Abstract
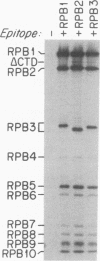
Mutations in the three largest subunits of yeast RNA polymerase II (RPB1, RPB2, and RPB3) were investigated for their effects on RNA polymerase II structure and assembly. Among 23 temperature-sensitive mutations, 6 mutations affected enzyme assembly, as assayed by immunoprecipitation of epitope-tagged subunits. In all six assembly mutants, RNA polymerase II subunits synthesized at the permissive temperature were incorporated into stably assembled, immunoprecipitable enzyme and remained stably associated when cells were shifted to the nonpermissive temperature, whereas subunits synthesized at the nonpermissive temperature were not incorporated into a completely assembled enzyme. The observation that subunit subcomplexes accumulated in assembly-mutant cells at the nonpermissive temperature led us to investigate whether these subcomplexes were assembly intermediates or merely byproducts of mutant enzyme instability. The time course of assembly of RPB1, RPB2, and RPB3 was investigated in wild-type cells and subsequently in mutant cells. Glycerol gradient fractionation of extracts of cells pulse-labeled for various times revealed that a subcomplex of RPB2 and RPB3 appears soon after subunit synthesis and can be chased into fully assembled enzyme. The RPB2-plus-RPB3 subcomplexes accumulated in all RPB1 assembly mutants at the nonpermissive temperature but not in an RPB2 or RPB3 assembly mutant. These data indicate that RPB2 and RPB3 form a complex that subsequently interacts with RPB1 during the assembly of RNA polymerase II.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allison L. A., Moyle M., Shales M., Ingles C. J. Extensive homology among the largest subunits of eukaryotic and prokaryotic RNA polymerases. Cell. 1985 Sep;42(2):599–610. doi: 10.1016/0092-8674(85)90117-5. [DOI] [PubMed] [Google Scholar]
- BEERS R. F., Jr, SIZER I. W. A spectrophotometric method for measuring the breakdown of hydrogen peroxide by catalase. J Biol Chem. 1952 Mar;195(1):133–140. [PubMed] [Google Scholar]
- Berghöfer B., Kröckel L., Körtner C., Truss M., Schallenberg J., Klein A. Relatedness of archaebacterial RNA polymerase core subunits to their eubacterial and eukaryotic equivalents. Nucleic Acids Res. 1988 Aug 25;16(16):8113–8128. doi: 10.1093/nar/16.16.8113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll S. B., Stollar B. D. Conservation of a DNA-binding site in the largest subunit of eukaryotic RNA polymerase II. J Mol Biol. 1983 Nov 5;170(3):777–790. doi: 10.1016/s0022-2836(83)80131-4. [DOI] [PubMed] [Google Scholar]
- Dayhoff M. O., Barker W. C., Hunt L. T. Establishing homologies in protein sequences. Methods Enzymol. 1983;91:524–545. doi: 10.1016/s0076-6879(83)91049-2. [DOI] [PubMed] [Google Scholar]
- Edwards A. M., Darst S. A., Feaver W. J., Thompson N. E., Burgess R. R., Kornberg R. D. Purification and lipid-layer crystallization of yeast RNA polymerase II. Proc Natl Acad Sci U S A. 1990 Mar;87(6):2122–2126. doi: 10.1073/pnas.87.6.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evan G. I., Lewis G. K., Ramsay G., Bishop J. M. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol Cell Biol. 1985 Dec;5(12):3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field J., Nikawa J., Broek D., MacDonald B., Rodgers L., Wilson I. A., Lerner R. A., Wigler M. Purification of a RAS-responsive adenylyl cyclase complex from Saccharomyces cerevisiae by use of an epitope addition method. Mol Cell Biol. 1988 May;8(5):2159–2165. doi: 10.1128/mcb.8.5.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda R., Ishihama A. Subunits of RNA polymerase in function and structure; Maturation in vitro of core enzyme from Escherichia coli. J Mol Biol. 1974 Aug 15;87(3):523–540. doi: 10.1016/0022-2836(74)90102-8. [DOI] [PubMed] [Google Scholar]
- Himmelfarb H. J., Simpson E. M., Friesen J. D. Isolation and characterization of temperature-sensitive RNA polymerase II mutants of Saccharomyces cerevisiae. Mol Cell Biol. 1987 Jun;7(6):2155–2164. doi: 10.1128/mcb.7.6.2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K., Iwakura Y., Ishihama A. Biosynthesis of RNA polymerase in Escherichia coli. III. Identification of intermediates in the assembly of RNA polymerase. J Mol Biol. 1975 Aug 5;96(2):257–271. doi: 10.1016/0022-2836(75)90347-2. [DOI] [PubMed] [Google Scholar]
- Julius D., Schekman R., Thorner J. Glycosylation and processing of prepro-alpha-factor through the yeast secretory pathway. Cell. 1984 Feb;36(2):309–318. doi: 10.1016/0092-8674(84)90224-1. [DOI] [PubMed] [Google Scholar]
- Kawakami K., Ishihama A. Defective assembly of ribonucleic acid polymerase subunits in a temperature-sensitive alpha-subunit mutant of Escherichia coli. Biochemistry. 1980 Jul 22;19(15):3491–3495. doi: 10.1021/bi00556a013. [DOI] [PubMed] [Google Scholar]
- Kolodziej P. A., Woychik N., Liao S. M., Young R. A. RNA polymerase II subunit composition, stoichiometry, and phosphorylation. Mol Cell Biol. 1990 May;10(5):1915–1920. doi: 10.1128/mcb.10.5.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodziej P. A., Young R. A. Epitope tagging and protein surveillance. Methods Enzymol. 1991;194:508–519. doi: 10.1016/0076-6879(91)94038-e. [DOI] [PubMed] [Google Scholar]
- Kolodziej P., Young R. A. RNA polymerase II subunit RPB3 is an essential component of the mRNA transcription apparatus. Mol Cell Biol. 1989 Dec;9(12):5387–5394. doi: 10.1128/mcb.9.12.5387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Kuo C. L., Campbell J. L. Cloning of Saccharomyces cerevisiae DNA replication genes: isolation of the CDC8 gene and two genes that compensate for the cdc8-1 mutation. Mol Cell Biol. 1983 Oct;3(10):1730–1737. doi: 10.1128/mcb.3.10.1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Mann C., Buhler J. M., Treich I., Sentenac A. RPC40, a unique gene for a subunit shared between yeast RNA polymerases A and C. Cell. 1987 Feb 27;48(4):627–637. doi: 10.1016/0092-8674(87)90241-8. [DOI] [PubMed] [Google Scholar]
- Martin C., Okamura S., Young R. Genetic exploration of interactive domains in RNA polymerase II subunits. Mol Cell Biol. 1990 May;10(5):1908–1914. doi: 10.1128/mcb.10.5.1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martindale D. W. A conjugation-specific gene (cnjC) from Tetrahymena encodes a protein homologous to yeast RNA polymerase subunits (RPB3, RPC40) and similar to a portion of the prokaryotic RNA polymerase alpha subunit (rpoA). Nucleic Acids Res. 1990 May 25;18(10):2953–2960. doi: 10.1093/nar/18.10.2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonet M., Scafe C., Sexton J., Young R. Eucaryotic RNA polymerase conditional mutant that rapidly ceases mRNA synthesis. Mol Cell Biol. 1987 May;7(5):1602–1611. doi: 10.1128/mcb.7.5.1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonet M., Sweetser D., Young R. A. Functional redundancy and structural polymorphism in the large subunit of RNA polymerase II. Cell. 1987 Sep 11;50(6):909–915. doi: 10.1016/0092-8674(87)90517-4. [DOI] [PubMed] [Google Scholar]
- Ovchinnikov Y. A., Lipkin V. M., Modyanov N. N., Chertov O. Y., Smirnov Y. V. Primary structure of alpha-subunit of DNA-dependent RNA polymerase from Escherichia coli. FEBS Lett. 1977 Apr 1;76(1):108–111. doi: 10.1016/0014-5793(77)80131-2. [DOI] [PubMed] [Google Scholar]
- Pati U. K., Weissman S. M. The amino acid sequence of the human RNA polymerase II 33-kDa subunit hRPB 33 is highly conserved among eukaryotes. J Biol Chem. 1990 May 25;265(15):8400–8403. [PubMed] [Google Scholar]
- Riva M., Schäffner A. R., Sentenac A., Hartmann G. R., Mustaev A. A., Zaychikov E. F., Grachev M. A. Active site labeling of the RNA polymerases A, B, and C from yeast. J Biol Chem. 1987 Oct 25;262(30):14377–14380. [PubMed] [Google Scholar]
- Saitoh T., Ishihama A. Subunits of RNA polymerase in function and structure. VI. Sequence of the assembly in vitro of Escherichia coli RNA polymerase. J Mol Biol. 1976 Jul 5;104(3):621–635. doi: 10.1016/0022-2836(76)90125-x. [DOI] [PubMed] [Google Scholar]
- Scafe C., Martin C., Nonet M., Podos S., Okamura S., Young R. A. Conditional mutations occur predominantly in highly conserved residues of RNA polymerase II subunits. Mol Cell Biol. 1990 Mar;10(3):1270–1275. doi: 10.1128/mcb.10.3.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scafe C., Nonet M., Young R. A. RNA polymerase II mutants defective in transcription of a subset of genes. Mol Cell Biol. 1990 Mar;10(3):1010–1016. doi: 10.1128/mcb.10.3.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sentenac A. Eukaryotic RNA polymerases. CRC Crit Rev Biochem. 1985;18(1):31–90. doi: 10.3109/10409238509082539. [DOI] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989 May;122(1):19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K., Hill D. E. Two related regulatory sequences are required for maximal induction of Saccharomyces cerevisiae his3 transcription. Mol Cell Biol. 1987 Jan;7(1):104–110. doi: 10.1128/mcb.7.1.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweetser D., Nonet M., Young R. A. Prokaryotic and eukaryotic RNA polymerases have homologous core subunits. Proc Natl Acad Sci U S A. 1987 Mar;84(5):1192–1196. doi: 10.1073/pnas.84.5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taketo M., Ishihama A. Biosynthesis of RNA polymerase in Escherichia coli. IV. Accumulation of intermediates in mutants defective in the subunit assembly. J Mol Biol. 1976 Apr 5;102(2):297–310. doi: 10.1016/s0022-2836(76)80055-1. [DOI] [PubMed] [Google Scholar]
- Taketo M., Ishihama A. Biosynthesis of RNA polymerase in Escherichia coli. V. Defects of the subunit assembly in a temperature-sensitive beta subunit mutant. J Mol Biol. 1977 May 5;112(1):65–74. doi: 10.1016/s0022-2836(77)80156-3. [DOI] [PubMed] [Google Scholar]
- Thompson N. E., Steinberg T. H., Aronson D. B., Burgess R. R. Inhibition of in vivo and in vitro transcription by monoclonal antibodies prepared against wheat germ RNA polymerase II that react with the heptapeptide repeat of eukaryotic RNA polymerase II. J Biol Chem. 1989 Jul 5;264(19):11511–11520. [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Young R. A. RNA polymerase II. Annu Rev Biochem. 1991;60:689–715. doi: 10.1146/annurev.bi.60.070191.003353. [DOI] [PubMed] [Google Scholar]
- Yura T., Ishihama A. Genetics of bacterial RNA polymerases. Annu Rev Genet. 1979;13:59–97. doi: 10.1146/annurev.ge.13.120179.000423. [DOI] [PubMed] [Google Scholar]