Abstract
CENPA/Cse4 assembles centromeric chromatin on diverse DNA. CENPA chromatin is epigenetically propagated on unique and different centromere DNA sequences in a pathogenic yeast Candida albicans. Formation of neocentromeres on DNA, nonhomologous to native centromeres, indicates a role of non-DNA sequence determinants in CENPA deposition. Neocentromeres have been shown to form at multiple loci in C. albicans when a native centromere was deleted. However, the process of site selection for CENPA deposition on native or neocentromeres in the absence of defined DNA sequences remains elusive. By systematic deletion of CENPA chromatin-containing regions of variable length of different chromosomes, followed by mapping of neocentromere loci in C. albicans and its related species Candida dubliniensis, which share similar centromere properties, we demonstrate that the chromosomal location is an evolutionarily conserved primary determinant of CENPA deposition. Neocentromeres on the altered chromosome are always formed close to the site which was once occupied by the native centromere. Interestingly, repositioning of CENPA chromatin from the neocentromere to the native centromere occurs by gene conversion in C. albicans.
A functional centromere, the chromosomal site where a kinetochore assembles and attaches to spindle microtubules to facilitate proper chromosome segregation, is formed only once on each monocentric chromosome in normal cells. Occasionally, dicentric chromosomes are formed in a diseased state such as in cancerous cells that show high genome instability (Shen 2011). The mechanism that assures the presence of only one functional centromere in a chromosome is largely unknown. While centromeres perform an indispensable function in chromosome segregation, underlying centromeric sequences are highly divergent (Henikoff et al. 2001). The presence of CENPA, a variant of histone H3, at centromeric chromatin is a conserved feature of all eukaryotic chromosomes (Stimpson and Sullivan 2010; Black and Cleveland 2011). However, the mechanism of CENPA recruitment in yeast to humans may not be universal (Sanyal 2012).
A landmark discovery of neocentromere formation on a locus nonhomologous to the native centromere in an altered “acentric” chromosome found in a boy with a learning disability provided strong evidence that non-DNA sequence determinants can play a role in centromere specification (Voullaire et al. 1993). Since the discovery of the first human neocentromere, several neocentromeres have been identified in humans, and the occurrence of neocentromeres has been reported in fungal, animal, and plant species as well (Marshall et al. 2008; Marshall and Choo 2009). Human neocentromeres also contain most other proteins required for proper kinetochore formation (Alonso et al. 2007). Thus neocentromere formation provides an excellent model for the study of the determinants of CENPA deposition that forms centromeric chromatin.
Neocentromere formation on a non-native locus takes place only in the absence of the functional native centromere in a normal cell (Ishii et al. 2008; Ketel et al. 2009). The mechanism that suppresses neocentromere loci in the presence of the functional native centromere remains largely unknown. In contrast to short point centromeres of budding yeast Saccharomyces cerevisiae that are genetically regulated, repetitive regional centromeres of most other eukaryotes including fission yeast Schizosaccharomyces pombe, Drosophila, plants, and humans are epigenetically regulated (Black and Cleveland 2011). Many epigenetic determinants of centromere function such as RNA interference, H3K9Me2-marked pericentric heterochromatin, and DNA methylation have been identified in several organisms (Lachner et al. 2001; Nakayama et al. 2001; Wong et al. 2006; Folco et al. 2008; Gopalakrishnan et al. 2009; Roy and Sanyal 2011). Two closely related human pathogenic budding yeasts, Candida albicans and Candida dubliniensis, carry 3- to 5-kb-long unique centromeres that lack specific sequence motifs, characteristics of point centromeres, as well as features associated with regional centromeres such as repeat elements, DNA methylation, H3K9Me2-mediated heterochromatin, or CENP-B proteins (Sanyal and Carbon 2002; Sanyal et al. 2004; Baum et al. 2006; Padmanabhan et al. 2008). These short regional centromeres of C. albicans have been shown to be regulated epigenetically (Baum et al. 2006). However, the nature of epigenetic factors remains to be determined. Neocentromere formation at multiple ectopic loci has been shown to be highly efficient in C. albicans (Ketel et al. 2009). Thus, the C. albicans neocentromere provides an attractive system for the study of enigmatic epigenetic regulatory factors involved in formation of CENPA chromatin.
More interestingly, centromeres of C. albicans have undergone rapid divergence from C. dubliniensis (they diverged from each other ∼20 million years ago) (Padmanabhan et al. 2008). Such rapid evolution of centromeres reveals a remarkable plasticity for DNA sequences in centromere function. A similar divergence in centromeric sequences has been reported in closely related fungal (Schizosaccahromyces sp.), plant (Oryza sativa and O. brachyantha), and animal (Muscaroli and other mouse species) species, and even in closely related lineages of the Saccharomyces paradoxus (Kipling et al. 1995; Lee et al. 2005; Bensasson et al. 2008; Rhind et al. 2011). Asymmetric female meiosis has been proposed to be the drive for selfish centromeres to gain a transmission advantage that leads to rapid centromeric evolution (Henikoff et al. 2001). However, the process responsible for the rapid evolution of CENPA-binding regions in lineages of the S. paradoxus with symmetric meiosis (Bensasson et al. 2008) and closely related C. albicans and C. dubliniensis which lack any detectable meiotic cycle remains unknown.
In the present study, a comprehensive analysis of neocentromere formation identified physical chromosomal location as a novel epigenetic factor that regulates CENPA deposition at Candida centromeres. A comparative analysis revealed that centromere location and the dynamics of neocentromere formation are evolutionarily conserved in C. albicans and C. dubliniensis. We demonstrate that the physical chromosomal location not only ensures CENPA deposition at only one locus, but also prevents its deposition at other potential centromere-forming loci in Candida. Most strikingly, we observed that gene conversion occurs at C. albicans centromeres. We speculate that gene conversion may contribute to both rapid evolution of centromere DNA and conservation of centromeric location in the same and other closely related species.
Results
Relative chromosomal position of CENPA-binding regions in C. albicans and C. dubliniensis remains evolutionarily conserved in spite of extensive chromosomal rearrangements
Two closely related species of Candida, C. albicans and C. dubliniensis, share a high degree of DNA sequence homology as they diverged ∼20 million years ago (Mishra et al. 2007). Centromere DNA sequences of C. albicans and C. dubliniensis share no sequence homology due to rapid evolution of centromere DNA sequences (Padmanabhan et al. 2008), although synteny of genes across CENPA-binding regions in most of the chromosomes is largely maintained in these two species. An SfiI macrorestriction map is now available for both C. albicans (1006 and white opaque switching strain WO-1; Chu et al. 1993) and C. dubliniensis (Strain Cd36; Magee et al. 2008). C. albicans WO-1 (http://www.broadinstitute.org/annotation/genome/candida_albicans/GenomesIndex.html?component=%24DirectLink&service=direct&session=T&sp=SCAW1) and C. dubliniensis (http://www.sanger.ac.uk/resources/downloads/fungi/candida-dubliniensis.html) have undergone various gross chromosomal rearrangements during the course of evolution, resulting in the rearrangement of SfiI fragments between different chromosomes (Magee and Magee 1997; Jackson et al. 2009). We aligned and compared the relative arrangement of SfiI fragments on orthologous chromosomes of C. albicans 1006, C. albicans WO-1, and C. dubliniensis (Supplemental Fig. S1). Despite the nature of the rearrangements that occurred being very different, orthologous centromeres of all the chromosomes involved in these rearrangement events are located at a similar distance from one end of the chromosome in two clinical isolates of C. albicans 1006 and WO-1 (Mishra et al. 2007) and C. dubliniensis (Padmanabhan et al. 2008). Thus, in spite of having dissimilar centromere DNA sequences, a high degree of conservation in centromere location suggests involvement of chromosomal locations in determining centromeres' identity in Candida.
Formation of centromeric chromatin on a DNA sequence is context-dependent
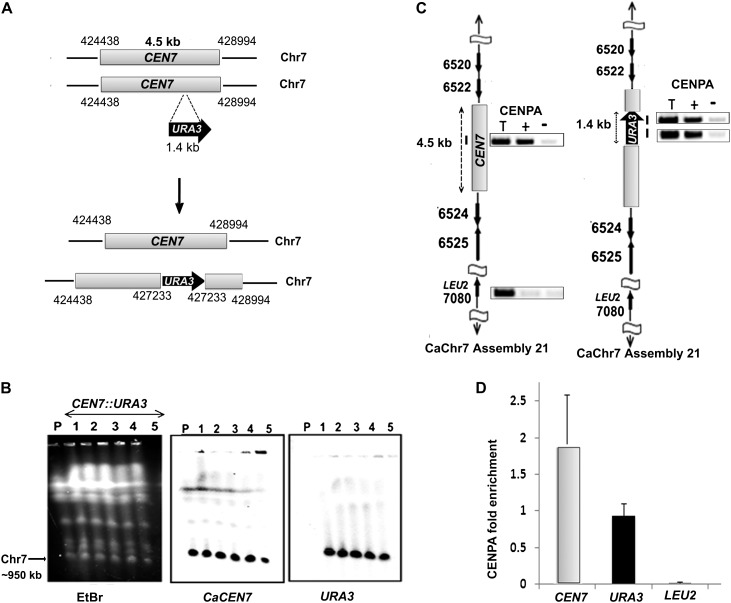
To test if a specific chromosomal location per se, irrespective of the DNA sequence present, could be an epigenetic determinant for centromere formation in Candida, a 1.4-kb URA3 gene sequence was integrated into CEN7 (coordinates: Assembly 21 CaChr7, 427233) in the C. albicans RM1000AH strain (Fig. 1A; Sanyal et al. 2004), and the expected integration was verified by Southern analysis (Supplemental Fig. S2A). Each homolog of chromosome 7 (Chr7) is marked with a unique marker ARG4 or HIS1 at the same location, unlinked to CEN7 and present ∼450 kb away from CEN7 on the right arm of Chr7 in RM1000AH. The frequency of chromosome loss was monitored in transformants carrying URA3 integrated at CEN7 by measuring the simultaneous loss of two markers: ARG4 and URA3 or HIS1 and URA3. We did not detect any chromosome loss (URA loss <1 in 103 cells) (Table 1) in such integrants, suggesting that the URA3-containing altered chromosome is mitotically stable. Subsequently, karyotypic analysis eliminated the possibility of chromosomal rearrangements in those RM1000AH (CEN7/CEN7∷URA3) transformants (Fig. 1B). Genomic DNA plugs prepared from five URA3 integrants were resolved on CHEF gels using conditions that separate all the chromosomes of C. albicans (see Methods), transferred to a membrane and probed with URA3 or CEN7. None of these integrants showed any observable chromosomal rearrangements (Fig. 1B). Interestingly, all the integrants (CEN7/CEN7∷URA3) exhibited reversible silencing of URA3 as evidenced by their growth both on 5-fluoro-orotidic acid (FOA) and complete medium lacking uridine (CM-Ura) plates (Supplemental Fig. S2B, left). Transcript levels were measured in one such integrant (J151) grown both on FOA and CM-Ura. Reverse transcriptase qPCR analysis revealed approximately fivefold repression in URA3 transcript levels on FOA as compared with CM-Ura (Supplemental Fig. S2B, right). It is important to note that C. albicans is a diploid organism, and thus one Chr7 homolog is unaltered while the other one has URA3 integrated at CEN7. Both ChIP-PCR and ChIP-qPCR analysis on CENPA/Cse4 ChIP DNA revealed that CENPA was recruited on URA3 of the altered homolog and on CEN7 of the unaltered homolog (Fig. 1C,D). These results strongly suggest that centromeric chromatin can assemble on noncentromeric DNA when placed at the native centromere, without disrupting functional centromere formation at the native locus in C. albicans.
Figure 1.
CENPA chromatin assembles at a specific chromosomal location. (A) Schematic showing the location of URA3 insertion at native CEN7 in one homolog of Chr7. Arrows indicate the gene location and the direction of transcription while the numbers represent coordinates on a specific chromosome based on Assembly 21 of C. albicans SC5314 genome. (B) An ethidium bromide (EtBr) stained CHEF gel image (left) showing no apparent alteration in the karyotype of RM1000AH-CEN7∷URA3 integrants (lanes 1–5) as compared with the parent RM1000AH (lane P). DNA was transferred on a membrane and Southern analysis was performed by probing the blot with either CaCEN7 (middle) or URA3 (right). (C) ChIP analysis with anti-Cse4 (CENPA) antibodies followed by PCR revealed recruitment of CENPA on URA3 of the altered homolog (left) as well as on native CEN7 of the unaltered homolog (right) of Chr7. Primer locations are marked with bars. LEU2 present on Chr7 but unlinked to CEN7 is used as a negative control for CENPA binding. (T) Total DNA; (+) with Ab; (−) without Ab (beads only). (D) Real-time qPCR analysis on CENPA ChIP DNA from RM1000AH-CEN7∷URA3/CEN7 strain J154 using primers from CEN7 and URA3 regions. Fold enrichment of CENPA binding was calculated as described in Methods.
Table 1.
Chromosome loss frequency in various CEN-deleted transformants

Replacement of the core CENPA-rich native centromere by URA3 is sufficient to induce centromeric chromatin relocation but depends on the transcriptional status of URA3
To examine whether CENPA-containing centromeric chromatin can also assemble on URA3 in the total absence of the core CENPA-rich region, we replaced the core 4.5-kb CENPA-rich CEN7 region (coordinates: Assembly 21 CaChr7, 424438–428994) by the 1.4-kb URA3 sequence in RM1000AH (CEN7/CEN7) (Supplemental Fig. S3A). We performed 15 independent transformation experiments. To ensure the independent nature of each transformant, only one correct transformant screened by Southern blot analysis (the strategy is described in the Supplemental Methods) was selected from each transformation experiment for subsequent analysis. The transformants with desired CEN7 deletion were further analyzed by CHEF gel followed by Southern analysis as described above. One out of 15 transformants showed chromosomal rearrangements of CEN7-deleted Chr7 (Supplemental Fig. S3B) and was excluded from further studies. One of the remaining 14 transformants exhibited a high rate of loss of Chr7 (URA loss 20%) and was also excluded from further analysis. In all other transformants, CEN7-deleted altered Chr7 was mitotically stable (URA loss <1 in 103 cells) (Table 1). To find out whether or not centromeric chromatin assembled on URA3 that replaced native CEN DNA, CENPA/Cse4 ChIP assays were performed in 11 stable RM1000AH-cen7Δ (CEN7/cen7Δ) transformants. In 10 out of 11 RM1000AH-cen7Δ transformants, PCR analysis using primers from different regions of URA3 revealed CENPA binding on one end of URA3. CENPA binding was found to be extended beyond URA3 until 2–3 kb toward the right arm of Chr7 spanning Orf19.6520 and Orf19.6522 (nCEN7-I) (Fig. 2; Supplemental Fig. S3C). Thus, a deletion spanning only a CENPA-rich region activates neocentromeres at a location situated next to the deleted region. When ChIP assays were performed on cells grown in FOA media, CENPA binding showed a shift away from Orf19.6520 and Orf19.6522 and encompassed the entire URA3 region, suggesting that transcriptional silencing favors CENPA binding in C. albicans.
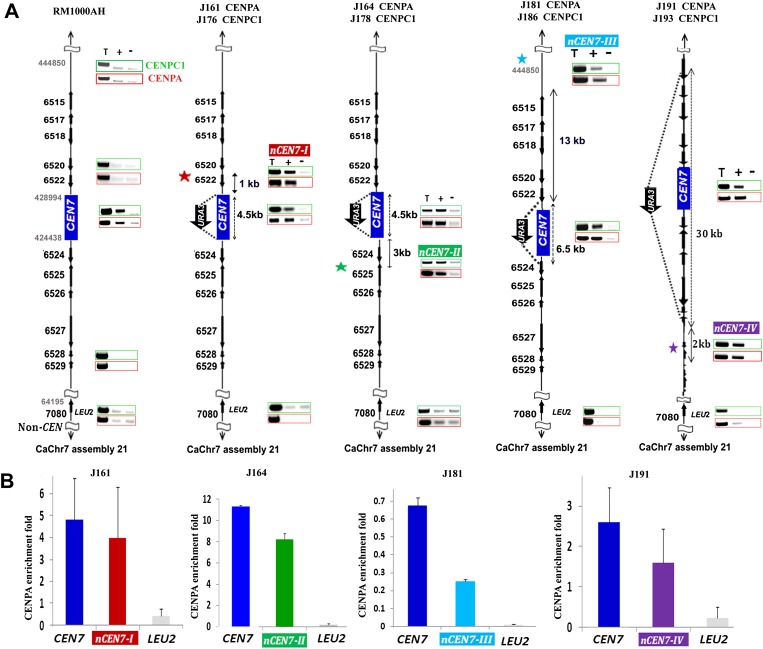
Figure 2.
Neocentromeres, like centromeres, preferentially form on specific chromosomal locations in a nonrandom fashion independent of the length of the deleted region in C. albicans. (A) CENPA and CENPC1 ChIP analyses in RM1000AH-cen7Δ, RM1000AH-cen7-6.5 kbΔ, or RM1000AH-cen7-30kbΔ clones mapped neocentromeres (nCENs) within 15 kb from the deleted region. C. albicans is a diploid yeast and carries two homologs of each chromosome. Only one homolog of Chr7 where CEN7 has been replaced by URA3 is shown. CENPA enrichment shown at the native CEN7 location is contributed by an unaltered homolog and is shown as a positive control for CENPA ChIP assays. Thick black arrows along with the Orf numbers show the gene arrangement and the direction of transcription. One representative clone from each class (see Supplemental Figs. S3C, S4B for other clones) is shown. Class I neocentromeres (nCEN7-I) formed on Orf19.6520 and Orf19.6522 (10/11 RM1000AH-cen7Δ and 4/5 RM1000AH-cen-6.5kbΔ transformants), Class II neocentromeres (nCEN7-II) mapped to Orf19.6526 and Orf19.6525 (1/11 RM1000AH-cen7Δ), Class III neocentromeres (nCEN7-III) mapped on an Orf-free region—Assembly 21, CaChr7 Coordinates 442000–445000 (1/5 RM1000AH-cen-6.5kbΔ transformant)—and nCEN7-IV mapped on Orf19.6531, Orf19.6532, Orf19.6533, Orf19.6534 (2/2 RM1000AH-cen7-30kbΔ transformants). Parent RM1000AH did not show CENPA or CENPC1 enrichment on these neocentromere sites. Coordinates of the control non-CEN (LEU2) region on Chr7 in Assembly 21 are 64195–64440 (Orf 19.7080). (T) Total DNA; (+) with Ab; (−) without Ab (beads only). CENPA ChIP profiles are shown in red boxes and the same for CENPC1 are shown in green boxes. Enrichment of CENPA at the URA3 location was not shown here but presented as a separate figure (Supplemental Fig. S5A). (B) CENPA ChIP qPCR analysis showing fold enrichment of CENPA on native and neocentromeres.
The physical chromosomal location of CENPA chromatin is an important determinant of centromere identity in Candida
Detailed PCR analysis on CENPA/Cse4 ChIP DNA from 11 RM1000AH-cen7Δ transformants, each of which carries a neocentromere-containing altered Chr7, using a set of primers covering a 20-kb region spanning CEN7, revealed that in addition to the native centromere of unaltered Chr7, a second neocentromere site (nCEN7-II, 1/11 transformant) was activated 3 kb away from URA3 on the left arm of the altered Chr7 spanning Orf19.6526 and Orf19.6525 (Fig. 2). To confirm the binding of another evolutionarily conserved kinetochore protein CENPC1/Mif2 on nCEN7-I and nCEN7-II, we functionally expressed Myc-tagged CENPC1/Mif2 (Methods) in RM1000AH-cen7Δ strains that carried a neocentric Chr7 (Supplemental Fig. S3D). PCR analysis on Myc (CENPC1/Mif2) ChIP DNA revealed CENPC1 enrichment on nCEN7-I and nCEN7-II consistent with CENPA/Cse4 binding (Fig. 2). The length of the CENPA-rich region on a neocentromere in each case was 3–4 kb including ∼1–2 kb of variation among transformants within a specific class of neocentromeres. If neocentromere formation occurs randomly anywhere on Chr7 (∼950 kb), the chance of its formation adjacent to a native centromere would be ∼1 in 300. Since all the transformants analyzed in this study formed neocentromeres adjacent to the native centromeres, we conclude that centromere proximal regions are the most preferred and thus biased for assembly of CENPA chromatin in the absence of the native core CEN7 sequence.
Neocentromere formation at centromere proximal regions is independent of the size of the pericentric region deleted
Pericentric regions that consist of inverted repeats and boundary elements facilitate CENPA deposition at 10- to 15-kb central domains (Takahashi et al. 2000; Allshire and Karpen 2008; Roy and Sanyal 2011) of the S. pombe centromere. In the absence of any boundary elements (such as tRNA genes in S. pombe; Kuhn et al. 1991; Takahashi et al. 1991), H3K9 methylation (found in pericentric chromatin of most regional centromere-containing organisms; Bernard et al. 2001; Nakayama et al. 2001; Rice et al. 2003) or common repeat elements (Fishel et al. 1988), the length of the pericentric region is difficult to ascertain in C. albicans (Sanyal 2012). Detailed bioinformatic analysis to find out pericentric repeats in our previous study (Padmanabhan et al. 2008) revealed that pericentric regions surrounding all the centromeres except CEN5 lack proper repeats and contain only cryptic (or remnants of) transposable elements or inverted repeats. Interestingly, degenerate retrotransposon-like repeat elements (e.g., Tcen, Tgl1, and Tgl2) are features of Neurospora crassa centromeres as well (Cambareri et al. 1998; Smith et al. 2011). Orthologous pericentric regions contain several short stretches of DNA sequences that are common in pericentric regions of some, but not all, C. albicans and C. dubliniensis chromosomes (Padmanabhan et al. 2008). The formation of neocentromeres almost invariably at centromere proximal locations prompted us to hypothesize that, in the absence of specific sequence motifs or repeats, the more conserved pericentric regions may facilitate recruitment of CENPA at these regions. We anticipated that residual CENPA present on CEN7 proximal regions may be responsible for seeding assembly of CENPA chromatin in these regions for neocentromere formation in the absence of the native centromere DNA.
To examine whether or not neocentromeres are formed randomly at multiple locations in the absence of pericentric regions, we replaced the entire 6.5-kb ORF-free region including CEN7 (coordinates: Assembly 21 CaChr7, 423450–429852) with the 1.4-kb URA3 sequence (Supplemental Fig. S4A, left). Five correct RM1000AH-cen7-6.5kbΔ transformants (Supplemental Fig. S4B, left) analyzed (each obtained from an independent transformation experiment) showed neither any rearrangement of Chr7 (Supplemental Fig. S4C, left) nor a significant chromosome loss (Table 1), suggesting neocentromere formation in each case. CENPA/Cse4 ChIP assays were performed in four RM1000AH-cen7-6.5kbΔ transformants to map neocentromere sites. The most prevalent site of neocentromere formation was mapped to the same hotspot identified when the CEN7 core was deleted (nCEN7-I). A new site (nCEN7-III) was mapped 13 kb away from the CEN7 sequence toward the right arm of Chr7 on an ORF-free region (Assembly 21, CaChr7 Coordinates 442000–445000) (Fig. 2A; Supplemental Fig. S4D). The nCEN7-III neocentromere location was further confirmed by co-localization of CENPA and CENPC1 binding on the neocentric chromosome as described above (Fig. 2A).
Attempts to construct a functional minichromosome in C. albicans have been unsuccessful so far; hence the precise boundary of a functional centromere or the pericentric region is undefined (Baum et al. 2006). It is possible that the pericentromeric module in C. albicans is longer than the ORF-free 6.5-kb region surrounding CEN7. Thus subsequently we deleted a 10-kb (data not shown) or a 30-kb region including CEN7 (coordinates: Assembly 21 CaChr7, 411320–440780) in RM1000AH (Supplemental Fig. S4, right panels). Due to the deletion of the 30-kb region, it was possible to separate the unaltered and altered Chr7 in RM1000AH-cen7-30kbΔ on a CHEF gel using a specific run condition (data not shown). We selected those independent RM1000AH-cen7-30kbΔ clones that were stable (URA loss <1 in 103 cells) (Table 1) and the altered chromosome moved faster on the CHEF gel (Supplemental Fig. S4C). The CENPA/Cse4 ChIP followed by PCR analysis using primers from Chr7 revealed that neocentromeres formed at a site adjacent to the deleted region on Orf19.6531, Orf19.6532, Orf19.6533, Orf19.6534 (nCEN7-IV, 1–2 kb away from URA3 toward the left arm of Chr7) on the altered chromosome, in addition to the native CEN7 of the unaltered Chr7 homolog in both the transformants (Fig. 2A; Supplemental Fig. S4D). To confirm the presence of a functional kinetochore at nCEN7-VI, we functionally expressed Myc-tagged CENPC1/Mif2 (see Methods) in RM1000AH-cen7-30kbΔ (J191) strains that carried a neocentric Chr7. CENPC1 ChIP assays in resulting strain J193 confirmed the co-localization with CENPA at nCEN7-VI (Fig. 2A). Together we conclude that neocentromeres always mapped to the centromere proximal regions when the core CENPA-rich region is deleted, irrespective of the length of the deleted region. Thus, the physical location, rather than the pericentric chromatin state, is a determinant of centromere/neocentromere identity in C. albicans.
Next we compared the strength of CENPA binding on native centromeres and neocentromeres by real-time qPCR analysis on CENPA ChIP DNA from neocentromere-containing strains. Real-time qPCR analysis revealed that native CEN7 is strongly enriched as compared with neocentromeres (Fig. 2B). Neocentromeres formed on the most prevalent site, nCEN7-I, which is closest to the native CEN7 location, showed ∼85% CENPA binding as compared with native CEN7, while neocentromeres nCEN7-II and nCEN7-IV that are formed 3–4 kb away from the deleted region showed ∼60% and ∼70% CENPA binding, respectively, as compared with that of the native centromeres (CEN7). As the distance of a neocentromere site from the deleted CEN7 region increased, CENPA binding progressively dropped and CENPA binding to nCEN7-III, which is located 13 kb away from the native centromere, showed ∼35% as compared with native CEN7 (Fig. 2B).
CENPA ChIP sequencing analysis confirms that CEN proximal neocentromeres are the unique enriched regions on the entire altered chromosome
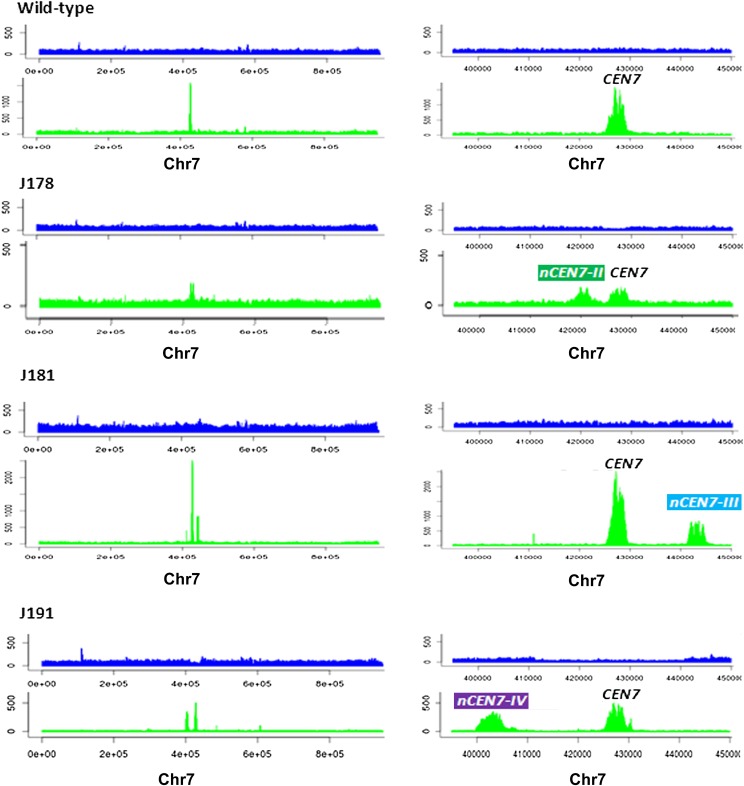
Since CENPA binding on nCEN7-II, nCEN7-III, and nCEN7-IV was significantly less (ranging from 35% to 70%) than the native CEN7, we wondered whether or not these sites are the only CENPA-rich sites present across the entire altered Chr7. In order to verify this we performed ChIP sequencing experiments (see Methods) on the CENPA/Cse4 or CENPC1/MycMif2 ChIP DNA fractions from transformants carrying active neocentric (nCEN7-II, nCEN7-III, and nCEN7-IV) chromosomes. All the sequence reads aligned to native CEN7, nCEN7-II, nCEN7-III, and nCEN7-IV sites over a region of 3–5 kb length (GEO accession number GSE42907). Exclusive enrichment of CENPA at native CEN7 (of unaltered Chr7 homolog) and the neocentromeres that we mapped (Fig. 3) excluded the possibility of the presence of ectopic CENPA-binding sites. Thus centromere-proximal CENPA-rich sites identified in this study are authentic neocentromeres.
Figure 3.
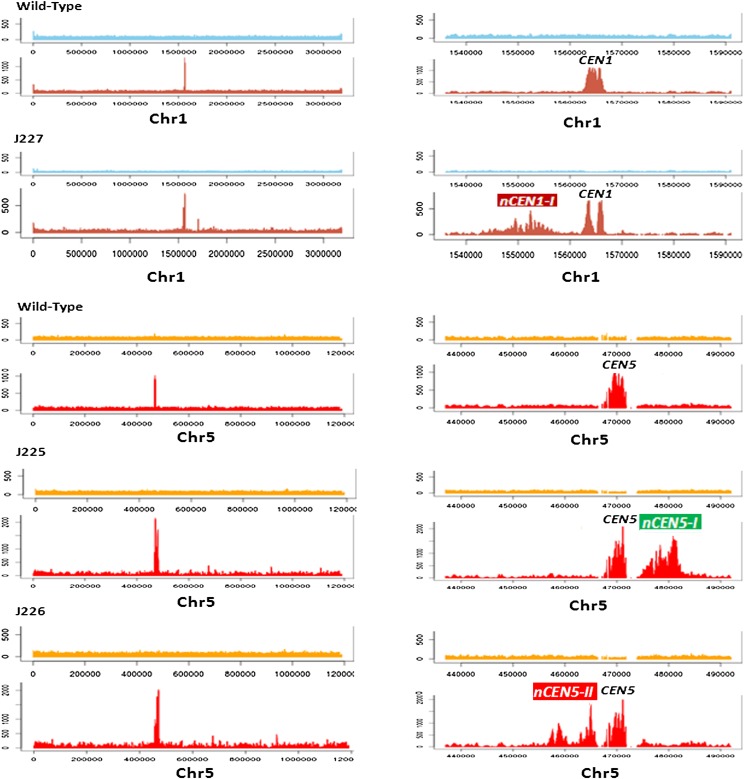
ChIP-seq analyses revealed that neocentromeres are always and exclusively formed at a close proximity to the native CEN. Relative number of sequence reads from the whole cell lysate (upper panels) or CENPA-Prot A or MycMif2/CENPC1 ChIP DNA (lower panels) in wild-type strain J200, Cen7Δ-deleted strains J178 (RM1000AH-cen7Δ), J181 (RM1000AH-cen7-6.5kbΔ), or J191 (RM1000AH-cen7-30kbΔ) mapped neocentromeres (nCENs) within 15 kb from the deleted region. (Left panels) Distribution of sequence reads across the entire chr7; (right panels) zoomed-in view of neocentromeric location with respect to the native CEN7 in each case. Chromosome coordinates are shown in the x-axis.
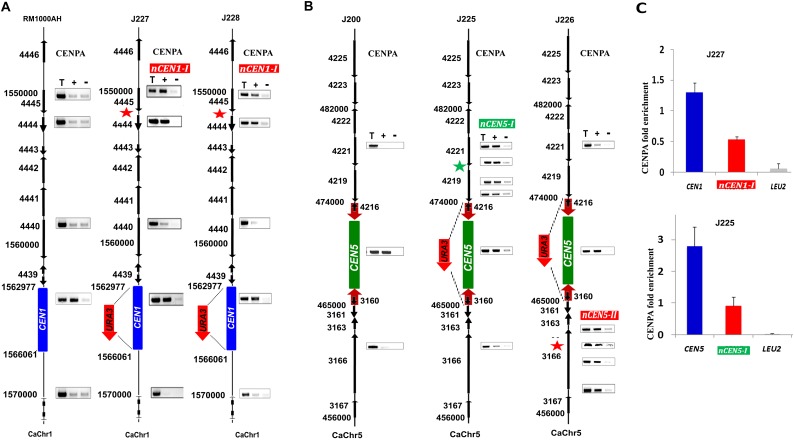
To examine if the dynamics of neocentromere formation is chromosome-specific, we extended our study to Chr1. The 4.2-kb CENPA/Cse4-binding region on Chr1 (coordinates: Assembly 21 CaChr1, 1562878–1567085) was replaced with the 1.4-kb URA3 sequence in RM1000AH. CENPA/Cse4 ChIP assays were performed in two transformants harboring stable URA3-containing Chr1 to map neocentromere locations. Again, both the neocentromeres formed at CEN1 proximal regions (coordinates: Assembly 21, CaChr1 1550000–1555000) (Fig. 4A), suggesting that similar to Chr7, CEN proximal regions are the most preferred sites of neocentromere formation on Chr1 as well as in C. albicans.
Figure 4.
Neocentromeres are preferentially formed on CEN adjacent regions on various chromosomes of C. albicans. (A) CENPA ChIP analyses in RM1000AH-cen1Δ clones mapped neocentromeres (nCENs) within 15 kb from the deleted region. (B) CENPA ChIP analyses in RM1000AH-cen5Δ clones also mapped neocentromeres (nCENs) within 15 kb from the deleted region. C. albicans carries two homologs of each chromosome. Only one homolog of Chr1 where CEN1 has been replaced by URA3 is shown. CENPA enrichment shown at the native CENs location is contributed by an unaltered homolog and is shown as a positive control for CENPA ChIP assays. Black arrows along with the Orf numbers show the gene arrangement and the direction of transcription. Red thick arrows on either side of CEN5 indicate inverted repeats. Neocentromeres on both chr1 (nCEN1-I: coordinates: Assembly 21, CaChr1 1544000–1556000) and Chr5 (nCEN5-I: coordinates: Assembly 21, CaChr5 456000–462000; nCEN5-II: coordinates: Assembly 21, CaChr5 474000–482000) formed at CEN1 proximal regions. Length of CENPA binding on Chr1 and Chr5 neocentromeres was longer (4–12 kb) than native centromeres (3–4 kb). Parent RM1000AH did not show CENPA enrichment on these neocentromere sites. (T) Total DNA; (+) with Ab; (−) without Ab (beads only). (C) CENPA ChIP qPCR analysis showing fold enrichment of CENPA on native centromeres and neocentromeres as compared with a noncentromeric region (LEU2). Enrichment of CENPA at the URA3 location was not shown here but presented as a separate figure (Supplemental Figure S5).
In contrast to our study where neocentromeres always formed on centromere proximal regions, a previous study had shown that neocentromere formation in C. albicans takes place at multiple loci on Chr5 even at a distance as far as 450 kb from the deleted CEN5 (Ketel et al. 2009). CEN5 is the only C. albicans centromere that contains a core CENPA-rich region surrounded by long inverted repeats. To reexamine whether the pattern of neocentromere formation is different or not on Chr5, we replaced the entire CEN5 spanning the core and the repeats (coordinates: Assembly 21, CaChr5 466279–473499) by the same 1.4-kb URA3 gene. We expressed Prot A-tagged CENPA in wild-type RM1000AH as well as in six randomly selected RM1000AH-cen5Δ transformants that exhibited no loss of the altered chromosome. ProtA (CENPA) ChIP-PCR analysis showed that CENPA was enriched at the native centromere of the unaltered Chr7 as well as on CEN-proximal regions (nCEN5-I: coordinates: Assembly 21, CaChr5 456000–462000; nCEN5-II: coordinates: Assembly 21, CaChr5 474000–482000) (Fig. 4B) to form neocentromeres. We observed that the length of neocentromeres was longer than native centromeres (3–5 kb) in Chr5 and Chr1 deletion experiments ranging from 6 to 12 kb (see the coordinates given above). These results, taken together, strongly suggest that CEN-proximal sites are the most preferred sites for centromere seeding in C. albicans. Real-time qPCR analysis revealed significant reduction in CENPA binding on neocentromeres formed on Chr1 and Chr5 as compared with their native counterparts (Fig. 4C). The presence of no other CENPA-binding region on these chromosomes was confirmed by ChIP-seq analysis on the CENPA/Cse4 or ProtA-Cse4 ChIP DNA fractions from transformants carrying neocentromeres (Fig. 5; GEO accession number GSE42907).
Figure 5.
ChIP-seq analyses show that CEN proximal neocentromeres are authentic neocentromeres in Chr1 and Chr5. Relative number of sequence reads from whole cell lysate (top panel) or CENPA-Prot A ChIP DNA (lower panels) in J200 (wild type), J227 (RM1000AH-cen1Δ), J225, or J226 (RM1000AH-cen5Δ) mapped neocentromeres (nCENs) within 15 kb from the deleted region. (Left panels) Distribution of sequence reads across the entire chromosome; (right panels) zoomed-in view of the neocentromeric location along with the native CEN in each case. Chromosome coordinates are shown in the x-axis.
Similarly to neocentromeres formed on Chr7, an additional CENPA-binding region apart from the native CEN confirms neocentromere sites on Chr5 identified by ChIP-PCR and ChIP-qPCR analysis. Interestingly, as reported in a previous study, a shift of CENPA binding from the original CEN5 proximal neocentric location to URA3 was observed when the strain was grown in FOA-containing media for ChIP analysis (Supplemental Fig. S5A,B).
The mechanism of neocentromere formation remains conserved in C. albicans and C. dubliniensis
Since the physical chromosomal location of centromeres is conserved in C. albicans and C. dubliniensis, we next examined the pattern of neocentromere formation in C. dubliniensis. We constructed a URA auxotrophic strain CdJ3 by transforming Cd36 using the MPA flipper cassette (Staib et al. 2001; see Methods). Next we replaced the CENPA-binding region of Chr7 (coordinates: CdChr7 434001–440600) of C. dubliniensis with the 1.4-kb URA3 sequence in the CdJ3 strain (Supplemental Fig. S6A). Replacement of CEN7 in one homolog of Chr7 in five independent transformants, each one obtained from an independent transformation experiment, was confirmed by PCR. Like C. albicans, the CdCEN7-deleted chromosome remains mitotically stable in each case (URA3 loss <1 in 103 cells) (Table 1). Of the six C. dubliniensis strains analyzed, no two showed identical karyotypes in a previous study (Magee et al. 2008). Thus, it is possible that CdCEN7-deleted chromosomes might have been fused to a natural centric chromosome rather than forming a neocentromere to become mitotically stable as frequently found in fission yeast (Ishii et al. 2008). Karyotypic analysis in five such C. dubliniensis transformants carrying a stable altered Chr7 exhibited no chromosomal rearrangements of URA3-containing CEN7-deleted Chr7 (Supplemental Fig. S6B), suggesting that neocentromere formation is highly efficient in C. dubliniensis as well. However, Southern analysis of the CHEF gel probed with the CdCEN7 sequence suggests additional rearrangements of chromosomes. Thus, we performed locus-specific Southern analysis of these clones to verify replacement of CdCEN7 with URA3. Only two CdJ3-cen7Δ transformants (J202 and J204) that exhibited desired bands, when EcoRV-digested DNA was probed with a sequence adjacent to CEN7, were taken further for mapping neocentromere locations (Supplemental Fig. S6C). We expressed Prot A-tagged CENPC1 (CdMif2-Prot A) in two strains, J202 and J204, and performed Prot A (CENPC1)-ChIP assays to identify neocentromere locations (Supplemental Fig. S6D) in resulting strains J212 and J216, respectively. Neocentromere locations were mapped between 1 and 10 kb away from the deleted CdCEN7 on the altered chromosome in each case (Supplemental Fig. S6E). Thus, similar to C. albicans, centromere proximal regions are the preferred sites of neocentromere formation in C. dubliniensis also.
Gene conversion occurs at C. albicans centromeres
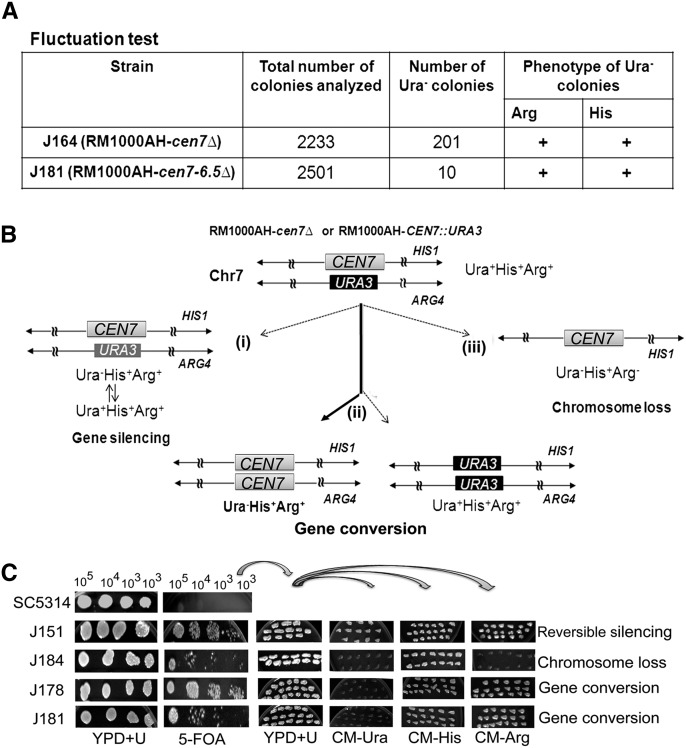
Replacement of chromosomal regions of various lengths (4.5–30 kb) spanning CEN7 by URA3 in C. albicans leads to stable propagation of the URA3-containing neocentric chromosome. During fluctuation assays to measure chromosome loss, some of the Ura− derivatives of neocentromere harboring strains J164 (RM1000AH-cen7Δ) and J181 (RM1000AH-cen7-6.5kbΔ) exhibited an unusual genotype: These segregants were prototroph for Arg and His markers indicating the presence of both the Chr7 homologs (Fig. 6A). These Ura− and FOA+ (resistant to FOA) segregants could not grow on CM-Ura media (Fig. 6B,C), suggesting that the URA3 gene was not reversibly silenced. Southern analysis of the above-mentioned Ura− colonies revealed the absence of the URA3 gene containing 7.6-kb BglII fragment that was originally present in their parent RM1000AH-cen7Δ (Supplemental Fig. S7A). CHEF gel analysis in these Ura− colonies revealed no detectable genomic rearrangement as compared with wild-type RM1000AH and parent RM1000AH-cen7Δ (Supplemental Fig. S7B). The presence of both the homologs of Chr7 and the absence of the URA3-containing BglII fragment confirmed that URA3 was nonreciprocally exchanged by the CEN7 sequence of the unaltered homolog of Chr7 through gene conversion. We detected similar gene conversion events in RM1000AH-cen7-6.5kbΔ transformants as well (Supplemental Fig. S7, right panels). Thus we conclude that gene conversion occurs at C. albicans centromeres.
Figure 6.
Gene conversion occurs at the C. albicans centromere. (A) Fluctuation tests (see Methods) in strain J164 (RM1000AH-cen7Δ) and J181 (RM1000AH-cen7-6.5kbΔ) identified Ura−Arg+His+ colonies (bottom panel), suggesting either silencing of URA3 or gene conversion at CEN7. (B) Schematic showing possible fates of a marker on a chromosome during segregation. The Ura− phenotype can arise due to (1) silencing of URA3 placed at the centromeric context, (2) loss of the chromosome (shown here is integration of URA3 into ARG4 containing Chr7 homolog), and (3) gene conversion, which results in replacement of URA3 by CEN7 from the unaltered Chr7 or vice versa. (C) Strains SC5314 (wild-type Ura+ strain), J151 (RM1000AH-CEN7∷URA3), J184 (RM1000AH-cen7-6.5kbΔ), J164 (RM1000AH-cen7Δ), and J181 (RM1000AH-cen7-6.5kbΔ) were counter-selected on FOA plates (lower panel). SC5314 failed to grow, while J151, J184, J178, and J181 could grow on 5-FOA-containing plates. To see whether the Ura− phenotype is due to silencing, chromosome loss, or gene conversion, FOA-resistant (FOA+) colonies were revived on YPDU plates and then transferred to plates lacking uridine, histidine, or arginine. Strains J151 (RM1000AH-CEN7∷URA3) and J184 (RM1000AH-cen7-6.5kbΔ) were used as positive controls for silencing and chromosome loss, respectively. Silencing at C. albicans CEN7 is reversible (Supplemental Fig. S1B). As expected, all FOA+ colonies of J151 were Ura+Arg+His+, confirming reversible silencing. FOA+ colonies of J184 were Ura−Arg−His+, which confirms a loss of the ARG4-containing homolog of Chr 7 (shown in Table 1). FOA+ colonies of J164 and J181, however, were Ura−Arg+His+, ruling out the occurrence of silencing (Ura+Arg+His+) or chromosome loss (Ura−Arg−His+), but indicative of gene conversion. Gene conversion in Ura−Arg+His+ derivatives of both J164 and J181 was subsequently confirmed by Southern (Supplemental Fig. S7) and PCR analysis (data not shown).
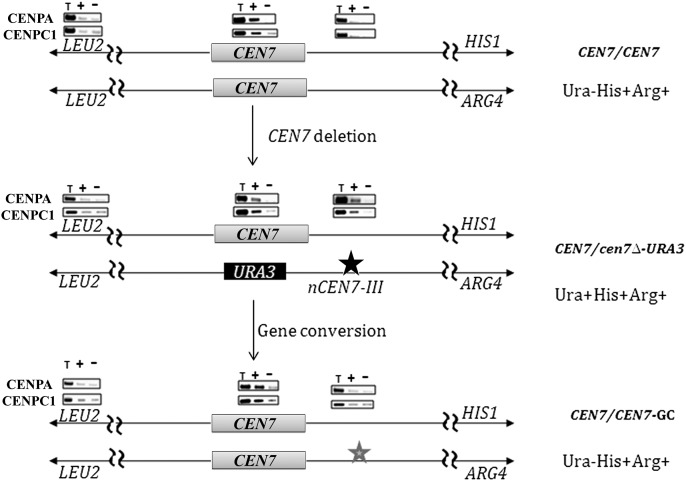
Centromere repositioning occurs during gene conversion
Strains J164 (RM1000AH-cen7Δ) and J181 (RM1000AH-cen7-6.5kbΔ) that exhibited gene conversion contained active neocentromeres on nCEN7-II (∼3 kb away from CEN7 toward the left arm of Chr7) and nCEN7-III (∼13 kb away from CEN7 toward the right arm of Chr7), respectively (Figs. 2, 3). If both the acquired CEN7 and the preexisting neocentromere in gene-converted derivatives are active in each case, Chr7 could become dicentric. We performed ChIP assays in gene-converted derivatives but no enrichment of CENPA/CENPC1 on neocentromeric location was observed (Fig. 7). Together these results suggest that neocentromeres previously established in parent RM1000AH-cen7Δ and RM1000AH-cen7-6.5kbΔ strains have been inactivated in their gene-converted derivatives which have acquired the native centromeric region from the unaltered Chr7 homolog. Thus, the active centromere has been repositioned (from the neocentromere location) to its original location.
Figure 7.
Centromere repositioning in C. albicans. PCR analysis of CENPA/Cse4 and CENPC1/Mif2 ChIP DNA in wild-type RM1000AH (upper), RM1000AH-cen7-6.5kbΔ (middle) harboring nCEN7-III (∼13 kb away from URA3 and marked by a star), and a gene-converted derivative of RM1000AH-cen7-6.5kbΔ (bottom). Absence of CENPA and CENPC1 binding at the nCEN7-III locus after gene conversion confirms neocentromere inactivation.
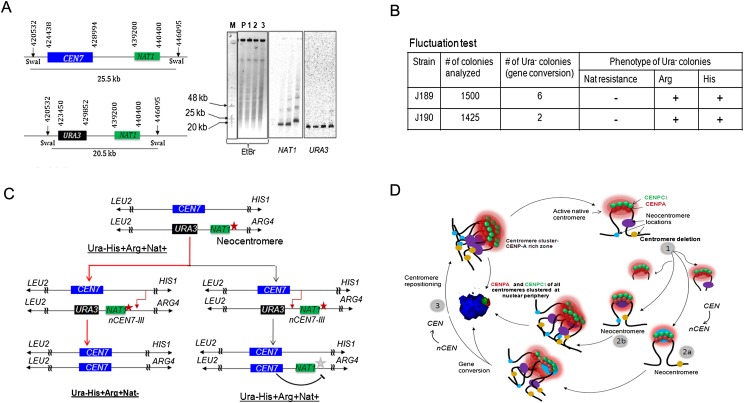
In meiosis, gene conversion tracts range from 1 to 2 kb (Borts and Haber 1989; Detloff and Petes 1992; Malone et al. 1992), while in mitosis, some gene conversion tracts range up to hundreds of kilobases (Judd and Petes 1988). Since gene conversion at C. albicans CEN7 described here is mitotic in nature, it can encompass a larger region and hence include the neocentromere located 13 kb away from the native CEN7 locus. To find out whether inactivation of the neocentromere in a gene-converted clone has occurred due to replacement of the neocentromere by homologous sequence during recombination or by an active suppression from the native centromere acquired from homologous chromosome, we sought to examine the possible length of the gene conversion tract. In strain J181(RM1000AH-cen7-6.5kbΔ), we replaced a 1.2-kb region (Assembly 21 CaChr7, 438440–439640) located next to the neocentromere (Assembly 21 CaChr7, 440500–444000) toward CEN7 with a dominant marker NAT1 (1.2 kb). Integration of the NAT1 marker on the neocentric chromosome was confirmed by Southern analysis (Fig. 8A). The resulting strain J189 is resistant to the drug nourseothricin. If the gene conversion tract encompasses the neocentromere, the NAT1 marker should be lost along with the neocentromere in its derivatives (Fig. 8B). Gene-converted Ura−Arg+His+ derivatives of two independent transformants of J189 were examined for the presence of the NAT1 marker. All Ura− gene-converted colonies were found to be sensitive to nourseothricin (Fig. 8C) as well. This indicates that the neocentromere has been replaced by a sequence from the homologous chromosome and thus inactivated by the native centromere. As a result, the centromere has been repositioned to its original place by gene conversion. Gene conversion can be an active mechanism to suppress neocentromeres to maintain the native centromere position conserved during evolution (Fig. 8D).
Figure 8.
Centromere repositioning occurs due to inactivation of neocentromeres by gene conversion. (A) Schematics showing a region across CEN7 and expected size of the SwaI fragment released either from the unaltered (25.5 kb) or CEN7-deleted (20.5 kb) homolog of Chr7 (left panel). SwaI-digested genomic DNA plugs from the parent (RM1000AH-cen7-6.5kbΔ), and three NAT1 integrants were separated on a CHEF gel (EtBr stained CHEF gel is shown here). Probing with NAT1 or URA3 sequences confirmed integration of NAT1 into CEN7-deleted neocentric Chr7 in these two integrants (J189 and J190). (B) A table summarizes the phenotypes of the markers of the segregants. Fluctuation test analysis in two independent NAT1 integrants (J189 and J190) of J181 (RM1000AH-cen7-6.5kbΔ) revealed the loss of NAT1 along with URA3. This result suggests that the neocentromere has been replaced along with URA3 by gene conversion. (C) Schematic showing possible fates of the segregants after gene conversion depending on the site of recombination initiation. (Red line) The observed event. (Black line) An alternative event. (Red star) Active neocentromere; (gray star) inactive neocentromere. (D) A possible mechanism of the maintenance of centromere function at the native physical chromosomal location. Similar to budding yeast S. cerevisiae, centromeres are always clustered toward the periphery of a nucleus throughout the cell cycle in C. albicans. A 3D reconstructed image showing peripheral localization of the kinetochore cluster immunostained with anti-CENPA/Cse4 (red) and anti-CENPC1/Mif2 (green) antibodies in a nucleus (blue, DAPI) in C. albicans. Clustering of kinetochores/centromeres is crucial for the integrity of the kinetochore ensemble as well as the fidelity of chromosome segregation in both S. cerevisiae and C. albicans (Anderson et al. 2009). We propose that clustering of all native centromeres into a specific 3D scaffold at the nuclear periphery creates a zone of high CENPA (and other kinetochore proteins including CENPC1) local concentration (pink). Potential neocentromere sites exist at centromere proximal regions (oval shapes of blue, brown, and violet colors) at least partly, due to their physical proximity to the CENPA-rich zone. Under normal conditions, the native centromere occupies the most preferred position in the centromere cluster and keeps adjacent potential neocentromere sites away from the CENPA-rich zone by providing steric hindrance. Removal of centromeric regions of various lengths brings centromere proximal regions in close proximity to the CENPA-rich zone (1). The new region now becomes a part of the CENPA-rich nuclear domain and assembles CENPA chromatin to form a neocentromere (2a and 2b). Gene conversion can inactivate an established neocentromere by replacing it with the homologous sequence that lacks the epigenetic information for CENPA assembly (3). This process is accompanied by acquisition of the native centromere from a homologous chromosome leading to repositioning of the centromere back to its original location.
Discussion
The exclusive presence of potential neocentromere-forming loci at centromere proximal regions in both Candida species demonstrates that the physical location of a centromere, once established, becomes evolutionarily conserved in spite of significant changes in the underlying sequence. A remarkably high conservation in relative centromere location in two independent clinical isolates, C. albicans 1006 and C. albicans WO-1, and in a related species, C. dubliniensis, strongly supports this hypothesis. We reason that although the native centromere is the strongest site for centromere formation, other potential sites (nCEN7-I–IV) are present at centromere proximal regions due to their similar relative location to that of the native centromere along the length of a chromosome. Interestingly, a comparative analysis (Fig. 2B) revealed that neocentromeres that are formed close to the native centromere (nCEN7-I) show CENPA binding comparable to that of the native centromeres (CEN7). As the distance of a neocentromere site from the deleted CEN7 region increased (nCEN7-II and nCEN7-III), CENPA binding progressively dropped. Previously we have shown that introduction of a CEN DNA sequence close to (∼7 kb away) a native CEN could not recruit CENPA chromatin (Baum et al. 2006). Neocentromere formation on a stable barley chromosome 7H derivative and Drosophila X minichromosome (γ238) occurred on sequences that were originally situated close to the native centromere in parent chromosomes (Williams et al. 1998; Nasuda et al. 2005), suggesting that proximity to the native centromere favors neocentromere formation. In light of these observations and our results from comprehensive analysis of neocentromere formation in C. albicans, we propose a model shown in Figure 8D.
The presence of neocentromere hotspots within the centromere proximal regions indicates that the nature of DNA sequences such as structural motifs or replication origins may play an important role in the assembly of CENPA on centromeres/neocentromeres (Sanyal 2012). Bending of DNA caused by centromeric binding proteins (CBF3 complex in S. cerevisiae and CENP-B in human) has been proposed to provide a proper geometry for kinetochore assembly in yeast and human (Anderson et al. 2009). A comparative analysis of structural conformation at functional centromeres/neocentromeres may provide information regarding structural blueprints that help centromere-forming sequences to be part of this specialized CENP-rich three-dimensional (3D) scaffold described in the model.
The previous report of the formation of neocentromeres at multiple locations on Chr5 in C. albicans depicts several features of neocentromeres: Neocentromere formation was seemingly efficient, random, and restricted primarily to intergenic regions when formed away from the native centromere (Ketel et al. 2009). Considering the conserved features of neocentromeres in various plant, animal, and fungal species, the presence of a combination of all these properties of neocentromeres together in C. albicans is particularly striking (Marshall and Choo 2009). Moreover, centromere location has been shown to be unchanged in a number of C. albicans clinical isolates analyzed in a previous study in spite of a considerable heterogeneity in centromere DNA sequence (Mishra et al. 2007). Our studies on the dynamics of neocentromere formation on Chr 1, Chr 5, and Chr 7 of C. albicans as well as on Chr7 of C. dubliniensis show some common properties reported to be associated with Chr5 neocentromeres. However, a total absence of ectopic centromere-distal neocentromeres from a large number of transformants analyzed in our study reveals a significant difference from the previous report (50% CEN5-deleted transformants formed neocentromeres at centromere-distal sites). Interestingly, CENPA enrichment was found to be very low in most of these distal neocentromeres (36%–42%, 8%–14%, 36%, and 60% in NeoCEN-1, NeoCEN-2, NeoCEN-3, and NeoCEN-4, respectively) (Ketel et al. 2009). A low-affinity binding of CENPA at several noncentromeric sites has been shown to occur in S. cerevisiae (Lefrançois et al. 2009). Co-localization of another kinetochore protein on such low-affinity CENPA-bound distal neocentromere sites should be done in order for them to be considered authentic neocentromeres.
Despite the rapid sequence divergence, centromere locations in C. albicans and C. dubliniensis remain unchanged. Thus, there must be factors or mechanisms that are responsible for conservation of physical locations of centromeres while underlying sequences are constantly changing. Centromeric regions were thought to be recombination deficient, but recently, mitotic recombination has been shown to be taking place at mammalian centromeres at a very high rate (Jaco et al. 2008). Moreover, gene conversion has been reported to occur at centromeres of S. cerevisiae and maize (Symington and Petes 1988; Shi et al. 2010). Gene conversion is a mechanism that repairs double-strand breaks by copying the homologous sequence. In the present study, we detected gene conversion at C. albicans centromeres. During gene conversion, an established neocentromere was nonreciprocally exchanged by its homologous sequence that originally lacked neocentromeric activity. This led to repositioning of CENPA from nCEN7-III to the native CEN7 locus. As discussed earlier, CENPA recruitment at neocentromeres progressively drops with an increase in distance from the native centromeres. Thus gene conversion might provide a selective advantage to a chromosome by repositioning centromeric activity from the neocentromeric location (less preferred) to the original centromeric location (more preferred). We speculate that this can be a possible mechanism responsible for conservation of the physical chromosomal location of centromeres in C. albicans and C. dubliniensis. Interestingly, some of neocentromere hotspots in humans are sites for centromere repositioning during evolution (Ventura et al. 2004). Recombination coupled replication during gene conversion has been shown to be a highly mutagenic process in budding yeast (Hicks et al. 2010). Recombination is required to bring about the mutations necessary to drive evolution. Thus, while keeping the physical chromosomal location unchanged, gene conversion might have brought about changes in DNA sequence by accumulating mutations over several million years, probably to drive speciation through functional incompatibility of centromeres in these two closely related asexually propagated yeasts.
Methods
Strain construction
Strains and primers are listed in Supplemental Tables S3 and S4, respectively.
Construction of centromeric deletion strains
The insert for deleting the core 4.5-kb CEN7 was released from pBSCR7Δ as described before (Sanyal et al. 2004). Other cassettes for deletion of various CEN7 regions, 4.2-kb CEN1 and 7.2-kb CEN5 sequences, were constructed as follows: CaURA3 was cloned in pBluescriptKSII (−) as a NotI and PstI fragment to generate the plasmid pBSURA3. Sequences upstream of and downstream from the region to be deleted were cloned in SacI/NotI and PstI/XhoI sites of pBSURA3. Coordinates of deleted regions, sequences upstream of and downstream from the region to be deleted that were used for the homologous recombination are given in Supplemental Table S1. Primer sequences are available in Supplemental Table S4. Inserts containing the deletion cassettes were released as SacI/XhoI fragments and transformed into the strain RM1000AH. Correct integrants were confirmed by Southern analysis (details of Southern analysis strategies are given in Supplemental Table S2).
Expression of ProtA-tagged CENP-A in C. albicans
The NAT1 sequence was amplified from the pGFPNAT plasmid using primers NAT-1 and NAT-2 and cloned into SmaI site of pBluescript to give rise to pBSNAT. A Cse4 TAP cassette was amplified from pCSE4TAP (Thakur and Sanyal 2012) using primer pair JTCse4SacII/CdMif2TAPNAT2 and cloned into end-filled SacII & SpeI sites of pBSNAT to construct pCaCse4TAPNAT. pCaCse4TAPNAT was partially digested with XhoI and transformed into RM1000AH and RM1000AH-cen5Δ clones. Expression and localization of Prot A-tagged CENPA/Cse4 in resulting strains were verified by indirect immunofluorescence using anti-Prot A antibodies. CENPA/Cse4 Prot A in all clones tested exhibited localization patterns similar to CENPA/Cse4 (data not shown).
Expression of MycMif2 (CENPC1) in C. albicans
To examine the binding of Mif2/CENPC1 at various neocentromeres (nCENI-IV) we expressed MycMif2 in RM1000AH, RM1000AH-CEN7∷URA3, RM1000AH-cen7-4.5kbΔ, RM1000AH-cen7-6.5kbΔ, RM1000AH-cen7-30kbΔ, RM1000AH-cen1-4.2kbΔ clones. A cassette containing PCK1prMycMIF2 was amplified from strain CAMB2 (Sanyal et al. 2004) using primers Mif2PckMycPst1-F and Mif2pck1SacII-R and subsequently cloned into PstI and SacII sites of pSF2A vector (Reuss et al. 2004) that contained NAT1, the nourseothricin (NAT) marker as described before (Thakur and Sanyal 2012). The resulting plasmid was linearized using HpaI and introduced into various neocentromere-containing transformants and their gene-converted derivatives. Correct integration in each case was confirmed using primes MycpckMif2pstF and Mif69248 (Supplemental Table S4). Expression and localization of Myc-tagged MycMif2/CENPC1 were verified by indirect immunofluorescence using anti-Myc antibodies. MycMif2/CENPC1 in all clones tested exhibited localization patterns similar to CENPA/Cse4 (Supplemental Figs. S1C, S3D), confirming functional expression of MycMif2/CENPC1.
Integration of the NAT1 marker gene to measure the length of gene conversion tracts
To replace a 1.2-kb region (Assembly 21 Chr7, 438440–439640) with NAT1 marker of the same length near nCEN7-III, upstream (Assembly 21 Chr7, 437700–438440, primer pair NATGC1 and NATGC2) and downstream (Assembly 21 Chr7, 439640–440300, primer pair NATGC3 and NATGC4) sequences were cloned into SacII/SpeI and ClaI/SacI sites of pBSNAT, respectively. The resulting plasmid, named pNATGC2, was used to amplify a fragment using NATGC1 and NATGC4 and used to transform J181 (RM1000AH-cen7-6.5kbΔ). Desired transformants were identified by Southern analysis for correct integration of NAT1 into the URA3-containing homolog of Chr7. In parent J181 (RM1000AH-cen7-6.5kbΔ), SwaI digestion releases two fragments of different sizes, a 25.5-kb fragment from the wild type and a 20.5-kb fragment from the CEN7-deleted homolog of Chr 7 (Fig. 8A). CHEF gel plugs were made from J181 and NAT1 integrants using the protocol described before. SwaI-digested genomic DNA plugs from the parent J181 and three NAT1 integrants of it were separated on a CHEF gel and transferred to a Zeta probe (Bio-Rad) membrane. Probing the membrane with NAT1 or URA3 sequence confirmed integration of NAT1 into the CEN7-deleted neocentric homolog in these two integrants (J189 and J190).
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation assays were performed as described previously (Thakur and Sanyal 2011, 2012).
Quantitative real-time PCR analysis
Total RNA was isolated from J151 grown in Complete-Ura and FOA-containing media using TRIzol Reagent from Invitrogen (Cat# 15596-026). cDNA was prepared using Reverse Transcriptase from Thermo Scientific (Cat# EP0441) and oligo (dT) primers (Sigma Cat# O4387). Real-time qPCR was performed using the SYBR Green supermix (BIO RAD Cat# 1708880). Reactions were run on the BIORAD real time machine (Bio-Rad). Three technical replicates were performed on RNA isolated from J151. DCt values were calculated from the mean of the technical replicates (Supplemental Fig. S2A).
Real-time analysis was performed on CENPA ChIP DNA from a representative clone from each class of neocentromere-containing strain. Three biological as well as technical replicates were performed for each experiment. Primers used are listed in Supplemental Table S4. The CENPA enrichment was determined by the percent input method. The Ct values for input were corrected for the dilution factor and then the percent of the input chromatin immunoprecipitated by the antibody was calculated as 100 × 2(Adjusted Input Ct− IP Ct).
ChIP sequencing and analysis of data
Enriched samples from ChIP experiments as well as the corresponding whole-cell extracts were quantified using Qubit before processing for Library preparation. Only samples with a minimum of 10 ng ds DNA (Qubit) were taken further for library preparation. Libraries for multiplex ChIP sequencing were constructed using NEXTflex ChIP-Seq Sample Preparation Kit protocol outlined in “Preparing Samples for ChIP Sequencing of DNA” (BIOO Scientific# IP-5143-01). Briefly, DNA was subjected to a series of enzymatic reactions that repair frayed ends and phosphorylate the fragments. The end repaired fragments were subjected to two rounds of SPRI clean up with Agencourt AMPURE XP beads (Beckman Coulter # A63881) for size selection of DNA inserts between 300 and 400 bp. The size-selected fragments were adenylated with a single nucleotide “A” overhang (BIOO Scientific# IP-5143-01) and adaptors were ligated (NEXT Flex adapters). The fragments with ligated adapters were enriched with 18 cycles of PCR. The prepared libraries were quantified using Qubit and validated for quality by running an aliquot on a High Sensitivity Bioanalyzer Chip (Agilent).
Sequencing was carried out on the Illumina Genome Analyzer IIx (GAIIx) using flowcell (ID-63867AAXX). ChIP-seq library fragments were diluted, denatured, and hybridized to a lawn of oligonucleotides immobilized on the flow cell surface. Hybridized library template was amplified using immobilized oligonucleotides as primers. Each hybridized template through the process of isothermal bridge amplification results in the formation of a cluster comprised of roughly 1000 clonal copies.
Sequencing was performed by synthesis (SBS) technology using four fluorescently labeled nucleotides to sequence every cluster on the flow cell surface in parallel. During each sequencing cycle, a single labeled deoxynucleotide triphosphate (dNTP) was added and clusters were imaged. The fluorescent dye and blocker was cleaved off and the next complementary base was added to the nucleic acid chain and imaged. Thirty-six such cycles were performed. Individual bases were called directly from signal intensity measurements during each cycle. Once sequencing was completed, the raw data was extracted from the server using the proprietary Illumina pipeline software to obtain FASTQ files. Quality check of raw data was performed using Seq QC and 7 to 30 million single-end 36-nt-long reads were generated for each sample. Raw reads were processed using SeqQC. Reads were aligned to the target C. albicans SC5314 genome Assembly 21 using Bowtie version 0.12.8 and using the parameters “-v 3–best -m 1” (Langmead and Salzberg 2012). A minimum of 2 million aligned reads were obtained per sample. Peak calling was performed using Homer v3.13 in “histone” mode using default parameters (Heinz et al. 2010). Chromosome-wise read distribution and fold-enrichment graphs were generated using R scripts.
CHEF analysis
C. albicans cells were grown in appropriate media (YPDU for RM1000AH and gene-converted strains and Complete-Ura for RM1000AH-cen7Δ) until OD600 = 1.0. Exponentially grown cells were pelleted down and washed with ice chilled 50 mM EDTA. Agarose plugs were made according to the instruction manual protocol (BioRad Kit, Cat No. 170-3593) with clean cut agarose (0.6%) and lyticase enzyme provided in the kit. Plugs were introduced into agarose gels in 0.5× TBE buffer (0.1 M Tris. 0.09 M boric acid, 0.01 M EDTA, pH 8), and electrophoresis was performed with a Bio-Rad CHEF-DRIII system. The gels were run in 0.5× TBE buffer and maintained at 14°C throughout the procedure. Electrophoresis settings were as follows (Magee et al. 2008):
For separation of all chromosomes: 0.6% Bio-Rad Pulsed Field Certified Agarose, in 0.5× TBE buffer (50 mMTris, 50 mM boric acid,1 mM EDTA, pH 8.3), 60–300 sec switch ramp, 24 h, 4.5 V/cm 120°; 720–900 sec ramp, 12 h, 2.0 V/cm;
For separation of smaller chromosomes: 0.9% agarose, 60–120 sec ramp 24 h, 6.0 V/cm; 120–360 sec ramp, 15h, 4.5 V/cm, 120°;
For separation of SwaI-digested DNA: 0.9% agarose, 0–10 sec ramp 19 h, 6.0 V/cm; 120°.
The gels were stained with ethidium bromide (EtBr) and analyzed by using the Quantity One software (Bio-Rad).
Chromosome loss assay
All the genetic manipulations in C. albicans were done in RM1000AH background (Sanyal et al. 2004). Each homolog of Chr7 is differentially marked with unique auxotrophic markers—ARG4 or HIS1 on the right arm. Chromosome loss in RM1000AH-cen7Δ, RM1000AH-cen7-6.5kbΔ, and RM1000AH-cen7-30kbΔ transformants was scored by simultaneous loss of URA3 and ARG4 or URA3 and HIS1 markers. Strains were grown overnight at 30°C in nonselective media (YPDU) and plated on nonselective plates. After two days of growth at 30°C, colonies were replica-plated on selective plates (CM-Ura, CM-Arg, and CM-His plates). Colonies that failed to grow on plates lacking uridine and arginine or uridine and histidine were counted as colonies that lost a chromosome. A similar fluctuation test analysis was performed in NAT1 integrants of RM1000AH-cen7-6.5kbΔ, but in addition to CM-Ura, CM-Arg, and CM-His plates, cells were transferred to YPDU +Nat plates as well.
Data access
The sequencing data used in this study have been submitted to the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE42907.
Acknowledgments
We thank Genotypic Technology Private Limited, Bangalore, for ChIP-seq library preparation, raw sequence data generation, and the ChIP-seq analysis reported in this publication. We thank Joachim Morschhauser for providing us plasmids pSFU2 and pSFU4 and the confocal facility of the Molecular Biology and Genetics Unit, JNCASR. This work was supported by grants from the Department of Science and Technology (DST) and the Department of Biotechnology (DBT), Government of India to K.S. and intramural support from JNCASR. J.T. was a DBT-SRF and is presently supported by a postdoctoral fellowship from JNCASR.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.141614.112.
References
- Allshire RC, Karpen GH 2008. Epigenetic regulation of centromeric chromatin: Old dogs, new tricks? Nat Rev Genet 9: 923–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso A, Fritz B, Hasson D, Abrusan G, Cheung F, Yoda K, Radlwimmer B, Ladurner AG, Warburton PE 2007. Co-localization of CENP-C and CENP-H to discontinuous domains of CENP-A chromatin at human neocentromeres. Genome Biol 8: R148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson M, Haase J, Yeh E, Bloom K 2009. Function and assembly of DNA looping, clustering, and microtubule attachment complexes within a eukaryotic kinetochore. Mol Biol Cell 20: 4131–4139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum M, Sanyal K, Mishra PK, Thaler N, Carbon J 2006. Formation of functional centromeric chromatin is specified epigenetically in Candida albicans. Proc Natl Acad Sci 103: 14877–14882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensasson D, Zarowiecki M, Burt A, Koufopanou V 2008. Rapid evolution of yeast centromeres in the absence of drive. Genetics 178: 2161–2167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard P, Maure JF, Partridge JF, Genier S, Javerzat JP, Allshire RC 2001. Requirement of heterochromatin for cohesion at centromeres. Science 294: 2539–2542 [DOI] [PubMed] [Google Scholar]
- Black BE, Cleveland DW 2011. Epigenetic centromere propagation and the nature of CENP-a nucleosomes. Cell 144: 471–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borts RH, Haber JE 1989. Length and distribution of meiotic gene conversion tracts and crossovers in Saccharomyces cerevisiae. Genetics 123: 69–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambareri EB, Aisner R, Carbon J 1998. Structure of the chromosome VII centromere region in Neurospora crassa: Degenerate transposons and simple repeats. Mol Cell Biol 18: 5465–5477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu WS, Magee BB, Magee PT 1993. Construction of an SfiI macrorestriction map of the Candida albicans genome. J Bacteriol 175: 6637–6651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detloff P, Petes TD 1992. Measurements of excision repair tracts formed during meiotic recombination in Saccharomyces cerevisiae. Mol Cell Biol 12: 1805–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishel B, Amstutz H, Baum M, Carbon J, Clarke L 1988. Structural organization and functional analysis of centromeric DNA in the fission yeast Schizosaccharomyces pombe. Mol Cell Biol 8: 754–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folco HD, Pidoux AL, Urano T, Allshire RC 2008. Heterochromatin and RNAi are required to establish CENP-A chromatin at centromeres. Science 319: 94–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalakrishnan S, Sullivan BA, Trazzi S, Della Valle G, Robertson KD 2009. DNMT3B interacts with constitutive centromere protein CENP-C to modulate DNA methylation and the histone code at centromeric regions. Hum Mol Genet 18: 3178–3193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38: 576–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Ahmad K, Malik HS 2001. The centromere paradox: Stable inheritance with rapidly evolving DNA. Science 293: 1098–1102 [DOI] [PubMed] [Google Scholar]
- Hicks WM, Kim M, Haber JE 2010. Increased mutagenesis and unique mutation signature associated with mitotic gene conversion. Science 329: 82–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii K, Ogiyama Y, Chikashige Y, Soejima S, Masuda F, Kakuma T, Hiraoka Y, Takahashi K 2008. Heterochromatin integrity affects chromosome reorganization after centromere dysfunction. Science 321: 1088–1091 [DOI] [PubMed] [Google Scholar]
- Jackson AP, Gamble JA, Yeomans T, Moran GP, Saunders D, Harris D, Aslett M, Barrell JF, Butler G, Citiulo F, et al. 2009. Comparative genomics of the fungal pathogens Candida dubliniensis and Candida albicans. Genome Res 19: 2231–2244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaco I, Canela A, Vera E, Blasco MA 2008. Centromere mitotic recombination in mammalian cells. J Cell Biol 181: 885–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judd SR, Petes TD 1988. Physical lengths of meiotic and mitotic gene conversion tracts in Saccharomyces cerevisiae. Genetics 118: 401–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketel C, Wang HS, McClellan M, Bouchonville K, Selmecki A, Lahav T, Gerami-Nejad M, Berman J 2009. Neocentromeres form efficiently at multiple possible loci in Candida albicans. PLoS Genet 5: e1000400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kipling D, Mitchell AR, Masumoto H, Wilson HE, Nicol L, Cooke HJ 1995. CENP-B binds a novel centromeric sequence in the Asian mouse Mus caroli. Mol Cell Biol 15: 4009–4020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn RM, Clarke L, Carbon J 1991. Clustered tRNA genes in Schizosaccharomyces pombe centromeric DNA sequence repeats. Proc Natl Acad Sci 88: 1306–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T 2001. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410: 116–120 [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HR, Zhang W, Langdon T, Jin W, Yan H, Cheng Z, Jiang J 2005. Chromatin immunoprecipitation cloning reveals rapid evolutionary patterns of centromeric DNA in Oryza species. Proc Natl Acad Sci 102: 11793–11798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefrançois P, Euskirchen GM, Auerbach RK, Rozowsky J, Gibson T, Yellman CM, Gerstein M, Snyder M 2009. Efficient yeast ChIP-Seq using multiplex short-read DNA sequencing. BMC Genomics 10: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magee BB, Magee PT 1997. WO-2, a stable aneuploid derivative of Candida albicans strain WO-1, can switch from white to opaque and form hyphae. Microbiology 143: 289–295 [DOI] [PubMed] [Google Scholar]
- Magee BB, Sanchez MD, Saunders D, Harris D, Berriman M, Magee PT 2008. Extensive chromosome rearrangements distinguish the karyotype of the hypovirulent species Candida dubliniensis from the virulent Candida albicans. Fungal Genet Biol 45: 338–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone RE, Bullard S, Lundquist S, Kim S, Tarkowski T 1992. A meiotic gene conversion gradient opposite to the direction of transcription. Nature 359: 154–155 [DOI] [PubMed] [Google Scholar]
- Marshall OJ, Choo KH 2009. Neocentromeres come of age. PLoS Genet 5: e1000370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall OJ, Chueh AC, Wong LH, Choo KH 2008. Neocentromeres: New insights into centromere structure, disease development, and karyotype evolution. Am J Hum Genet 82: 261–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra PK, Baum M, Carbon J 2007. Centromere size and position in Candida albicans are evolutionarily conserved independent of DNA sequence heterogeneity. Mol Genet Genomics 278: 455–465 [DOI] [PubMed] [Google Scholar]
- Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI 2001. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292: 110–113 [DOI] [PubMed] [Google Scholar]
- Nasuda S, Hudakova S, Schubert I, Houben A, Endo TR 2005. Stable barley chromosomes without centromeric repeats. Proc Natl Acad Sci 102: 9842–9847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan S, Thakur J, Siddharthan R, Sanyal K 2008. Rapid evolution of Cse4p-rich centromeric DNA sequences in closely related pathogenic yeasts, Candida albicans and Candida dubliniensis. Proc Natl Acad Sci 105: 19797–19802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuss O, Vik A, Kolter R, Morschhauser J 2004. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene 341: 119–127 [DOI] [PubMed] [Google Scholar]
- Rhind N, Chen Z, Yassour M, Thompson DA, Haas BJ, Habib N, Wapinski I, Roy S, Lin MF, Heiman DI, et al. 2011. Comparative functional genomics of the fission yeasts. Science 332: 930–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice JC, Briggs SD, Ueberheide B, Barber CM, Shabanowitz J, Hunt DF, Shinkai Y, Allis CD 2003. Histone methyltransferases direct different degrees of methylation to define distinct chromatin domains. Mol Cell 12: 1591–1598 [DOI] [PubMed] [Google Scholar]
- Roy B, Sanyal K 2011. Diversity in requirement of genetic and epigenetic factors for centromere function in fungi. Eukaryot Cell 10: 1384–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal K 2012. How do microbial pathogens make CENs? PLoS Pathog 8: e1002463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal K, Carbon J 2002. The CENP-A homolog CaCse4p in the pathogenic yeast Candida albicans is a centromere protein essential for chromosome transmission. Proc Natl Acad Sci 99: 12969–12974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal K, Baum M, Carbon J 2004. Centromeric DNA sequences in the pathogenic yeast Candida albicans are all different and unique. Proc Natl Acad Sci 101: 11374–11379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Z 2011. Genomic instability and cancer: An introduction. J Mol Cell Biol 3: 1–3 [DOI] [PubMed] [Google Scholar]
- Shi J, Wolf SE, Burke JM, Presting GG, Ross-Ibarra J, Dawe RK 2010. Widespread gene conversion in centromere cores. PLoS Biol 8: e1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KM, Phatale PA, Sullivan CM, Pomraning KR, Freitag M 2011. Heterochromatin is required for normal distribution of Neurospora crassa CenH3. Mol Cell Biol 31: 2528–2542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staib P, Moran GP, Sullivan DJ, Coleman DC, Morschhäuser J 2001. Isogenic strain construction and gene targeting in Candida dubliniensis. J Bacteriol 183: 2859–2865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stimpson KM, Sullivan BA 2010. Epigenomics of centromere assembly and function. Curr Opin Cell Biol 22: 772–780 [DOI] [PubMed] [Google Scholar]
- Symington LS, Petes TD 1988. Meiotic recombination within the centromere of a yeast chromosome. Cell 52: 237–240 [DOI] [PubMed] [Google Scholar]
- Takahashi K, Murakami S, Chikashige Y, Niwa O, Yanagida M 1991. A large number of tRNA genes are symmetrically located in fission yeast centromeres. J Mol Biol 218: 13–17 [DOI] [PubMed] [Google Scholar]
- Takahashi K, Chen ES, Yanagida M 2000. Requirement of Mis6 centromere connector for localizing a CENP-A-like protein in fission yeast. Science 288: 2215–2219 [DOI] [PubMed] [Google Scholar]
- Thakur J, Sanyal K 2011. The essentiality of the fungus-specific Dam1 complex is correlated with a one-kinetochore-one-microtubule interaction present throughout the cell cycle, independent of the nature of a centromere. Eukaryot Cell 10: 1295–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur J, Sanyal K 2012. A coordinated interdependent protein circuitry stabilizes the kinetochore ensemble to protect CENP-A in the human pathogenic yeast Candida albicans. PLoS Genet 8: e1002661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura M, Weigl S, Carbone L, Cardone MF, Misceo D, Teti M, D'Addabbo P, Wandall A, Björck E, de Jong PJ, et al. 2004. Recurrent sites for new centromere seeding. Genome Res 14: 1696–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voullaire LE, Slater HR, Petrovic V, Choo KH 1993. A functional marker centromere with no detectable α-satellite, satellite III, or CENP-B protein: Activation of a latent centromere? Am J Hum Genet 52: 1153–1163 [PMC free article] [PubMed] [Google Scholar]
- Williams BC, Murphy TD, Goldberg ML, Karpen GH 1998. Neocentromere activity of structurally acentric mini-chromosomes in Drosophila. Nat Genet 18: 30–37 [DOI] [PubMed] [Google Scholar]
- Wong NC, Wong LH, Quach JM, Canham P, Craig JM, Song JZ, Clark SJ, Choo KH 2006. Permissive transcriptional activity at the centromere through pockets of DNA hypomethylation. PLoS Genet 2: e17. [DOI] [PMC free article] [PubMed] [Google Scholar]