Abstract
Structured water molecules near redox cofactors were found recently to accelerate electron-transfer (ET) kinetics in several systems. Theoretical study of interprotein electron transfer across an aqueous interface reveals three distinctive electronic coupling mechanisms that we describe here: (i) a protein-mediated regime when the two proteins are in van der Waals contact; (ii) a structured water-mediated regime featuring anomalously weak distance decay at relatively close protein-protein contact distances; and (iii) a bulk water–mediated regime at large distances. Our analysis explains a range of otherwise puzzling biological ET kinetic data and provides a framework for including explicit water-mediated tunneling effects on ET kinetics.
Protein ET reactions play a critical role in biologically vital processes in living cells, most notably photosynthesis and respiration (1). Describing the structure dependence of intermolecular ET reactions is particularly challenging because of the wide range of the accessible docking geometries, and several studies have addressed these reaction mechanisms (2–8). The factors that control unimolecular ET rates, namely the donor-to-acceptor (D to A) distance and energies, the structure of the ET-mediating protein matrix, and the thermal atomic motion, have been extensively explored both experimentally (4–6) and theoretically (9–14).
Intermolecular ET reactions, however, remain a challenge. In addition to the above factors, the rate depends on the D-to-A docking geometry, as well as on the structure and thermal motion of the solvent (2–7). The number of structural degrees of freedom makes quantitatively reliable theoretical calculations extremely difficult. We show that the intervening water structure leads to one of three distinctly different ET tunneling regimes, in contrast to the common assumption of single-exponential distance decay (2, 5–7). The identification of these three regimes provides a framework for understanding the mechanisms that underlie several unexplained and seemingly unrelated water-mediated biological ET rate processes (7, 15–19), as well as providing a strategy for making theoretical estimates of bimolecular rates that take these water-mediation effects into account.
Water can influence the ET reaction rates by mediating ET coupling pathways, as well as by controlling activation free energies (5, 9). In the past decade, the distance dependence of water-mediated ET reaction rates has become the focus of intensive experimental (4–7, 13, 15–17, 20, 21) and theoretical (18, 19, 22–24) investigation. Until recently, experimental and theoretical analysis suggested a single-exponential decay of the ET rates with distance through water, with a characteristic decay constant of about 1.6 to 1.7 Å−1 (5, 20, 21). In comparison with proteins that exhibit decay constants of about 1.0 to 1.2 Å−1 (5), water appeared to be a rather poor ET mediator because of extensive through-space links in tunneling pathways (20).
In the past few years, however, a number of important experimental observations emerged that are inconsistent with the single-exponential decay model for water and with the generality of rapid distance decay for ET through water molecules. In crystals, ET across thin aqueous interfaces was found to be facile (7). In covalently cross-linked azurin complexes, water dimers that formed between the redox centers appeared to increase the ET rate substantially (15). In DNA, the influence of water (and counterions) may be even more pronounced, and fluctuations in hydration were proposed to gate ET (16). In small water clusters in gas phase or on TiO2 surfaces, hydrated electrons were suggested to facilitate water-mediated ET reactions (25, 26). These recent experiments all suggest the need for a deeper unifying theoretical framework to describe the distance and structure dependence of water-mediated ET reaction rates.
Here, we use computation to explore how the aqueous protein environment influences ET rates as a function of distance. Coupling interactions and rates were computed for the trypsin-solubilized bovine liver cytochrome b5 [(27), Protein Data Bank (PDB) entry 1CYO] self-exchange ET reaction (27–33), which represents a broad class of solvent-mediated ET reactions involving α-helical redox proteins. To explore the influence of protein orientation, we constrained the cytochrome b5 heme-heme angles to 0°, 45°, or 90°.
For each orientation, the molecules were also constrained to a specified distance between the porphyrin-ring edges between 6 and 16 Å. For each of the geometries, the protein molecules were solvated in the TIP3 (34) water; Na+ and Cl− counterions were added to establish an ionic strength of 0.2 M, and the system was equilibrated for 200 ps using the Charmm27 forcefield, constant pressure (NpT) ensemble, periodic boundary conditions, and particle-mesh Ewald full electrostatics (35). After equilibration, another 100-ps molecular dynamics (MD) simulation was performed, and the system conformation was saved every 1 ps, yielding 100 “snapshots.” The D-to-A electronic coupling TDA was computed for each snapshot with an extended–Hückel-level (XH) electronic Hamiltonian; the ET rate was estimated from the mean-square electronic coupling 〈 〉 in the non-adiabatic Fermi’s golden rule expression (10, 11). The TDA calculations included the protein atoms and the interfacial water molecules. For comparison, these calculations were repeated without the interfacial water (34). This computational approach provides a reasonable qualitative estimate of the D-A couplings in proteins and small molecules (10, 11, 36, 37). The coupling was also estimated using the empirical Pathways model (38) and atomic packing density analysis (13). Finally, bimolecular self-exchange ET rates were estimated by using the computed mean square donor-acceptor interactions (as a function of distance) in Brownian dynamics simulations (28, 29).
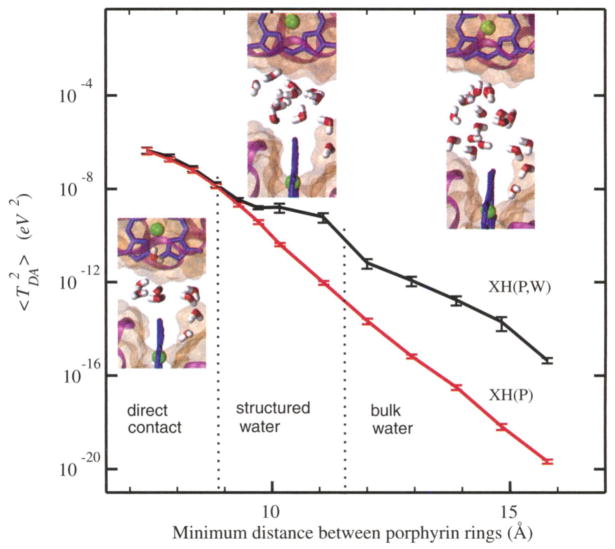
The dependence of 〈 〉 on the distance between porphyrin-ring edges is essentially orientation independent (35); Fig. 1 shows the distance dependence for the 90° orientation between the porphyrin rings. Most important, rather than finding single-exponential decay with distance, the XH results reveal three distinct ET regimes at distances of 7 to 9 Å, 9 to 12 Å, and >12 Å, respectively. At 7 to 9 Å, the distance decay is similar to that for intramolecular ET (5); that is, the tunneling is facilitated primarily by the protein atoms, and water has little effect on the electronic coupling. In the distance range from 9 to 12 Å, the coupling decay is anomalously weak, and water is the principal mediator of tunneling across the protein-protein interface (i.e., the plateau regime). At distances >12 Å, the distance decay is consistent with tunneling through liquid water.
Fig. 1.
The mean-square electronic coupling dependence on the distance between the two porphyrin rings (90° angle between porphyrin rings): XH(P,W), proteins and water included in the coupling calculation at the extended-Hückel level; XH(P), only protein mediation is included in the extended-Hückel coupling calculations. The three distance ranges correspond to the direct contact regime, structured water–mediated regime, and bulk water–mediated regime, respectively. The insets show configurations of the interfacial water molecules typical for each regime. The error bars show the sampling error estimated using a renormalization group-based method (39).
The empirical Pathways-level analysis of the 〈 〉 distance dependence also captures the qualitative features of these three regimes (fig. S1). The atomic packing density method does not resolve the three regimes because there is little change in the overall packing density over the entire distance range. Although the XH electronic structure method is a qualitative one, we believe that the existence of the three coupling regimes is a robust characteristic of the solvated protein environment.
The structural origin of these three different electronic coupling regimes is explained by the interfacial water structure at difference distances, as shown in Fig. 1. At small distances, there are only a few water molecules at the interface, and they are positioned outside of the dominant ET coupling pathways. At intermediate distances, a small number of water molecules penetrate the space between the hemes, establishing strongly coupled tunneling pathways. Figure 2 shows the evolution of 〈 〉 in the coupling plateau region for time snapshots taken from an MD trajectory. In a number of conformations, ET is mediated by either a single water molecule (snapshot A) or by several water molecules that provide multiple ET pathways that interfere constructively (snapshot B). These constructively interfering geometries control the value of 〈 〉, and the relative abundance of these conformations (as compared to conformations with destructively interfering pathways, snapshot C) is characteristic of the plateau regime. This behavior arises from the confined space between proteins, combined with strong protein surface-water interactions (electrostatic and van der Waals interactions), in the plateau regime. At larger protein-protein distances, the water configurations that lead to strong TDA values occur much less frequently than in the structured water regime. The electronic coupling between proteins in the shortest-distance regime, where the gap between proteins is too small for water to enter the dominant coupling pathways, is mediated by through-space tunneling. The through-space mechanism is known to decay rapidly with distance (12, 38). These three ET regimes are essentially independent of protein orientation (figs. S1 and S2). We refer to the three regimes as the direct contact regime (short distances), the structured water-mediated regime (intermediate distances), and the bulk water–mediated regime (larger distances). Although the extended-Hückel method may introduce systematic errors, we expect it to capture the essential features of the bridge-mediated coupling and interference effects.
Fig. 2.

A typical time series of computed values in the structured water (plateau) regime. The relatively rare conformations that dominate the D-to-A coupling feature either a single dominant bridging water molecule (snapshot A), or several constructively interfering water-mediated pathways (snapshot B). In most of the time snapshots, the hydrogen-bonding network provides multiple ET pathways with mixed constructive and destructive interference, resulting in relatively small D-to-A coupling (snapshot C). The electrostatic and van der Waals interactions between the water and protein surfaces increase the probability of generating water-molecule configurations that provide large D-to-A coupling leading to the smooth distance dependence of the ET rate in the plateau regime. The large peak at 94 ps arises from a constructively interfering coupling network established by three water molecules.
We have tested the consistency of a three-coupling regime model with ET self-exchange rates using a Brownian dynamics framework for computing the intermolecular ET rates (28, 29, 35). Table 1 shows that the computed ET rates are in good qualitative agreement with experimental self-exchange rates (28). Earlier simulation of Northrup and co-workers (28) based on single-exponent models with two adjustable parameters was also consistent with the data. The results of the present study lack adjustable TDA parameters, although the quantitative accuracy of the extended-Hückel approach to electronic coupling calculation is certainly dependent on its parameterization.
Table 1.
Experimental (28) and theoretically estimated (Brownian dynamics) bimolecular rate constants k2 (M−1 s−1) for the cytochrome b5 self-exchange ET as a function of ionic strength μ.
| μ(M) | k2 (M−1 s−1)
|
|
|---|---|---|
| Experiment | Theory | |
| 0.1 | 2.6 × 103 | 1.0 × 103 |
| 0.3 | 4.6 × 103 | 2.4 × 104 |
| 0.6 | 1.6 × 104 | 7.6 × 104 |
| 1.0 | 2.8 × 104 | 1.1 × 105 |
| 1.5 | 4.5 × 104 | 1.7 × 105 |
The existence of multiple tunneling regimes also provides insight into several recent (and otherwise puzzling) experimental and theoretical observations in biological ET reaction kinetics. Winkler, Gray, and co-workers found that ET across protein-protein interfaces in protein crystals mediated by three water molecules is nearly as rapid as unimolecular ET is over the same distance (7). Canters and co-workers showed that water dimers between covalently cross-linked azurin complexes could substantially enhance the intermolecular ET kinetics (15). Similarly, Klinman and co-workers investigated the copper-to-copper ET over about 7 Å in the hydroxylating domain of peptidylglycine α-amidylating monooxygenase and found an unusually large electronic coupling mediated, apparently, by water rather than by the protein or substrate (17). Using Pathways-level analysis, Onuchic and co-workers found that water molecules mediate the dominant ET coupling routes between cytochrome c2 and the photosynthetic reaction center (18). Cave and co-workers showed that water molecules between model D and A pairs substantially enhance intermolecular ET rates as well (19). All of these recent observations support our conclusion that a small number of structured water molecules interposed between the donor and the acceptor cofactors can substantially enhance ET rates.
The influence of aqueous tunneling pathways on interprotein ET kinetics has remained a key open issue in biological ET for some time. Single-exponential decay models fail to describe water-mediated ET reactions properly. The existence of multiple tunneling mediating regimes identified above is evinced by a body of recent experimental and theoretical observations. Most importantly, the structured water coupling regime may provide an important mechanism to facilitate ET reactions in the critical near-contact distance range relevant to biological ET kinetics. We hypothesize that water may be a particularly strong tunneling mediator when it occupies a sterically constrained space between redox cofactors with strong organizing forces that favor constructively interfering coupling pathways. It will be particularly interesting to use both theory and experiment to explore how the water-mediated coupling between proteins varies with protein-protein shape complementarity, surface charge and polarity, and dynamical fluctuations of the proteins and of the organized water at the interface.
Supplementary Material
Acknowledgments
This work was supported by the NIH (GM-048043). We are grateful to H. B. Gray and J. R. Winkler for stimulating discussions. We thank T. Kawatsu for providing code for the XH calculations, I. V. Kurnikov for providing code for the Pathways-based and atomic packing density-based calculations, and M. A. Pasquinelli for assistance with the Brownian dynamics calculations.
Footnotes
References and Notes
- 1.Berg JM, Stryer L, Tymoczko JL. Biochemistry. 5. Freeman; New York: 2002. [Google Scholar]
- 2.Gray HB, Winkler JR. Proc Natl Acad Sci USA. 2005;102:3534. doi: 10.1073/pnas.0408029102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liang ZX, et al. J Am Chem Soc. 2004;126:2785. doi: 10.1021/ja038163l. [DOI] [PubMed] [Google Scholar]
- 4.Osyczka A, Moser CC, Daldal F, Dutton PL. Nature. 2004;427:607. doi: 10.1038/nature02242. [DOI] [PubMed] [Google Scholar]
- 5.Gray HB, Winkler JR. Q Rev Biophys. 2003;36:341. doi: 10.1017/s0033583503003913. [DOI] [PubMed] [Google Scholar]
- 6.Blankenship RE. Nat Struct Biol. 2001;8:94. doi: 10.1038/84191. [DOI] [PubMed] [Google Scholar]
- 7.Tezcan FA, Crane BR, Winkler JR, Gray HB. Proc Natl Acad Sci USA. 2001;98:5002. doi: 10.1073/pnas.081072898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McLendon G, Hake R. Chem Rev. 1992;92:481. [Google Scholar]
- 9.Marcus RA, Sutin N. Biochim Biophys Acta. 1985;811:265. [Google Scholar]
- 10.Skourtis SS, Balabin IA, Kawatsu T, Beratan DN. Proc Natl Acad Sci USA. 2005;102:3552. doi: 10.1073/pnas.0409047102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Balabin IA, Onuchic JN. Science. 2000;290:114. doi: 10.1126/science.290.5489.114. [DOI] [PubMed] [Google Scholar]
- 12.Jones M, Kurnikov IV, Beratan DN. J Phys Chem A. 2002;106:2002. [Google Scholar]
- 13.Page CC, Moser CC, Chen XX, Dutton PL. Nature. 1999;402:47. doi: 10.1038/46972. [DOI] [PubMed] [Google Scholar]
- 14.Tan M-L, Balabin IA, Onuchic JN. Biophys J. 2004;86:1813. doi: 10.1016/S0006-3495(04)74248-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Amsterdam IMC, et al. Nat Struct Biol. 2002;9:48. doi: 10.1038/nsb736. [DOI] [PubMed] [Google Scholar]
- 16.Barnett RN, Cleveland CL, Landman U, Schuster GB. Science. 2001;294:567. doi: 10.1126/science.1062864. [DOI] [PubMed] [Google Scholar]
- 17.Francisco WA, Wille G, Smith AJ, Merkler DJ, Klinman JP. J Am Chem Soc. 2004;126:13168. doi: 10.1021/ja046888z. [DOI] [PubMed] [Google Scholar]
- 18.Miyashita O, Okamura MY, Onuchic JN. Proc Natl Acad Sci USA. 2005;102:3558. doi: 10.1073/pnas.0409600102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller NE, Wander MC, Cave RJ. J Phys Chem A. 1999;103:1084. [Google Scholar]
- 20.Wenger OS, Leigh BS, Villahermosa RM, Gray HB, Winkler JR. Science. 2005;307:99. doi: 10.1126/science.1103818. [DOI] [PubMed] [Google Scholar]
- 21.Ponce A, Gray HB, Winkler JR. J Am Chem Soc. 2000;122:8187. [Google Scholar]
- 22.Benjamin I, Evans D, Nitzan A. J Chem Phys. 1997;106:6647. [Google Scholar]
- 23.Newton MD. J Electroanal Chem. 1997;438:3. [Google Scholar]
- 24.Larsson S. J Phys Chem. 1984;88:1321. [Google Scholar]
- 25.Verlet JRR, Bragg AE, Kammrath A, Cheshnovsky O, Neumark DM. Science. 2004;307:93. doi: 10.1126/science.1106719. [DOI] [PubMed] [Google Scholar]
- 26.Onda K, et al. Science. 2005;308:1154. doi: 10.1126/science.1109366. [DOI] [PubMed] [Google Scholar]
- 27.Durley RCE, Mathews FS. Acta Crystallogr. 1996;D52:65. doi: 10.1107/S0907444995007827. [DOI] [PubMed] [Google Scholar]
- 28.Andrew SM, Thomasson KA, Northrup SH. J Am Chem Soc. 1993;115:5516. [Google Scholar]
- 29.Northrup SH, Boles JO, Reynolds JCL. Science. 1988;241:67. doi: 10.1126/science.2838904. [DOI] [PubMed] [Google Scholar]
- 30.Strittmatter P, et al. Proc Natl Acad Sci USA. 1974;71:4565. doi: 10.1073/pnas.71.11.4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hegesh E, Hegesh J, Kaftory AN. N Engl J Med. 1986;314:757. doi: 10.1056/NEJM198603203141206. [DOI] [PubMed] [Google Scholar]
- 32.Utecht RE, Kurtz DM., Jr Biochim Biophys Acta. 1988;953:164. doi: 10.1016/0167-4838(88)90021-0. [DOI] [PubMed] [Google Scholar]
- 33.Dixon DW, Hong X, Woehler SE, Mauk AG, Sishta BP. J Am Chem Soc. 1990;112:1082. [Google Scholar]
- 34.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. J Chem Phys. 1983;79:926. [Google Scholar]
- 35.Details of system setup, electronic structure, molecular dynamics, and Brownian dynamics simulations are available on Science Online.
- 36.Kawatsu T, Kakitani T, Yamato T. J Phys Chem B. 2002;106:5068. [Google Scholar]
- 37.Siddarth P, Marcus RA. J Phys Chem. 1993;97:1308. [Google Scholar]
- 38.Beratan DN, Betts JN, Onuchic JN. Science. 1991;252:1285. doi: 10.1126/science.1656523. [DOI] [PubMed] [Google Scholar]
- 39.Flyvbjerg H, Petersen HG. J Chem Phys. 1989;91:461. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.