional steps in transcription initiation and reinitiation from the major late promoter in a HeLa nuclear extract. J Biol Chem. 1987 Mar 15;262(8):3452–3461. [PubMed] [Google Scholar]
- Hawley D. K., Roeder R. G. Separation and partial characterization of three functional steps in transcription initiation by human RNA polymerase II. J Biol Chem. 1985 Jul 5;260(13):8163–8172. [PubMed] [Google Scholar]
- Hay N., Skolnik-David H., Aloni Y. Attenuation in the control of SV40 gene expression. Cell. 1982 May;29(1):183–193. doi: 10.1016/0092-8674(82)90102-7. [DOI] [PubMed] [Google Scholar]
- Hernandez N. Formation of the 3' end of U1 snRNA is directed by a conserved sequence located downstream of the coding region. EMBO J. 1985 Jul;4(7):1827–1837. doi: 10.1002/j.1460-2075.1985.tb03857.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez N., Weiner A. M. Formation of the 3' end of U1 snRNA requires compatible snRNA promoter elements. Cell. 1986 Oct 24;47(2):249–258. doi: 10.1016/0092-8674(86)90447-2. [DOI] [PubMed] [Google Scholar]
- Horikoshi M., Sekimizu K., Natori S. Analysis of the stimulatory factor of RNA polymerase II in the initiation and elongation complex. J Biol Chem. 1984 Jan 10;259(1):608–611. [PubMed] [Google Scholar]
- Kao S. Y., Calman A. F., Luciw P. A., Peterlin B. M. Anti-termination of transcription within the long terminal repeat of HIV-1 by tat gene product. Nature. 1987 Dec 3;330(6147):489–493. doi: 10.1038/330489a0. [DOI] [PubMed] [Google Scholar]
- Kerppola T. K., Kane C. M. Analysis of the signals for transcription termination by purified RNA polymerase II. Biochemistry. 1990 Jan 9;29(1):269–278. doi: 10.1021/bi00453a037. [DOI] [PubMed] [Google Scholar]
- Kessler M., Ben-Asher E., Aloni Y. Elements modulating the block of transcription elongation at the adenovirus 2 attenuation site. J Biol Chem. 1989 Jun 15;264(17):9785–9790. [PubMed] [Google Scholar]
- Landick R., Yanofsky C. Stability of an RNA secondary structure affects in vitro transcription pausing in the trp operon leader region. J Biol Chem. 1984 Sep 25;259(18):11550–11555. [PubMed] [Google Scholar]
- Lattier D. L., States J. C., Hutton J. J., Wiginton D. A. Cell type-specific transcriptional regulation of the human adenosine deaminase gene. Nucleic Acids Res. 1989 Feb 11;17(3):1061–1076. doi: 10.1093/nar/17.3.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laybourn P. J., Dahmus M. E. Transcription-dependent structural changes in the C-terminal domain of mammalian RNA polymerase subunit IIa/o. J Biol Chem. 1989 Apr 25;264(12):6693–6698. [PubMed] [Google Scholar]
- Maderious A., Chen-Kiang S. Pausing and premature termination of human RNA polymerase II during transcription of adenovirus in vivo and in vitro. Proc Natl Acad Sci U S A. 1984 Oct;81(19):5931–5935. doi: 10.1073/pnas.81.19.5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T., Segall J., Weil P. A., Roeder R. G. Multiple factors required for accurate initiation of transcription by purified RNA polymerase II. J Biol Chem. 1980 Dec 25;255(24):11992–11996. [PubMed] [Google Scholar]
- Mok M., Maderious A., Chen-Kiang S. Premature termination by human RNA polymerase II occurs temporally in the adenovirus major late transcriptional unit. Mol Cell Biol. 1984 Oct;4(10):2031–2040. doi: 10.1128/mcb.4.10.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers R. M., Tilly K., Maniatis T. Fine structure genetic analysis of a beta-globin promoter. Science. 1986 May 2;232(4750):613–618. doi: 10.1126/science.3457470. [DOI] [PubMed] [Google Scholar]
- Rappaport J., Reinberg D., Zandomeni R., Weinmann R. Purification and functional characterization of transcription factor SII from calf thymus. Role in RNA polymerase II elongation. J Biol Chem. 1987 Apr 15;262(11):5227–5232. [PubMed] [Google Scholar]
- Reinberg D., Roeder R. G. Factors involved in specific transcription by mammalian RNA polymerase II. Transcription factor IIS stimulates elongation of RNA chains. J Biol Chem. 1987 Mar 5;262(7):3331–3337. [PubMed] [Google Scholar]
- Reines D., Chamberlin M. J., Kane C. M. Transcription elongation factor SII (TFIIS) enables RNA polymerase II to elongate through a block to transcription in a human gene in vitro. J Biol Chem. 1989 Jun 25;264(18):10799–10809. [PubMed] [Google Scholar]
- Resnekov O., Aloni Y. RNA polymerase II is capable of pausing and prematurely terminating transcription at a precise location in vivo and in vitro. Proc Natl Acad Sci U S A. 1989 Jan;86(1):12–16. doi: 10.1073/pnas.86.1.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnekov O., Ben-Asher E., Bengal E., Choder M., Hay N., Kessler M., Ragimov N., Seiberg M., Skolnik-David H., Aloni Y. Transcription termination in animal viruses and cells. Gene. 1988 Dec 10;72(1-2):91–104. doi: 10.1016/0378-1119(88)90130-8. [DOI] [PubMed] [Google Scholar]
- Resnekov O., Kessler M., Aloni Y. RNA secondary structure is an integral part of the in vitro mechanism of attenuation in simian virus 40. J Biol Chem. 1989 Jun 15;264(17):9953–9959. [PubMed] [Google Scholar]
- Rougvie A. E., Lis J. T. The RNA polymerase II molecule at the 5' end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell. 1988 Sep 9;54(6):795–804. doi: 10.1016/s0092-8674(88)91087-2. [DOI] [PubMed] [Google Scholar]
- Samuels M., Fire A., Sharp P. A. Separation and characterization of factors mediating accurate transcription by RNA polymerase II. J Biol Chem. 1982 Dec 10;257(23):14419–14427. [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawadogo M., Lescure B., Sentenac A., Fromageot P. Native deoxyribonucleic acid transcription by yeast RNA polymerase--P37 complex. Biochemistry. 1981 Jun 9;20(12):3542–3547. doi: 10.1021/bi00515a037. [DOI] [PubMed] [Google Scholar]
- Sawadogo M., Roeder R. G. Factors involved in specific transcription by human RNA polymerase II: analysis by a rapid and quantitative in vitro assay. Proc Natl Acad Sci U S A. 1985 Jul;82(13):4394–4398. doi: 10.1073/pnas.82.13.4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawadogo M., Sentenac A., Fromageot P. Interaction of a new polypeptide with yeast RNA polymerase B. J Biol Chem. 1980 Jan 10;255(1):12–15. [PubMed] [Google Scholar]
- Schmidt M. C., Zhou Q., Berk A. J. Sp1 activates transcription without enhancing DNA-binding activity of the TATA box factor. Mol Cell Biol. 1989 Aug;9(8):3299–3307. doi: 10.1128/mcb.9.8.3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skarnes W. C., Tessier D. C., Acheson N. H. RNA polymerases stall and/or prematurely terminate nearby both early and late promoters on polyomavirus DNA. J Mol Biol. 1988 Sep 5;203(1):153–171. doi: 10.1016/0022-2836(88)90099-x. [DOI] [PubMed] [Google Scholar]
- Sopta M., Carthew R. W., Greenblatt J. Isolation of three proteins that bind to mammalian RNA polymerase II. J Biol Chem. 1985 Aug 25;260(18):10353–10360. [PubMed] [Google Scholar]
- Spencer C. A., Groudine M. Transcription elongation and eukaryotic gene regulation. Oncogene. 1990 Jun;5(6):777–785. [PubMed] [Google Scholar]
- Spencer C. A., LeStrange R. C., Novak U., Hayward W. S., Groudine M. The block to transcription elongation is promoter dependent in normal and Burkitt's lymphoma c-myc alleles. Genes Dev. 1990 Jan;4(1):75–88. doi: 10.1101/gad.4.1.75. [DOI] [PubMed] [Google Scholar]
- Yang X. J., Hart C. M., Grayhack E. J., Roberts J. W. Transcription antitermination by phage lambda gene Q protein requires a DNA segment spanning the RNA start site. Genes Dev. 1987 May;1(3):217–226. doi: 10.1101/gad.1.3.217. [DOI] [PubMed] [Google Scholar]
- Yuo C. Y., Ares M., Jr, Weiner A. M. Sequences required for 3' end formation of human U2 small nuclear RNA. Cell. 1985 Aug;42(1):193–202. doi: 10.1016/s0092-8674(85)80115-x. [DOI] [PubMed] [Google Scholar]
- de Vegvar H. E., Lund E., Dahlberg J. E. 3' end formation of U1 snRNA precursors is coupled to transcription from snRNA promoters. Cell. 1986 Oct 24;47(2):259–266. doi: 10.1016/0092-8674(86)90448-4. [DOI] [PubMed] [Google Scholar]